1. Introduction
Mango is a tropical fruit with global significance, generating over USD 30 billion in annual revenue and supporting the livelihoods of more than 20 million farmers worldwide [
1,
2]. Despite its economic and nutritional importance, mango cultivation faces serious challenges due to leaf diseases such as Anthracnose, Powdery Mildew, and Bacterial Canker, which can lead to yield losses of up to 30% [
3]. In India alone, these diseases result in annual financial losses ranging from USD 50 million to USD 100 million [
4], while on a global scale, plant diseases cause over USD 5 billion in losses each year [
5]. The implications for food security are also concerning. Mangoes contribute significantly to local diets by providing essential vitamins and minerals. Consequently, a decline in mango production not only diminishes dietary diversity but also threatens food supply in regions where agriculture is a primary source of income.
Traditional methods of detecting plant diseases rely on experts who conduct manual visual inspections. Research shows that the accuracy of these assessments for early-stage mango leaf diseases can fall to 65% [
6]. This decline in accuracy is primarily due to the inherent subjectivity and variability in human judgment. Additionally, evaluations by different experts often produce inconsistent results, leading to unreliable diagnoses. On average, an expert can spend up to 30 minutes inspecting a single tree [
7]. As a result, they can examine only about 20 trees per day, which is insufficient in regions with large mango plantations. This slow inspection process not only delays the identification of disease outbreaks but also hinders timely intervention. The heavy reliance on expert knowledge also limits the scalability of traditional methods. In many rural areas, fewer than 37% of smallholder farms have regular access to trained agricultural diagnosticians [
8]. This lack of access increases the risk of undetected diseases, contributing to additional yield losses estimated at around 10–15% over the growing season [
9].
The automated classification of mango leaf diseases through DL and ensemble methods has become the focus of several research investigations [
10,
11]. However, each of these approaches has its limitations. CNNs excel at extracting features from images and can achieve high accuracy rates. However, they require large annotated datasets for training, which are difficult to gather in agriculture. Additionally, training these models demands significant computational resources and time, making them less accessible in low-resource settings. Transfer learning employs pre-trained models to reduce the need for extensive data and expedite training. This is particularly useful when labeled data are scarce, allowing for more efficient adaptation in detecting mango leaf diseases [
