1. Introduction
Cloud computing is now essential for real-time business applications. However, its widespread use has also led to several high-profile data breaches and losses. Cloud computing [
1] enables the protection of business applications’ data against system vulnerabilities and security attacks, promoting business continuity. One of the most significant security challenges in using cloud computing is protecting data through cloud-based systems, given the nature of the model (XaaS). The growing number of cloud computing service providers only increases the complexity of selecting a vendor to provide data storage services that can reliably protect sensitive data. This growing complexity has also increased the likelihood of an attack, leading to unauthorized access to this data when cloud-based services are deployed and used. As all cloud applications and data are stored in third-party environments via the Internet, protecting cloud data is paramount. Suppose an attacker successfully gains access to sensitive information stored in the cloud. In that case, the cloud user may suffer significant repercussions, including unauthorized access and the complete loss of that sensitive data.
To protect confidential information in transit over the cloud, a strong data confidentiality mechanism must be employed. The purpose of cryptographic algorithms is to encrypt data in transit using a mathematical transformation, with the additional requirement that such data be decrypted at the destination. A significant factor affecting the efficiency of cryptographic algorithms is proper key management. As more businesses of all sizes adopt cloud computing, there will be an increasing reliance on real-time applications that require continuous data transfer between cloud service providers and end-users. Therefore, the secure transmission framework for the proposed cloud services incorporated within the Subject Service will follow this model to emphasize the need for robust [
2], encryption of data during transmission. Researchers are beginning to investigate DNA-based cryptosystems as a new method to increase throughput, enhance security, and maintain confidentiality by increasing the amount of information stored digitally. Additionally, DNA computing offers a different approach to securely managing data in cloud computing environments, opening new areas of cryptographic design that are not currently addressed by existing technologies [
3].
Proper key management and robust data confidentiality practices can mitigate the risk of customer data breaches when utilising cloud computing. The growing number of potential customer breaches has heightened the risk in cloud computing. Recent incidents (Arby’s, OneLogin, ESEA, America’s Job Link, Verizon) demonstrate the inadequacy of some current practices in using cloud storage. An array of companies have repeatedly suffered major data breaches due to incorrectly configured systems or insufficient protection for sensitive data. There is an increased need for businesses to develop better ways to safeguard the privacy of their sensitive data as they continue to use cloud-based services to interact with their customers [
4]. Cloud service providers (CSPs) such as Amazon, Microsoft, and Google use the strongest and most reliable encryption technology available: typically, secret key encryption systems to encrypt user data and Public Key Systems (PKS) to distribute the secret keys. However, as threats to CSPs continually evolve and the number of security breaches increases, traditional methods for encrypting user data and distributing keys need to be enhanced to better protect users’ stored data [
5].
In contrast to the NDNA framework [
6], which uses a static DNA encoding table and a predetermined intron-ordered sequence, the proposed Secure DNA Cryptosystem (SDNA) will employ dynamic encoding-table creation, session-based key diversification, and biologically inspired intron randomization. These enhancements significantly increase the randomness of the encoding, minimize correlations in the ciphertext, and provide vastly superior performance for both encryption and decryption. Strong data confidentiality in cloud computing environments is necessary, but using traditional cryptography can impose high computational overhead, scalability challenges, and complex key management in lightweight/high-volume environments. These limitations have led to increased interest in exploring DNA-based cryptography, which offers the advantages of large key spaces, automatically generated random keys, and biologically inspired encoding. The use of the genetic code (DNA sequences) as a method for generating dynamic tables for symmetric encryption offers a potential alternative to traditional cryptographic methods, enhancing security and reducing computational overhead in modern cloud computing systems.
Novelty and Contributions
The proposed Secure DNA Cryptographic System (SDNA) provides several advancements over the prior NDNA design and current DNA cryptography systems as follows:
- (a)
Dynamic Encoding Tables: Unlike NDNA’s static encoding lookup table, SDNA generates the encoding lookup table per session using a Pseudorandom Number Generator (PRNG). As such, no mapping is reused, thereby reducing statement correlation.
- (b)
Randomized Intron Generation: NDNA used a fixed intron structure while SDNA utilizes a PRNG-seeded intron structure comprising six randomized components (uppercase, lowercase, odd, even, square, and multiple), resulting in greater entropy and session variability.
- (c)
Multiple Parallel Processing Blocks: SDNA also provides encodable processes that may be carried out concurrently during the encryption/decryption process while maintaining the security properties offered by the DNA-based approaches.
- (d)
Hybrid Cloud-Oriented Architecture: SDNA separates the DNA-based symmetric Encryption from asymmetric key exchange (RSA/ElGamal), improving the scalability, efficiency, and capabilities of key distribution.
The rest of the paper has been organized as follows:
Section 2 covers related work on alterations in the DNA cryptosystem,
Section 3 provides preliminaries,
Section 4 presents a secure DNA cryptosystem for cloud as an example,
Section 5 includes the results of the implementation of a secure DNA cryptosystem, and
Section 6 comprises a security analysis of the secure DNA cryptosystem.
2. Literature Review
Cryptographic algorithms can be classified as (i) single key cryptosystems (i.e, symmetric key, such as DES, AES, Blowfish, etc.) and (ii) public key cryptosystems (i.e, asymmetric key, such as RSA, ElGamal, etc.) In cloud services, three entities are considered primary roles: the Data Owner (DO), Data User (DU), and Cloud Storage Server (CSS). The DO can generate, modify, store, retrieve, and delete the application data. The DU can read and write the application data based on its privileges. The CSS can provide storage services for cloud users. In symmetric key cryptography in the cloud, the DO must generate a secret key and share it with the DU over a secure channel. Then, the DO generates the data and shares it with the CSS, encrypting it with a symmetric key cryptographic algorithm. The DU will decrypt the contents using the same key and the same symmetric key cryptographic algorithm. In the case of asymmetric key cryptosystem implementation in the cloud, the DO and DU generate public and private keys via a certificate authority. If the DO needs to share data with the DU, the DO retrieves the DU’s public key from a certificate authority and encrypts the data using the DU’s public key in an asymmetric cryptosystem. Then, the DO transfers the encrypted data to the DU in the cloud, and the DU decrypts it using its own private key in an asymmetric cryptosystem. The security of the asymmetric cryptosystem relies on key length and concepts from number theory. The security of the symmetric cryptosystem relies on confusion, diffusion, and key length. If the key length increases by a certain number of bits, the algorithm’s cryptanalysis appears to become more difficult. In terms of performance, throughput is high, and power consumption is lower with symmetric cryptosystems. To improve the implementation of cryptosystems in cloud environments, researchers have proposed modifications to DNA cryptosystems, which are analyzed in terms of security and the properties of an efficient DNA cryptosystem.
Table 1 summarizes the security-related properties of DNA cryptosystems that inform the comparative analysis of the reviewed works.
