Abstract
Age-related ocular conditions like macular degeneration (AMD), diabetic retinopathy (DR), and glaucoma are leading causes of irreversible vision loss globally. Optical coherence tomography (OCT) provides essential non-invasive visualization of retinal structures for early diagnosis, but manual analysis of these images is labor-intensive and prone to variability. Deep learning (DL) techniques have emerged as powerful tools for automating the segmentation of the retinal layer in OCT scans, potentially improving diagnostic efficiency and consistency. This review systematically evaluates the state of the art in DL-based retinal layer segmentation using the PRISMA methodology. We analyze various architectures (including CNNs, U-Net variants, GANs, and transformers), examine the characteristics and availability of datasets, discuss common preprocessing and data augmentation strategies, identify frequently targeted retinal layers, and compare performance evaluation metrics across studies. Our synthesis highlights significant progress, particularly with U-Net-based models, which often achieve Dice scores exceeding 0.90 for well-defined layers, such as the retinal pigment epithelium (RPE). However, it also identifies ongoing challenges, including dataset heterogeneity, inconsistent evaluation protocols, difficulties in segmenting specific layers (e.g., OPL, RNFL), and the need for improved clinical integration. This review provides a comprehensive overview of current strengths, limitations, and future directions to guide research towards more robust and clinically applicable automated segmentation tools for enhanced ocular disease diagnosis.
1. Introduction
Ocular diseases such as glaucoma, diabetic retinopathy, and age-related macular degeneration remain among the leading causes of vision loss and blindness worldwide. Early diagnosis and precise monitoring of these conditions are critical to prevent irreversible damage, and OCT imaging has become a cornerstone modality for non-invasive visualization of the retinal structures. However, the manual segmentation of retinal layers in OCT scans is labor-intensive, time-consuming, and subject to inter-observer variability, limiting its scalability and consistency in clinical settings. To address these challenges, DL techniques have emerged as powerful tools for automating retinal layer segmentation in OCT images. These methods have demonstrated remarkable capabilities in extracting retinal boundaries, identifying pathological alterations, and even predicting disease progression. Nevertheless, despite the growing number of publications in this area, the field lacks a recent and comprehensive synthesis of the advances, trends, and applications of DL in retinal layer segmentation. This review responds to several critical gaps in current literature. First, while numerous studies report individual deep learning models or techniques, there is no up-to-date review that systematically compares these approaches or analyzes their architectural trends and use cases across various ocular conditions. Second, the diversity in datasets used, many of which are private, sparsely labeled, or poorly described, has created a significant barrier to reproducibility and benchmarking. No previous works provide a consolidated overview of the available OCT datasets annotated for retinal layer segmentation, which is essential for both model development and validation. Third, there is currently no consensus on standard evaluation protocols or metrics. Studies employ a wide range of criteria, such as the Dice similarity coefficient, mean absolute error, and Hausdorff distance, often without clarifying how these metrics relate to clinical utility. This inconsistency hinders cross-study comparisons and obfuscates real-world performance assessments. Lastly, there is a lack of analysis on how artificial intelligence is being applied across different clinical contexts or patient populations, leaving researchers and clinicians without guidance on which models are most applicable to specific diagnostic tasks or settings. This review aims to bridge these gaps by (i) categorizing and analyzing recent deep learning techniques for OCT-based retinal layer segmentation, (ii) compiling the available datasets and their characteristics, (iii) examining the performance metrics and evaluation trends, and (iv) identifying current limitations and future directions. By doing so, we provide a structured and critical overview to support both academic research and clinical innovation in AI-assisted ophthalmology.
1.1. Glaucoma
Glaucoma is the leading cause of irreversible blindness globally [1]. In 2002, the WHO estimated that 4.4 million people (12.3% of the global blind population) were blind due to glaucoma. A significant portion of glaucoma cases worldwide still go undiagnosed [2], and it is projected that about 111.8 million people will be affected by this disease by 2040 [3]. Significant changes in the retinal layer thickness has been observed in patients with different levels of glaucoma, for the macular retinal nerve fiber layer outer inferior sector, the ganglion cell layer outer temporal sector, and for the inner plexiform layer outer temporal sector [4].
The symptoms of glaucoma described by [5] indicate that the principal sign is a raised intraocular pressure, which can exceed 70 mm Hg and cause symptoms like halos around lights and a cloudy vision due to the cornea being waterlogged by the sudden pressure increase. The increase in the intraocular pressure causes damage on the retinal nerve fibers, resulting in visual field loss, being the characteristic tunnel vision, with a gradual increase that even the patient might not notice until the disease is in an advanced stage, and optic disc changes that result in a cupping of the optic nerve. Additionally, elevated intraocular pressure can obstruct blood flow in the venous system, raising the risk of retinal vein occlusion, and even causing other uncommon signs, such as pain and enlargement of the eye in children under 3 years old.
1.1.1. Primary Open-Angle Glaucoma
POAG is the most common form of open-angle glaucoma and a leading cause of blindness worldwide, with approximately 6 million cases of blindness in 2020 [6]. While most patients with POAG do not go blind, the disease often causes significant visual function limitations and reduced quality of life. The pathophysiology of POAG remains poorly understood, but early damage to RGC and their axons, which eventually form the optic nerve, disrupts the visual pathway and leads to vision loss. Risk factors include elevated IOP, older age, black race, family history, and thin central cornea, though the disease can occur with normal IOP. POAG management focuses on lowering IOP, the only modifiable risk factor, through medical or surgical interventions. However, many patients continue to experience progressive vision loss despite treatment, prompting ongoing research into neuroprotective strategies and regenerative therapies for RGC axons [6,7,8].
1.1.2. Acute Angle-Closure Glaucoma
AACG is an ophthalmic emergency characterized by a sudden rise in IOP due to the closure of the iridocorneal angle, which obstructs the drainage of aqueous humor. This condition typically occurs when the lens blocks the flow of aqueous from the posterior to the anterior chamber, leading to a rapid increase in IOP. Symptoms include severe ocular pain, headache, blurred vision, nausea, and the appearance of halos around lights. Hypermetropic individuals and older women are at higher risk due to their shallower anterior chambers. Without timely intervention, AACG can lead to permanent vision loss. Management focuses on immediate reduction of IOP using medications such as miotics, beta-blockers, and osmotic diuretics, followed by definitive treatments like laser peripheral iridotomy or surgery. Delayed treatment can result in irreversible damage to the trabecular meshwork, requiring further surgical intervention [5,9].
1.2. Age-Related Macular Degeneration
AMD is a progressive and chronic condition affecting the central retina, and it stands as another primary cause of vision loss globally. Vision impairment primarily occurs during the advanced stages of the disease, which manifests in two forms: neovascular (“wet”) AMD and geographic atrophy (“late dry”) AMD. In the neovascular form, abnormal blood vessels from the choroid invade the neural retina, leading to fluid accumulation, lipid deposits, and bleeding, eventually resulting in scar tissue formation. On the other hand, geographic atrophy is characterized by the gradual degeneration of the retinal pigment epithelium, the choriocapillaris, and photoreceptors. These advanced stages of AMD are responsible for the most significant visual deterioration. A decade ago, treatment options for AMD were limited. However, the introduction of pharmaceuticals that inhibit vascular endothelial growth factor (VEGF) has significantly improved disease management, preventing vision loss in nearly 95% of patients, and improving vision in 40% [10].
1.3. Diabetic Retinopathy
Diabetic retinopathy is the most frequent cause of new blindness cases in adults aged 20–74 [11]. Diabetic retinopathy progresses from mild nonproliferative changes that are characterized by an increased vascular permeability, to moderate and severe NPDR, which is characterized by vascular closure, and eventually to PDR, distinguished by the growth of new blood vessels on the retina and the posterior surface of the vitreous. Macular edema, defined by retinal swelling due to leaky blood vessels, can occur at any stage of retinopathy. Nearly all patients with type 1 diabetes develop retinopathy within the following two decades. Up to 21% of individuals with type 2 diabetes already have retinopathy at the time of their diabetes diagnosis, and many go on to develop some level of retinopathy over time [11].
1.4. OCT-Aided Ocular Diseases Diagnosis
Optical coherence tomography is an imaging technology that enables noninvasive optical medical diagnosis by collecting optical backscattering signal for cross-sectional and three-dimensional images for visualization of the internal microstructure in living tissue [12,13].
1.5. Retinal Layers
The retina is composed of six types of cells organized into ten separate layers, each contributing to the processing and transmission of visual information. The retina is a structure composed of ten layers of interconnected neurons (Figure 1), which are as follows.
The inner limiting membrane (ILM) is the retina’s innermost layer, formed by the expanded endfeet of Müller cells and serving as a basement membrane between the retina and the vitreous [14,15].
The nerve fiber layer (NFL) is composed of ganglion cell axons that converge at the optic disc to form the optic nerve. It is thickest near the optic disc and thins toward the periphery. Axons from the macula form the papillomacular bundle, which is essential for central vision. The NFL also contains astrocytes, Müller cell processes, and superficial retinal vessels. Its integrity is crucial for visual function, and thinning of the NFL is an early marker of diseases like glaucoma, optic neuritis, and other neurodegenerative or vascular conditions. Flame-shaped hemorrhages in this layer are common in diabetic and hypertensive retinopathies [14,15].
The ganglion cell layer (GCL) contains around 1.2 million ganglion cell bodies, along with displaced amacrine cells, astrocytes, endothelial cells, and pericytes. These ganglion cells send their axons to form the optic nerve. The GCL is thickest in the perifoveal macula, with 8–10 rows of cells, and thins to a single row outside the macula, disappearing at the foveola. Most ganglion cells are midget or parasol types, preserving the center-surround organization of visual signals [15,16].
The inner plexiform layer (IPL) is the second layer involved in retinal signal processing. It is where bipolar cells connect with ganglion cells, and amacrine cells help regulate this connection. The IPL is divided into six sublayers, which allow for parallel processing of visual information through organized interactions between bipolar, amacrine, and ganglion cells [15,16].
The inner nuclear layer (INL) contains the cell bodies of bipolar, horizontal, and amacrine cells, along with Müller glial cells. Bipolar cells relay signals from photoreceptors to ganglion cells, with rod and cone subtypes supporting scotopic and photopic vision, respectively. Horizontal cells modulate glutamatergic input from photoreceptors [16,17].
The outer plexiform layer (OPL) is where visual signals are transmitted through synapses between photoreceptors and the dendrites of over twelve types of bipolar cells, as well as the dendrites and axon terminals of horizontal cells. This layer contains synaptic connections between rods, cones, and the footplates of horizontal cells, and it also houses capillaries that primarily run through the region [18,19].
The outer nuclear layer (ONL) houses the nuclei of rod and cone photoreceptors. Damage to this cellular machinery leads to irreversible photoreceptor loss and impaired visual function. ONL thickness has become a key anatomical marker for studies of retinal degeneration. Monitoring of ONL thickness allows researchers to track disease progression [20].
The external limiting membrane (ELM) is a distinct, less prominent hyper-reflective line in the outer retina, located just above the inner segment–outer segment (IS-OS) junction [21,22]. Functionally, this linear confluence of zonular adherents between Müller cells and photoreceptor cell bodies acts as a selective barrier against macromolecules and provides structural support for photoreceptor alignment [23].
The photoreceptor layer (layer of rods and cones) is the region containing the inner and outer segments of the photoreceptors, the first-order neurons in the visual pathway, that convert light into electrical signals. Rods enable low-light vision, while cones support color and high-acuity vision. These cells have high energy demands, especially cones, making energy metabolism vital for their function and survival. Degeneration of photoreceptors, particularly cones, is a key factor in many retinal diseases causing vision loss [24].
The retinal pigment epithelium (RPE) is a single layer of pigmented cells between Bruch’s membrane and the photoreceptors, playing key roles in nutrient transport, waste removal, and photoreceptor support. Its structural features aid in substance exchange and phagocytosis, while tight junctions help form the blood-retinal barrier. Melanin protects against UV and oxidative damage. RPE dysfunction is linked to retinal diseases like retinitis pigmentosa, age-related macular degeneration, and Stargardt disease, highlighting its importance in vision and potential therapies [25].