12]. However, differences between the pre-trained datasets and actual mango leaf images can lead to performance issues. Fine-tuning is necessary to accurately capture the specific features of these diseases. Their “black-box” nature can also impede understanding and trust in their predictions [
13]. ensemble methods have also been proposed to enhance classification scores by combining predictions from multiple models. However, managing multiple models increases computational complexity [
14], making real-time deployment challenging in rural areas. ViTs have become a promising alternative to these models for image classification, as they effectively capture long-range dependencies and global context. They use self-attention mechanisms to evaluate relationships across the entire image and can improve the detection of subtle disease patterns on plant leaves. However, ViTs have not been widely adopted for mango leaf disease classification due to the lack of large annotated datasets and their higher computational requirements.
Several research gaps and challenges remain in the field of mango leaf disease classification. Firstly, many studies are limited by datasets that are not only small in size but also lack diversity. This often leads to issues like severe domain shifts and overfitting when models are trained on region-specific data. As a result, their ability to generalize across different agroecological zones is restricted [
15]. Furthermore, there is a prevalent problem of class imbalance, where certain disease types are underrepresented in the datasets. This skewed distribution negatively impacts recall and precision, especially for rare disease categories [
16]. The ambiguity of transfer learning architectures adds to the challenge of interpretability. The absence of intrinsic explainability mechanisms limits their usefulness in critical agricultural decision-making processes where trust and clarity are essential [
17,
18]. Moreover, while ViTs provide enhanced contextual feature representation, their high computational demands and significant inference latency hinder real-time deployment in resource-constrained field environments [
19]. Lastly, although some mobile-based diagnostic applications have been created, they often lack model interpretability and do not incorporate rigorous post hoc explainability techniques. Consequently, there is an urgent need for a reliable diagnostic tool in precision agriculture.