Aich et al. [
7] proposed a symmetric key cryptosystem based on DNA sequences. This approach uses a one-time password (OTP) to both encrypt and decrypt the message. Here, two encryption stages are performed before the data is sent to the channel. Then, a random number is generated and considered the original key (OTP). To produce the ciphertext for the first stage, an XOR operation is carried out between the original key (OTP) and the plaintext. During encryption, the translation table is applied. Here, the key serves as an OTP, a random number that is difficult to predict. Gupta et al. [
8] propose that the plaintext is transformed into artificial DNA sequences. The DNA sequence is then converted to bits using a binary coding scheme. Hussain et al. [
9] propose a new encryption scheme based on DNA cryptography, which satisfies the properties inherent to DNA. To fulfill the DNA properties, dynamic encoding tables are employed. The encoding tables are constructed by assigning the alphabet and symbols to the DNA sequence. The mapping process and the biological process of DNA are performed to obtain the ciphertext [
10]. Researchers’ contributions [
11] towards DNA cryptosystems highlight that DNA molecules can be used to scramble and transform confidential data into a meaningless form, thereby protecting privacy. The DNA cryptosystem [
12] works by identifying six properties related to security measures. Gugnani et al. [
13] proposed DNA cryptography using XML SOAP file encryption. Sensitive data is extracted and transformed into binary values and then into DNA bases with complementary pair rules. Then, the reference string is combined with the DNA sequences, yielding ciphertext as position values. The process does not involve generating character encodings specific to reference sequence bases. During the encryption phase, confusion techniques are employed, including shift and inverse shift operations [
14]. Paul et al. [
15] proposed an encryption technique based on an XOR operation with a one-time pad DNA sequence. The mirror image of the XOR operation ensures the unbroken result. The time complexity is analyzed. Marwan et al. [
16] propose a DNA-based data-hiding technique. The data is hidden in genomic DNA using a new, improved method of DNA steganography. Here, security depends entirely on the key. Hossain et al. [
17] proposed a DNA cryptosystem that relies on a generated sequence table and random ASCII characters. The OTP method is applied to modify the plain-text encoding. The DNA sequence table is generated through iterations, and the amino acid mapping table is also generated to facilitate randomness and diffusion of the ciphertext. Logical operations, such as XOR and XNOR, are performed. The proposed technique provides greater security than other systems and is protected against attacks such as brute-force and collision attacks. Earlier, variants of DNA cryptosystems [
6,
18] were proposed to maintain data privacy during storage and retrieval by incorporating DNA properties into security measures.
Khashan et al. [
19] introduce OutFS, a user-side encrypted file system that provides transparent encryption for stored and shared outsourced data. OutFS uses a hybrid encryption scheme that combines symmetric and asymmetric methods. The key management is designed for convenience. To enhance data-sharing security, an identity-based encryption (IBE) scheme is employed. OutFS is intended to preserve the integrity of outsourced file data and the file system’s data structure. The research analyzes performance and experimental results, which indicate that OutFS is efficient. OutFS achieves an average throughput of 8.8 MB/s and, for writing and reading outsourced files, the throughput is between 8.7 and 10.5 MB/s. Security analysis indicates that the OutFS system is very secure and robust against attacks such as brute-force, eavesdropping, man-in-the-middle, and offline dictionary attacks.
Namasudra et al. [
20] proposed a novel, secure, and fast DNA-computing-based Access Control Model (ACM). In the proposed scheme, the Cloud Service Provider (CSP) maintains a table or list for quick data access. A 1024-bit random key is generated from the user’s secret information and used to encrypt data. A theoretical analysis and numerous experimental results are presented, demonstrating the efficiency and effectiveness of the proposed access control model compared to other existing models. Pavithran et al. [
21] proposed a novel cryptosystem based on finite automata theory and deoxyribonucleic acid (DNA) cryptography. Three components make up the system: a sender, a receiver, and a key pair generator. Using the receiver’s characteristics, the sender generates a 256-bit secret key based on DNA, which is then used to encrypt the data. The DNA sequence is then coded using a randomly generated Mealy machine, increasing the security of the ciphertext. Numerous security threats, including brute-force, known-plaintext, differential cryptanalysis, ciphertext-only, man-in-the-middle, and phishing attacks, can be thwarted by the proposed technique. The findings and conversations demonstrate that the proposed plan is more secure and effective than the current ones.
He et al. [
22] presented a highly effective ciphertext retrieval scheme based on homomorphic encryption for multiple data owners in a hybrid cloud, known as CRHM, in which a public cloud server and a private cloud server work together to enable ciphertext retrieval. The research design included an encrypted balanced binary index tree structure and a homomorphic encryption scheme based on large integer operations in CRHM to support the “multiple owners” mode, and multi-keyword ranked retrieval. Security analysis shows that CRHM can effectively protect user files and privacy during retrieval. Performance evaluation shows that CRHM is highly efficient in index generation and retrieval compared to related schemes, while maintaining relatively high retrieval accuracy. Pavithran et al. [
23] propose a new encryption method based on a Moore machine, a hyper-chaotic system, and deoxyribonucleic acid (DNA) cryptography. The hyperchaotic system generates four pseudo-random number sequences utilized in DNA-based processes. By changing the DNA sequence, the Moore machine increases the system’s security. The suggested method can defend a system against various attacks, including brute-force, known-plaintext, ciphertext-only, man-in-the-middle, and differential cryptanalysis. With an average avalanche impact of 54.75%, the suggested strategy ensures high robustness. Additionally, experimental results demonstrate that the proposed scheme outperforms existing schemes in both efficiency and security.
Sohal et al. [
24] introduced a new cryptographic method that encrypts data before uploading it to the cloud using client-side data encryption. Based on DNA cryptography, this is a multifold symmetric key cryptography approach. In addition to outlining the specifics of the strategy, the research contrasted it with the current symmetric key algorithms (DNA, AES, DES, and Blowfish). The experimental findings show that, in terms of ciphertext size, encryption time, and throughput, our proposed technique outperforms these conventional algorithms. Therefore, the recently suggested method is more effective and provides superior results. Kumar et al. [
25] propose an encryption strategy that uses ECC (Elliptic Curve Cryptography) in conjunction with AES (Advanced Encryption Standard) to efficiently protect sensitive data in the cloud and, more importantly, safeguard the user’s personal information against adversaries. This new method is demonstrated to be viable, yielding superior, more effective results.
Rao et al. [
26] outline a public cloud security framework based on Hybrid Elliptic Curve Cryptography (HECC). The proposed method generates keys using the lightweight structure of Edwards curves. The author’s identity-based encryption varies the produced private keys. The author’s proposed key-reduction technique minimizes key length to speed up the Advanced Encryption Standard (AES) encryption process. The Diffie–Hellman exchange would then be used to exchange public keys. Throughput and key generation, encryption, and decryption times are used to assess the proposed model’s performance. The proposed model outperformed all existing models in all aspects. The proposed model’s key generation time is 0.000025 s, yielding an encryption time of 0.00349 s for the resultant ciphertext. The throughput achieved is 693.10 kB/s. Beggas et al. [
27] introduced a new method for generating unpredictable, random keys for symmetric OTP (One-Time Pad) cryptosystems. A self-assembly structure, computational processes, an entropy source, and a chaotic function are utilized in the OTP key generation process to enhance the unpredictability of the generated OTP keys. This process involves creating and reassembling OTP keys of different lengths, each of which is less than 1 MB in size. The proposed two-stage secure transmission method offers a high level of security. In the first stage, secret parameters are sent over a public channel using an OTP-based encryption scheme. In the second stage, a very short secret key is transmitted via an asymmetric method. This approach also minimizes and optimizes the public and secure communication channels. The advantages of a secure key exchange strategy are discussed, and it is recommended to optimize and mitigate both public and secure key exchange channels. This method does not include an analysis of encryption and decryption.