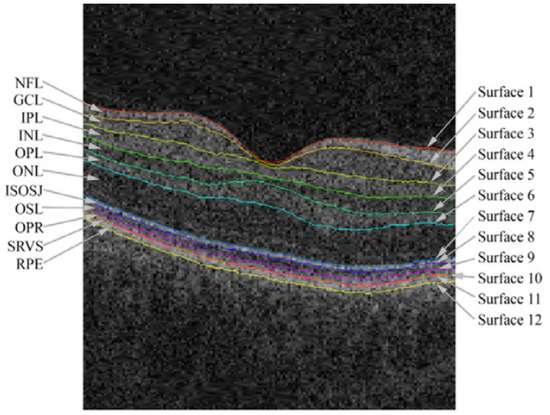
Figure 1.
Retinal layer boundaries annotated from an OCT image from the macular region of the retina, taken from [26].
As an early indicator of glaucoma, changes in the retinal layers, specifically in the NFL, can be measured as early as 6 years before any detectable vision loss symptoms, indicating damage to the optic nerve head [27]. In addition, asymmetry of the retinal layers can be an indicator of glaucoma, especially GCL, which is a promising early indicator for glaucoma [28].
2. Background Concepts
Deep learning enables computational models to learn data representations at various levels of abstraction. These techniques have significantly advanced the performance in areas such as natural language processing, computer vision, and many other fields. By using the backpropagation algorithm, deep learning uncovers complex patterns in large datasets. Deep convolutional networks have led to major advancements in processing different data types, such as visual data (images, video) and audio [29].
2.1. Convolutional Neural Networks
CNNs are a specialized type of neural network designed to process data with a grid-like structure, such as images or time-series data, by taking advantage of the relationships between nearby data points. Unlike fully connected neural networks, where every unit is connected to all units in the next layer, CNNs utilize a local connectivity structure that reflects the spatial arrangement of input data. This is achieved through the convolution operation (Figure 2), which replaces the general matrix multiplication used in traditional neural networks. Convolution enables CNNs to capture local patterns and features, such as edges and textures, by performing dot-product operations between grid-structured weights and input data [30,31,32]. Initially inspired by the biological visual cortex, CNNs have proven highly effective in tasks like image classification, achieving groundbreaking results in competitions like ImageNet [33]. Their hierarchical structure allows for the extraction of increasingly abstract features at deeper layers, making CNNs one of the foundational architectures in modern deep learning.
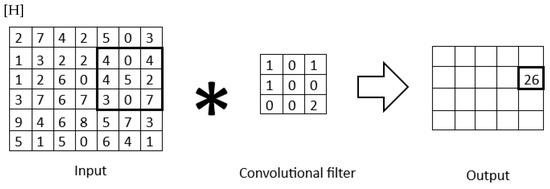
Figure 2.
Convolutional filter applied to a 2D matrix.
U-Net
U-Net is a convolutional neural network architecture designed for biomedical image segmentation. Introduced by Ronneberger et al. in 2015 [34], it is widely used in medical imaging applications due to its ability to produce accurate segmentations with limited annotated data. The architecture features a symmetric encoder-decoder structure, as shown in Figure 3, where the encoder captures high-level contextual information, and the decoder reconstructs spatial details to generate accurate segmentation maps. Skip connections between corresponding layers in the encoder and decoder help preserve detailed information features. U-Net has shown strong performance in various applications, including retinal layer segmentation in OCT images, through a variety of architecture adaptations.
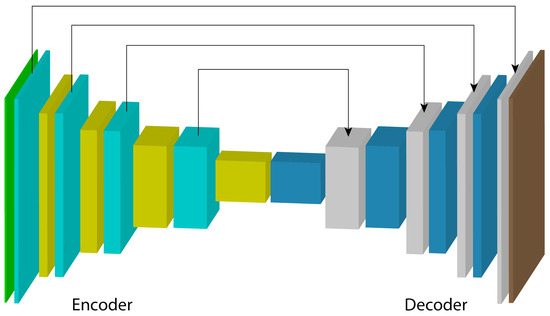
Figure 3.
Encoder–decoder-based U-Net architecture.
2.2. Generative Adversarial Networks
GANs are architectures composed of two models: a generator G and a discriminator D, with parameters and , respectively. G learns a mapping from a random noise vector z to an output map (image) x, represented by , while the task of D is to distinguish between examples from the dataset and examples generated by G, denoted as [35].
The model’s function is represented by a min-max game, where D is trained to maximize the probability of correctly labeling real (from the training set) and fake (generated by G) examples, while G tries to minimize this by generating examples that are indistinguishable from the real ones, as shown in Equation (1):
The goal of the model is for the discriminator to learn the properties of the training dataset, considering these as the real examples, while treating those generated by the generator as fake. Based on the discriminator’s classification, the generator is forced to produce better results that are increasingly closer to being indistinguishable from the training set.
The structure of a generative adversarial network is shown in Figure 4. A noise map is introduced into the generator model G, which produces an output example with the characteristics of the data distribution it was trained on. Regardless of how closely it resembles a real example from the dataset, it is assigned the label fake to train the discriminator model D with these examples. By complementing the training with the real dataset, D becomes capable of extracting features that belong to and do not belong to the data distribution of the training set.
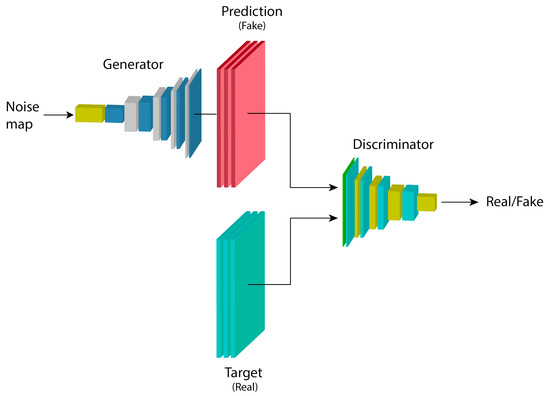
Figure 4.
Basic architecture of a generative adversarial network.
2.3. Conditional Generative Adversarial Networks
cGANs, such as the image-to-image translation proposed by [36], add the property to GANs of having an input example alongside the noise map z as extra information, in order to condition the model to generate examples based on this input. This is highly useful for a variety of applications, such as image segmentation, where the model’s output map is relatively similar to the input, and thus an entirely different image from the input is not necessary. The objective function for these models is as shown in Equation (2).
Figure 5 shows the architecture of a conditional generative adversarial network. The previously described maps are fed into the generator model G, which makes a prediction to obtain an output map . The maps generated by G’s inference, given the parameters , are used to train the parameters of the discriminator model, with an output label equal to 0 since they belong to the “fake” example set. In the opposite case, where the output label equals 1, the D model is trained directly with the “real” data distribution set. The loss obtained in D from this training process with both sets is useful for updating the parameters and , as previously shown in the equation describing the conditional model.
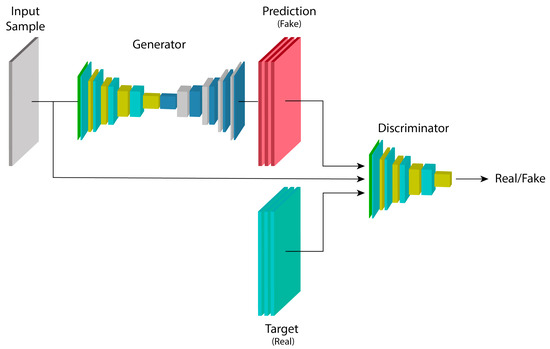
Figure 5.
Architecture of a conditional generative adversarial network.
2.4. Transformers
Based on the recurrent architecture, transformers are proposed in [37], where its architecture is based on attention blocks, entirely removing recurrence and convolutions from the architecture itself. This model is based on two submodules: an encoder and a decoder.
The encoder maps an input sequence of symbolic representations to a continuous representation , which is fed into the decoder. The decoder generates an output sequence of symbols one at a time, with each output being fed back into the model to generate the next output. This architecture is shown in Figure 6.
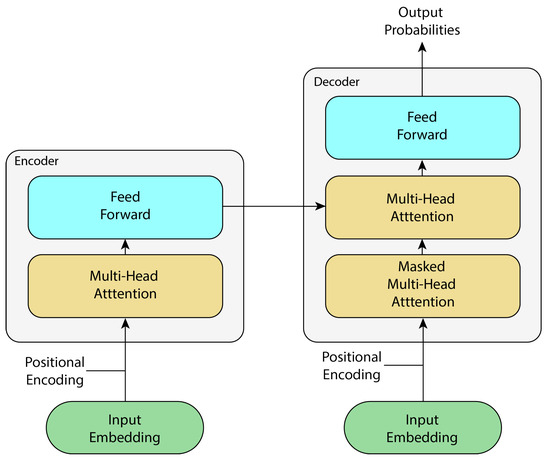
Figure 6.
Simplified transformer architecture, adapted from [37].
While originally designed for sequence-to-sequence tasks in natural language processing, the direct application of transformers to computer vision (e.g., vision transformer or ViT) faced challenges, particularly the quadratic computational cost of global self-attention with high-resolution images [38]. To overcome this, several efficient variants have been developed that are highly relevant for dense prediction tasks like segmentation. Swin transformer, for example, introduces a hierarchical structure and computes self-attention within local, non-overlapping windows that are shifted across layers to facilitate cross-window connections, where approach achieves linear complexity relative to image size [39]. Similarly, the CSWin transformer [40] employs a cross-shaped window self-attention mechanism to capture a wider receptive field more efficiently by processing horizontal and vertical stripes in parallel. Furthermore, hybrid CNN–transformer models have emerged as a powerful paradigm. These architectures typically use a convolutional backbone to extract rich, low-level feature maps and then employ a transformer encoder to model long-range spatial dependencies, effectively combining the local feature extraction strengths of CNNs with the global context modeling capabilities of transformers.
Attention Block
Attention is described in [37] as a mapping of a query Q and a set of keys K and values to an output V, where the queries, keys, and values are all vectors. The attention block, known as “Scaled Dot-Product Attention” (Figure 7), consists of queries and keys with dimension , and values with dimension . Then, using the softmax activation function, attention is calculated as follows:
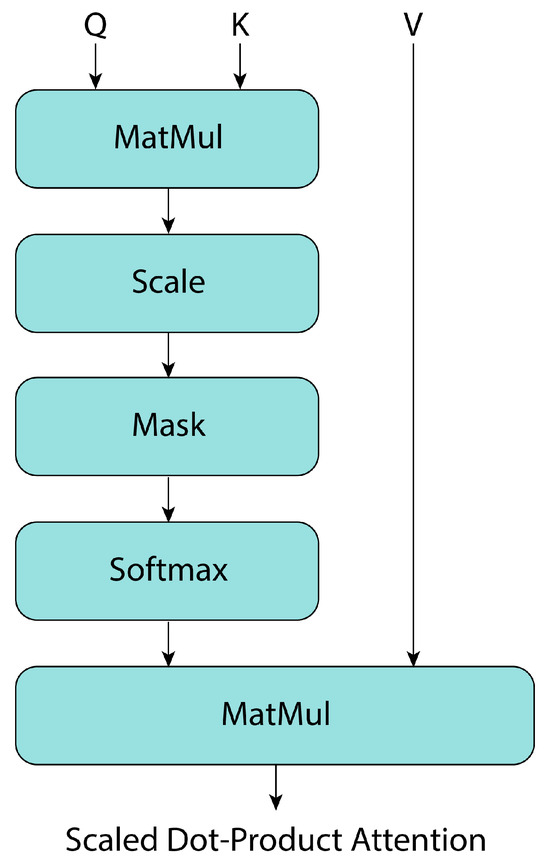
Figure 7.