This study aims to develop an interpretable and computationally efficient DL model for mango leaf disease classification, ensuring high accuracy, transparency, and real-time deployment. The primary objectives of this study are as follows:
Develop a transformer-based classification system that combines convolutional layers and self-attention to effectively identify different mango leaf diseases.
Improve the discriminatory power of the model by integrating statistical texture descriptors derived from the GLCM.
Embed explainable features into the model to provide visual explanations for predictions, enhancing user trust and supporting decision-making in agriculture.
Address the limitations of single-dataset training by validating the model on heterogeneous, multi-regional datasets to ensure robust generalization and adaptability to varying agroecological conditions.
Integrate the XAI-enabled transformer model within a web application for an accessible and interpretable tool for real-time mango leaf disease detection.
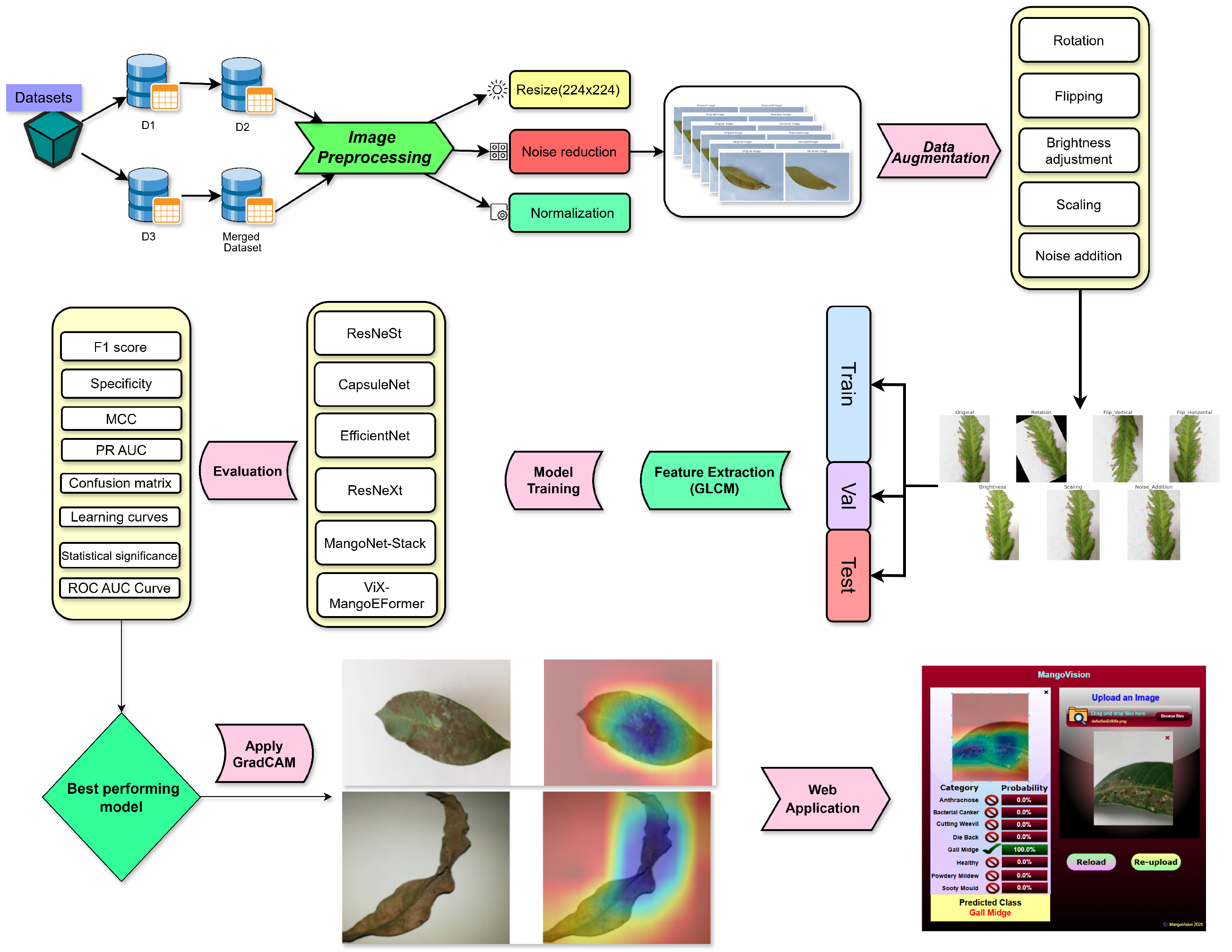
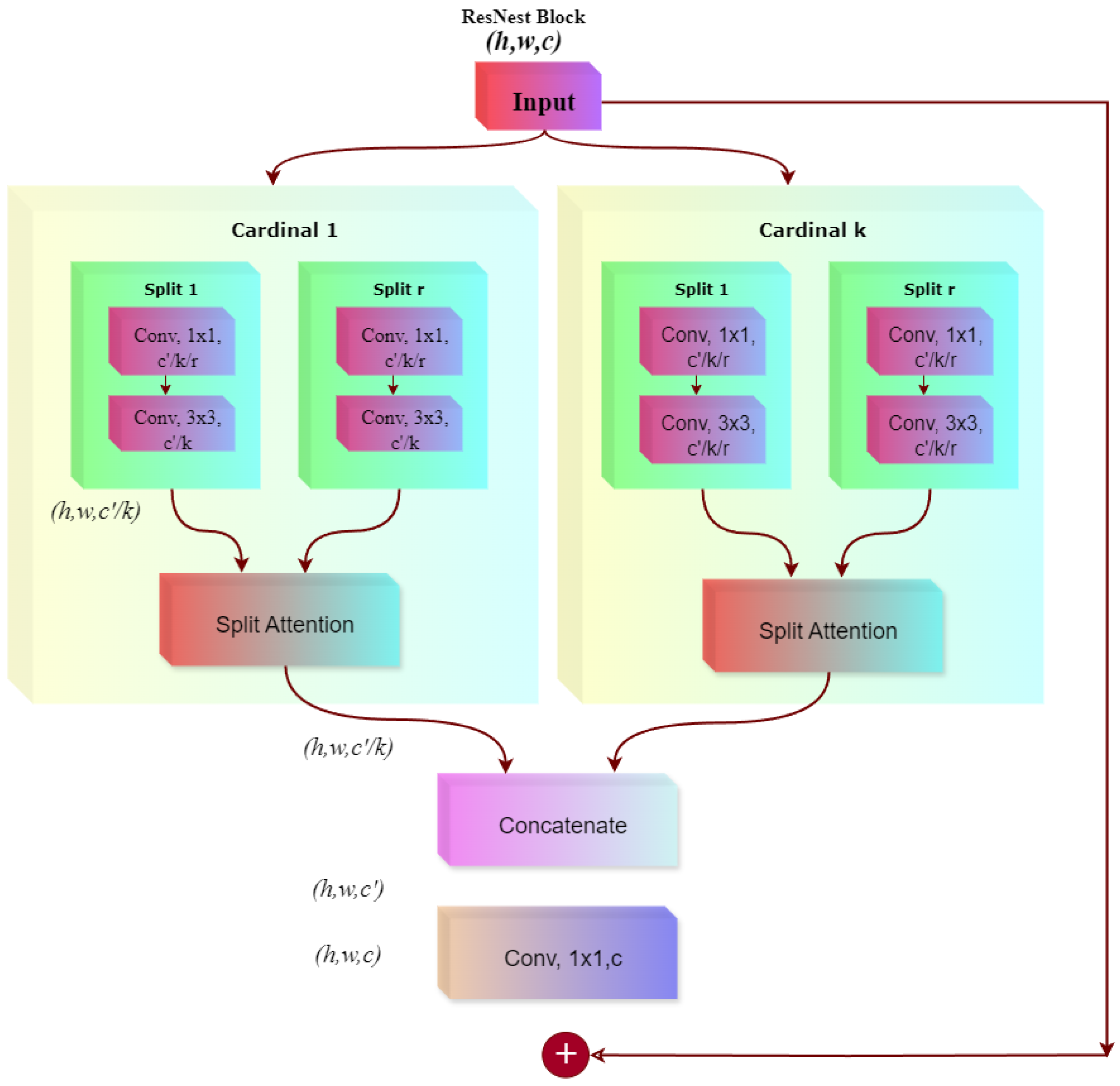
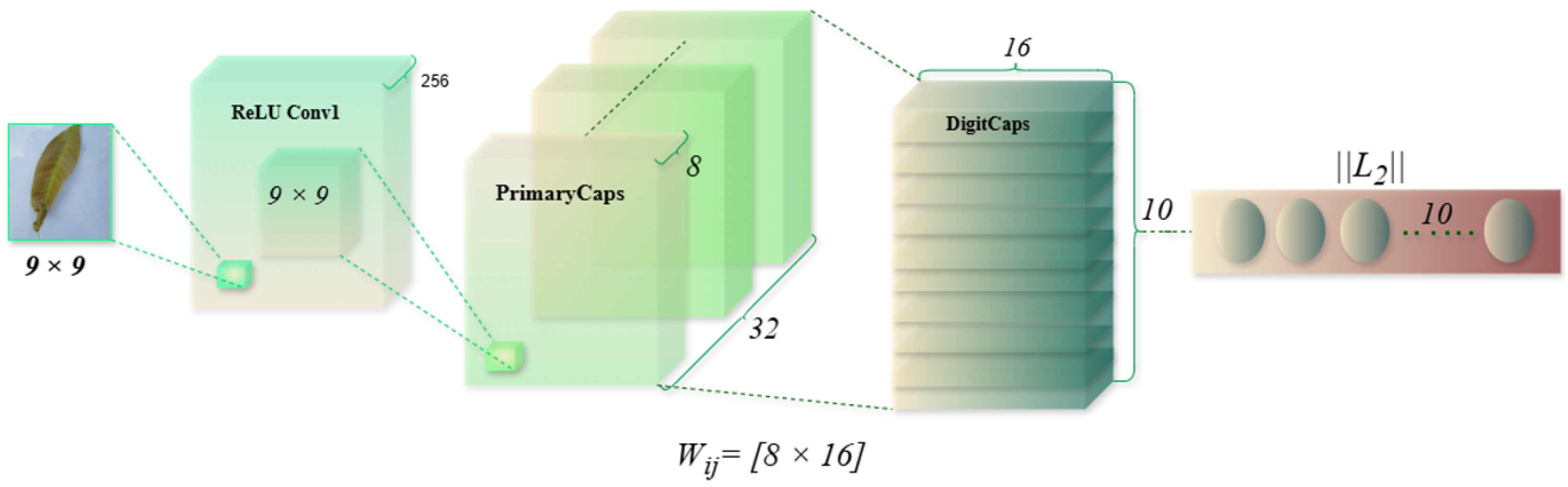
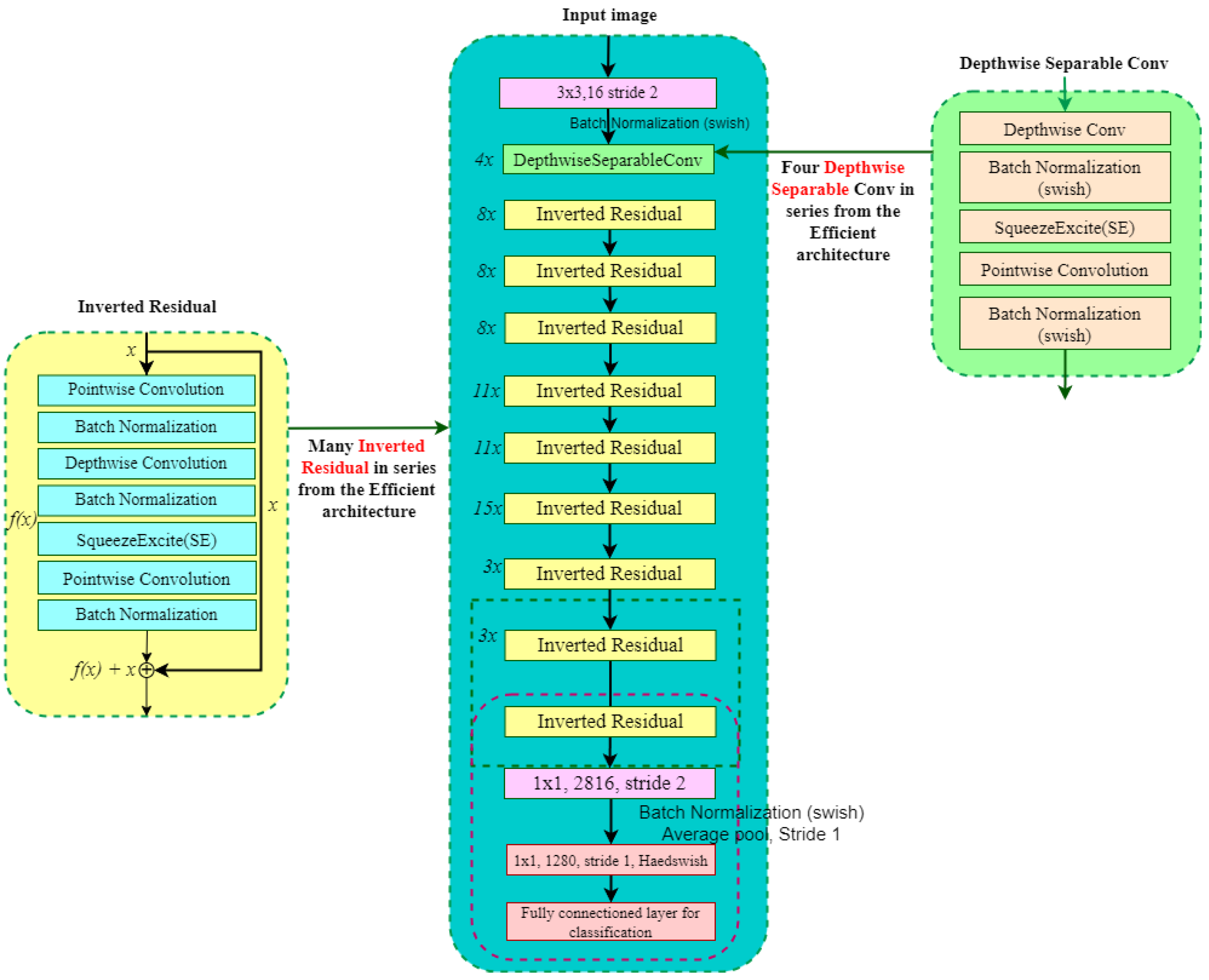
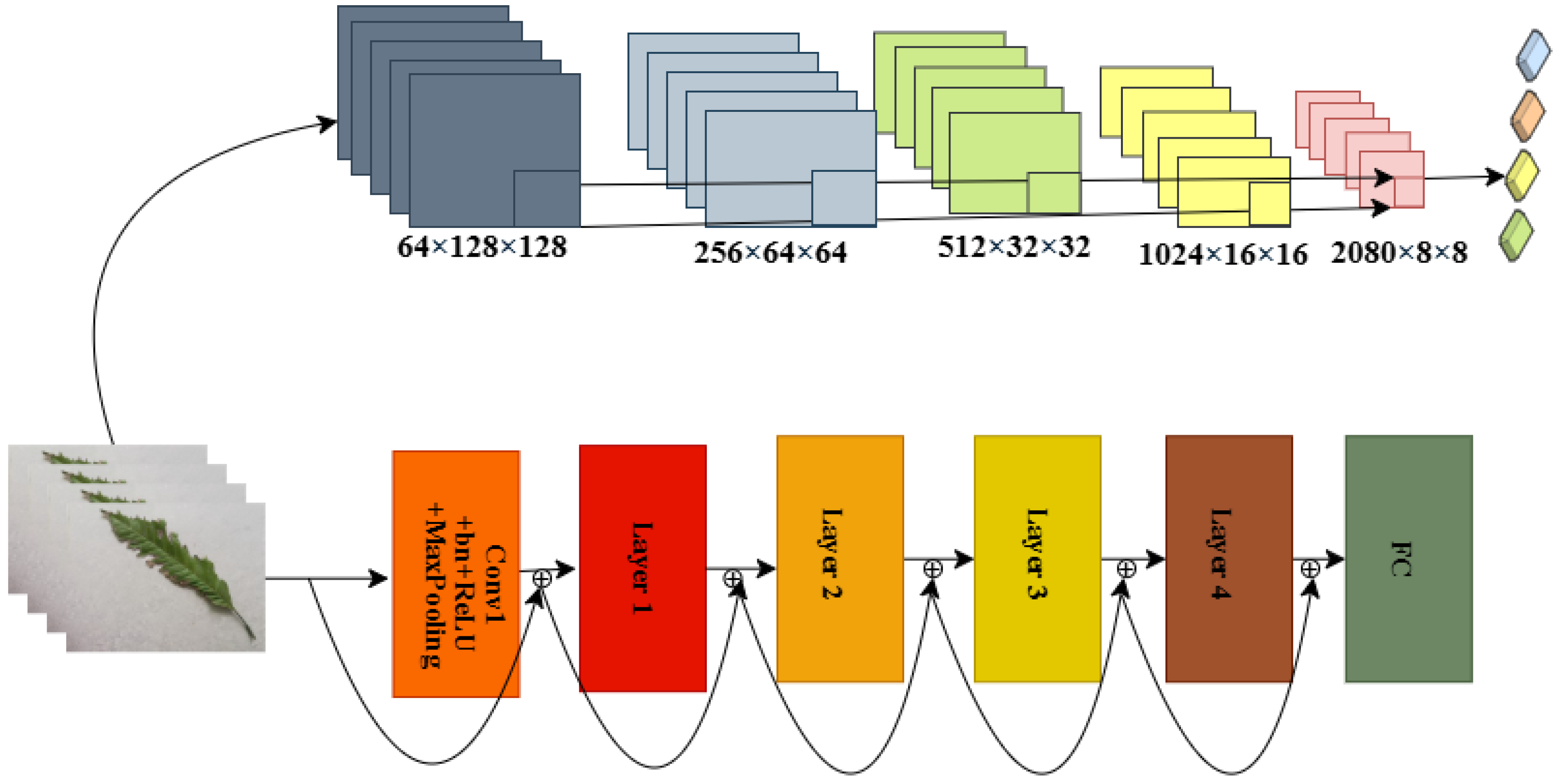
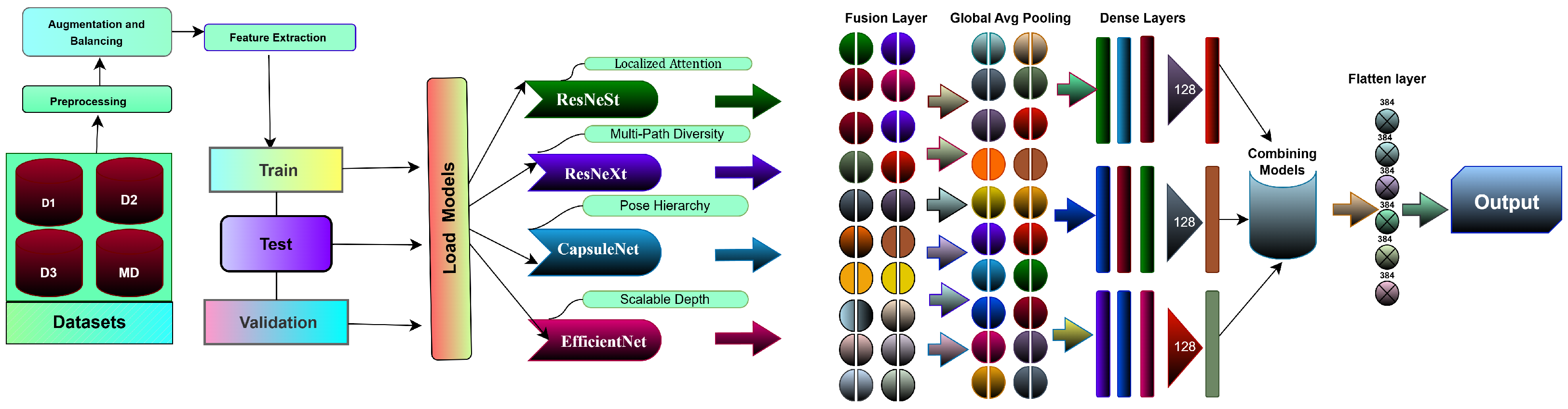
An overview of the proposed pipeline is presented in
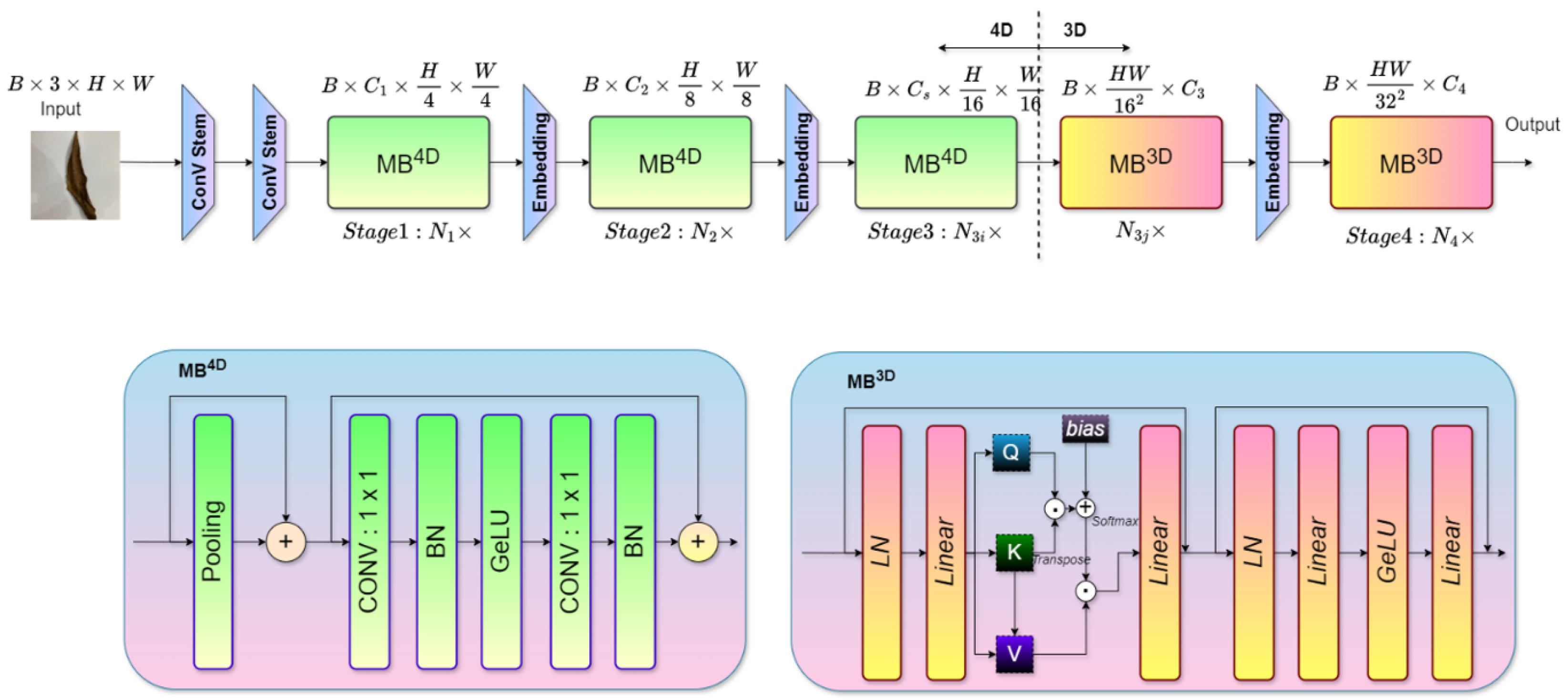
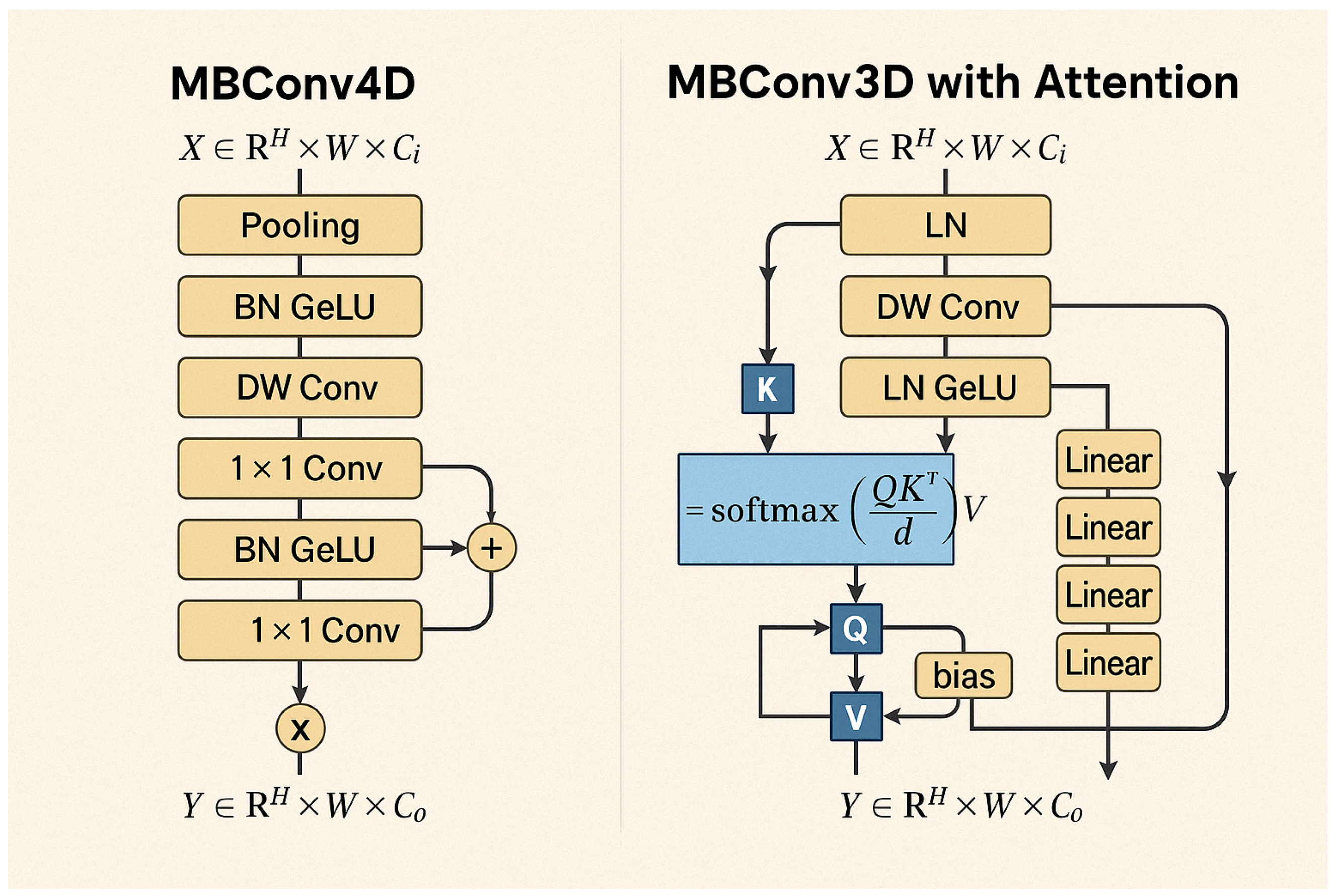
Figure 1. The approach utilizes four datasets—two balanced and two imbalanced—to ensure diversity. Preprocessing involves anisotropic diffusion filtering, followed by data augmentation to improve generalization. GLCM-based texture features are fused with high-level features extracted by transfer learning models and the proposed ViX-MangoEFormer architecture. The MangoNet-Stack ensemble learning is implemented by utilizing base learners such as ResNest, ResNeXt, CapsuleNet, and EfficientNet-B8. Features from these models are pooled and concatenated, then passed to a Flatten layer functioning as the meta-learner. All the models are compared to our proposed ViX-MangoEFormer model, a transformer-based architecture that uses MBConv4D and MBConv3D modules to capture local and global features in diseased leaf images. Multi-Head Self-Attention (MHSA) was used to improve the model’s capability to learn long-range dependencies. Finally, fully connected layers map the extracted feature embeddings to the corresponding mango leaf disease classes. Grad-CAM was used to provide visual explanations that enhance transparency and build trust among end-users. The best-performing model is integrated into a web platform that outputs disease classes, prediction probabilities, and Grad-CAM heatmaps for transparent interpretation.