Rahul et al. [
28] present an efficient, well-performed image encryption scheme based on dynamic DNA encoding and chaotic maps with relatively simple structures and high chaotic behaviour, such as the Logistic map, Henon map, and Lorenz system, which provide much stronger security for digital images. In addition, the scheme uses the SHA-256 hash and zig-zag traversal to further help in covering the scheme. This research also proposes an improved scheme for encoding the DNA of four bits at once rather than two. The scheme also generates unique keys for each encryption and decryption session. The proposed scheme possesses several good features, including a low processing cost, high randomness, ample key space, flexible parameter space, high sensitivity for both keys and plaintext, and fast speed. Consequently, this scheme effectively protects sensitive digital images against a variety of cryptographic attacks. Various evaluations indicate that the scheme is more secure and efficient than state-of-the-art methods against a wide range of cryptographic attacks. Vaishali et al. [
29] proposed a novel approach to protect data during communication using bioinformatics and the Diffie–Hellman Key exchange. The cryptosystem proposes an encryption and decryption technique that utilizes the full Central Dogma of Molecular Biology (CDMB), which describes the process by which DNA is transformed into proteins. The Hellman–Diffie algorithm generates keys via a key-exchange approach and incorporates several additional security enhancements. Even on large datasets, the proposed bio-inspired cryptosystem demonstrates a cryptographic efficiency superior to that of existing systems. Furthermore, it develops a highly secure, fast cryptosystem that protects data against various internet-based threats.
Selvakumaer et al. [
30] described a cryptographic algorithm (encryption and decryption) that uses Huffman coding and DNA cryptography to securely communicate private digital healthcare data. The intriguing feature is that the size of the cipher produced by the technique is the same as the size of the cipher created using the character set of the provided data. To demonstrate the security of data when stored and transferred to the cloud, a security analysis is provided. The requirements for cryptography, key space analysis, sensitivity of keys and plain text, sensitivity and specificity, sensitive score analysis, optimal threshold, randomness analysis, uniqueness of implementation, entropies of binary bits, DNAbases, DNAbases with Huffman code, Huffman encoded binary bits, and the risk of cloud service providers are all examined. When the suggested method is contrasted with other cryptographic techniques, it is found to be more robust and secure. Vadladi et al. [
31] developed an ECC-based authentication and integrity-checking architecture using an internal error-correcting code technique. It generates the DNA code and adds an encrypted message, making the authentication procedure more robust. Here, ECC is utilized to encrypt the original plaintext, providing increased security for IoT device authentication while using less memory, space, and power.
Zitouni et al. [
32] propose a new lightweight, energy-efficient block cipher based on DNA cryptography, called “LWBC_DNA”. LWBC_DNA combines DNA cryptography and lightweight cryptography, and its architecture features a hybrid substitution-permutation network and a Feistel network. The LWBC_DNA cipher encrypts blocks of 64 bits, uses a 16-bit key, and performs 16 iterative rounds of simple operations, including concatenation, XOR, and XNOR, to produce a 32-bit ciphertext. An evaluation of performance and security has shown that the LWBC_DNA cipher provides excellent protection and meets IoMT device requirements for simplicity, storage space, and energy consumption. Furthermore, the security analysis confirms that the LWBC_DNA scheme is highly resistant to a variety of cryptographic attacks. Kairi et al. [
33] proposed a hybrid approach that provides robust cloud data security by combining machine learning with DNA-based cryptography encoding. The proposed approach presents an adaptive model that utilizes supervised machine learning methods to optimize DNA cryptographic operations, including complementary rules, XOR operations, and DNA encoding. These techniques leverage performance metrics and anomaly detection to dynamically enhance encryption and decryption. Without sacrificing processing speed, experimental results on benchmark cloud datasets demonstrate notable improvements in encryption strength, key management, and defense against frequent attacks.
In Djaa et al. [
34], SDEAP is a novel, lightweight, symmetric DNA encryption technique inspired by protein synthesis. It draws on the fundamental principles of molecular biology and leverages the randomness of DNA to generate a powerful OTP key. To create an algorithm with straightforward stages and intricate security levels that make the resulting ciphertext difficult to decipher, the research leveraged characteristics of protein synthesis. Both the key and the plaintext are transformed into proteins. To create a ciphertext in protein form, a fresh XOR operation between proteins is then carried out. In contrast to previous efforts, the research proposes an additional encryption level to securely transmit the produced keys along with the ciphertext in a message of the ideal size. SDEAP is simulated in an IoT environment using the Cooja simulator, available in Contiki OS. The results demonstrate that SDEAP is more effective than SIMON and PRESENT in terms of time and power usage, with reductions of 60% and 93%, respectively. Jero et al. [
35] aim to safeguard data from various risks and minimize overlapping possibilities by developing an enhanced Cloud Computing Security (CSS) model. The purpose of SHA-512/256 is to leverage user-identifying information to generate a fixed-length hash. Additionally, the data is compressed using Deflate, reducing the byte size and saving storage space. The data is then transferred to the encryption processing stage, where hybrid Chaotic-DNA (CDNA) encryption is applied. The Chaotic-DNA process encrypts data using a chaotic workflow and provides DNA-based key generation for an efficient security procedure. A trusted cloud center creates the key for the hybrid encryption technique based on the data’s sensitivity. The user must overcome four authentication hurdles to access the stored data from the cloud system: password, user ID, OTP, and fingerprint. The proposed model achieves 98% security and a 2 s authentication time, as assessed across various data types, including text, integers, and images. Data in that image takes 90 s to encrypt and 0.18 s to decrypt, text data takes 149 s to encrypt and 0.69 s to decrypt, and integer data takes 101 s to encrypt and 1.4 s to decrypt. This experimental investigation demonstrates that the proposed security approach effectively minimizes overlap and utilizes space more efficiently.
Selvi et al. [
36] proposed a unique approach for secure cloud-based healthcare solutions, structured into user, doctor, and cloud modules to manage patient data and generate treatment suggestions. This model addresses inefficiencies in current encryption implementations by enabling high-speed cryptographic processing with strong security, mitigating constraints on the real-time storage and retrieval of medical data. Statistical analysis has demonstrated that it will outperform existing cryptosystems by 25–40% in terms of operating overhead. In addition, it presents the integration of machine learning (ML)-based depression detection as effective in an encrypted environment for privacy-preserving analyses. The results indicate sufficient promise for the significant uptake of SymECCipher in healthcare settings, as it can provide a scalable, quantum-secure, and blockchain-compatible encryption framework. Future work can be extended by integrating lattice-based cryptography to improve quantum security and by expanding SymECCipher beyond healthcare to wearable health devices and telemedicine platforms. The method of Kumaran et al. [
37] provides a new hybrid encryption system that combines DNA cryptography with Elliptic Curve Cryptography (ECC). While DNA-based coding shows high randomness and equality, ECC provides significant security and confidentiality. The proposed method employs DNA encoding and secure key generation to obtain the medical image. The hybridization of these two methods addresses many of the main limitations of existing methods by increasing security and computational efficiency, making it suitable for real-time medical applications. The experimental analysis was performed using various parameters, including histogram analysis, correlation coefficient, Chi-square, MSE, PSNR, and entropy. The results show that the proposed method outperforms state-of-the-art methods, achieving an entropy value of 7.9981, a correlation coefficient of 0.0019, and a PSNR of 53.97. In addition, the proposed method was tested for runtime, memory usage, and security.
Comparative Analysis of Existing DNA Cryptosystems
Table 2 summarizes potential algorithms, including techniques, encryption and decryption methods, results, limitations, and year of publication.
Our contribution is to enhance the DNA cryptosystem for cloud data storage and retrieval, improving performance and security, reducing computational time, enabling dynamic operations, and facilitating effective key generation.