Transformer’s attention block, adapted from [37].
From this, multi-head attention blocks are a generalization of “Scaled Dot-Product Attention” (Figure 8), where multiple attention blocks are stacked, and their outputs are concatenated. This allows the architecture to process multiple input vectors in parallel, providing more context information at each propagation step of the feature maps. The mathematical representation of these blocks is as follows, where W denotes the model weights [37]:
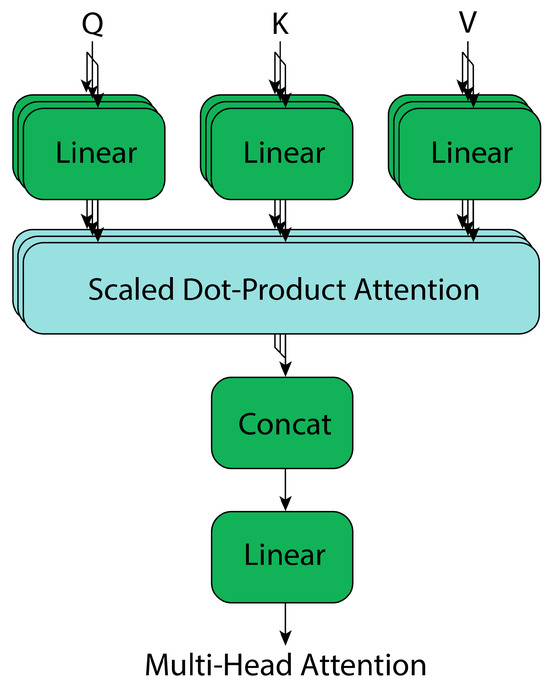
Figure 8.
Transformer’s multi-head attention, adapted from [37].
In the transformer architecture [37], masking plays a critical role in the attention mechanisms. The encoder’s self-attention primarily makes use of a padding mask to discard irrelevant padded tokens, ensuring focus on actual sequence content. This is achieved by setting the logits corresponding to padded positions to −∞ before the softmax function, effectively making their attention weights zero. The decoder’s masked multi-head attention similarly uses a causal mask, which prevents attention to subsequent tokens during generation, and therefore, maintaining the auto-regressive property essential for tasks like machine translation. For this, future positions in the sequence are also set to −∞ in the attention logits, ensuring they receive no attention.
3. Materials and Methods
In this review, the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) methodology [41] is used to ensure a comprehensive and systematic evaluation of the literature on deep learning techniques for retinal layer segmentation in OCT images, as shown in Figure 9. The PRISMA framework provides a structured approach for identifying, selecting, and synthesizing relevant studies.
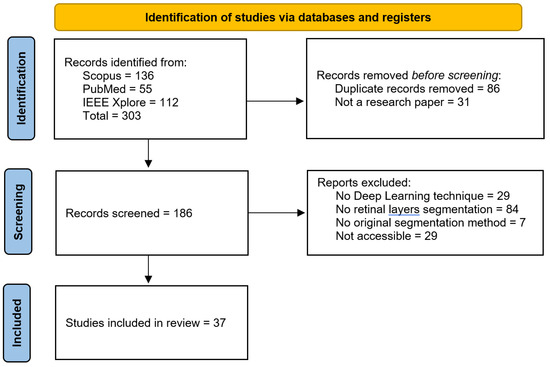
Figure 9.
PRISMA methodology followed to identify key state-of-the-art methods to analyze in this review.
3.1. Eligibility Criteria
The scope in the scientific articles to search considers the different terminologies of the techniques of interest, the search was defined for the following keywords that cover deep learning techniques for the segmentation of the retinal layers applied to the diagnosis of the three key eye diseases in this review:
- Optical Coherence Tomography OCT-based segmentation;
- Retinal layer(s);
- Segmentation/Detection;
- Deep learning/Neural network(s)/Machine learning/Artificial intelligence;
- Glaucoma/Diabetic retinopathy/Macular degeneration.
The articles retrieved using the specified keywords from Scopus, PubMed, and IEEE Xplore span from 2014 to October 2024, the date on which the search was conducted, as shown in Figure 10.
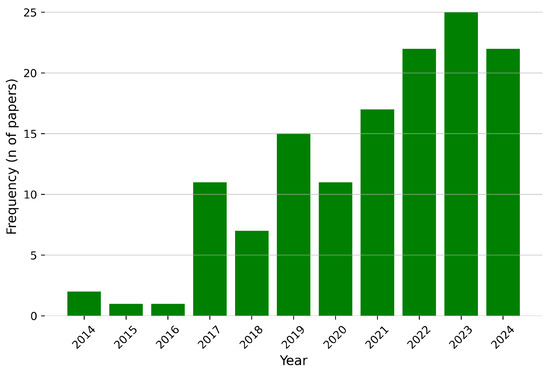
Figure 10.
Publication year distribution for the identified articles for this review.
3.2. Study Selection
A total of 303 articles were identified, of which 86 were duplicates and 31 were not original research articles (e.g., databases or reviews). From the remaining 186 articles, 119 were excluded based on three filters. The first filter required the use of deep learning-based image processing techniques. The second filter focused on the area of interest for segmentation or detection; many articles referenced the retinal layers but aimed to identify fluid or abnormalities rather than the retinal layers themselves. The third filter involved a full-text review to ensure that the proposed segmentation method was both original and developed by the authors. After applying these filters, 66 articles were selected from which 37 articles were accessible by the authors and included in this review (Figure 9).
3.3. Data Extraction
The information extracted from the included articles was divided into four main categories: data acquisition and processing, segmentation method, detected retinal layers, and model performance, as shown in Figure 11 and reviewed in the following sections of this review.
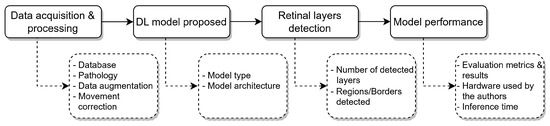
Figure 11.
Overview of the data extraction framework used in this review. The selected articles were analyzed across four main categories: data acquisition and processing, segmentation method, detected retinal layers, and model performance. This structure facilitated a consistent and comparative evaluation of DL techniques applied to retinal layer segmentation in OCT images.
Data acquisition and processing. Aiming to understand the data distribution, the database used in each article is reviewed to point out relevant factors, such as the dataset availability, distribution of the population (healthy individuals and patients with ocular diseases), data augmentation/preprocessing and if any technique for movement correction was implemented prior to the retinal layer segmentation process.
Segmentation method. Training strategy for the proposed model (supervised, semi-supervised learning) and its architecture, to understand the trends and effectiveness of the deep learning models used for the retinal layer segmentation.
Retinal layer detection. To compare the scope of each one of the included articles, comparing the number of retinal layers detected, which might be conditioned by the annotated data available, and which ones are being segmented, with the goal of supporting the medical diagnosis.
Model performance. Parameters such as evaluation metrics for the segmentation or detection of the retinal layers are retrieved, together with the information about the hardware requirements for the training of the proposed models and the inference time for the OCT images or volumes.
3.4. Risk-of-Bias Assessment
To ensure the reliability and validity of the selected studies, a risk-of-bias assessment was conducted. The evaluation focused on key aspects that may influence the performance and reported outcomes of deep learning models for retinal layer segmentation. These include the quality and availability of annotated data, transparency in methodology, reproducibility of results, and completeness of evaluation metrics.
Studies that lacked detailed descriptions of dataset characteristics, annotation procedures, or preprocessing steps were considered to have a higher risk of bias. In addition, articles relying on private datasets with no access or insufficient annotation transparency were scrutinized more critically. Similarly, models evaluated solely on a limited number of metrics, or without external validation, were flagged for potential overfitting or inflated performance estimates.
To mitigate the impact of such biases, only studies with clearly stated segmentation objectives, well-documented datasets (preferably public or semi-public), and the use of standardized performance metrics (e.g., Dice score, MAE, RMSE) were prioritized in the analysis. This approach helps ensure a balanced overview of the current landscape and facilitates meaningful comparisons across the reviewed models.
4. Results
This section synthesizes the findings from the selected studies, organized into four key categories from the data extraction process: data acquisition and preprocessing, deep learning segmentation methods, retinal layers detected, and model performance evaluation. The goal is to identify trends, assess methodological strengths, and highlight areas for improvement. The analysis begins with an overview of the datasets used, considering accessibility, demographic diversity, and labeling quality. It then examines the deep learning architectures applied, including CNNs, U-Net variants, and generative or attention-based models. Next, it reviews the scope and specificity of the segmented retinal layers. Finally, it discusses the evaluation metrics used to quantify model performance, emphasizing their clinical and technical relevance.
4.1. Data Acquisition and Processing
The foundation of any deep learning approach relies on the quality and characteristics of the data used for training and evaluation. In the context of retinal layer segmentation, access to high-resolution, accurately labeled OCT images is essential. This section reviews the datasets utilized in the selected studies, their acquisition parameters, and the preprocessing techniques applied to enhance model performance and generalizability.
OCT Databases
A critical step in image analysis is the acquisition of a dataset that includes the features needed for the study, such as the image modality in the medical area, the focus in the tissues of interest, a well-calibrated scanner, and a demography for the subjects to study, to name a few.
Acquiring the data is complicated due to a variety of reasons, including the resource allocation (budget, specialists, subjects), but most importantly, a correct data labeling is crucial to train any supervised deep learning-based model. Labeling data is very time-consuming and requires an expert in the area to properly perform such a task.
Access to medical equipment might also be a challenge, as top hospitals and universities might have access to all the aforementioned resources, but it might become a major obstacle for most researchers around the world.
To overcome this issue and build the foundation blocks for the scientific community, several research centers have made their data freely available for everyone to train and test their models. For the articles included in this review, the publicly available datasets for the detection of retinal layers in OCT images were identified, and their information is comprised in Table 1, comparing the data acquisition variables (OCT scanner brand, medical conditions and demography of subjects) and the properties of the datasets (number of samples, OCT image resolution, retinal layers labeled), being the following:
- AFIO database [42]. The OCT and fundus images database obtained from the Armed Forces Institute of Ophthalmology (AFIO) contains ONH-centered OCT and fundus images for glaucoma detection. It includes images from healthy and glaucomatous subjects, and manual annotations for the RPE and ILM retinal layers, along with the CDR values annotated by glaucoma specialists.
- U. of Miami [43]. The OCT data from the University of Miami was obtained at the Bascom Palmer Eye Institute, which contains OCT images from subjects with diabetes. The database contains the annotations for 11 retinal boundaries (10 layers): PRS-NFL, NFL-GCL, GCL-IPL, IPL-INL, INL-OPL, OPL-HFLONL, HFLONL-ELMMYZ, ELMMYZ-ELZOS, ELZOS-IDZ, IDZ-RPE, RPE-CRC.
- Duke University-AMD [44]. The data was collected in four clinics: Devers Eye Institute, Duke Eye Center, Emory Eye Center, and National Eye Institute, containing OCT images from healthy subjects and others with intermediate and advanced AMD, aged between 50 and 85 years old. The annotations made by the authors correspond to the ILM, inner RPEDC, and outer Bruch’s membrane.
- OCT MS and Healthy Controls Data [45]. The dataset collected in the Johns Hopkins Hospital contains OCT images including healthy subjects and patients with multiple sclerosis. The dataset includes manual delineations for the following retinal layers: RNFL, GCL+IPL, INL, OPL, ONL, IS, OS, RPE.
- Duke University-DME [46]. The OCT images were acquired from patients identified in the Duke Eye Center Medical Retina with DME, for a posterior manual segmentation to segment fluid and eight retinal boundaries that result in seven regions: NFL, GCL-IPL, INL, OPL, ONL-ISM, ISE, OS-RPE.
- AROI [47]. The Annotated Retinal Optical Coherence Tomography Images (AROI) database collected at the University Hospital Center, Croatia, contains OCT images from 60-year-old subjects and older diagnosed with nAMD. The annotated retinal layers in the data include ILM, IPL-INL, RPE, and BM, along with three fluid classes.