While Vision Transformers have shown promise in agricultural image classification tasks [
20,
21,
22], most existing models face challenges, including high computational demands, inadequate fine-grained texture modeling, and limited applicability for real-time use. Previous approaches [
19,
23,
24] often require large-scale training data, are difficult to interpret, and tend to overlook the integration of handcrafted domain-relevant features. Unlike existing transformer-based studies that consider explainability as an afterthought [
25,
26,
27], our MangoEFormer model features native Grad-CAM integration within a web interface. This ensures that disease prediction outputs are not only accurate and fast but also transparent and actionable. Furthermore, our system demonstrated superior adaptability by validating performance across multi-source datasets, effectively addressing the domain shift. Our key contributions are as follows:
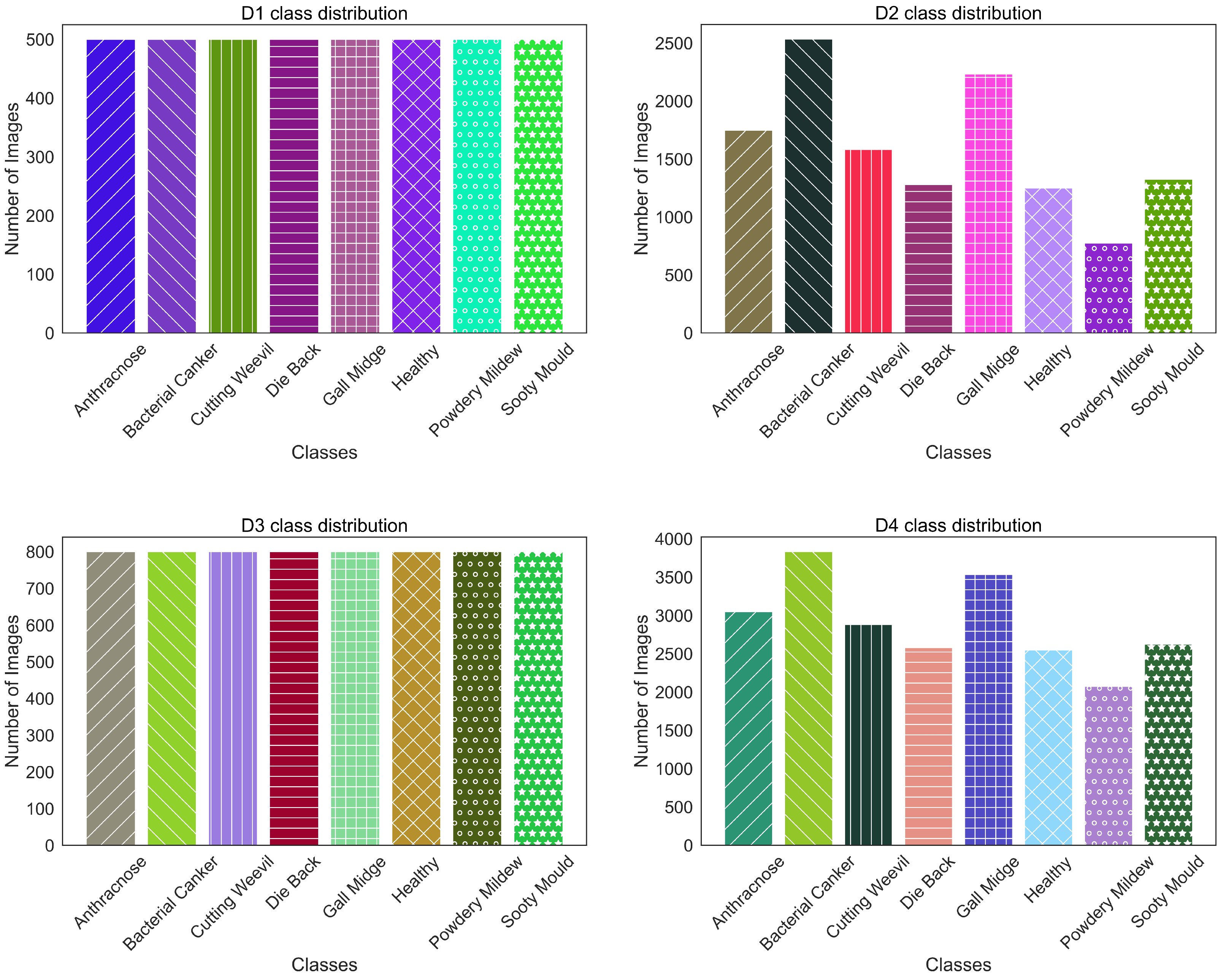
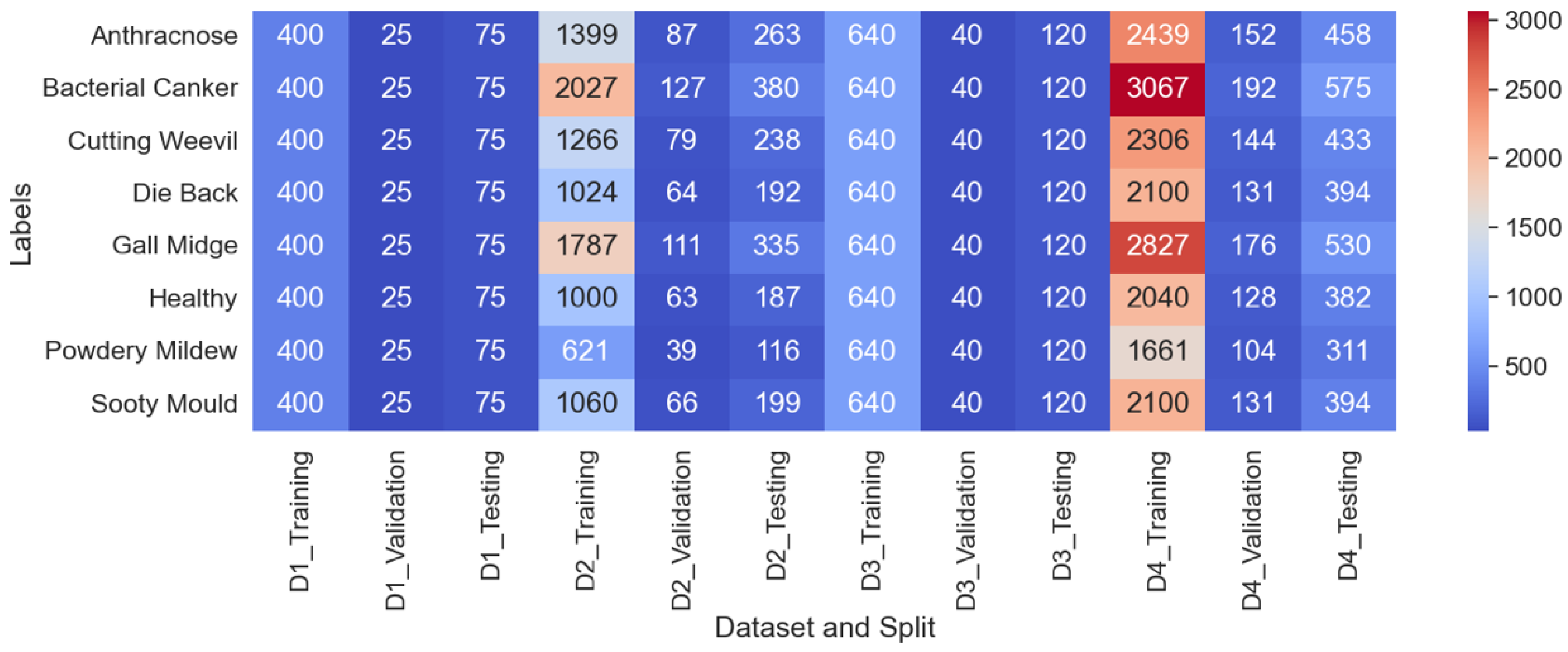
Developed an extensive mango leaf disease classification dataset comprising 25,530 high-quality images across eight distinct classes by combining four publicly available datasets. Its improved data diversity and sample size helped our study achieve state-of-the-art performance over prior studies.
Proposed a dual approach featuring a transfer learning-based ensemble MangoNet-Stack and a transformer-based ViX-MangoEFormer model, addressing key limitations in previous studies and achieving superior feature extraction and classification performance.
Compared the performance of all models on both augmented and non-augmented training sets, and on balanced versus imbalanced datasets, to thoroughly assess the impact of data augmentation and distribution on model accuracy.
Conducted class-wise error analysis to identify and mitigate misclassification issues and validated the optimized model on different crop leaves exhibiting similar disease classes as mango.
Deployed the optimized model via a web interface that delivers real-time disease detection along with probability scores and visual explanations.
The rest of the paper is organized as follows:
Section 2 reviews the research on using AI for classifying mango leaf diseases.
Section 3 outlines the methodology, including details about the dataset, feature extraction methods, and model architecture.
Section 4 presents the experimental results, evaluation metrics, and performance comparisons.
Section 5 discusses the findings, their implications, and practical applications. It also addresses the study’s limitations and suggests directions for future research. Finally,
Section 6 concludes with key findings and final thoughts.
