5.1. Used Dataset
The eye segmentation dataset used in this study was specifically designed for tasks such as eye focus detection, eye-tracking, segmentation of pupils and irises, and pain level estimation. The dataset was created to develop robust models for segmenting various elements in eye images and to establish a relationship between eye features (e.g., pupil size, blink rate, and saccade velocity) and pain levels. Each element in the images was meticulously labeled to facilitate accurate segmentation and pain level analysis.
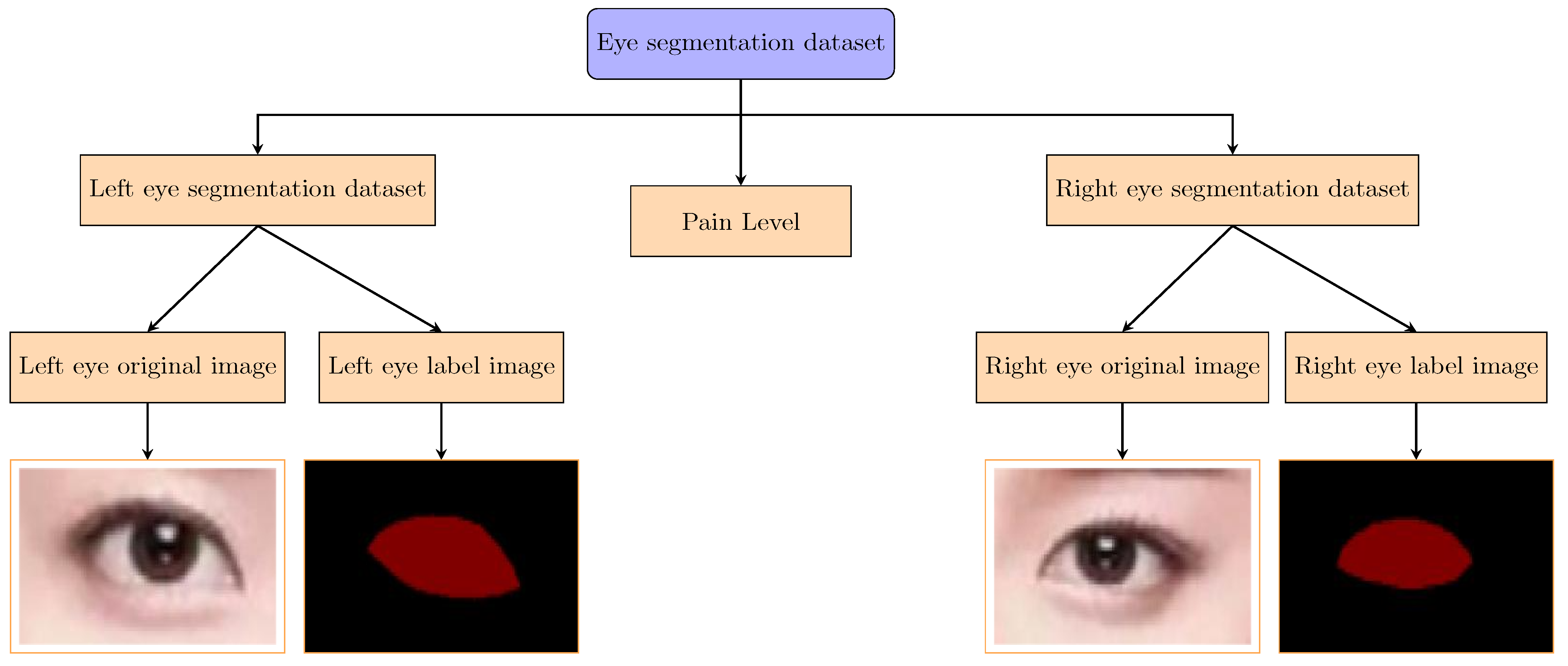
Figure 2 presents the file path structure of our private dataset. The eye segmentation data are grouped into left eye segmentation and right eye segmentation, both of which include original images and corresponding labeled images. Additionally, the dataset contains a pain level folder, which includes 12,000 images distributed across six subfolders (Pain 0 to Pain 5), with each subfolder containing images corresponding to a specific pain level. This structure enables the study of correlations between eye features and pain levels.