- OCTID [48]. The Optical Coherence Tomography Image Database (OCTID) contains fovea-centered images from healthy subjects and with different diseases, such as AMD, CSR, DR, and MH, collected at Sankara Nethralaya (SN) Eye Hospital, India. The authors of the dataset include also a GUI to perform/refine the annotations for the samples. Additionally, He et al. [49] performed the layers labeling for the NFL, GCL + IPL, INL, OPL, ONL, ELM + IS, OS, and RPE retinal layers, available upon request to the authors.
- IOVS [50]. The Investigative Ophthalmology and Visual Science database includes OCT images from subjects with AMD, collected in 4 different clinics (Devers Eye Institute, Duke Eye Center, Emory Eye Center, and the National Eye Institute), and includes annotations for the ILM, RPE + drusen complex, and Bruch’s membrane structures.
4.2. Deep Learning Methods for Retinal Layers Detection
The challenge of retinal layer segmentation has been addressed using a range of DL techniques, from foundational convolutional networks to more specialized architectures. This section explores these key methodologies.
4.2.1. Convolutional Neural Networks
CNNs have proven to be a powerful tool for segmenting retinal layers in OCT images, as they can learn complex features directly from training data. Unlike traditional methods, which depend on handcrafted, application-specific transformations, such as cost functions, constraints, and model parameters, deep learning approaches automatically learn both the features and the model from the data itself. Traditional methods often rely on expert-designed, application-specific transforms, including cost functions, constraints and model parameters, whereas deep learning-based methods are able to directly learn the model and features from training data [51,52]. However, these methods are often dependent on specific rules and may perform poorly with artifacts. Classic machine learning algorithms, such as support vector machines, have also been used [52]. In recent years, CNN-based methods have been extensively studied, showcasing substantial advancements in both accuracy and robustness through a series of techniques, such as the following.
Direct Segmentation: Some methods use CNNs to directly segment retinal surfaces or layers. For instance, a CNN can be trained to simultaneously segment multiple retinal surfaces in one pass. These methods can classify each voxel as belonging to a certain class. Some CNNs are designed to learn local surface profiles for both normal and diseased images [51,52].

Table 1.
Databases for OCT retinal layer segmentation.
Table 1.
Databases for OCT retinal layer segmentation.
| Database | Layers Labeled | Number of Subjects | Demography | Subjects Conditions | Scanner | Number of Samples | Sample Resolution (pixels) | Voxel Resolution (μm/pixel) |
|---|---|---|---|---|---|---|---|---|
| AFIO [42] | ILM, RPE | 26 | Male and female. Different age groups | 32 glaucomatous, 18 healthy | TOPCON 3D OCT-1000 (Topcon Corporation, Tokio, Japan) | 50 images | 951 × 456 | - |
| U. of Miami [43] | PRS, NFL, GCL, IPL, INL, OPL, HFLONL, ELMMYZ, ELZOS, IDZ, RPE, CRC | 10 | 6 male and 4 female aged 53 ± 6 years old | All diabetic subjects | Spectralis SD-OCT (Heidelberg Engineering, Heidelberg, Germany) | 10 volumes | 768 × 61 × 496 | 11.11 × 3.87 |
| Duke University-AMD [44] | ILM, RPEDC, Outer Bruch’s membrane | 384 | Normal subjects aged 51 to 83 years old. Subjects with AMD aged 51 to 87 years old | 269 intermediate AMD, 115 healthy | SD-OCT imaging systems from Bioptigen, Inc. Research Triangle Park, NC, USA | 38,400 images | 100 × 1000 (resampled to 1001 × 1001) | 6.70 × 3.24 |
| OCT MS and Healthy Controls Data [45] | RNFL, GCL + IPL, INL, OPL, ONL, IS, OS, RPE | 35 | 6 male and 29 female aged 39.49 mean, 10.94 SD years old | 21 multiple sclerosis, 14 healthy | Spectralis OCT system (Heidelberg Engineering, Heidelberg, Germany) | 35 volumes | 49 × 496 × 1024 | 5.8 (±0.2) × 3.9 (±0.0) × 123.6 (±3.6) |
| Duke University-DME [46] | NFL, GCL-IPL, INL, OPL, ONL-ISM, ISE, OS-RPE, BM | 10 | - | All subjects with DME | Spectralis (Heidelberg Engineering, Heidelberg, Germany) | 10 volumes | 496 × 768 × 61 | 3.87 × 11.07–11.59 × 118–128 |
| AROI [47] | ILM, IPL-INL, RPE, BM | 24 | Subjects aged 60 and older | All subjects with nAMD | Zeiss Cirrus HD OCT 4000 device (Zeiss, Oberkochen, Germany) | 3072 images (1136 labelled) | 1024 × 512 | 1.96 × 11.74 × 47.24 |
| OCTID [48] | NFL, GCL + IPL, INL, OPL, ONL, ELM + IS, OS, RPE | - | - | 102 MH, 55 AMD, CSR, 107 DR, 206 healthy | Cirrus HD-OCT machine (US Ophthalmic, Miami, US) | 470 images (25 labelled) | 500 × 750 | 5 × 15 |
| IOVS [50] | ILM, RPEDC, Bruch’s membrane | 20 | - | All subjects with AMD | SD-OCT imaging systems from Bioptigen, Inc. (Research Triangle Park, NC, USA) | 25 volumes | 1000 × 512 × 100 | 3.06–3.24 × 6.50–6.60 × 65.0–69.8 |
Cascaded CNNs: A cascaded CNN architecture can use intermediate tasks to contribute to the final segmentation by incorporating shape-related information. For instance, FourierNet employs a cascaded fully convolutional network (FCN) design, where the intermediate task of estimating Fourier descriptors contributes to improving the final segmentation [53].
Multi-Scale CNNs: Some CNN architectures utilize multi-scale features to obtain more abundant contextual information. For example, a multi-scale convolutional neural network architecture can automatically extract multi-scale features using multiple patch sizes. These networks can improve the identification of samples on both clear and fuzzy boundaries [54].
Attention Mechanisms: Attention mechanisms can be used to improve segmentation accuracy. For example, spatial and channel-attention gates can be leveraged at multiple scales for fine-grained segmentation. A depth-efficient attention module can also be incorporated to improve feature utilization [49,55].
Joint Segmentation and Classification: Some models perform joint segmentation and classification. For example, a model can simultaneously predict retinal layers and boundaries and then use this information for eye diseases diagnosis [56].
Fully Convolutional Networks: FCNs can be used to directly predict pixel-wise labels of retinal layers. They are efficient because they can process entire images, rather than patches. Some models fuse information from retinal boundaries with the OCT image to facilitate layer segmentation [56,57].
4.2.2. U-Net-Based Architectures
The U-Net architecture and its variants have become a cornerstone in the field of medical image segmentation, particularly for retinal layer segmentation in OCT images. These networks are favored for their ability to learn complex spatial features and provide accurate pixel-wise segmentations [58,59,60,61]. At its base, the U-Net consists of a contracting (encoder) path and an expansive (decoder) path. The encoder extracts features from the input image through convolutional and pooling layers, whereas the decoder reconstructs the segmentation by upsampling feature maps from the encoder and applying convolutional layers. Skip connections between corresponding layers of the encoder and decoder are crucial for preserving spatial information that might be lost during downsampling [60,61,62,63].
4.2.3. U-Net Modifications for Retinal Layer Segmentation
Several modifications and enhancements to the basic U-Net architecture have been proposed to address specific challenges in retinal OCT segmentation. These include the following.
Two-Dimensional U-Net for Direct Segmentation: Many studies employ a 2D U-Net to directly segment retinal layers from individual B-scans [60,61,64]. These approaches treat layer segmentation as a pixel-wise classification problem, assigning each pixel to a specific retinal layer, background, or fluid region [59,60,64,65,66].
U-Net with Pre-trained Encoders: The pre-trained encoders, such as ResNet, within the U-Net architecture has been useful to leverage the knowledge gained from large image datasets such as ImageNet. Transfer learning can lead to improved segmentation performance, especially when training data for retinal layers is limited [60,61].
Multi-Task Learning: Some approaches incorporate multi-task learning, using a single U-Net to perform different but related tasks like layer segmentation and fluid detection. For example, some networks jointly segment retinal layers and detect retinal fluids such as IRF, SRF and PED [59,61,64,66,67].
Three-Dimensional U-Net for Volumetric Segmentation: To incorporate the spatial information between B-scans, some studies have utilized 3D U-Nets, allowing the network to learn features from the entire OCT volume, improving segmentation accuracy and spatial coherence [63,67,68]. Some approaches use blocks of adjacent B-scans as input [63,67].
Shape-Based Regression: Instead of direct pixel classification, some methods employ shape-based regression, where U-Nets are trained to predict signed distance maps (SDMs) representing the distance to the next object contour. This approach preserves topology and can be used to produce smooth, topologically correct segmentations [60,62].
Attention Mechanisms: To improve segmentation in regions with blurry boundaries or complex pathologies, some studies incorporate attention mechanisms into the U-Net architecture. This allows the network to focus on the most relevant features for accurate segmentation [66].
Cascaded U-Nets: Some approaches use cascaded U-Nets, in which the first U-Net identifies the region of interest (e.g., the retina), and a second U-Net refines the segmentation of the retinal layers within that region. This can improve segmentation accuracy, especially in cases of severe pathologies [60,63].
U-Net with Dynamic Programming: The integration of the U-Net with a constrained differentiable dynamic programming module to explicitly enforce surface smoothness, utilizing feedback from the downstream model optimization module to guide feature learning [68].
Loss Functions: Different loss functions have been explored to train the U-Net models. Common options include:
- Dice Loss: Based on the Dice coefficient, this loss is used for mask segmentation, maximizing the overlap between the predicted and ground truth masks. It is particularly useful in cases of class imbalance [58,59,60,61]. The Dice loss is defined as shown in Equation (5), where represents the predicted value for pixel i, is the corresponding ground truth value, and N is the total number of pixels in the mask.
- Mean Squared Error (MSE): This loss function is used in shape regression methods to predict signed distance maps (SDMs), representing the distance to object boundaries. It is also used to directly predict the positions of retinal layer surfaces [60,62,63,64]. The MSE Loss is mathematically defined as shown in Equation (6), where and denote the ground truth and predicted values for pixel i, respectively, and N is the total number of pixels.
- Cross-Entropy Loss: This loss is used for pixel-wise classification, where each pixel is assigned to a layer, background, or fluid region. It calculates the difference between the predicted and actual probability distribution. Some studies modify or combine it with other functions [58,61,62,66]. For a binary classification problem, the cross-entropy loss is expressed as shown in Equation (7), where is the ground truth label for pixel i, is the predicted probability for the positive class, and N is the total number of pixels.
4.3. Graph-Based Methods
Traditional methods for retinal layer segmentation have utilized graph theory to formulate the segmentation problem as finding an optimal path or surface within a graph, where nodes represent pixels or voxels and edges represent the relationships between them [69,70,71,72].
Graph search algorithms use graph theory to identify optimal paths for segmentation, often incorporating shape and context prior knowledge to penalize deviations and surface changes. Graph-based multi-surface segmentation methods utilize trained hard and soft constraints on various retinal layers to construct segmentation graphs [69], while shortest path algorithms determine the segmentation by identifying the shortest path through the graph. To enhance accuracy, these shortest path approaches frequently integrate graph edge weights with probability maps generated by deep learning networks [73].
Graph convolutional networks (GCNs) are used to leverage structural information in retinal layers. GCNs can be incorporated into U-shape networks to conduct spatial reasoning. This approach is especially useful for the peripapillary region where the optic disc is present [72,74]. Different methods combine the strengths of both graph theory and deep learning for improved retinal layer segmentation.
CNN-GS: These methods use a CNN to extract features and generate probability maps for layer boundaries, followed by a graph search to find the final boundaries [70,75].