3. Preliminaries
The DNA cryptosystem is a groundbreaking approach to encrypting data using DNA sequences. Compared with existing cryptosystems, the DNA cryptosystem offers a significantly higher storage capacity. In one single gram of DNA, 700 terabytes of data can be stored. On Earth, the availability of DNA base pairs has been estimated to be 5.0 × 1037. DNA molecules can be used to solve computational problems because they contain genetic information related to the development and growth of living organisms. DNA, or deoxyribonucleic acid, is the hereditary material in almost all organisms, including humans. In the development, growth, reproduction, and functioning of all living organisms, DNA forms a coiled double helix consisting of two antiparallel polynucleotide strands. The two strands (i.e., polynucleotides) are a collection of monomer units known as nucleotides. Each nucleotide comprises four nucleobases: A (Adenine), C (Cytosine), G (Guanine) and T (Thymine). The base pairing rules for the two separate polynucleotide strands are C with G and A with T (via hydrogen bonds). In DNA cryptography, base pairs form an information carrier. The DNA cryptosystem encrypts the user’s data and outputs the results as a DNA sequence (A, C, T, G). In the first phase, data are considered to be text, image, and audio. The second phase involves binary conversion, which is stored as binary data or any other numerical system, depending on the researcher. The nucleotides are associated with binary values (i.e, A—00, C—10, T—11, and G—01) for the transformation of DNA sequences. DNA encryption is the next phase, which encrypts the binary data. The encryption process varies from author to author. This is performed through biological operations, including DNA bimolecular, One-Time Pad (OTP), DNA chip technology, DNA fragmentation, and Polymerase Chain Reaction (PCR). It can also be achieved through logical operations, such as XOR and XNOR. The next phase is DNA conversion. Amino acids in protein synthesis and a defined character set are used to generate intermediate results in the DNA cryptosystem. The ciphertext will be completely different from the plaintext, and it is not easy for an intruder to create the plaintext from the given ciphertext.
Terminology and Notation
- (a)
intron sequence (inseq): This is a portion of non-coding DNA that is added to a data sequence as a means to increase security while generating tables by providing randomness.
- (b)
collate_character (cc): A collection of pre-defined characters used to align or map the output of amino acids to cipher characters used in constructing the encoding tables.
- (c)
collate_amino (ca): An ordered list of the amino acids used to make the mapping space larger (for example, cs64 → cs256) and create a codon-to-amino-acid lookup table.
- (d)
D4/D64: A term used for DNA-derived codon matrices. The D4 version refers to the first 4 × 4 table created from two tRNA sequences, while the D64 version expands that to a 64 × 4 table used for encoding.
- (e)
NDNA Previously a DNA-based cryptosystem model using static encoding tables and fixed intron sequences to encrypt data.
- (f)
DO1t/DO2t/DO1m/DO2m: Intermediate biological transformations of the two data-owner DNA sequences. DO1m and DO2m are mRNA versions of these sequences, while DO1t and DO2t are tRNA complements of this information, which are used to generate D4 and D64, respectively.
4. Proposed Work
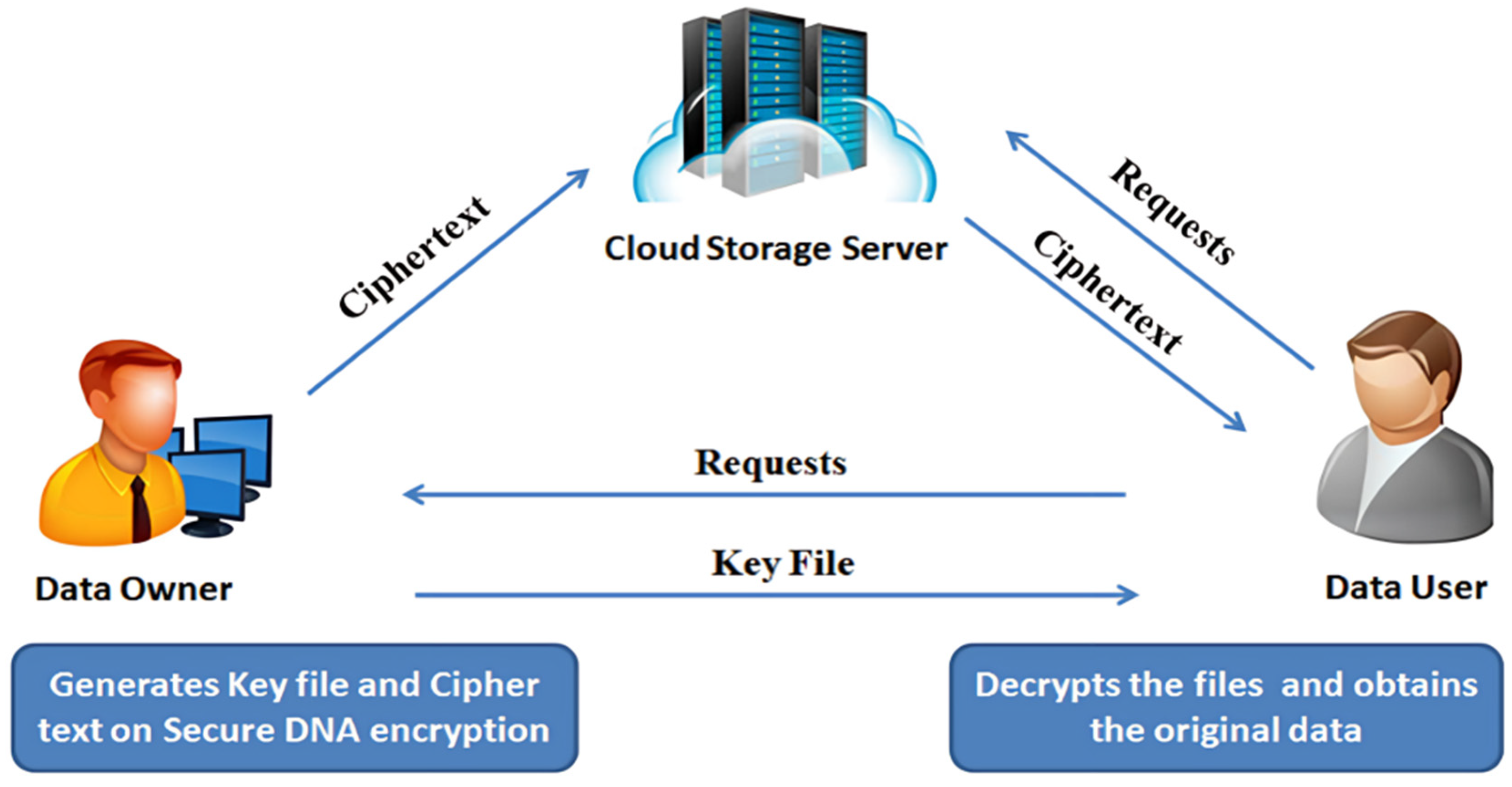
The proposed scheme provides a secure cloud data framework with specific roles for each user performing data operations. In the proposed scheme framework, the DO is responsible for encrypting and storing data in the cloud. At the same time, the DU retrieves and decrypts the data using a key shared by the DO via a secure channel. The framework users are as follows:
DO—stores the data on Cloud Storage Server (CSS);
DU—retrieves the data from the Cloud Storage Server (CSS);
CSS—handles DO and DU requests.