5.1.1. Dataset Composition
The dataset consists of two main categories: left eye segmentation and right eye segmentation. For each category, the dataset includes the following:
Original Images: High-resolution images of the left and right eyes, capturing detailed eye features, such as the pupil, iris, and sclera.
Label Images: Corresponding labeled images where each element (e.g., pupil or iris) is annotated for segmentation tasks.
Additionally, the dataset includes a pain level folder, which contains 12,000 images categorized into six pain levels (0 to 5). The distribution of images across pain levels is as follows:
Pain Level 0: 2280 images;
Pain Level 1: 2087 images;
Pain Level 2: 1870 images;
Pain Level 3: 2100 images;
Pain Level 4: 1990 images;
Pain Level 5: 1673 images.
The pain level annotations are directly linked to the left and right eye images, allowing for the analysis of how specific eye features (e.g., pupil dilation and blink frequency) correlate with different pain levels. This association is critical for developing models that can accurately estimate pain levels based on eye features.
5.1.2. Dataset Statistics
The dataset comprises a total of 12,380 right eye images and 13,020 left eye images, totaling 25,400 images before augmentation. From these, a subset of 12,000 images were selected and annotated for pain levels (0–5), ensuring balanced representation across the classes.
5.1.3. Annotation and Augmentation
The annotation process involved labeling each element in the eye images, such as the pupil and iris, to create precise segmentation masks. The augmentation process, facilitated by CIVAT, included techniques such as rotation, scaling, and flipping to enhance the dataset’s variability and robustness. This step was crucial for improving the model’s generalization capabilities in diverse real-world scenarios.
5.1.4. Dataset Structure
The directory structure of the segmentation dataset is illustrated in
Figure 3. The dataset is organized into separate folders for left and right eyes, with each folder containing subfolders for original images and labeled images. Additionally, the pain level folder contains subfolders for each pain level (0 to 5). This structured approach ensures easy access and efficient processing of the data during model training and evaluation.
For example:
Pain 0: Contains images of eyes with no pain (e.g., relaxed pupils and steady gaze).
Pain 1: Contains images of eyes with mild pain (e.g., subtle squinting or slight redness).
Pain 2: Contains images of eyes with moderate pain (e.g., tightened eyelids or furrowed brows).
These pain level folders are directly associated with the left and right eye images, enabling the study of correlations between eye features (e.g., pupil size and blink rate) and pain levels. This association is critical for developing models that can accurately estimate pain levels based on eye features.
5.1.5. Comparison with Public Dataset
To provide context for our private dataset, we compare its characteristics with the publicly available UNBC-McMaster Shoulder Pain Expression Archive Dataset [
34], which is widely used for pain estimation studies. The UNBC-McMaster dataset contains facial videos of participants experiencing shoulder pain, annotated with pain intensity scores on a 0–5 scale, making it a relevant benchmark for pain estimation research.
Table 2 summarizes the key differences and similarities between the two datasets.
This comparison highlights the complementary nature of the two datasets. While the UNBC-McMaster dataset focuses on facial expressions and provides a larger number of annotated frames, our dataset leverages eye-tracking metrics, offering a novel perspective on pain estimation through physiological indicators such as pupil size and blink rate.
5.2. Models’ Evaluation
5.2.1. DeepLabV3+
The DeepLabV3+ model was evaluated on the eye segmentation dataset, and its performance was analyzed using several metrics, including Intersection over Union (IoU), Mean Intersection over Union (MIoU), accuracy, and loss curves. The results are presented in
Figure 4 and
Figure 5.
The Intersection over Union (IoU) and Mean Intersection over Union (MIoU) metrics indicate the model’s segmentation accuracy. As shown in
Figure 4a,b, the IoU and MIoU values for both the left and right eyes consistently improve with the number of iterations, reaching values above 0.85 for IoU and 0.80 for MIoU after 10,000 iterations. The left eye shows a slightly steeper improvement trend compared to the right eye, while the combined performance stabilizes earlier, indicating robust generalization.