4. Result Analysis
4.1. Comparative Performance Analysis
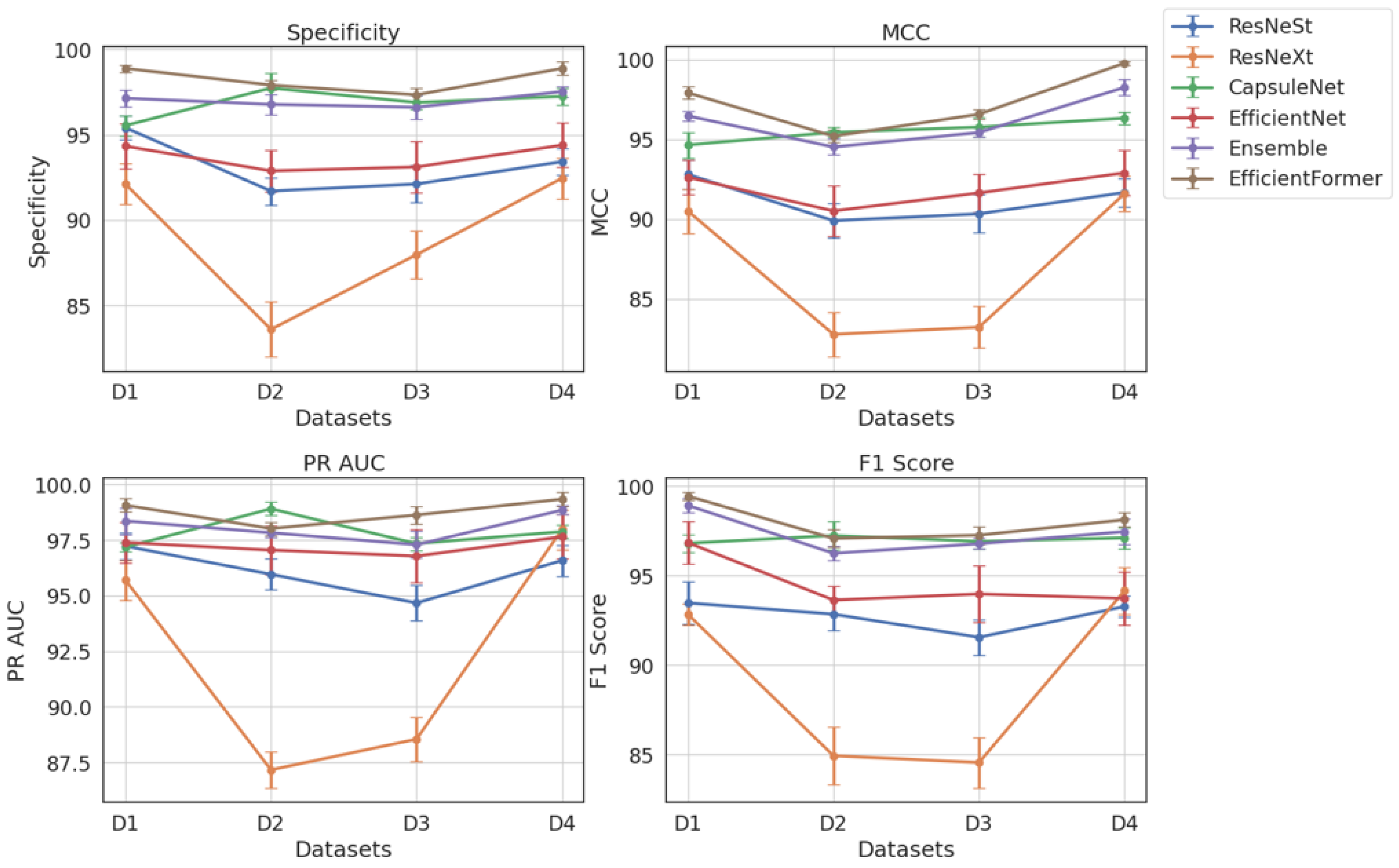
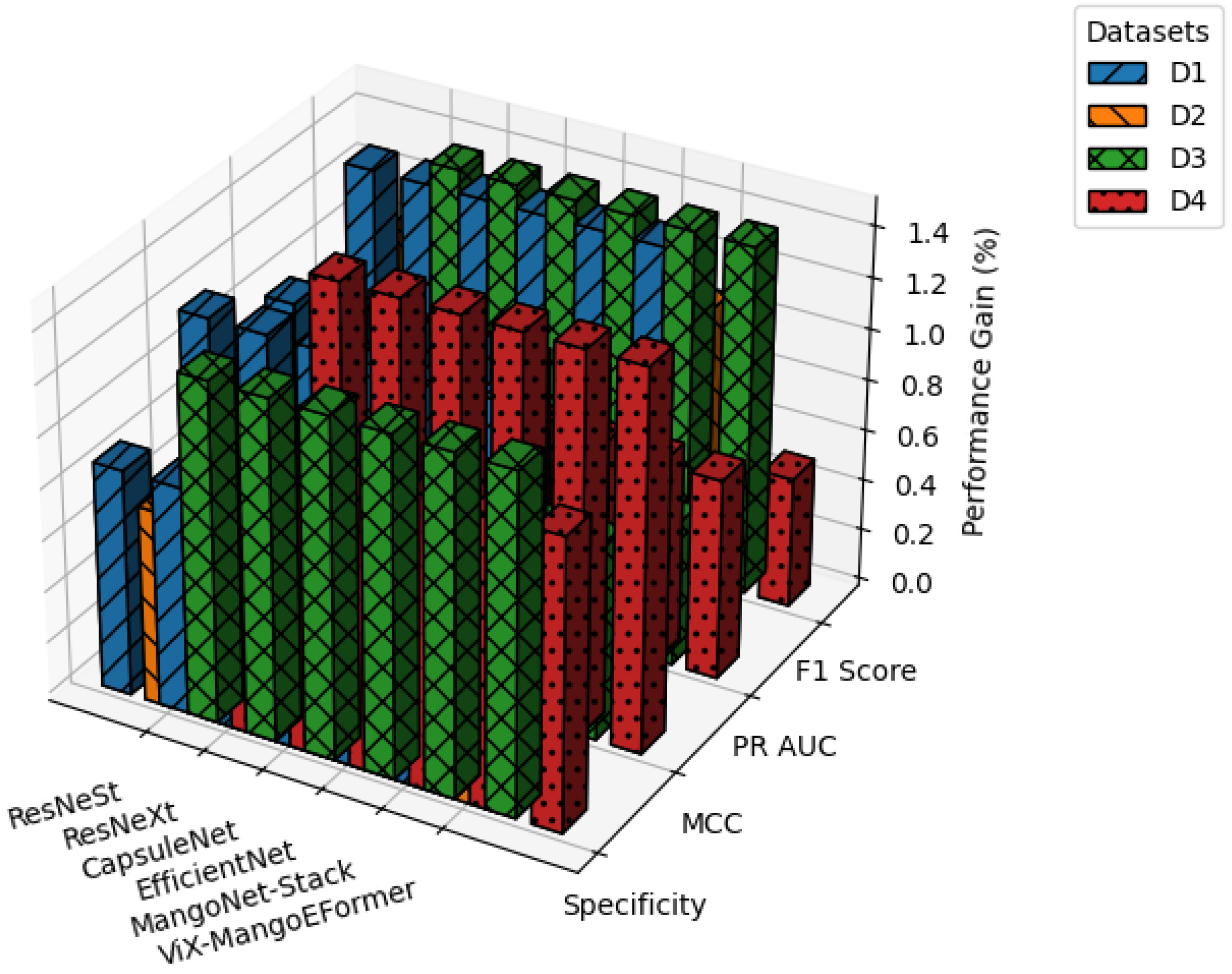
Table 4 presents a comparative evaluation of all models across balanced datasets (D1–D4). As shown in
Figure 17, ViX-MangoEFormer consistently ranks highest, securing the top position in three out of four datasets. In D1, it leads with a Specificity of 98.89 and an F1 Score of 99.43, outperforming both MangoNet-Stack (with scores of 97.14 and 98.91) and CapsuleNet (with scores of 95.54 and 96.80). ViX-MangoEFormer also demonstrates remarkable stability, with the lowest deviation in F1 Score (0.2), while ResNeXt shows greater variability, with a deviation of 1.2 in Specificity. In the more competitive D2 setting, CapsuleNet achieves the highest MCC of 95.44 and a PR AUC of 98.91, while ViX-MangoEFormer leads in Specificity and F1 Score. The differences between these top models are minimal, as both exhibit low variability (≤0.5). MangoNet-Stack remains a strong contender but does not surpass either model in any of the metrics. ResNeXt again displays instability, particularly in Specificity (1.6).
In D3, ViX-MangoEFormer maintains its lead, with a Specificity of 97.34 and a PR AUC of 98.63, closely followed by CapsuleNet and MangoNet-Stack. The proposed model exhibits consistent precision, with deviations in PR AUC and F1 Score of ≤0.5. In contrast, EfficientNet and ResNeXt show higher standard deviations (≥1.2), indicating less reliable predictions. Finally, in D4, ViX-MangoEFormer again dominates with the highest Specificity and MCC. Although MangoNet-Stack (with scores of 97.53 and 98.26) and CapsuleNet (with scores of 97.25 and 96.33) perform well, neither of them surpasses the proposed model. Notably, ViX-MangoEFormer maintains one of the lowest variances. Meanwhile, ResNeXt continues to demonstrate instability, with a variation in MCC reaching 1.3.
Analyzing the performance gaps in the non-augmented training datasets (see
Table 5) confirms the dominance of the ViX-MangoEFormer and MangoNet-Stack models. ViX-MangoEFormer consistently leads across most metrics, with its F1 Score in D1 surpassing the MangoNet-Stack’s by a noticeable margin. However, the MangoNet-Stack model remains competitive. The gap between MangoNet-Stack and CapsuleNet is more significant, particularly in D4, where MangoNet-Stack outperforms CapsuleNet in MCC. In D2, CapsuleNet performs best, with slightly higher MCC and F1 Score than ViX-MangoEFormer, but the difference falls within the standard deviation range.
Analyzing model performance on imbalanced data shows that ViX-MangoEFormer consistently outperforms the others, especially in D1 and D4, with scores of 98.71 in F1 Score and 98.23 in Specificity for D4 and the lowest variance. MangoNet-Stack is a strong second, particularly in D1 (95.52 MCC, 97.45 F1 Score) and D3 (95.56 MCC, 95.68 F1 Score), maintaining low variability. In D2, CapsuleNet leads in MCC and F1 Score, but ViX-MangoEFormer is better in Specificity. The close scores indicate comparable performance. CapsuleNet also competes well in D3, achieving 95.78 Specificity and 95.44 F1 Score, but it falls short in D1 and D4. ResNeXt consistently underperformed, with D2 and D3 metrics dropping to 81.46 MCC and 83.92 F1 Score, and showed high variability (e.g., ±2.3 Specificity in D2). ResNeSt and EfficientNet yield moderate results, with ResNeSt achieving 94.53 Specificity in D1 and EfficientNet a PR AUC of 96.10, but both fall behind top-tier models. ViX-MangoEFormer and MangoNet-Stack provide reliable predictions with low metric deviations, while CapsuleNet shows slightly higher variability. ResNeXt and ResNeSt exhibit the greatest fluctuations.
Figure 18 illustrates how data augmentation boosts performance across all datasets. Improvements are most notable in F1 Score and MCC of D1 and D3. D2 shows marginal gains, suggesting sufficient inherent diversity, while D4 sees moderate improvements in ViX-MangoEFormer and MangoNet-Stack. These results confirm that augmentation benefits hybrid ViT and ensemble models the most, whereas traditional CNNs like ResNeXt and ResNeSt exhibit limited responsiveness.
4.2. Performance Validation
The misclassification pattern of ViX-MangoEFormer and MangoNet-Stack on balanced and imbalanced datasets are summarized in
Table 6 and
Table 7, respectively. ViX-MangoEFormer and MangoNet-Stack performed similarly overall, but a detailed class-wise evaluation revealed that ViX-MangoEFormer consistently outperforms MangoNet-Stack, particularly in class imbalance situations. In balanced datasets (D1, D3), both models generally performed well on common classes like Healthy, Bacterial Canker, and Cutting Weevil. However, ViX-MangoEFormer achieved higher MCC scores (97.5–98.0) and PR AUCs (up to 99.5), while MangoNet-Stack scored lower (MCC 96.2–97.5). Minor misclassifications in Anthracnose and Gall Midge for MangoNet-Stack resulted in F1 Scores that were 0.2–0.3 points less, mainly due to visual similarities that ViX-MangoEFormer’s attention-based architecture managed to handle better.
In imbalanced datasets (D2, D4), ViX-MangoEFormer showed a stable performance, with MCCs above 97.0 and PR AUCs of 99.2–99.4. In contrast, MangoNet-Stack experienced significant drops in performance, especially for rare classes like Sooty Mould and Powdery Mildew. For example, in D4, MangoNet-Stack’s MCC for Sooty Mould dropped to 96.4, compared to 97.3 for ViX-MangoEFormer. The misclassifications mainly resulted in false negatives where rare classes were incorrectly identified as more common diseases, highlighting MangoNet-Stack’s bias in skewed distributions. Also, both Die Back and Gall Midge showed moderate performance declines moving from balanced to imbalanced datasets, with MangoNet-Stack struggling more with visually overlapping classes. Overall, misclassification patterns indicate that ViX-MangoEFormer is more resilient to distribution shifts, with fewer false positives and negatives across all datasets. In contrast, MangoNet-Stack, while effective on balanced data, shows degraded performance under class imbalance.
The paired
t-test results (see
Table 8 and
Table 9) indicate that ViX-MangoEFormer significantly outperforms other models across all datasets. It shows substantial improvements in MCC and F1 Score, with
p-values often below 0.005. On dataset D2, ResNeXt has
p-values ranging from 0.0001 (MCC and PR AUC) to 0.0002 (Specificity and F1 Score), maintaining similar low values on D3. ResNeSt performs slightly better than ResNeXt but remains inferior, with
p-values between 0.0001 and 0.0008 on D4 and 0.0036 to 0.0047 on D1. CapsuleNet is more competitive, with higher
p-values on D2 showing no significant difference. However, on D3, it has lower
p-values (0.0147 for MCC and 0.0175 for F1 Score), indicating better performance. EfficientNet is also a strong contender but is outperformed statistically, with
p-values decreasing from D1 to D4, suggesting it lacks the context modeling for highly imbalanced data. MangoNet-Stack is the next closest competitor, with
p-values near the significance threshold. In D3, it has
p-values for MCC and F1 Score, just above the significance level. However, in D2, ViX-MangoEFormer outperforms it, with
p-values of 0.0125 for MCC and 0.0138 for F1 Score, highlighting that while MangoNet-Stack is strong, its ensemble approach does not match the attention-driven representation of ViX-MangoEFormer.