Initially, the DO selects two random DNA sequences, referred to as DO Sequence 1 and DO Sequence 2. Next, the DO generates a secure DNA-encoding table from the sequences. Then, the DO encrypts the data using the secure DNA encryption algorithm, producing a ciphertext file and a key file. The ciphertext file is placed in the cloud storage. The DU places requests for the ciphertext file and retrieves it from CSS. Then, the key file is requested from the DO. The key file is shared by encrypting it with asymmetric key cryptosystems over a secure channel [
26]. Then, the DU decrypts the key file using the same asymmetric key cryptosystem and the secure DNA decryption algorithm, producing a plaintext file.
Figure 1 shows the overall workflow of the proposed SDNA scheme for cloud data storage and retrieval. Thus, the proposed scheme secure cloud data framework includes two types of cryptosystems: (i) symmetric key cryptosystem—encrypt and decrypt cloud data using SDNA; (ii) asymmetric key cryptosystem—share the key files between the DO and DU in a secure channel using existing algorithms like RSA, ElGamal, etc.
The SDNA ensures data secrecy in the cloud with unparalleled security. The security of a DNA cryptosystem lies in the biological processes, randomness, and dynamism of its encryption, decryption, and table-generation processes. The inclusion of DNA cryptosystem properties (
Table 1) enhances the security of the cryptosystem and the cloud data storage and retrieval framework.
The SDNA requires less computation time for storage and retrieval processes but requires more time for cryptanalysis. This essential feature is partially achieved by many traditional cryptographic algorithms, underscoring the importance of the proposed scheme, the secure DNA cryptosystem. The proposed scheme, SDNA, consists of three algorithms:
- (i)
Secure DNA encoding table generation algorithm;
- (ii)
Secure DNA encryption algorithm;
- (iii)
Novel DNA decryption algorithm.
4.1. Comparison with NDNA Baseline
The proposed SDNA algorithm contains multiple improvements compared to the prior NDNA [
6] design to increase its efficiency and scalability. The most significant changes are dynamic DNA encoding, parallelized multi-block processing, and a streamlined lookup structure. Each of these enhancements can reduce encryption time and resource consumption while maintaining the same level of security. Indeed, all upgrades were made while retaining the same level of security, which is better for efficient computing and parallelization.
Table 3 compares NDNA [
6] and SDNA, highlighting design improvements, performance impacts, and security considerations.
Table 4 summarizes the significant symbols used in the SDNA cryptosystem and defines their roles in the encoding process. Elements DO1 and DO2 represent the random DNA inputs for a session that will be encoded into mRNA (DO1m, DO2m) and tRNA (DO1t, DO2t). Intermediate DNA matrices D4 and D64 form the building blocks of codon expansion, while the character set of cs64 and cs256 determines which character is assigned to each codon. The descriptor terms ELT, ET, and AT describe the data structures used to look up and index codons and to produce characters during encryption. The last item lists the specific random string inseq that is used to increase the security of the session.
The SDNA method incorporates randomness into several aspects of security. DNA sequences DO1 and DO2 are generated at random from the nucleotide set {A, C, G, T} using a cryptographically seeded PRNG, thereby preventing predictable session initialization. The intron inseq is generated similarly by selecting a plurality of uppercase letters, lowercase letters, symbols, and timestamp characters (OD, ED, M) from their respective sets using PRNG. The mapping from the expanded D64 codon matrix to the cs256 character set is permuted using a PRNG-generated permutation to achieve randomness and is thus unique for each session encoding structure.