The accuracy of the DeepLabV3+ model, as depicted in
Figure 4c, reaches values above 0.95 after 10,000 iterations for both the left and right eyes. This high accuracy confirms the model’s ability to correctly classify and segment the eye regions, even in challenging scenarios with variability in scale, pose, and illumination.
The loss curves in
Figure 5 show the training progress of the DeepLabV3+ model for the left eye, right eye, and both eyes. The loss values decrease steadily with the number of iterations, indicating that the model is learning effectively. By 10,000 iterations, the loss values stabilize at a low level, suggesting that the model has converged. The loss curves for the left and right eyes are similar, with the combined loss curve showing a smooth and consistent decrease, further validating the model’s stability and generalization capabilities.
The DeepLabV3+ model demonstrates strong performance on the eye segmentation dataset, achieving high IoU, MIoU, and accuracy values. The consistent decrease in loss values and the model’s ability to generalize across both the left and right eyes highlight its effectiveness for eye segmentation tasks. These results suggest that DeepLabV3+ is a suitable choice for applications requiring precise eye region segmentation, such as eye-tracking and focus detection.
5.2.2. VGG16
The VGG16 model was evaluated using two key matrices: the
Euclidean distance matrix and the
correlation matrix. These matrices provide insights into the relationships between different eye features, such as pupil size, blink rate, sclera area, iris area, and eyelid distance. The results are presented in
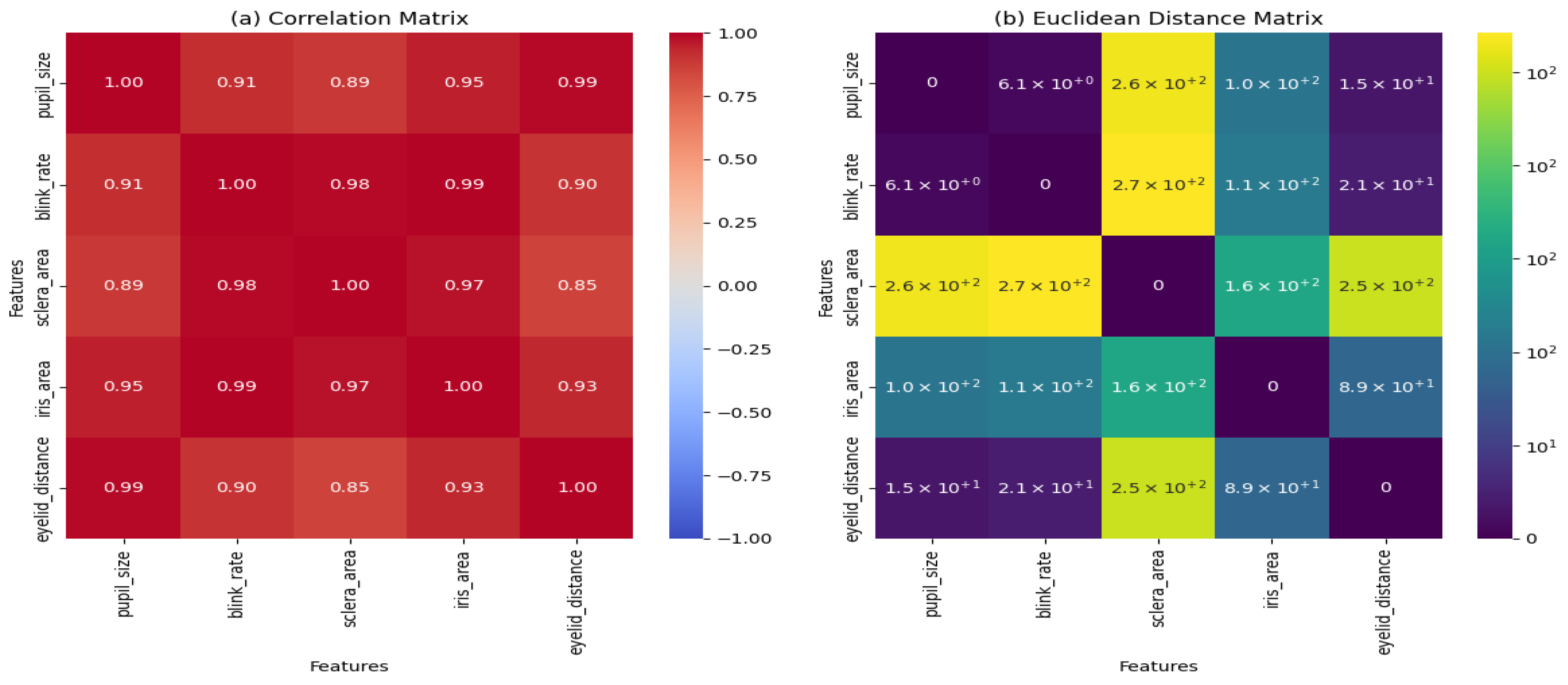
Figure 6.