To assess the generalizability and adaptability of ViX-MangoEFormer beyond mango leaf disease classification, the model was evaluated on datasets from Guava, Tomato, Grape, Apple, and Jute crops. These crops include the leaf disease category common in our dataset. The results in
Table 10 show that ViX-MangoEFormer demonstrates strong versatility and accuracy in handling diverse plant species and their disease categories. It excels in identifying Guava leaf diseases, with a Specificity of 98.51% and an MCC of 97.85%. Its MCC for Jute is even higher, indicating variability in symptoms among different species. However, the model faces challenges with Tomato leaf classification, achieving its lowest MCC at 95.95% and a PR AUC of 96.47%, showing difficulty in distinguishing similar symptoms. In contrast, Grape leaf disease classification yields the highest PR AUC of 99.58% and an F1 Score of 98.91%, demonstrating excellent accuracy. For Apple leaf diseases, the model records an MCC of 96.89% and an F1 Score of 97.43%, which are slightly lower than Grape due to subtler symptom variations. Notably, in Jute leaf classification, it achieves an F1 Score of 99.01% and a Specificity of 99.15%, showcasing its ability to reduce false positives.
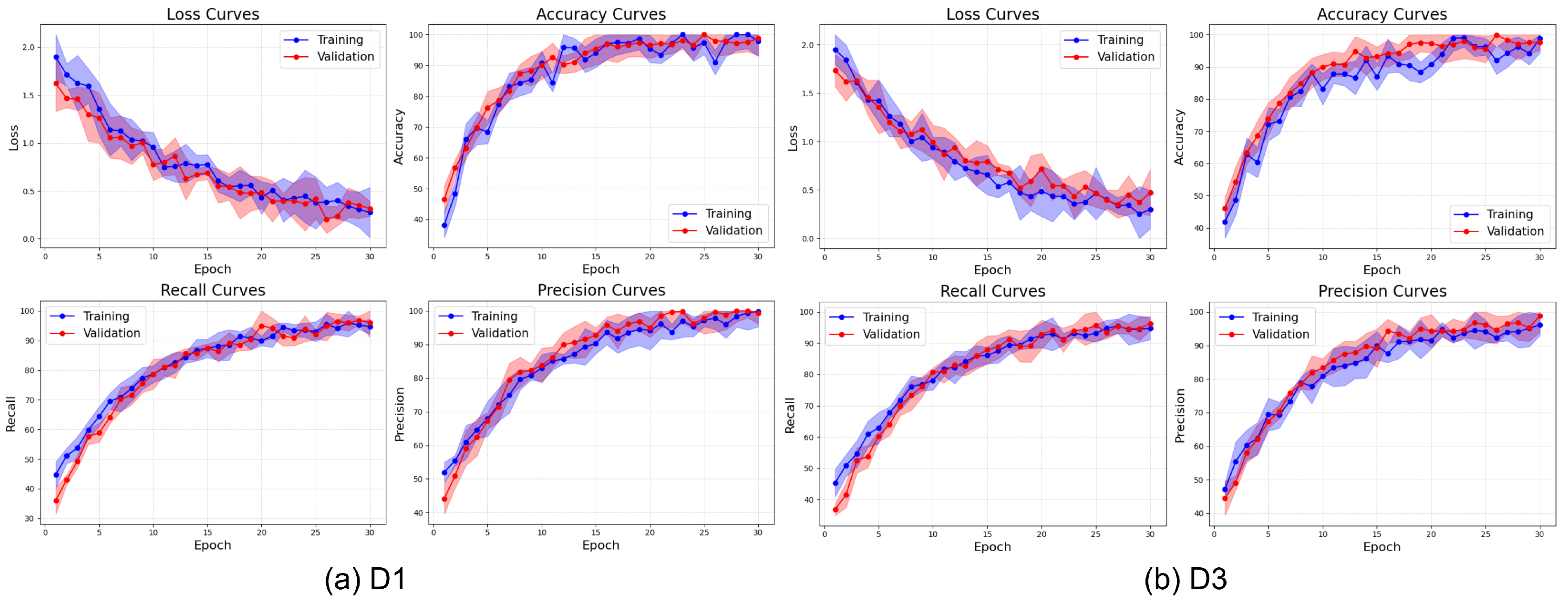
The learning curves presented in
Figure 19 illustrate the training behavior of ViX-MangoEFormer on the balanced datasets. The loss curves show a steep decline during the initial 10 epochs, followed by a gradual stabilization near zero, indicating effective convergence. D3 achieves a slightly lower final loss compared to D1. The close alignment between training and validation losses in both datasets indicates minimal overfitting. The accuracy curves also demonstrate rapid increases during the early epochs, with accuracy surpassing 98% after 15 to 20 epochs. Notably, D3 reaches peak accuracy at around 99% by epoch 10, while D1 takes more epochs to converge. This implies that ViX-MangoEFormer learns more efficiently from D3, possibly due to clearer class separability. The recall curves show the model’s capacity to identify true positives over epochs. By epoch 20, recall approaches 99% for both datasets. D1 has some fluctuation in validation recall during early training but starts with higher validation recall than training recall, indicating strong performance on unseen data. Precision also improves steadily, stabilizing between 98% and 99%. D3 achieves faster and smoother convergence, while D1 shows mild early variations. These trends suggest that ViX-MangoEFormer extracts features more effectively from D3, leading to more stable training results.
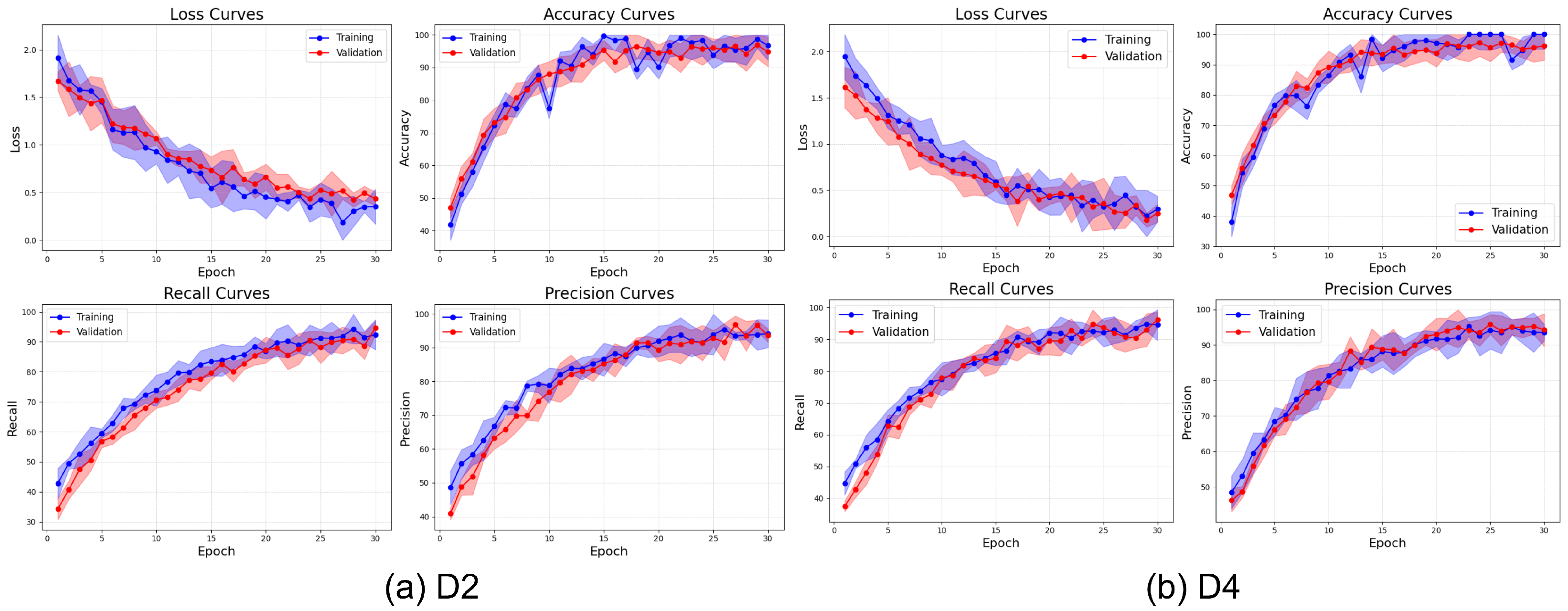
Figure 20 illustrates the learning behavior of ViX-MangoEFormer on the imbalanced D2 and D4 datasets. Both datasets show a rapid reduction in loss during the first 10 epochs; however, D4 converges more smoothly and achieves a lower final loss, indicating more efficient learning. The close alignment of training and validation loss in both cases suggests strong regularization and minimal overfitting. The accuracy curves exhibit similar trends, with both datasets surpassing 98% accuracy by epoch 15. However, D4 stabilizes earlier, implying that the model adapts more quickly when the class distributions are less skewed. In contrast, D2 takes more epochs to converge and demonstrates instability, particularly in the early recall and precision curves. D4’s steady progression in recall and precision reflects a balanced representation across classes, while D2’s variability, especially early in the training process, indicates that the model struggles to consistently detect minority classes. Although precision in D2 improves, it fluctuates more, reinforcing the model’s difficulty in establishing clear decision boundaries. These observations reveal that while ViX-MangoEFormer is generally robust, it performs more confidently and effectively on datasets with less severe class skew, such as D4.