GCN-assisted U-shape networks: Graph reasoning blocks are inserted between the encoder and decoder of a U-shape network for spatial reasoning and segmentation of retinal layers, which captures long-range contextual information [72,74].
Motion Correction with 3D Segmentation: Neural networks are used to combine motion correction with 3D segmentation. Motion correction is performed prior to a graph-assisted 3D neural network which utilizes 3D convolution and a novel graph pyramid structure [76].
4.4. Generative Models
Traditional methods for retinal layer segmentation are often time-consuming and vulnerable to noise and distortions in OCT images. Recently, CNNs have shown great promise in automatically segmenting retinal layers from OCT images. However, these methods often rely on large amounts of annotated data, which can be difficult and resource-intensive to obtain. Semi-supervised and generative models offer an alternative by leveraging unlabeled data to enhance segmentation performance.
The method proposed by [77] used a Pix2Pix GAN-based approach for segmenting peripapillary retinal layers, utilizing a U-Net generator and a PatchGAN discriminator. To address limited labeled data, a semi-supervised strategy was employed: the GAN was first trained on labeled data, then fine-tuned using a mix of labeled and pseudo-labeled images, with only the generator’s weights being updated for unlabeled data.
Another modern technique for image processing is the transformer architecture, where in [78] a retinal layer segmentation network based on the CSWin transformer architecture was proposed. The authors combine convolution and attention mechanisms for improved segmentation accuracy. The network features a U-shaped structure with an encoder that converts images into token sequences processed by CSWin transformer blocks, and a decoder that upsamples features using patch expanding layers and convolution. To enhance edge detection, a BADice loss function was proposed, focusing on boundary areas and combined with the Dice loss for training.
4.5. Retinal Layers Detection
The detection of retinal layers using deep learning (DL) models is strongly influenced by the datasets employed for training, particularly the availability and quality of annotations defining the layer boundaries as ground truth data. The focus on specific pathologies often dictates which retinal layers are prioritized for analysis by physicians, aligning with their role in supporting medical diagnosis. This clinical emphasis shapes current research trends, highlighting the most relevant layers for detection and segmentation.
In the analysis conducted for this review, the retinal regions segmented by the different approaches show that the most frequently reported retinal layers are the RPE, RNFL, OPL, INL, and ONL (Figure 12). This is no coincidence, as the RPE layer is a biomarker for diabetic retinopathy diagnosis [79], whereas the RNFL is for glaucoma [80]. Different databases offer different limits or borders to the regions of interest, in which cases some retinal layers are combined to form a single region, or even primarily focusing on the total thickness of the retina, by providing delineation for the upper and lower layers and a few in between. A full specification of the regions of interest to segment by the analyzed publications can be found in Table 2, showing also the pathology present in the segmented OCT data, together with the metrics used to evaluate the segmentation performance, which will be discussed in the following section.
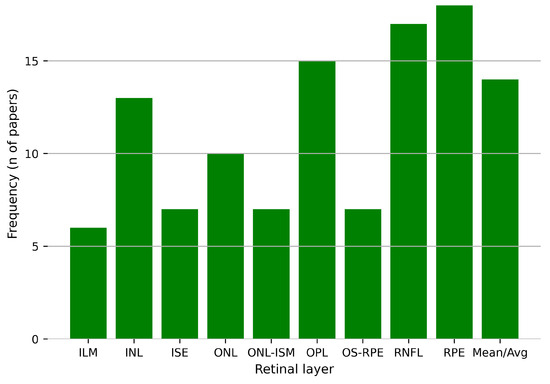
Figure 12.
Frequency of the retinal layers segmented by the models proposed in the selected studies for this review.

Table 2.
Databases, retinal layers detected, and metrics evaluated for each article.
4.5.1. Evaluation Metrics
To evaluate the performance of deep learning models for retinal layer segmentation, a variety of evaluation metrics are commonly used in the articles analyzed in this review. These metrics serve as standardized tools to quantitatively measure how well a model’s predictions align with the ground truth annotations made by experts, offering insights into different aspects of performance, including accuracy, precision, robustness, and boundary alignment across different retinal regions. Given the clinical significance of accurate retinal layer segmentation, these metrics not only evaluate the overall quality of the segmentation but also highlight specific strengths and weaknesses of the models. Furthermore, the choice of evaluation metrics often depends on the objective of the study, whereas the focus is on global accuracy, boundary precision, or the ability to handle pathological variations. The most frequently reported metrics in the literature included for this study are shown in Figure 13. Each of these metrics emphasizes different aspects of model performance, contributing to both the evaluation and optimization of training. Authors select various metrics to demonstrate the effectiveness and competitiveness of their models. The most commonly used metrics are described below to provide a clearer understanding of their definitions and how they reflect model behavior in image segmentation tasks.
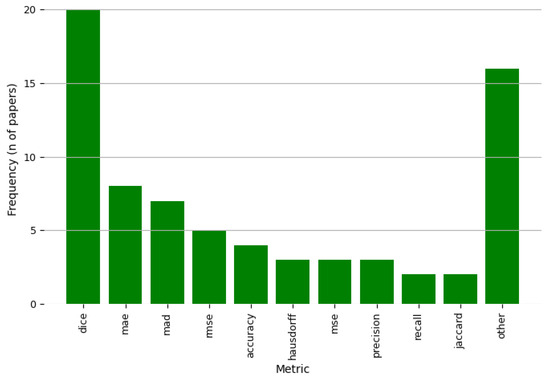
Figure 13.
Frequency of the metrics evaluated in the selected studies for this review.
Dice Similarity Coefficient (Dice Score): The Dice score (Equation (8)) is a widely used metric for measuring the overlap between two binary sets. It quantifies the similarity between the predicted and ground truth segmentation by computing the ratio of the intersection to the total number of elements. A higher Dice score indicates better agreement between the predicted and true layers.
Mean Absolute Error (MAE): MAE (Equation (9)) measures the average absolute difference between the predicted and actual values of the layer boundaries. This metric is useful for assessing the accuracy of boundary predictions, with lower values indicating better performance.
Median Absolute Deviation (MAD): MAD (Equation (10)) is a robust measure of the spread of prediction errors, calculated as the median of the absolute deviations from the median of the predictions. This metric is less sensitive to outliers compared to MAE and provides a more stable estimate of prediction variability.
Root Mean Squared Error (RMSE): RMSE (Equation (11)) calculates the square root of the average squared differences between predicted and true values. This metric penalizes larger errors more significantly than MAE and is useful for evaluating the overall model performance.
Accuracy: Accuracy (Equation (12)) measures the proportion of correctly predicted pixels in the image, with a higher value indicating that a larger portion of the image was accurately segmented. While commonly used, it may not be as informative for imbalanced datasets or small structural features, such as the thinnest retinal layers where the background composes the majority of the image area.
Hausdorff Distance: The Hausdorff distance (Equation (13)) measures the greatest distance between two sets of points, in this case, the predicted and ground truth layer boundaries. This metric is particularly useful for evaluating the spatial alignment of boundaries, with lower values indicating better boundary detection.
4.5.2. Data Preprocessing
Data augmentation plays a key role in training DL models, and the medical field is no exception. Data scarcity, particularly labeled data, remains a significant challenge due to the time-intensive process required from physicians for annotation. To mitigate this constraint, data augmentation techniques have been widely employed in the state-of-the-art approaches for retinal layer segmentation. These techniques artificially increase the diversity and size of datasets, enhancing model performance and generalization.
In the analyzed studies, various data augmentation techniques have been employed to enhance the diversity and robustness of the training datasets, as shown in Table 3. These techniques can be grouped into the following categories based on their primary transformation type.

Table 3.
Deep learning methods implemented for retinal layer segmentation in OCT images for the reviewed research articles.
Geometric Transformations:
- Rotation: Slightly rotates images to simulate variability in acquisition angles.
- Translation: Shifts images horizontally or vertically to reduce positional bias.
- Horizontal Flip: Mirrors images horizontally to increase sample diversity.
- Vertical Crop: Extracts specific vertical sections of the image to focus on relevant regions.
- Random Crop: Selects random sections of the image to enhance spatial variability.
- Random Shearing: Applies slight angular distortions to simulate realistic deformations.
Noise Injection:
- Gaussian Noise: Adds subtle pixel-level noise to simulate sensor variability and reduce overfitting.
- Salt-and-Pepper Noise: Introduces random white and black pixel noise to improve robustness against artifacts.
- Additive Blur: Slightly blurs the image to mimic imperfections during image acquisition.
Intensity and Contrast Adjustments:
- Contrast Adjustment: Modifies image contrast to account for lighting variability during acquisition.
These techniques are often applied individually or in combination, depending on the dataset characteristics and the specific properties intended for enhancement, aiming to improve generalization and segmentation performance.
Figure 14 shows the distribution of Dice score percentages for the segmentation of the most reported retinal layers using deep learning techniques. Each boxplot represents the reported Dice scores for a specific retinal layer, highlighting both the central tendency and the variability observed across the included studies. Please note that several articles publish their results testing their models in multiple databases, so this plot comprises all the results provided by the authors. Even though each database might differ from the others in terms of the characteristics of the collected OCT images, it is still a good reference to compare the individual segmentation performance for the proposed DL methods to offer a quick glance of the open challenges in such task.
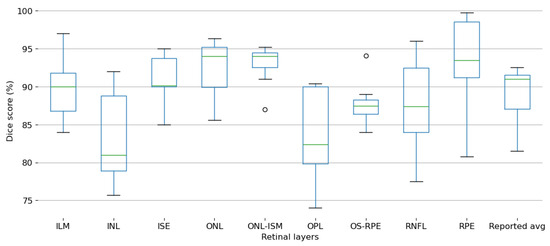
Figure 14.
Box and whisker plot summarizing the Dice Score values reported across all selected studies for retinal layer segmentation.
The ILM layer demonstrates relatively high Dice scores with moderate variability, suggesting consistent segmentation performance across studies. In contrast, the inner nuclear layer (INL) exhibits a broader range of Dice scores, indicating greater variability and potential challenges in achieving reliable segmentation accuracy.
ISE and ONL layers show consistently high Dice scores with minimal variability, reflecting robust segmentation performance. Similarly, the ONL-ISM layer maintains high accuracy, with only minor outliers observed.
The OPL displays a noticeable drop in Dice scores accompanied by significant variability, suggesting inherent difficulties in segmenting this layer accurately. The OS-RPE also exhibits moderate segmentation performance, with little variability across the reported results.
The RNFL shows substantial variability in Dice scores, with both high and low performances reported, highlighting inconsistencies in segmentation outcomes. In contrast, the retinal pigment epithelium (RPE) achieves the highest Dice scores among all layers, indicating excellent segmentation accuracy and consistency across studies. Both of these layers are the most common target for segmentation in the reported studies.
Lastly, the reported average boxplot aggregates the Dice scores across all retinal layers, offering an overall view of the segmentation performance reported in the analyzed literature.
These findings suggest that while certain retinal layers, such as RPE and ONL, are segmented with high precision and consistency, others, like the INL, OPL, and RNFL, remain challenging due to their structural complexity and varying imaging quality across datasets. This variability underscores the need for continued refinement of deep learning models tailored for retinal layer segmentation.
While Figure 13 illustrates the frequency with which various evaluation metrics are reported, and Figure 14 shows performance based on the Dice score, directly comparing methods remains challenging due to the heterogeneity of datasets and evaluation protocols. To enable a more standardized comparison, Table 4 summarizes the performance of the reviewed methods on the publicly available Duke University–DME dataset [46], using the Dice score, being the most commonly used dataset and metric combination in the reviewed studies. This consistent benchmark highlights the relative performance of different architectures and provides insight into the current state of the art in pathological retina segmentation.

Table 4.
Comparison of Dice scores for the reviewed methods evaluated on the Duke University-DME dataset [46].