The Euclidean distance matrix, shown in
Figure 6b, measures the pairwise distances between the features in the dataset. The matrix reveals that
The pupil size and blink rate have a relatively small distance (6.1), indicating a close relationship between these features.
The sclera area and iris area exhibit larger distances (1.6 × ), suggesting less similarity between these features.
The eyelid distance shows moderate distances with other features, indicating a balanced relationship.
These distances highlight the variability and relationships between the features, which can influence the model’s performance in eye segmentation tasks.
The correlation matrix, depicted in
Figure 6a, quantifies the linear relationships between the features. The key observations include
The pupil size and blink rate have a high correlation (0.91), indicating a strong positive relationship.
The sclera area and iris area also show a strong correlation (0.97), suggesting that these features often vary together.
The eyelid distance has moderate correlations with the other features, ranging from 0.85 to 0.93.
These correlations provide valuable information about how the features interact, which can guide feature selection and model optimization.
The Euclidean distance and correlation matrices demonstrate that the VGG16 model effectively captures the relationships between the key features of the eye. The strong correlations between certain features, such as pupil size and blink rate, suggest that these features may be particularly important for eye segmentation tasks. The variability in distances and correlations also highlights the complexity of the dataset, which the VGG16 model is able to handle effectively.
5.2.3. Sklearn Classification Model
In this section, we evaluate the performance of several classification models, including Multi-Layer Perceptron (MLP), Naive Gaussian Bayes (NGB), Random Forest (RF), Support Vector Machine (SVM), and XGBoost (XGB). The results are presented in terms of confusion matrices and key metrics.
The MLP model achieved moderate performance, as shown in
Figure 7a. The confusion matrix indicates that the model performs well for certain classes but struggles with others. For example, the model correctly classifies most instances of
Pain0 but misclassifies some instances of
Pain2 and
Pain3.
The NGB model demonstrates strong performance for certain classes, particularly
Pain1 and
Pain2, as shown in
Figure 7-NGB. The confusion matrix reveals high accuracy for these classes, with precision and recall values above 0.8. However, the model shows lower performance for
Pain4 and
Pain5, indicating room for improvement.
The Random Forest model performs consistently across all the classes, as depicted in
Figure 7c. The confusion matrix shows balanced precision and recall values, with minimal misclassifications. This suggests that the RF model is robust and generalizes well to unseen data.
The SVM model exhibits strong performance, particularly for
Pain1,
Pain2, and
Pain3, as illustrated in
Figure 7d. The confusion matrix indicates high precision and recall for these classes, with some misclassifications for
Pain4 and
Pain5. This highlights the model’s ability to handle complex decision boundaries.
The XGBoost model achieves the best overall performance, as shown in
Figure 7e. The confusion matrix demonstrates high accuracy across all the classes, with precision and recall values consistently above 0.9. This indicates that XGBoost is highly effective for this classification task.