The three critical stages in the randomized data system establish session-level security through randomization. First, the random data system creates a pair of session DNA sequences using a cryptographically seeded pseudo-random number generator (PRNG) to generate two random nucleotide sequences (i.e., DO1 and DO2). Then, the random data system generates an intron sequence (i.e., inseq) by randomly selecting from character sets that include upper case (UC), lower case (LC), symbols (S), other delimiters/separator characters (OD), other delimiters/separator characters (ED), and whitespace characters (M) via a PRNG. Finally, the ELT is generated by applying a pseudo-random permutation algorithm to create a unique mapping of D64 codons to cs256 characters for each session.
4.2. Secure DNA Encoding Table Generation
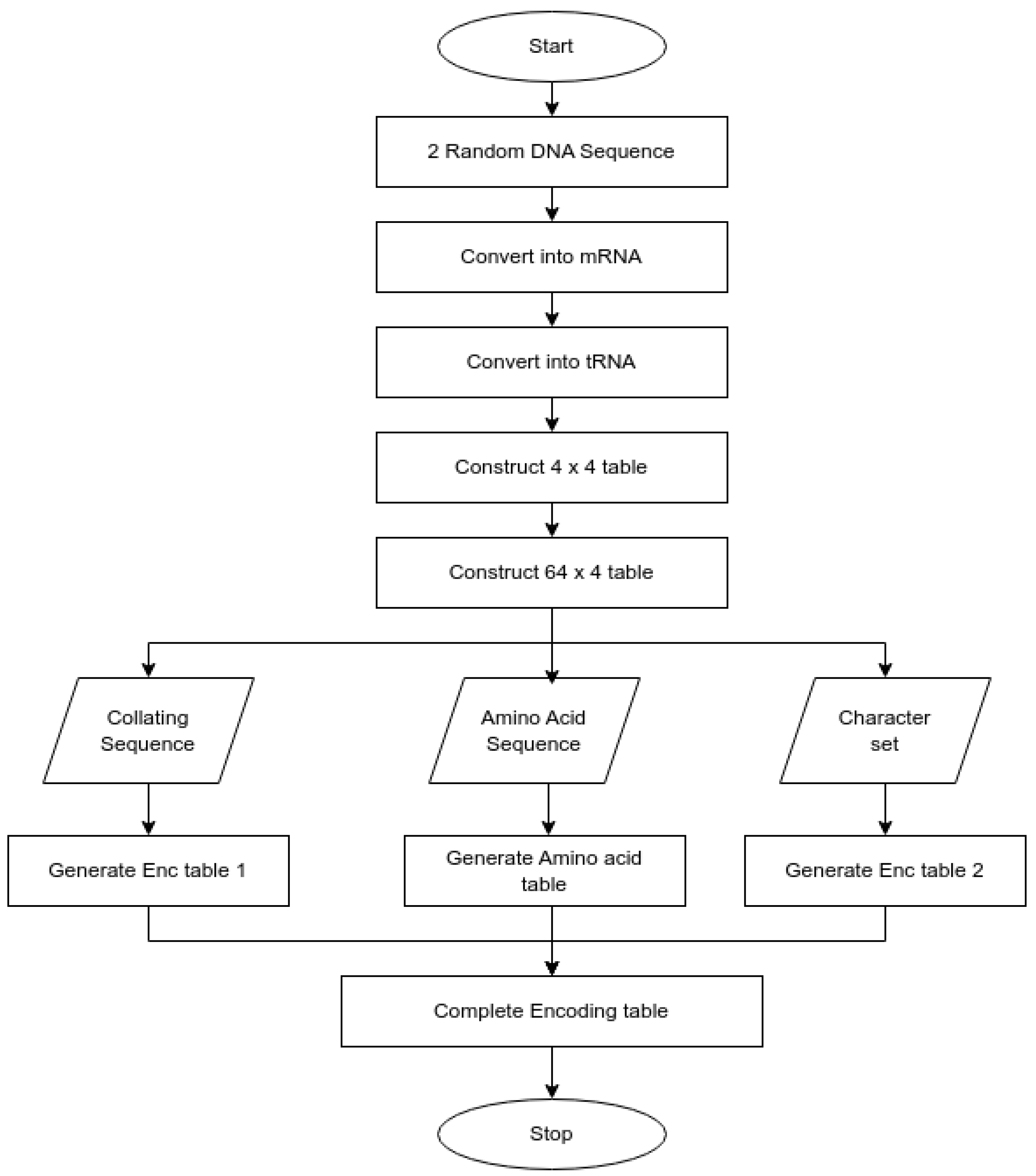
The generation of the SDNA encoding table is based on the amino acids encoded by DNA, a process related to protein synthesis. The SDNA encoding table generation algorithm is initiated by the DU and the DO, as shown in
Figure 2. In the pseudocode below, DO1t and DO2t represent the tRNA sequences derived from the DO’s two random DNA sequences. D64 refers to the 64 × 4 matrix generated by combining codon pairs. The pseudocode provides a complete and reproducible description of the algorithmic steps. The corresponding implementation was developed and validated in a controlled environment. The SDNA encoding table generation pseudocode 1 is as follows:
Pseudocode 1:
DNA_Encoding_table (DO1, DO2, cc, ca) |
| Inputs: Data Owner Sequence 1 DO1, Data Owner Sequence 2 DO2, collate_character cc, collate_amino ca |
| Output: Encoding lookup Table ELT |
| Method Variables: tRNA sequence of DO1 DO1t, tRNA sequence of DO2 DO2t, mRNA sequence of DO1 DO1m, mRNA sequence of DO2 DO2m. The product of DO1t and DO2t forms the D4 matrix. This matrix is further expanded into a 64 × 4 matrix, referred to as D64, character set cs64, character set cs256 |
| Procedure: |
| Convert DO1, DO2 into mRNA sequence DO1m, DO2m |
| Convert DO1m, DO2m to tRNA sequence DO1t, DO2t |
| Compute D4, Compute D64 & collate D64 through ca |
| Produce cs64 and expand to cs256 |
| Collate cs256 with cc |
| map D64 and cs256 to form ELT |
To generate the final ciphertext and key file by merging the amino-acid mapping and encoding tables, the process involves successively transforming and mapping two random DNA sequences. Each DNA sequence is first converted to an mRNA sequence, then to a tRNA sequence. These combined RNA sequences will form the basis of two codon tables: D4 and D64, for amino acids and characters, respectively (cs64 and cs256). The amino-acid codon tables and character codon tables are clustered together under a single logical heading (ELT) such that each codon in the amino-acid tables directly corresponds to a specific character in the character codon tables. After generation, the textual input is converted to its binary representation for encryption. This encrypted binary text is encoded as DNA. As such, a key file contains only the minimum number of key components: the hash of the DNA sequence used to generate the encoded text, the random seed(s) utilized to generate/encode the key, and references to the encoding table(s) used to create the encoded text that are essential to enable a deterministic reconstruction of these tables to facilitate text decryption. During final mapping, each tRNA triplet is matched with a unique amino acid and corresponding character in the encoding table. The concatenated outputs form the ciphertext, while the intron and mapping parameters form the session key file.
Figure 2 presents the generation of a DNA-based encoding table for encryption. Two random DNA sequences are first converted to mRNA and then to tRNA. A 4 × 4 table is built to represent amino acid transmission for each frame, with each frame expanded to a 64 × 4 table to describe combinations of codons. These increments also provide character sets and collating sequences. The resulting DNA sequences, tables, intervals, and frames each create two encryption tables and an amino acid table. These, together with the created tables, constitute the complete encoding table for securely encrypting the intended data.
4.3. Secure DNA Encryption
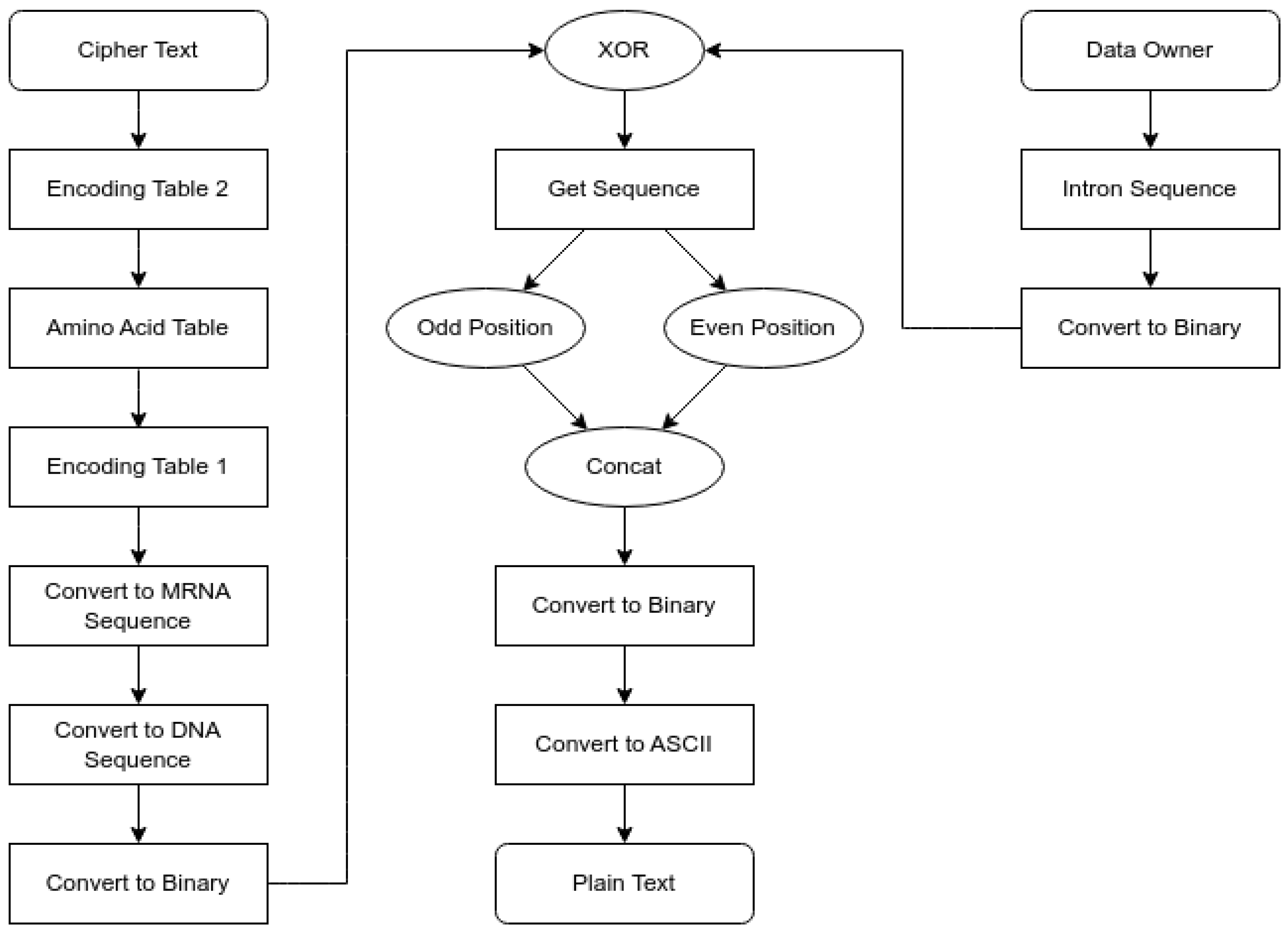
The Data Owner performs the secure DNA encryption process shown in
Figure 3.
Figure 3 depicts a DNA-based encryption process in which the Data Owner preprocesses and encodes the data into a binary format. The binary bits will be divided into odd and even positions, joined together, and XORed to increase data diffusion.