4.3. Time Complexity and Runtime Analysis
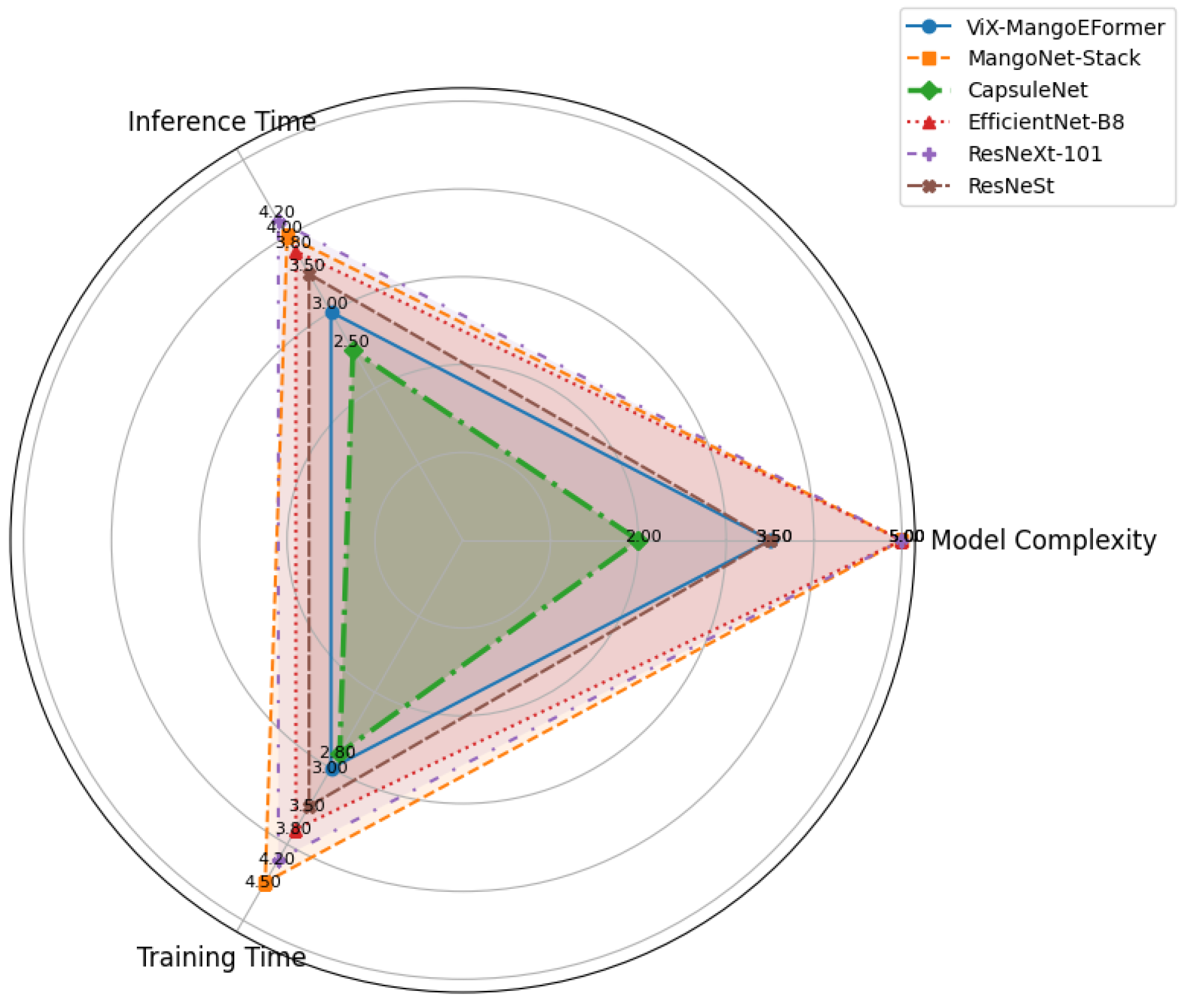
In addition to evaluating classification performance, we analyzed each model’s computational overhead by measuring training duration, model complexity (number of trainable parameters), and inference speed per image.
Figure 21 visually compares these aspects among the models. MangoNet-Stack and EfficientNet-B8 have the highest complexity rating of 5.0, indicating they are heavy architectures. ViX-MangoEFormer has a moderate complexity rating of 3.5, balancing performance and efficiency. CapsuleNet is the simplest model at 2.0, offering faster execution but limited capacity. In terms of inference time, CapsuleNet and ViX-MangoEFormer are the fastest, while MangoNet-Stack and ResNeXt-101 are the slowest, making ViX-MangoEFormer suitable for real-time applications. For training, CapsuleNet and ViX-MangoEFormer require less time compared to MangoNet-Stack and EfficientNet-B8, which demand more computational resources. In summary, ViX-MangoEFormer offers the best trade-off between model complexity, inference speed, and training time, making it particularly well-suited for efficient, real-time mango disease classification in practical settings.
Overall, ViX-MangoEFormer effectively balances speed, size, and accuracy, outperforming the ensemble-based MangoNet-Stack. Although it has more parameters than standard CNNs like ResNeXt-101 and EfficientNet-B8, its enhanced performance justifies the additional overhead. For large-scale or real-time mango leaf disease detection, ViX-MangoEFormer offers a practical trade-off between high accuracy and manageable computational costs. On the other hand, while MangoNet-Stack delivers superior overall accuracy, it requires more resources for training and inference. These insights can aid researchers and agricultural practitioners in selecting the best model based on their performance needs.
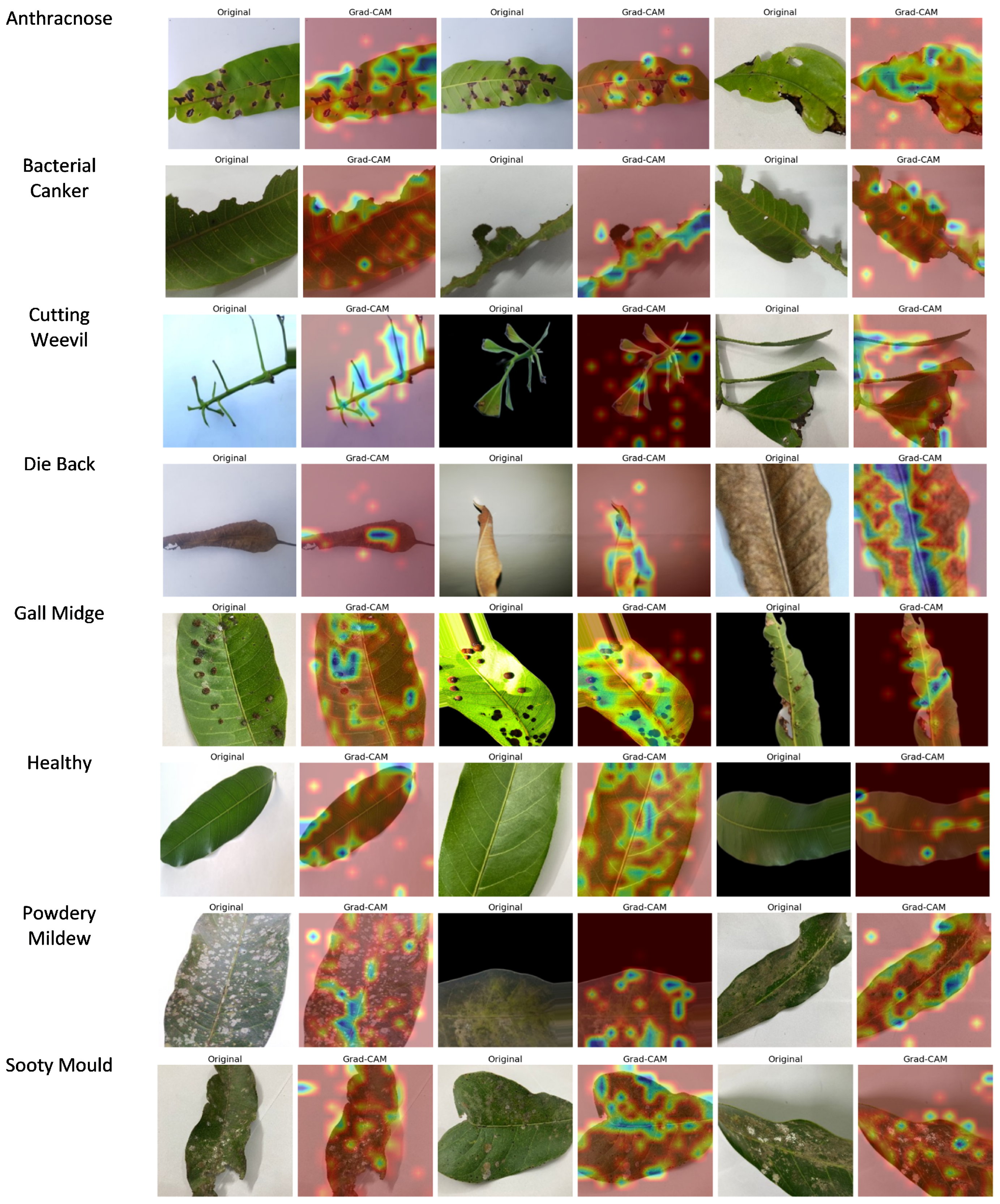
4.4. Grad-CAM Prediction Analysis
Grad-CAM was applied to the final convolutional layer of ViX-MangoEFormer to generate heatmaps that highlight critical areas in the input images of mango leaves. These heatmaps visually indicate which regions of the leaf were most influential in predicting disease presence or in classifying a leaf as healthy. By overlaying the heatmaps on the original images, professionals can verify whether the model correctly identified disease-affected areas or mistakenly focused on irrelevant regions.
Figure 22 presents class-wise Grad-CAM visualizations for ViX-MangoEFormer’s predictions on the D4 dataset. These output show that the model successfully highlighted regions that align with recognizable expert symptoms.
The Grad-CAM prediction for Anthracnose highlights dark, irregular necrotic spots on the leaves, indicating a focus on the characteristics of fungal infection. This suggests that the model effectively recognizes the severity and distribution of lesions, while ignoring background distractions. For Bacterial Canker, attention is drawn to brownish areas with yellowish halos, particularly near the midrib and veins. These features correlate with sites of bacterial infection and internal tissue degradation. The model successfully distinguishes this condition from other necrosis-based diseases, such as Anthracnose and Die Back, by concentrating on damage around the veins. In the case of Cutting Weevil, highlighted regions exhibit sharp, torn edges and missing tissue, reflecting the feeding behavior of the insect. The model accurately identifies these structural issues, demonstrating its understanding of the disease based on shape rather than color. With Die Back, Grad-CAM focuses darkened regions of the petiole and withered leaf tips, which are signs of nutrient deprivation and tissue death. This indicates that the model has developed a spatial understanding of how the disease progresses from the stem to the tip of the leaf.
For Gall Midge, predictions highlight small, round bumps on the leaf caused by insect larvae activity. The model focuses on the size, distribution, and surface distortion of these bumps, effectively distinguishing them from other diseases like Powdery Mildew by analyzing their 3D structure rather than just 2D visuals. In healthy leaves, there is low-intensity activation around the midrib, indicating that there are no disease features present and reinforcing confidence in the model’s classification abilities. For Powdery Mildew, heatmaps reveal white, chalky patches characteristic of the infection, confirming the model’s proficiency in detecting low-contrast textures. This class requires detailed texture learning, and the visualizations support the model’s capabilities. In the Sooty Mould class, activations appear on the black deposits found on leaves, which are linked to fungal growth resulting from insect honeydew. The model successfully differentiates these deposits from shadows or darker textures, showcasing its ability to recognize the subtle, unique features of Sooty Mould. These visual explanations are crucial for validating the model and serve as practical tools for agronomists and plant pathologists in diagnostic work.