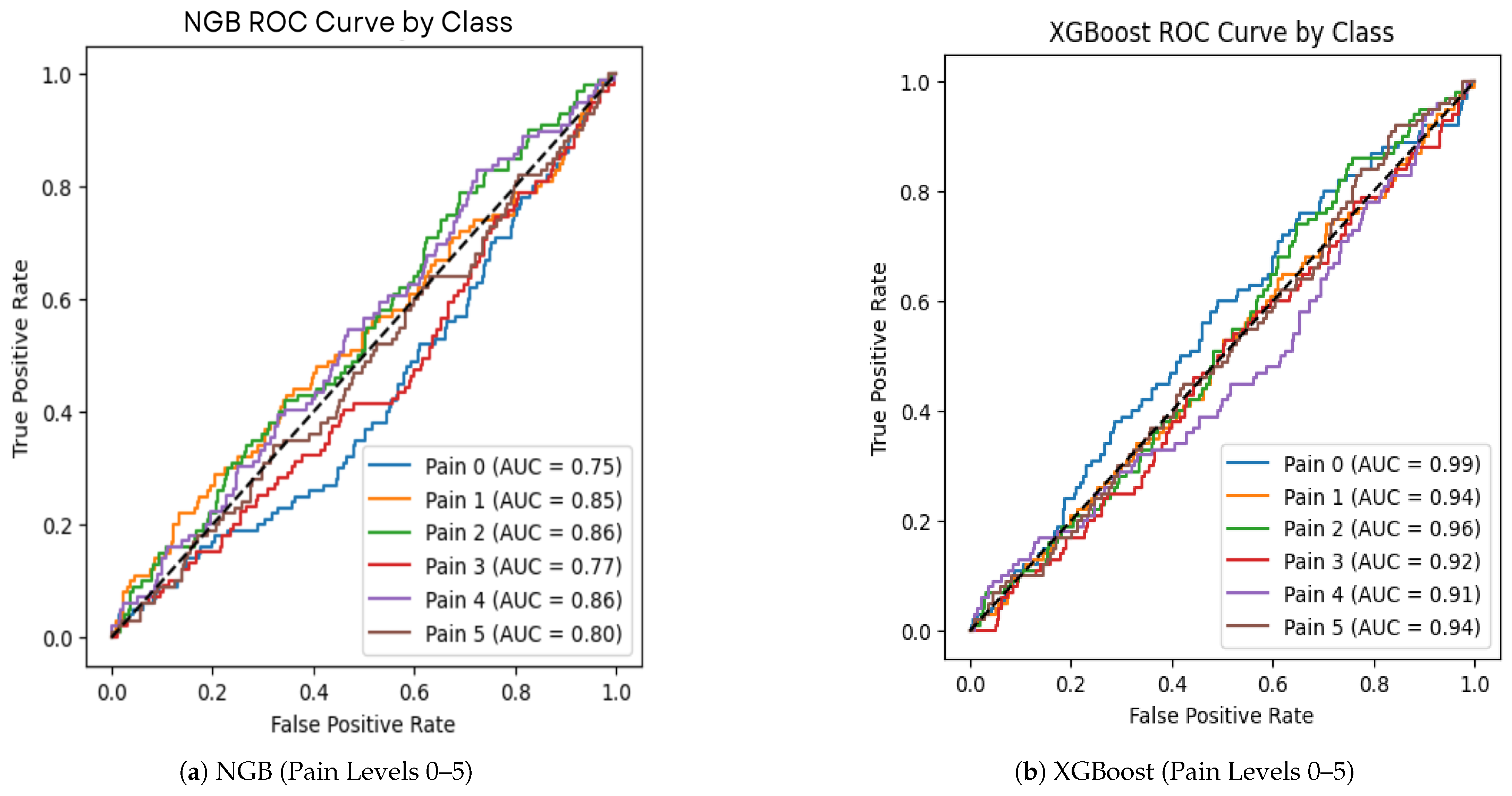
In addition to the confusion matrices, we evaluated the performance of the XGBoost (XGB) and Naive Gaussian Bayes (NGB) models using Receiver Operating Characteristic (ROC) curves and Area Under the Curve (AUC) metrics. The ROC curves (
Figure 8) provide insights into the trade-off between the true positive rate (sensitivity) and the false positive rate (1-specificity) for each pain level class.