The newly created sequence is converted into a DNA, mRNA, and tRNA sequence. At the same time, two random DNA sequences generate encoding and amino acid tables, which are used to convert the preprocessed data into ciphertext for transmission.
Intermediate data representations will be included in the pseudocode. These will consist of intermediate representations of binary plaintext, intron representations, and computational representations (e.g., DNA, mRNA, and tRNA) to create an encoding table for generating the ciphertext. In addition, aspects of biology, such as letter casing (upper- and lowercase letters), odd/even letter numbers, and the month of the year, will create additional dynamic introns for creating intermediate representations of intermediate data. This provides greater complexity and security throughout the encryption process, including all stages of encryption and secure communication. The SDNA encryption process strengthens the diffusion and randomness properties by applying several transformation steps to the plaintext stream. In the first transformation step, the plaintext bits stream is separated into two distinct streams, one containing all even-positioned bits and the other containing all odd-positioned bits. The two streams are then independently mapped to the four DNA bases (A, C, G, T), yielding two DNA representations of the plaintext. The DNA representations are subsequently transcribed into mRNA (messenger RNA), which is then translated into tRNA (transfer RNA). tRNA codons triplets, which represent a unique combination of the four bases of DNA, are then used to look up and encode the plaintext via the Encoding Lookup Table (ELT). The final cipher block is generated by XOR’ing the session-specific intron sequence (which also provides a layer of randomness) with the encoding process. By doing so, each time the encryption session occurs, a unique ciphertext will be produced. The secure DNA encryption process pseudocode 2 is as follows.
Pseudocode 2:
DNA_Encryption (pt, DNA seq1, DNA seq2, inseq) |
| Inputs: plaintext pt, two random DNA sequences 1 DNA seq1, DNA sequence 2 DNA seq2, and intron sequence inseq. |
| Output: ciphertext ct, keyfile cl |
| Method Variables: plaintext in binary ptb, altered plaintext in binary aptb, intron sequence inseq, intron sequence in binary inseqb, DNA sequence DNAseq, mRNA sequence mRNAseq, tRNA sequence tRNAseq, one upper case UC, one lower case LC, first letter of odd day OD, first letter of even day ED, symbol S, first letter of month M, odd position OP, even position EP, encoding table ET, amino acid table AT, encoding lookup table ELT. |
| Procedure: |
| Generate ASCII values from pt. |
| Convert ASCII values to plaintext in binary ptb |
| SplitBits(ptb) to OP and EP |
| Concatenate OP and EP to aptb |
| Convert aptb to DNAseq. |
| generate inseq with UC, OD, ED, S, M, LC |
| Convert inseq as ASCII values. |
| Convert ASCII values as an intron sequence in binary inseqb |
| XOR (inseqb, aptb) |
| Convert to DNAseq & then convert to mRNAseq |
| Convert to tRNAseq |
| Map in ET, AT, ELT & convert into ct and generate cl |
4.4. Secure DNA Decryption Algorithm
The reverse process of secure DNA encryption is used to retrieve the original plaintext, as shown in
Figure 4.
Figure 4 illustrates the process of decrypting ciphertext using the DNA method, in which the ciphertext is decoded sequentially using a DNA encoding table and an amino acid table. The information is translated from mRNA (DNA) to binary. The binary string is split into odd- and even-position bits, then re-XORed and converted back to ASCII to recreate the plaintext. The pseudocode in this section outlines the exact operational logic verified through internal implementation. The described steps enable reproduction in any standard programming environment (e.g., Python, C++, MATLAB).
The pseudocode used to decrypt the ciphertext, as defined in this document, uses variables reconstructed from the output values and the DNA encoding table(s), as well as from all other biological DNA sequence types. DNA encoding table(s) for the regenerated units of mRNA, tRNA, and introns are part of the process of recovering the binary form of the original plaintext from the reconstructed biological component sequences. The remaining variables used for decryption include the odd or even position of the decoded sequences and the value of the XORed sequence. The Secure DNA decryption process pseudocode 3 is as follows.
Pseudocode 3:
DNA_Decryption (ct, cl, DU, ET) |
| Inputs: ciphertext ct, Key file cl. |
| Output: Plaintext pt |
| Method Variables: Binary plaintext ptb, intron sequence inseq, intron sequence in binary inseqb, DNA sequence DNAseq, mRNA sequence mRNAseq, tRNA sequence tRNAseq, encoding table ET, amino acid table AT, encoding lookup table ELT, odd position OP, even position EP |
| Procedure: |
| Using cl, generate DNA encoding tables. |
| Map ct with ELT, AT, ET. |
| Convert to tRNAseq & then convert to mRNAseq |
| Convert to DNAseq |
| Convert the DNAseq into binary to get the inseq from DO. |
| Convert inseq into ASCII values. |
| Convert ASCII values to intron sequence in binary inseqb |
| XOR inseqb with the binary sequence to form the XOR seq |
| Split XORseq into OP and EV |
| Concatenate OP and EP |
| Convert to plaintext pt. |
The pseudocode and worked example enable the reader to produce an exhaustive statement for the SDNA system, facilitating high-level programming or scripting implementations in languages such as Python, C++, Java, etc. They are a valuable way to convey the full definition of SDNA at a very high level.
According to the SDNA system, plaintext is divided into “blocks” that are fixed in size, and each block will go through the whole transformation process (as described under the full transformation process) as separate blocks (i.e., they do not depend on previous blocks). The same session parameters (ELT, Intron, and PRNG seeds) will be used to encrypt all blocks, enabling parallel encryption without requiring CBC or other chaining methods. The final ciphertext is created by concatenating all ciphertext blocks, which were generated independently, allowing users to achieve maximum throughput and the very effective parallel processing of data.
4.5. Detailed Example of Secure DNA Cryptosystem
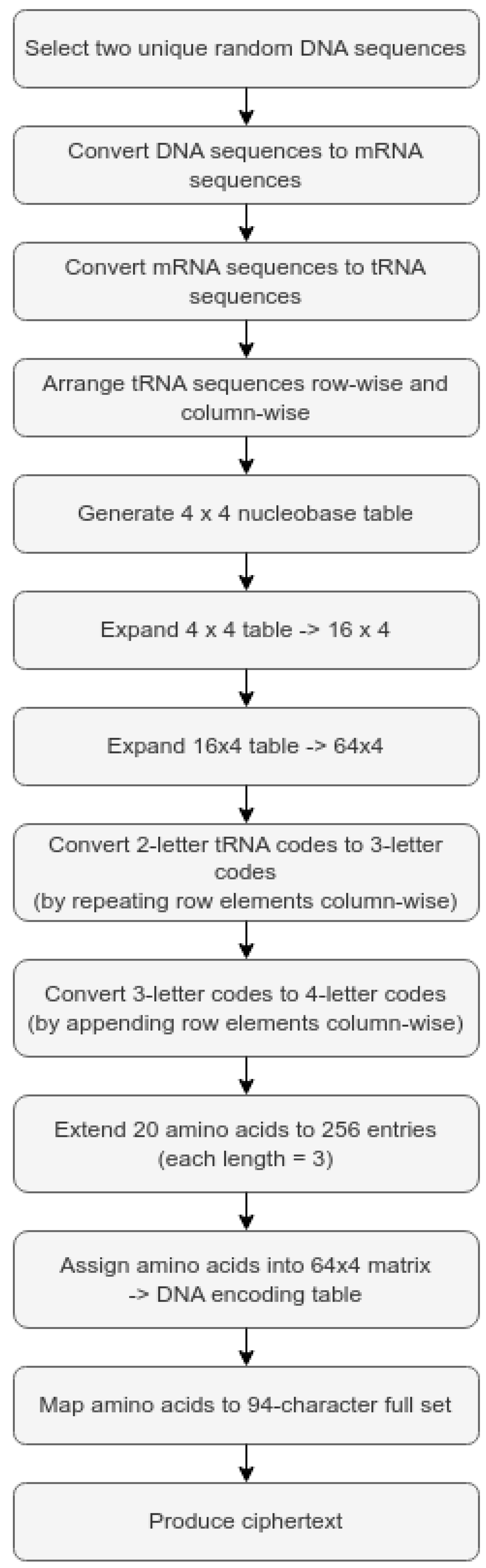
The plaintext is taken as “HELLO”. The DO choose the random sequence as GTAC and ATGC. (Let the random values be B1 and 39.) Using
Table 3, random values are converted to DNA sequences as GTAC and ATGC, respectively.