4.6. Model Performance
The computational efficiency of DL models for retinal layer segmentation is crucial for their real-world applicability in clinical areas. Key aspects include the hardware used during training and inference, and the associated computational costs, such as inference time. Table 5 summarizes the hardware specifications and inference times reported in the analyzed studies, together with the model dimensionality, to identify whether a model takes images or volumetric data to infer the segmentation maps.

Table 5.
Deep learning methods implemented for retinal layer segmentation in OCT images for the reviewed research articles.
Most studies reported using high-performance GPUs for both training and inference, with the NVIDIA GeForce series (GTX and RTX models) being the most commonly employed. Advanced setups included multi-GPU configurations, such as the use of several NVIDIA GPUs, enabling parallel processing of larger datasets. In some cases, studies utilized integrated systems, such as the cluster from Jiao Tong University, demonstrating the scalability of segmentation tasks to larger datasets and distributed computing environments.
Inference time varied significantly depending on the model architecture, dataset size, and hardware configuration. Reported times ranged from as low as 0.04 s per image to more than 2 min per OCT volume.
The vast majority of the reviewed methods operate on a 2D, slice-by-slice basis. As shown in Table 4, these 2D methods typically report the fastest inference times, some achieving processing speeds of milliseconds per image (e.g., Shen et al. [69] at 0.04 s/image). This high speed makes them well-suited for applications where computational resources are limited. However, as noted in our analysis, this speed advantage comes with a significant compromise: by processing each B-scan independently, 2D methods may fail to enforce spatial coherence across the volume. This can result in anatomically implausible discontinuities between adjacent slices, limiting their utility for precise volumetric analysis.
In contrast, a smaller group of studies employs 3D models, where these methods process entire OCT volumes or sub-volumes at once, typically using 3D convolutions. Their primary advantage is the inherent preservation of spatial coherence, which results in smoother and more anatomically consistent segmentation surfaces. The trade-off, however, is a substantially higher computational cost, requiring more powerful GPUs with greater VRAM, which are more complex to implement. This computational demand can be a significant barrier to their integration into fast-paced clinical workflows.
The variability in reported hardware specifications and inference times highlights the trade-offs between computational efficiency and segmentation accuracy. Future research should aim to optimize DL architectures to achieve faster inference times without sacrificing segmentation quality, particularly for clinical environments with limited computational resources.
5. Discussion
Deep learning techniques have transformed and improved the approaches for retinal layer segmentation in OCT images, offering pathways for early diagnosis and monitoring of ocular diseases, critical to prevent their progression and improve people’s quality of life. This review highlights the remarkable progress made in algorithm development, dataset utilization, and clinical applicability. However, several challenges remain, requiring continued exploration to fully take advantage of these techniques in real-world clinical environments.
5.1. Strengths and Innovations
The reviewed studies demonstrate significant advancements in segmentation performance, with architectures such as U-Net and its variants (e.g., attention-enhanced U-Nets, multi-scale U-Nets) achieving high Dice coefficients and robust boundary delineation for critical retinal layers. These methods emphasize the adaptability of deep learning techniques in capturing the smallest and most complicated features of OCT images, challenging even for experts, which are essential for diagnosing diseases like AMD, glaucoma, and diabetic retinopathy.
Attention mechanisms and hybrid models incorporating both convolutional and transformer-based architectures have further enhanced the capacity of segmentation models. Specifically, the adoption of efficient vision transformers, such as the Swin or CSWin transformers, and the development of hybrid models that make use of a CNN’s feature extraction power with a transformer’s ability to model global context, have been proven critical for handling variability in image quality and pathological features. cGANs and semi-supervised approaches have proven particularly effective in addressing the scarcity of labeled data in the medical field given the complexity and the level of expertise needed for such task, enabling the generation of synthetic annotations that improve model generalization, which is critical to tackle the variability across different pathologies, patients, scanners, and even the conditions in which the data was acquired can impact the quality of the images.
5.2. Challenges and Limitations
Even though there are several advancements, the field faces several challenges. First, the variability in image acquisition protocols and dataset characteristics (e.g., resolution, patient demographics) makes training DL models more difficult. Many studies rely on private or institution-specific datasets, which limits cross-study comparisons and the applicability of models to diverse clinical populations. Publicly available datasets such as those from Duke University and AFIO have provided a foundation, but additional efforts are needed to standardize annotations and expand dataset diversity.
The demographic homogeneity of many training datasets raises ethical concerns related to bias and fairness. AI models developed using data from a narrow population, often lacking detailed information on race, ethnicity, or socioeconomic background, may show reduced accuracy for underrepresented groups. Variations in retinal morphology or disease patterns across populations can further amplify this effect, leading to the already existing health disparities. To mitigate these risks, future research must emphasize the development of diverse and representative datasets, enforce transparent reporting of population characteristics, and include fairness in validation protocols. Ensuring equitable model performance is critical to realizing the full potential of AI in improving healthcare for all.
Second, certain retinal layers, such as the OPL and RNFL, exhibit significant variability in segmentation accuracy across studies. The structural complexity of these layers, combined with their thinner profiles and susceptibility to noise, presents a formidable challenge for existing models. Furthermore, the use of evaluation metrics like Dice and MAE highlights the trade-offs between global segmentation accuracy and boundary precision, underscoring the need for task-specific optimization strategies.
Finally, the computational demands of deep learning models remain a concern for their widespread adoption in clinical settings. Although most studies utilize high-performance GPUs for training and inference, the scalability of these approaches for real-time clinical applications, especially in resource-limited environments, requires further investigation in model optimization, improving inference time without sacrificing accuracy, given the critical implications in the diagnosis support for specialists.
5.3. Clinical Relevance and Impact
Accurate segmentation of retinal layers holds immense clinical significance. The ability to detect early biomarkers, such as RNFL thinning in glaucoma or RPE abnormalities in diabetic retinopathy, enables timely interventions that prevent vision loss. Furthermore, automated segmentation tools can alleviate the burden on physicians by enhancing the analysis of high-volume OCT scans, improving both diagnostic efficiency and consistency.
The integration of these models into clinical workflows is promising, as evidenced by their capacity to simultaneously segment layers and detect pathological features (e.g., fluid accumulations, drusen deposits). However, it raises a critical and complex ethical question: who is responsible for diagnostic errors? The accountability for an error made by a Deep Learning model cannot be assigned to a single entity. In our view, it is a shared responsibility distributed among several key stakeholders. Model developers and researchers have the responsibility for building robust, transparent, and rigorously validated algorithms and models, while being explicit about their limitations and intended use cases. Regulatory entities are tasked with establishing stringent approval processes to ensure these tools meet high standards of safety and efficacy before clinical deployment. Healthcare institutions must implement clear usage protocols and ensure that clinical staff receive adequate training in how to interpret and apply AI-generated outputs.
Despite the huge recent advancement, these systems are intended to support—not replace—clinical decision-making. As such, final diagnostic and treatment decisions must remain in the hands of qualified clinicians, who are tasked with interpreting AI outputs in the context of patient history, clinical findings, and their own professional judgment. A well-defined legal and ethical framework that delineates these responsibilities is essential for ensuring the safe, fair, and effective integration of AI into ophthalmology and the broader medical practice.
5.4. Future Research Directions
To address the identified limitations and further enhance the clinical utility of deep learning-based segmentation models, future research should prioritize the following areas:
- Dataset expansion and standardization: Collaborative efforts to develop large, diverse, and publicly available datasets with standardized annotations will improve model training and evaluation. Beyond simple collection, this includes the use of generative models to create realistic synthetic OCT images. This is particularly valuable for augmenting datasets with rare pathologies or simulating variations from different scanner types, improving model robustness without compromising patient privacy. Furthermore, advanced augmentation techniques, such as elastic deformations and random nonlinear transformations, can create more challenging and realistic training scenarios.
- Improved generalization: Incorporating domain adaptation techniques and augmenting datasets with diverse pathological cases can improve the robustness of models across different populations and imaging systems. This can be achieved through unsupervised methods that learn to align feature distributions from different domains (e.g., scanners or hospitals) or by using data normalization strategies that standardize intensity profiles and reduce scanner-specific artifacts before model training.
- Hybrid architectures: Using the strengths of hybrid models that combine convolutional and attention-based mechanisms remains a promising direction. Future work could also explore vision-language models that integrate textual clinical reports with OCT images to provide contextual priors, potentially improving segmentation accuracy in ambiguous cases.
- Enhanced segmentation of challenging layers: To improve accuracy for complex layers like OPL and RNFL, research should move beyond standard loss functions. This involves exploring advanced, boundary-aware loss functions that heavily penalize errors at layer interfaces, or topology-aware losses that explicitly preserve the structural integrity and continuity of thin layers.
- Real-Time Implementation: Efforts to optimize model efficiency for deployment on edge devices or cloud-based platforms can facilitate their integration into clinical workflows, especially in under-resourced environments.
6. Conclusions
Deep learning models have revolutionized retinal layer segmentation, achieving unprecedented accuracy and enabling early detection of vision-threatening diseases. These advancements have enabled segmentation techniques to identify subtle changes in the retinal structure, providing critical insights into the early stages of conditions such as glaucoma and diabetic retinopathy. However, challenges such as dataset variability, computational demands, and layer-specific segmentation accuracy remain significant hurdles. Addressing these issues will require not only technical innovations but also interdisciplinary collaboration to ensure models meet the diverse needs of real-world clinical environments. Innovations in model design, including hybrid architectures and domain-specific optimizations, continue to strengthen the foundation for future breakthroughs. By overcoming these challenges, deep learning tools can fully transform ophthalmic diagnostics, improving both clinical workflows and patient outcomes globally.
Author Contributions
Conceptualization, O.J.Q.-Q. and S.T.-A.; methodology, O.J.Q.-Q.; software, O.J.Q.-Q.; validation, O.J.Q.-Q. and S.T.-A.; formal analysis, O.J.Q.-Q.; investigation, O.J.Q.-Q.; resources, S.T.-A.; data curation, O.J.Q.-Q.; writing—original draft preparation, O.J.Q.-Q.; writing—review and editing, S.T.-A., M.A.A.-F., J.C.P.-O., and G.A.-F.; visualization, O.J.Q.-Q.; supervision, S.T.-A., J.C.P.-O., and M.A.A.-F.; project administration, S.T.-A.; funding acquisition, S.T.-A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
No new data were created by the work presented in this study.