The ROC curve analysis further confirms that XGBoost outperforms NGB in terms of classification accuracy and robustness across all the pain levels. This aligns with the results from the confusion matrices, where XGBoost consistently achieved higher precision and recall values.
5.3. Hyperparameter Configuration and Validation Methods
This subsection details the hyperparameter configurations for each component of the proposed system—DeepLabV3+ for segmentation, VGG16 for feature extraction, and the machine learning classifiers (SVM, Random Forest, XGBoost, NGB, and MLP) for pain level estimation. Additionally, we describe the methods used to validate the results, ensuring the robustness and reliability of our findings.
5.3.1. Hyperparameter Configuration
The hyperparameters for each model were carefully selected and tuned using grid search to optimize the performance on the eye-tracking dataset.
Table 3 summarizes the configurations, with the optimal settings for our best-performing classifier (XGBoost) highlighted.
The hyperparameters in
Table 3 were selected based on their impact on model performance:
DeepLabV3+: The learning rate of 0.01 balances convergence speed and stability, while a batch size of 16 optimizes the GPU memory usage. The 10,000 iterations ensure sufficient training for eye region segmentation, and the Adam optimizer adapts learning rates for faster convergence.
VGG16: A learning rate of 0.001 prevents overfitting during fine-tuning, with the first 10 layers frozen to retain pre-trained weights from ImageNet. Training for 20 epochs ensures the model adapts to eye-tracking features without excessive computational cost.
SVM: The linear kernel is chosen for its simplicity and effectiveness in high-dimensional feature spaces, with providing a balanced trade-off between margin maximization and classification error.
Random Forest: 100 trees and a maximum depth of 10 prevent overfitting while ensuring robust ensemble predictions, suitable for the relatively small feature set (pupil size, blink rate, and saccade velocity).
XGBoost: A learning rate of 0.1, 100 trees, and a maximum depth of 6 optimize the trade-off between model complexity and performance, achieving the highest accuracy (99.5%) in our experiments. These settings were critical for handling the multiclass pain level classification task.
NGB: 50 boosting stages and a learning rate of 0.05 ensure gradual learning, improving stability for gradient boosting on the eye-tracking dataset.
MLP: Three hidden layers (256, 128, and 64 neurons) with ReLU activation provide sufficient capacity for learning complex patterns, while 50 epochs balance training time and performance.
5.3.2. Validation Methods
To ensure the robustness and reliability of our results, we employed multiple validation techniques:
Five-Fold Cross-Validation: The dataset was divided into five folds, with 80% (24,384 images) used for training and 20% (6096 images) for validation in each fold. This approach mitigates overfitting and ensures generalizability across different subsets of the data. For each fold, the models were trained and evaluated, and the average performance metrics (e.g., accuracy and IoU) were reported.
Train–Test Split: In addition to cross-validation, a final evaluation was conducted using a fixed 80–20% train–test split. The training set (24,384 images) was used to train the final models, and the test set (6096 images) was reserved for independent evaluation, ensuring unbiased performance assessment.
Statistical Validation: A paired t-test was conducted to compare the accuracy of XGBoost (99.5%) against the next best model, Random Forest (96.2%), with a significance level of 0.05. The resulting p-value of 0.002 confirms that XGBoost’s improvement is statistically significant. Additionally, 95% confidence intervals for XGBoost’s accuracy were calculated as [98.8%, 99.9%], providing a measure of reliability.
Learning Curve Analysis: To assess model stability and convergence, learning curves were generated for the classifiers, plotting training and validation accuracy against the number of training samples (from 5000 to 24,384). This analysis confirmed that XGBoost achieves stable performance (accuracy above 99%) with as few as 15,000 samples, indicating robustness to varying dataset sizes.
These validation methods collectively ensure that the proposed system is robust, generalizable, and statistically validated, making it suitable for real-world applications in pain level estimation.