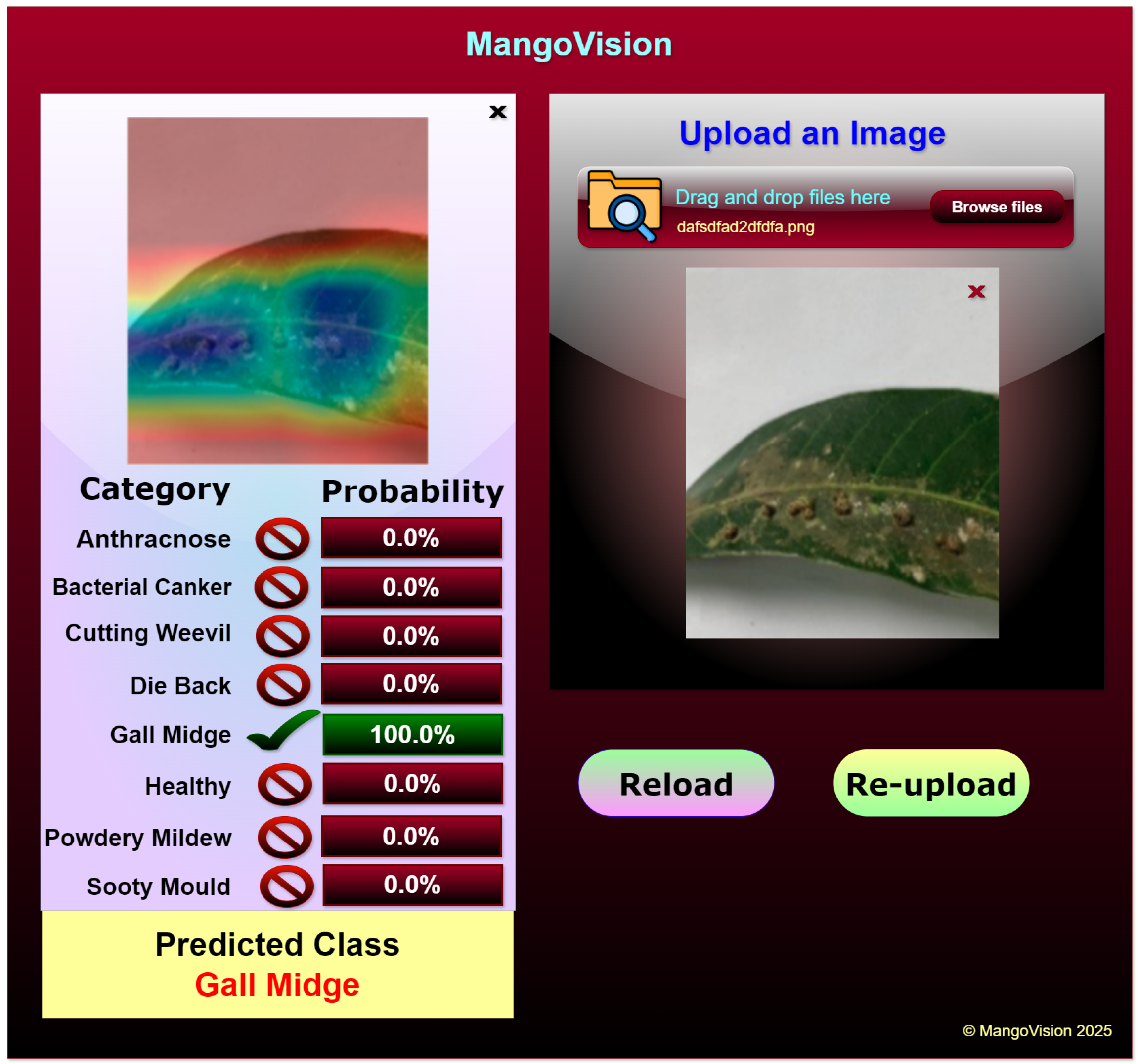
4.5. Web Application
The web application utilizes the Grad-CAM-enabled ViX-MangoEFormer model to classify diseases in mango leaves. It is specifically designed to assist farmers, agricultural experts, and researchers by allowing them to upload images of mango leaves for immediate disease detection. Once an image is uploaded, the model processes it and provides detailed classification results. The prediction results are displayed in a structured format, showing the probability scores for each disease category. For instance, in
Figure 23 the application accurately classifies the uploaded leaf image as Gall Midge with 100% confidence, while all other categories show a 0% probability, indicating the model’s certainty in its prediction. The left section of the interface displays a Grad-CAM-generated heatmap, which visually highlights the most influential areas in the input image that contributed to the classification. In this case, the activation map focuses on the gall formations along the leaf surface, confirming that the model correctly identifies the affected regions. This feature enhances the explainability of the decision-making process, allowing users to visually verify the areas that influenced the diagnosis. Its user-friendly interface allows easy reloading of images, enabling the testing of multiple samples and comparison of results.
In future, features such as real-time disease monitoring, predictive analytics, and integrated crop health systems could be added to expand its capabilities. For example, IoT-enabled sensors could be used to continuously monitor environmental factors like temperature, humidity, and soil moisture, all of which are crucial to plant health. These sensors could send data to the cloud via Bluetooth Low Energy (BLE) or Wi-Fi, where advanced predictive models could analyze the information and identify correlations with disease outbreaks. The application could also be linked to agricultural data platforms or government databases to create region-specific disease prediction maps. Notifications or alerts, delivered via Firebase Cloud Messaging (FCM) or SMS gateways, could inform farmers of potential risks, enabling them to take preventive measures. Integrating geographic information systems (GISs) could allow large-scale monitoring, providing policymakers with actionable insights for regional agricultural planning. Incorporating edge computing capabilities would further enhance the application’s performance by enabling real-time data processing with minimal latency. Such developments would make the application even more efficient and adaptable to various field conditions. With its scalable and modular design, the application bridges the gap between disease classification technology and practical agricultural applications.
4.6. State-of-the-Art Comparison
The analysis presented in
Table 11 shows that our ViX-MangoEFormer model outperforms existing models in classifying mango leaf diseases. It achieves this by combining high accuracy, comprehensive dataset coverage, explainability, and practical applicability. Previous studies, including those using InceptionV3 [
28], EfficientNetV2L [
29], and DeiT [
46], reported competitive results. However, they did not incorporate XAI or develop web applications. Additionally, these studies primarily assessed models on single datasets. While the ViT-B/16 model achieved a perfect accuracy of 100%, this result was based on a smaller dataset and focused only on binary classification. In contrast, our ViX-MangoEFormer model was trained on a much larger dataset consisting of 25,530 images, significantly surpassing the typical dataset sizes in prior studies, which usually ranged from 4000 to 6000 images. This extensive dataset is a key factor in the superior performance and robustness of ViX-MangoEFormer, ensuring its reliability across various scenarios.
ViX-MangoEFormer stands out for maintaining high accuracy across various datasets, achieving 99.43% on 4000 images, 98.02% on 12,730 images, and 98.63% on 6400 images. Its strong generalization capability makes it suitable for real-world applications. The model incorporates XAI techniques—this is a significant advantage over many earlier models that lack these features. Unlike most previous studies, which remain experimental, ViX-MangoEFormer has practical applications, making it accessible to farmers and agricultural experts. While other models, such as ConvNeXtXLarge [
41] and Swin Transformer [
44], are large and resource-intensive, ViX-MangoEFormer strikes a balance between complexity and efficiency, achieving top performance without excessive resource demands, making it ideal for real-time use.
5. Discussion
This study shows that data augmentation effectively reduces overfitting and improves classification performance across all tested models and datasets. By increasing training data while maintaining class labels, augmentation acts as a regularization strategy that enhances generalization, especially on imbalanced datasets (D2 and D4). It particularly helped underrepresented classes like Sooty Mould and Powdery Mildew, resulting in better metrics like MCC and PR AUC. The ViX-MangoEFormer model consistently outperformed all baselines, owing to its hybrid architecture that integrates convolutional modules for local feature extraction with transformer-based self-attention for long-range contextual modeling. This dual design allows the model to capture subtle lesion textures while attending to broader discoloration patterns. Moreover, its memory-efficient inference design, based on MBConv layers and sparse attention, further ensures low latency without sacrificing depth or resolution.
While ensemble learning has previously shown strong performance in leaf disease classification, ViX-MangoEFormer outperformed traditional CNN ensembles by effectively capturing long-range spatial correlations. Its transformer modules reweighted feature importance, allowing for better identification of subtle disease patterns. Additionally, integrating GLCM features improved the model’s sensitivity to fine texture statistics, which is essential for differentiating similar diseases like Sooty Mould and Powdery Mildew. By utilizing second-order intensity co-occurrence patterns, the model enhanced its discrimination capabilities. Furthermore, incorporating explainability mechanisms enhances the model’s trustworthiness. By integrating GLCM-based texture features with Grad-CAM visualizations, users can see which areas of the leaf were considered during classification. This transparency is crucial in agriculture as it helps build user confidence and supports regulatory compliance. Visual indicators like mold textures, leaf speckling, and vascular damage can be easily interpreted, allowing experts to validate model decisions and take action when necessary.
The implementation of ViX-MangoEFormer in a web application demonstrates its practical use. Designed for accessibility on laptops and smartphones, the application allows farmers and agronomists to conduct on-site disease diagnosis. The application is modular and can be extended to support additional crop types, disease categories, or geographic zones. This enables rapid decision-making regarding pesticide use, plant isolation, and irrigation adjustments. It features an interactive interface for real-time feedback and collaboration among agricultural extension teams. Despite its deep architecture, ViX-MangoEFormer achieves inference speeds of less than 120 milliseconds per image on mid-tier GPUs by optimizing attention computation and utilizing compact convolutional blocks. However, training the model remains resource-intensive, requiring approximately 18.7 GB of GPU memory and 42 GB of RAM over a span of 9.5 h on the merged dataset. While inference is optimized for edge devices such as the NVIDIA Jetson Xavier NX and T4 GPUs, the training pipeline currently relies on access to high-performance hardware.
However, there are limitations to consider. While the model has achieved high performance across various datasets, most leaf images were collected under consistent lighting conditions and clean backgrounds. This does not accurately reflect the variability encountered in real-world agricultural environments, where differences in lighting, occlusion, background clutter, and camera resolution can significantly alter the data distribution. To enhance generalizability, we plan to expand our dataset to include images captured in diverse field conditions and incorporate domain adaptation methods such as CycleGAN-based style transfer, test-time adaptation, and domain adversarial learning. These strategies aim to bridge the gap between laboratory conditions and practical deployments.
Although ViX-MangoEFormer is optimized for low-memory inference, its training complexity presents a challenge for institutions that lack high-end GPUs. The inclusion of GLCM-based feature fusion is advantageous for texture modeling but increases the computational load. To address this, future versions will explore model compression techniques such as quantization-aware training, structured pruning, and knowledge distillation. We are also considering the use of learnable texture encoders to replace handcrafted GLCM modules and to apply texture extraction selectively to disease-prone regions, thereby reducing overhead. Furthermore, integrating adaptive inference mechanisms, such as early exit branches, may enable dynamic computation in latency-sensitive scenarios.
Regarding interpretability, our Grad-CAM visualizations effectively localized critical disease areas, including necrotic zones, marginal blight, and chlorotic speckling. While this provides spatial insights, it does not fully elucidate the internal decision-making process of transformer modules. We propose incorporating advanced explainable AI (XAI) techniques, such as layer-wise relevance propagation (LRP) and concept-based interpretation, to achieve greater transparency. Additionally, using multispectral imaging, hyperspectral sensors, or RGB–NIR fusion can help differentiate similar disease types in complex conditions.
From a deployment and policy perspective, practical scalability requires a clear understanding of implementation costs, training logistics, and maintenance protocols. Anticipated implementation costs include GPU requirements, cloud hosting, and data storage infrastructure for large-scale adoption. Moreover, technical training for end-users and periodic model updates are essential to ensure ongoing accuracy and system relevance in dynamic agricultural environments. While formal usability testing has not yet been conducted, future studies will include a comprehensive cost-benefit analysis. This analysis will address model deployment, investments in cloud and edge hardware, training requirements, and the anticipated return on investment (ROI) in terms of yield improvement and pesticide reduction. Additionally, we plan to create a low-resource training pipeline that requires minimal hardware, allowing field officers to fine-tune models without needing extensive technical expertise. Lastly, pilot deployments across various agricultural zones will evaluate the framework’s scalability, adaptability, and stakeholder engagement, ensuring that the system can effectively influence real-world decision-making at scale.