Acknowledgments
During the preparation of this study, the authors used ChatGPT and Gemini for the purposes of proofreading/editing. The authors have reviewed and edited the output and take full responsibility for the content of this publication.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AACG | Acute Angle-Closure Glaucoma |
| AMD | Age-related Macular Degeneration |
| ASSD | Average Symmetric Surface Distance |
| CDR | Cup to disc ration |
| CSR | Central Serous Retinopathy |
| cGAN | Conditional Generative Adversarial Networks |
| CNN | Convolutional Neural Network |
| GAN | Generative Adversarial Network |
| DL | Deep Learning |
| DR | Diabetic Retinopathy |
| FCN | Fully Convolutional Network |
| HD | Hausdorff Distance |
| IOU | Intersection over Union |
| IOP | Intraocular pressure |
| IRF | Intraretinal fluid |
| MH | Macular Hole |
| MAD | Mean Absolute Deviation |
| MAE | Mean Absolute Error |
| MASD | Mean Absolute Surface Distance |
| mIOU | Mean Intersection over Union |
| mPA | Mean Pixel Accuracy |
| MSE | Mean Squared Error |
| MUE | Mean Unsigned Error |
| NPDR | Nonproliferative Diabetic Retinopathy |
| OCT | Optical Coherence Tomography |
| ONH | Optical nerve head |
| PED | Pigment epithelial detachment |
| PDR | Proliferative Diabetic Retinopathy |
| POAG | Primary Open-Angle Glaucoma |
| RGC | Retinal Ganglion Cells |
| RMSE | Root Mean Square Error |
| SRF | Subretinal fluid |
| UASSD | Uniform Average Symmetric Surface Distance |
| UMSP | Uniform Mean Surface Position |
| UMSPE | Uniform Mean Surface Position Error |
| USPE | Uniform Surface Position Error |
| WHO | World Health Organization |
References
- Resnikoff, S.; Pascolini, D.; Etya’ale, D.; Kocur, I.; Pararajasegaram, R.; Pokharel, G.P.; Mariotti, S.P. Global data on visual impairment in the year 2002. Bull. World Health Organ. 2004, 82, 844–851. [Google Scholar]
- Shaarawy, T.M.; Sherwood, M.B.; Hitchings, R.A.; Crowston, J.G. Glaucoma, 2nd ed.; Elsevier Health Sciences: Amsterdam, The Netherlands, 2015. [Google Scholar] [CrossRef]
- Tham, Y.C.; Li, X.; Wong, T.Y.; Quigley, H.A.; Aung, T.; Cheng, C.Y. Global Prevalence of Glaucoma and Projections of Glaucoma Burden through 2040. Ophthalmology 2014, 121, 2081–2090. [Google Scholar] [CrossRef]
- Cifuentes-Canorea, P.; Ruiz-Medrano, J.; Gutierrez-Bonet, R.; Peña-Garcia, P.; Saenz-Frances, F.; Garcia-Feijoo, J.; Martinez-de-la Casa, J.M. Analysis of inner and outer retinal layers using spectral domain optical coherence tomography automated segmentation software in ocular hypertensive and glaucoma patients. PLoS ONE 2018, 13, e0196112. [Google Scholar] [CrossRef]
- Khaw, P.; Elkington, A. Glaucoma—1: Diagnosis. BMJ 2004, 328, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Weinreb, R.N.; Leung, C.K.; Crowston, J.G.; Medeiros, F.A.; Friedman, D.S.; Wiggs, J.L.; Martin, K.R. Primary open-angle glaucoma. Nat. Rev. Dis. Prim. 2016, 2, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.H.; Fingert, J.H.; Kuehn, M.H.; Alward, W.L. Primary open-angle glaucoma. N. Engl. J. Med. 2009, 360, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Weinreb, R.N.; Khaw, P.T. Primary open-angle glaucoma. Lancet 2004, 363, 1711–1720. [Google Scholar] [CrossRef]
- Choong, Y.F.; Irfan, S.; Menage, M.J. Acute angle closure glaucoma: An evaluation of a protocol for acute treatment. Eye 1999, 13, 613–616. [Google Scholar] [CrossRef]
- Lim, L.S.; Mitchell, P.; Seddon, J.M.; Holz, F.G.; Wong, T.Y. Age-related macular degeneration. Lancet 2012, 379, 1728–1738. [Google Scholar] [CrossRef]
- Fong, D.S.; Aiello, L.; Gardner, T.W.; King, G.L.; Blankenship, G.; Cavallerano, J.D.; Ferris, F.L.; Klein, R.; Association, A.D. Retinopathy in Diabetes. Diabetes Care 2004, 27, s84–s87. [Google Scholar] [CrossRef]
- Drexler, W.; Fujimoto, J.G. (Eds.) Optical Coherence Tomography: Technology and Applications, 2nd ed.; Springer International Publishing: Cham, Switzerland, 2015. [Google Scholar] [CrossRef]
- Huang, D.; Swanson, E.A.; Lin, C.P.; Schuman, J.S.; Stinson, W.G.; Chang, W.; Hee, M.R.; Flotte, T.; Gregory, K.; Puliafito, C.A.; et al. Optical Coherence Tomography. Science 1991, 254, 1178–1181. [Google Scholar] [CrossRef]
- Mashige, K.P.; Oduntan, O.A. A review of the human retina with emphasis on nerve fibre layer and macula thicknesses. Afr. Vis. Eye Health 2016, 75, 1–8. [Google Scholar] [CrossRef]
- Hildebrand, G.D.; Fielder, A.R. Anatomy and physiology of the retina. In Pediatric Retina; Springer: Berlin/Heidelberg, Germany, 2010; pp. 39–65. [Google Scholar]
- Nguyen, K.; Patel, B.; Tadi, P. Anatomy, Head and Neck: Eye Retina. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Masri, R.A.; Weltzien, F.; Purushothuman, S.; Lee, S.C.; Martin, P.R.; Grünert, U. Composition of the inner nuclear layer in human retina. Investig. Ophthalmol. Vis. Sci. 2021, 62, 22. [Google Scholar] [CrossRef]
- Kim, S.Y.; Park, C.H.; Moon, B.H.; Seabold, G.K. Murine Retina Outer Plexiform Layer Development and Transcriptome Analysis of Pre-Synapses in Photoreceptors. Life 2024, 14, 1103. [Google Scholar] [CrossRef]
- Mahabadi, N.; Al Khalili, Y. Neuroanatomy, Retina. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Lujan, B.J.; Roorda, A.; Croskrey, J.A.; Dubis, A.M.; Cooper, R.F.; Bayabo, J.K.; Duncan, J.L.; Antony, B.J.; Carroll, J. Directional optical coherence tomography provides accurate outer nuclear layer and Henle fiber layer measurements. Retina 2015, 35, 1511–1520. [Google Scholar] [CrossRef]
- Landa, G.; Gentile, R.; Garcia, P.; Muldoon, T.; Rosen, R. External limiting membrane and visual outcome in macular hole repair: Spectral domain OCT analysis. Eye 2012, 26, 61–69. [Google Scholar] [CrossRef]
- Saxena, S.; Akduman, L.; Meyer, C.H. External limiting membrane: Retinal structural barrier in diabetic macular edema. Int. J. Retin. Vitr. 2021, 7, 1–3. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, L.; Sohn, E.H.; Lee, K.; Niemeijer, M.; Chen, J.; Sonka, M.; Abramoff, M.D. Quantification of external limiting membrane disruption caused by diabetic macular edema from SD-OCT. Investig. Ophthalmol. Vis. Sci. 2012, 53, 8042–8048. [Google Scholar] [CrossRef] [PubMed]
- Narayan, D.S.; Chidlow, G.; Wood, J.P.; Casson, R.J. Glucose metabolism in mammalian photoreceptor inner and outer segments. Clin. Exp. Ophthalmol. 2017, 45, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhou, J.; Li, D. Functions and diseases of the retinal pigment epithelium. Front. Pharmacol. 2021, 12, 727870. [Google Scholar] [CrossRef] [PubMed]
- Sonka, M.; Abràmoff, M.D. Quantitative analysis of retinal OCT. Med. Image Anal. 2016, 33, 165–169. [Google Scholar] [CrossRef]
- Jeoung, J.W.; Park, K.H. Comparison of Cirrus OCT and Stratus OCT on the Ability to Detect Localized Retinal Nerve Fiber Layer Defects in Preperimetric Glaucoma. Investig. Opthalmology Vis. Sci. 2010, 51, 938. [Google Scholar] [CrossRef]
- Yamada, H.; Hangai, M.; Nakano, N.; Takayama, K.; Kimura, Y.; Miyake, M.; Akagi, T.; Ikeda, H.O.; Noma, H.; Yoshimura, N. Asymmetry Analysis of Macular Inner Retinal Layers for Glaucoma Diagnosis. Am. J. Ophthalmol. 2014, 158, 1318–1329.e3. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, C.C. Neural Networks and Deep Learning: A Textbook; Springer International Publishing: Cham, Switzerland, 2018. [Google Scholar] [CrossRef]
- Bengio, Y.; Goodfellow, I.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2017; Volume 1. [Google Scholar]
- Charniak, E. Introduction to Deep Learning; Cambridge University Press: Cambridge, UK, 2019; Volume 53, pp. 1689–1699. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; proceedings, part III 18. Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. In Proceedings of the Advances in Neural Information Processing Systems; Ghahramani, Z., Welling, M., Cortes, C., Lawrence, N., Weinberger, K., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2014; Volume 27. [Google Scholar]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-Image Translation with Conditional Adversarial Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Vaswani, A. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. arXiv 2021, arXiv:2010.11929. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021; pp. 9992–10002. [Google Scholar]
- Dong, X.; Bao, J.; Chen, D.; Zhang, W.; Yu, N.; Yuan, L.; Chen, D.; Guo, B. CSWin Transformer: A General Vision Transformer Backbone with Cross-Shaped Windows. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 12114–12124. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Raja, H.; Akram, M.U.; Khawaja, S.G.; Arslan, M.; Ramzan, A.; Nazir, N. Data on OCT and fundus images for the detection of glaucoma. Data Brief 2020, 29, 105342. [Google Scholar] [CrossRef]
- Tian, J.; Varga, B.; Tatrai, E.; Fanni, P.; Somfai, G.M.; Smiddy, W.E.; Debuc, D.C. Performance evaluation of automated segmentation software on optical coherence tomography volume data. J. Biophotonics 2016, 9, 478–489. [Google Scholar] [CrossRef]
- Farsiu, S.; Chiu, S.J.; O’Connell, R.V.; Folgar, F.A.; Yuan, E.; Izatt, J.A.; Toth, C.A. Quantitative Classification of Eyes with and without Intermediate Age-related Macular Degeneration Using Optical Coherence Tomography. Ophthalmology 2014, 121, 162–172. [Google Scholar] [CrossRef]
- He, Y.; Carass, A.; Solomon, S.D.; Saidha, S.; Calabresi, P.A.; Prince, J.L. Retinal layer parcellation of optical coherence tomography images: Data resource for multiple sclerosis and healthy controls. Data Brief 2019, 22, 601–604. [Google Scholar] [CrossRef] [PubMed]
- Chiu, S.J.; Allingham, M.J.; Mettu, P.S.; Cousins, S.W.; Izatt, J.A.; Farsiu, S. Kernel regression based segmentation of optical coherence tomography images with diabetic macular edema. Biomed. Opt. Express 2015, 6, 1172. [Google Scholar] [CrossRef] [PubMed]
- Melinščak, M.; Radmilović, M.; Vatavuk, Z.; Lončarić, S. Annotated retinal optical coherence tomography images (AROI) database for joint retinal layer and fluid segmentation. Automatika 2021, 62, 375–385. [Google Scholar] [CrossRef]
- Gholami, P.; Roy, P.; Parthasarathy, M.K.; Lakshminarayanan, V. OCTID: Optical coherence tomography image database. Comput. Electr. Eng. 2020, 81, 106532. [Google Scholar] [CrossRef]
- He, X.; Wang, Y.; Poiesi, F.; Song, W.; Xu, Q.; Feng, Z.; Wan, Y. Exploiting multi-granularity visual features for retinal layer segmentation in human eyes. Front. Bioeng. Biotechnol. 2023, 11, 1191803. [Google Scholar] [CrossRef]
- Chiu, S.J.; Izatt, J.A.; O’Connell, R.V.; Winter, K.P.; Toth, C.A.; Farsiu, S. Validated Automatic Segmentation of AMD Pathology Including Drusen and Geographic Atrophy in SD-OCT Images. Investig. Opthalmology Vis. Sci. 2012, 53, 53. [Google Scholar] [CrossRef]