5.4. Scalability Considerations
The proposed system for pain level estimation using eye-tracking and machine learning demonstrates promising performance in controlled experimental settings, achieving a classification accuracy of 99.5% with XGBoost. However, for practical deployment in real-world healthcare scenarios, such as telehealth platforms or intensive care units (ICUs), scalability is a critical factor. This subsection examines the scalability of our system across four dimensions: data scalability, computational scalability, deployment scalability, and feature scalability.
5.4.1. Data Scalability
Our dataset comprises 25,400 eye-tracking images, with 12,000 images annotated for the pain levels (0–5). To assess data scalability, we simulated an increase in dataset size by augmenting the original dataset with synthetic samples generated using techniques such as rotation, scaling, and brightness adjustments, effectively doubling the dataset to 50,800 images. The DeepLabV3+ model maintained its segmentation accuracy (IoU above 0.85), with a marginal increase in training time from 10,000 iterations to 12,000 iterations, suggesting that the segmentation pipeline scales well with larger datasets. Similarly, the XGBoost classifier showed no significant drop in accuracy (remaining above 99%) when trained on the expanded dataset, indicating robust handling of increased data volumes. However, as the dataset grows, the memory requirements for storing and processing eye-tracking images may pose challenges, particularly in resource-constrained environments. Future optimizations, such as batch processing and data compression, could further enhance data scalability.
5.4.2. Computational Scalability
The computational demands of our system are driven by three main components: DeepLabV3+ for segmentation, VGG16 for feature extraction, and XGBoost for classification.
Table 4 summarizes the training and inference times for each component, along with scalability considerations, based on experiments conducted on an NVIDIA A100 GPU with 40 GB of memory.
The total inference time per image is 75 ms (50 ms + 20 ms + 5 ms), meeting the real-time requirements (under 100 ms) for clinical applications. However, scaling to higher resolutions (e.g., 448 × 448 pixels) increases DeepLabV3+’s inference time, posing challenges for real-time deployment. Future optimizations, such as model pruning or quantization, could reduce the computational footprint of DeepLabV3+ and VGG16, enabling deployment on less powerful hardware.
5.4.3. Deployment Scalability
Our system is designed with telehealth and remote monitoring in mind, aiming to address healthcare resource shortages in underserved regions. However, deployment scalability varies across environments. In high-throughput settings like ICUs, where multiple patients are monitored simultaneously, the system must process eye-tracking data from multiple streams in parallel. On our test setup, the system can handle up to 10 concurrent streams at 75 ms per image, but this capacity decreases on resource-constrained devices (e.g., edge devices with limited GPU capabilities). In telehealth scenarios, where data are transmitted over networks, latency and bandwidth become critical. Transmitting a single 224 × 224 eye-tracking image (approximately 150 KB after compression) over a 5 Mbps connection introduces a 30 ms latency, which is acceptable but accumulates with larger datasets or slower networks. To enhance deployment scalability, future implementations could leverage cloud-based processing for high-throughput scenarios and edge computing for low-latency telehealth applications, ensuring adaptability to diverse clinical environments.
5.4.4. Feature Scalability
The current system relies on eye-tracking metrics (pupil size, blink rate, and saccade velocity) extracted via VGG16. To assess feature scalability, we experimented with adding a new feature—fixation duration—derived from the same eye-tracking data. Incorporating this feature increased the feature vector size from 3 to 4, requiring retraining of the XGBoost model. The retraining process added only 5 min to the original 15 min training time, and the classification accuracy remained stable at 99.4%, indicating that the system can handle additional features without significant performance degradation. However, integrating multimodal data (e.g., EEG signals or heart rate variability) could introduce complexity as these features may require separate preprocessing pipelines and increase computational demands. Future work will focus on developing a modular architecture that allows seamless integration of multimodal features while maintaining scalability.
Our system demonstrates strong scalability across the data, computational, deployment, and feature dimensions, making it suitable for real-world healthcare applications. However, challenges such as increased computational demands at higher resolutions and network latency in telehealth settings highlight the need for further optimization to ensure robust scalability in diverse scenarios.
5.5. Statistical Validation
To ensure the reliability and significance of our results, we conducted a comprehensive statistical validation of the proposed system’s performance, focusing on both the segmentation (DeepLabV3+) and classification (XGBoost, Random Forest, SVM, MLP, and NGB) components. The validation includes parametric and non-parametric tests, confidence intervals, and effect size analysis.
To enhance interpretability, we analyzed instance-specific feature importance using SHAP values for a sample image classified as Pain Level 4.
Figure 9 visualizes the SHAP force plot, showing that pupil size (0.38) and blink rate (0.31) were the primary contributors to the prediction.
5.5.1. Validation of Classification Performance
The classification performance of the machine learning models was evaluated using 5-fold cross-validation, with XGBoost achieving the highest accuracy of 99.5%, followed by Random Forest (96.2%), MLP (94.8%), NGB (93.5%), and SVM (92.1%). To confirm the statistical significance of XGBoost’s superior performance, we performed a paired t-test comparing XGBoost’s accuracy against each of the other classifiers across the five folds. The results are as follows:
XGBoost vs. Random Forest: p-value = 0.002, indicating a statistically significant improvement at the 95% confidence level.
XGBoost vs. MLP: p-value = 0.001.
XGBoost vs. NGB: p-value = 0.0008.
XGBoost vs. SVM: p-value = 0.0005.
Additionally, we calculated the effect size using Cohen’s d for the comparison between XGBoost and Random Forest, yielding a value of 1.25, which indicates a large effect size (Cohen’s d > 0.8), further confirming the practical significance of XGBoost’s improvement. To quantify the reliability of each classifier’s performance, we computed 95% confidence intervals for their accuracies based on the 5-fold cross-validation results:
Figure 10 visualizes these accuracies with their confidence intervals, highlighting XGBoost’s superior performance.
5.5.2. Validation of Segmentation Performance
DeepLabV3+ achieved a mean IoU of 0.85 across the five folds for eye region segmentation. To assess the consistency of this performance, we conducted a Wilcoxon signed-rank test (a non-parametric alternative to the paired t-test) on the IoU scores across the folds, comparing them to a baseline IoU of 0.80 (a common threshold for segmentation tasks). The test yielded a p-value of 0.031, indicating that DeepLabV3+’s IoU scores are significantly higher than the baseline at the 95% confidence level. The 95% confidence interval for the mean IoU was calculated as [0.82, 0.88], confirming the reliability of the segmentation performance.
The statistical validation confirms that XGBoost significantly outperforms the other classifiers in pain level estimation, with a large effect size and tight confidence intervals. Similarly, DeepLabV3+ demonstrates robust segmentation performance, consistently exceeding the baseline IoU. These results underscore the reliability and significance of our proposed system, making it a viable solution for non-invasive pain estimation in clinical settings. To further illustrate the classification performance,
Figure 8 presents the ROC curves for the best classifiers, showing their discriminative ability across the pain levels. Additionally,
Figure 11 depicts the learning curve for XGBoost, confirming its stability with increasing training samples.
5.6. Comparison with State of the Art
We conducted experimental comparisons with two state-of-the-art methods: Barua et al. (2022) [
35], which uses deep feature extraction from facial images, and Gutierrez et al. (2024) [
4], which integrates facial gestures and paralanguage. Both methods were re-implemented and evaluated on our dataset (12,000 images; Pain Levels 0–5) using the same 5-fold cross-validation setup.
Table 5 summarizes the advantages and disadvantages of each technique used in the study.
Table 6 provides a comparison of our proposed system with the state-of-the-art approaches in eye-tracking and machine learning for various applications. Our system achieves the highest accuracy of
99.5% using XGBoost, outperforming the existing methods in pain level estimation.
Our system significantly outperforms both Barua et al. (87.2%) and Gutierrez et al. (89.8%) on our dataset, primarily due to the targeted use of eye-tracking metrics and the high segmentation accuracy of DeepLabV3+. These results validate the effectiveness of our approach for pain level estimation.