- Shah, A.; Zhou, L.; Abrámoff, M.D.; Wu, X. Multiple surface segmentation using convolution neural nets: Application to retinal layer segmentation in OCT images. Biomed. Opt. Express 2018, 9, 4509–4526. [Google Scholar] [CrossRef]
- Raja, H.; Akram, M.U.; Shaukat, A.; Khan, S.A.; Alghamdi, N.; Khawaja, S.G.; Nazir, N. Extraction of Retinal Layers Through Convolution Neural Network (CNN) in an OCT Image for Glaucoma Diagnosis. J. Digit. Imaging 2020, 33, 1428–1442. [Google Scholar] [CrossRef]
- Cansiz, S.; Kesim, C.; Bektas, S.N.; Kulali, Z.; Hasanreisoglu, M.; Gunduz-Demir, C. FourierNet: Shape-Preserving Network for Henle’s Fiber Layer Segmentation in Optical Coherence Tomography Images. IEEE J. Biomed. Health Inform. 2023, 27, 1036–1047. [Google Scholar] [CrossRef]
- Hu, K.; Shen, B.; Zhang, Y.; Cao, C.; Xiao, F.; Gao, X. Automatic segmentation of retinal layer boundaries in OCT images using multiscale convolutional neural network and graph search. Neurocomputing 2019, 365, 302–313. [Google Scholar] [CrossRef]
- Cazañas-Gordón, A.; Cruz, L.A.d.S. Multiscale Attention Gated Network (MAGNet) for Retinal Layer and Macular Cystoid Edema Segmentation. IEEE Access 2022, 10, 3198657. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Z.; Li, F.; Qu, G.; Qiao, Y.; Lv, H.; Zhang, X. Joint retina segmentation and classification for early glaucoma diagnosis. Biomed. Opt. Express 2019, 10, 2639–2656. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Carass, A.; Liu, Y.; Jedynak, B.M.; Solomon, S.D.; Saidha, S.; Calabresi, P.A.; Prince, J.L. Structured layer surface segmentation for retina OCT using fully convolutional regression networks. Med. Image Anal. 2021, 68, 101856. [Google Scholar] [CrossRef] [PubMed]
- Fazekas, B.; Aresta, G.; Lachinov, D.; Riedl, S.; Mai, J.; Schmidt-Erfurth, U.; Bogunović, H. SD-LayerNet: Semi-supervised Retinal Layer Segmentation in OCT Using Disentangled Representation with Anatomical Priors. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2022; Springer: Cham, Switzerland, 2022; Volume 13438. [Google Scholar] [CrossRef]
- Cao, J.; Liu, X.; Zhang, Y.; Wang, M. A Multi-task Framework for Topology-guaranteed Retinal Layer Segmentation in OCT Images. In Proceedings of the 2020 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Toronto, ON, Canada, 11–14 October 2020. [Google Scholar] [CrossRef]
- Vázquez, E.R.; Rodríguez, M.N.B.; López-Varela, E.; Penedo, M.G. Deep Learning for Segmentation of Optic Disc and Retinal Layers in Peripapillary Optical Coherence Tomography Images. In Proceedings of the Fifteenth International Conference on Machine Vision (ICMV 2022), Rome, Italy, 18–20 November 2023; Volume 12701. [Google Scholar] [CrossRef]
- Matovinovic, I.Z.; Loncaric, S.; Lo, J.; Heisler, M.; Sarunic, M. Transfer learning with U-net type model for automatic segmentation of three retinal layers in optical coherence tomography images. In Proceedings of the 2019 11th International Symposium on Image and Signal Processing and Analysis, Dubrovnik, Croatia, 23–25 September 2019; Volume 2019. [Google Scholar] [CrossRef]
- Kepp, T.; Ehrhardt, J.; Heinrich, M.P.; Hüttmann, G.; Handels, H. Topology-Preserving Shape-Based Regression Of Retinal Layers In Oct Image Data Using Convolutional Neural Networks. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, H.; Linton, E.F.; Johnson, B.A.; Choi, Y.J.; Kupersmith, M.J.; Sonka, M.; Garvin, M.K.; Kardon, R.H.; Wang, J.K. Hybrid deep learning and optimal graph search method for optical coherence tomography layer segmentation in diseases affecting the optic nerve. Biomed. Opt. Express 2024, 15, 3681–3698. [Google Scholar] [CrossRef]
- Ndipenoch, N.; Miron, A.; Wang, Z.; Li, Y. Simultaneous Segmentation of Layers and Fluids in Retinal OCT Images. In Proceedings of the 2022 15th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Beijing, China, 5–7 November 2022. [Google Scholar] [CrossRef]
- Moradi, M.; Chen, Y.; Du, X.; Seddon, J.M. Deep ensemble learning for automated non-advanced AMD classification using optimized retinal layer segmentation and SD-OCT scans. Comput. Biol. Med. 2023, 154, 106512. [Google Scholar] [CrossRef]
- Zhilin, Z.; Yan, W.; Zeyu, P.; Yunqing, Z.; Rugang, Y.; Guogang, C. Dual Attention Network for Retinal Layer and Fluid Segmentation in OCT. In Proceedings of the 2023 8th International Conference on Intelligent Informatics and Biomedical Sciences (ICIIBMS), Okinawa, Japan, 23–25 November 2023; Volume 8. [Google Scholar] [CrossRef]
- Mukherjee, S.; De Silva, T.; Grisso, P.; Wiley, H.; Tiarnan, D.K.; Thavikulwat, A.T.; Chew, E.; Cukras, C. Retinal layer segmentation in optical coherence tomography (OCT) using a 3D deep-convolutional regression network for patients with age-related macular degeneration. Biomed. Opt. Express 2022, 13, 3195–3210. [Google Scholar] [CrossRef]
- Xie, H.; Xu, W.; Wang, Y.X.; Wu, X. Deep learning network with differentiable dynamic programming for retina OCT surface segmentation. Biomed. Opt. Express 2023, 14, 3190–3202. [Google Scholar] [CrossRef]
- Shen, Y.; Li, J.; Zhu, W.; Yu, K.; Wang, M.; Peng, Y.; Zhou, Y.; Guan, L.; Chen, X. Graph Attention U-Net for Retinal Layer Surface Detection and Choroid Neovascularization Segmentation in OCT Images. IEEE Trans. Med. Imaging 2023, 42, 3140–3154. [Google Scholar] [CrossRef]
- Hu, K.; Liu, D.; Chen, Z.; Li, X.; Zhang, Y.; Gao, X. Embedded Residual Recurrent Network and Graph Search for the Segmentation of Retinal Layer Boundaries in Optical Coherence Tomography. IEEE Trans. Instrum. Meas. 2021, 70, 1–17. [Google Scholar] [CrossRef]
- Kugelman, J.; Alonso-Caneiro, D.; Read, S.A.; Vincent, S.J.; Collins, M.J. Automatic segmentation of OCT retinal boundaries using recurrent neural networks and graph search. Biomed. Opt. Express 2018, 9, 5759–5777. [Google Scholar] [CrossRef]
- Li, J.; Jin, P.; Zhu, J.; Zou, H.; Xu, X.; Tang, M.; Zhou, M.; Gan, Y.; He, J.; Ling, Y.; et al. Multi-scale GCN-assisted two-stage network for joint segmentation of retinal layers and discs in peripapillary OCT images. Biomed. Opt. Express 2021, 12, 2204–2220. [Google Scholar] [CrossRef] [PubMed]
- Mishra, Z.; Ganegoda, A.; Selicha, J.; Wang, Z.; Sadda, S.R.; Hu, Z. Automated Retinal Layer Segmentation Using Graph-based Algorithm Incorporating Deep-learning-derived Information. Sci. Rep. 2020, 10, 9541. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ling, Y.; He, J.; Jin, P.; Zhu, J.; Zou, H.; Xu, X.; Shao, S.; Gan, Y.; Su, Y. A GCN-assisted deep learning method for peripapillary retinal layer segmentation in OCT images. In Proceedings of the Optical Coherence Tomography and Coherence Domain Optical Methods in Biomedicine XXV, Online, 6–11 March 2021; Volume 11630. [Google Scholar] [CrossRef]
- Fang, L.; Cunefare, D.; Wang, C.; Guymer, R.H.; Li, S.; Farsiu, S. Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search. Biomed. Opt. Express 2017, 8, 2732–2744. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Galang, C.; Freeman, W.R.; Warter, A.; Heinke, A.; Bartsch, D.U.G.; Nguyen, T.Q.; An, C. Retinal OCT Layer Segmentation via Joint Motion Correction and Graph-Assisted 3D Neural Network. IEEE Access 2023, 11, 103319–103332. [Google Scholar] [CrossRef]
- Heisler, M.; Bhalla, M.; Lo, J.; Mammo, Z.; Lee, S.; Ju, M.J.; Beg, M.F.; Sarunic, M.V. Semi-supervised deep learning based 3D analysis of the peripapillary region. Biomed. Opt. Express 2020, 11, 3843–3856. [Google Scholar] [CrossRef]
- Xue, S.; Wang, H.; Guo, X.; Sun, M.; Song, K.; Shao, Y.; Zhang, H.; Zhang, T. CTS-Net: A Segmentation Network for Glaucoma Optical Coherence Tomography Retinal Layer Images. Bioengineering 2023, 10, 230. [Google Scholar] [CrossRef]
- Tonade, D.; Kern, T.S. Photoreceptor cells and RPE contribute to the development of diabetic retinopathy. Prog. Retin. Eye Res. 2021, 83, 100919. [Google Scholar] [CrossRef]
- Wheat, J.L.; Rangaswamy, N.V.; Harwerth, R.S. Correlating RNFL thickness by OCT with perimetric sensitivity in glaucoma patients. J. Glaucoma 2012, 21, 95–101. [Google Scholar] [CrossRef]
- Morelle, O.; Wintergerst, M.W.; Finger, R.P.; Schultz, T. Accurate drusen segmentation in optical coherence tomography via order-constrained regression of retinal layer heights. Sci. Rep. 2023, 13, 8162. [Google Scholar] [CrossRef]
- Baidu AI Studio Competition: Retinal OCT Layer Segmentation. 2022. Available online: https://aistudio.baidu.com/aistudio/competition/detail/783/0/introduction (accessed on 18 December 2024).
- He, Y.; Carass, A.; Liu, Y.; Calabresi, P.A.; Saidha, S.; Prince, J.L. Longitudinal deep network for consistent OCT layer segmentation. Biomed. Opt. Express 2023, 14, 1874–1893. [Google Scholar] [CrossRef]
- Sousa, J.A.; Paiva, A.; Silva, A.; Almeida, J.D.; Braz, J.G.; Diniz, J.O.; Figueredo, W.K.; Gattass, M. Automatic segmentation of retinal layers in OCT images with intermediate age-related macular degeneration using U-Net and DexiNed. PLoS ONE 2021, 16, e0251591. [Google Scholar] [CrossRef]
- Gende, M.; de Moura, J.; Fernández-Vigo, J.I.; Martínez-de-la-Casa, J.M.; García-Feijóo, J.; Novo, J.; Ortega, M. Robust multi-view approaches for retinal layer segmentation in glaucoma patients via transfer learning. Quant. Imaging Med. Surg. 2023, 13, 2846. [Google Scholar] [CrossRef]
- Dodo, B.I.; Li, Y.; Eltayef, K.; Liu, X. Automatic annotation of retinal layers in optical coherence tomography images. J. Med. Syst. 2019, 43, 336. [Google Scholar] [CrossRef]
- Konno, T.; Ninomiya, T.; Miura, K.; Ito, K.; Himori, N.; Sharma, P.; Nakazawa, T.; Aoki, T. Retinal Layer Segmentation from Oct Images Using 2D-3D Hybrid Network with Multi-Scale Loss and Refinement Module. In Proceedings of the 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI), Cartagena, Colombia, 18–21 April 2023. [Google Scholar] [CrossRef]
- Hassan, T.; Usman, A.; Akram, M.U.; Masood, M.F.; Yasin, U. Deep Learning Based Automated Extraction of Intra-Retinal Layers for Analyzing Retinal Abnormalities. In Proceedings of the 2018 IEEE 20th International Conference on e-Health Networking, Applications and Services (Healthcom), Ostrava, Czech Republic, 17–20 September 2018. [Google Scholar] [CrossRef]
- Liu, X.; Cao, J.; Wang, S.; Zhang, Y.; Wang, M. Confidence-Guided Topology-Preserving Layer Segmentation for Optical Coherence Tomography Images with Focus-Column Module. IEEE Trans. Instrum. Meas. 2020, 70, 5005612. [Google Scholar] [CrossRef]
- Wei, H.; Peng, P. The Segmentation of Retinal Layer and Fluid in SD-OCT Images Using Mutex Dice Loss Based Fully Convolutional Networks. IEEE Access 2020, 8, 60929–60939. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).