Figure 5 depicts the steps that comprise the construction of the DNA-based encoding table used for the encryption of genetic material. The first step is to select two random DNA sequences. These sequences will be converted to mRNA and then translated into tRNA before being arranged into a 4 × 4 codon table. After creating the 4 × 4 table, the author will make a 16 × 4 table and a 64 × 4 table that include both the original codons and the new codons. The codons will then be expanded from two-letter to three-letter codons, and then to four-letter codons. The next step is to create an amino acid set of 256 entries, which will be mapped to the 64 × 4 matrix to produce the DNA encoding table. Finally, to complete the encoding process, each amino acid in the DNA encoding table will be mapped to a character set, allowing for the generation of ciphertext from encoded codons.
4.5.1. Secure DNA Encoding Table Generation—Example
seq1: GTAC seq2: ATGC
Step 2: Transform the DNA sequence into an mRNA sequence.
Step 3: Transform the mRNA sequence into a tRNA sequence.
Step 4: Assign the two tRNA sequences randomly, row-wise and column-wise. A 4 × 4 table is then generated by multiplying the tRNA nucleobases. This is shown in
Table 5.
Step 5: The 4 × 4 table is expanded to a 16 × 4 table, then to a 64 × 4 table.
Step 6: Then, the two-letter tRNA nucleobases are converted into three-letter tRNA nucleobases in every matrix element by repeating the row matrix elements four times in a column-wise manner.
Step 7: Then, the three-letter tRNA nucleobases are converted into four-letter tRNA nucleobases in every matrix element by appending the row matrix elements column-wise.
Step 8: The 256 amino acid entries are extended from 20 amino acids. Each amino acid is three atoms long.
Step 9: To produce the DNA-encoding table in
Table 4, amino acids are assigned to a 64 × 4 matrix. The 94 elements of the complete character set are taken to generate this table.
Step 10: The amino acids are associated with the entire character set, as shown in
Table 6, to generate the ciphertext.
4.5.2. Secure DNA Encryption Algorithm—Example
For ease of understanding, the DO’s input file is assumed to contain “HELLO”.
Step 1: The input file is converted from ASCII to binary.
Append ‘0’ to make it as even in length, for example, eight digits.
Odd position group: 00100000001000100011
Even position group: 10001011101010101011
Concatenated binary value: 0010000000100010001110001011101010101011
Step 3: The intron sequence is generated using the values shown in
Table 7.
Intron sequence: Tsw#ad
Step 4: Each intron sequence character is converted to ASCII, then to binary.
Intron sequence binary value:
0101010001110011011101110010001101100001 01100100
011101000101000101001111100110011100101010011011
Note: While performing the XOR operation, the length of the binary values is balanced by appending ‘1’ as a suffix.
Step 6: The resultant binary value is transformed into a DNA sequence, as shown into
Table 8.
TGTA TTAT TAGG CTCT GACC CTCG
mRNA sequence: UGUA UUAU UAGG CUCU GACC CUCG
tRNA sequence: ACAU AAUA AUCC GAGA CUGG GAGC
Step 7: The above tRNA sequence is associated with the DNA encoding table (
Table 1) to obtain the unique index term. These unique index terms are associated with the amino acid table to obtain the amino acid sequence. The values are
P0$S1#T2*G1#V3*G2#
Step 8: The row index and the last character in the amino acid sequence are associated with the final DNA encoding
Table 4 to generate the ciphertext wsB~p`.
The corresponding column header is appended to each ciphertext character. Finally, the DO can send the ciphertext and the key file:
4.5.3. Secure DNA Decryption Algorithm—Example
Ciphertext: $w#s*B#~*p#`
P0$ S1# T2* G1# V3* G2#
Step 2: The obtained amino acid sequences are matched against the amino acid table to obtain the unique index term. Then, the index terms are associated with the encoding
Table 1 to obtain the corresponding tRNA sequences, as follows:
ACAU AAUA AUCC GAGA CUGG GAGC
mRNA sequence: UGUA UUAU UAGG CUCU GACC CUCG
DNA sequence: TGTA TTAT TAGG CTCT GACC CTCG
Step 3: The DNA sequence is transformed into binary values using
Table 1, as follows. DNA Binary sequence:
011101000101000101001111100110011100101010011011
Intron sequence: Tsw#ad
Step 5: Each intron sequence character is converted into ASCII and further transformed into binary:
Intron sequence binary value: 0101010001110011011101110010001101100001 01100100
001000000010001000111000101110101010101111111111
Group1: 00100000001000100011.
Group2: 10001011101010101011.
Note: Concatenate the bits from Group1 (first bit) and Group2 (first bit), then concatenate the bits from Group1 (second bit) and Group2 (second bit), and so on till the nth bit.
Concatenated group: 0100100001000101010011000100110001001111
Binary: 01001000 01000101 01001100 01001100 01001111.
ASCII: 72 69 76 76 79
Plaintext: HELLO
4.6. Relevance in Cloud Computing
The proposed scheme ensures that the SDNA cryptosystem is an excellent choice for cloud computing environments that require lightweight, biologically inspired, or hybrid encryption methods, e.g., healthcare datastores, IoT-based medical systems, or multi-tenant cloud computing environments that require frequent key updates and/or changes. The computational cost of SDNA will usually be lower than that of traditional methods such as AES and DES, which are more resource-intensive for smaller datasets and also provide session-level flexibility and knowledge for managing symmetric–asymmetric key integration. While AES is the industry standard, SDNA is a complementary methodology better suited for privacy-preserving cloud environments that require session-level encryption with low-cost key distribution and key changes.
By separating data protection from key distribution, the proposed design specifies a symmetric approach to encrypting data using a DNA-based encryption system and an asymmetric approach to distributing the key, ensuring maximum efficiency in the overall system of data and key management. A reduction in the encryption system’s computational burden is achieved by leveraging DNA’s lightweight properties, enabling faster key exchange. Since key regeneration occurs only during the initial setup of a session key, it requires less time and effort to regenerate each session key than to often exchange multiple keys across the network (environmentally). Other issues formerly associated with DNA-based cryptosystems were addressed by clearly distinguishing between symmetric encryption for protecting data against attacks and asymmetric encryption for distributing keys across the network (environmentally). This distinction leads to a significant increase in processing speed and ease of key rotation, as well as greater scalability for future use in cloud-based and IoT environments.
4.7. Preserve Security Properties of NDNA in SDNA
Table 9 summarizes the different types of cryptographic security mechanisms used by the original NDNA framework. The NDNA security mechanism’s primary cryptographic strength (e.g., static character mapping) is shifted to the dynamic mapping of characters within a given session and randomly generated keys based on a multi-component Pseudo-Random Number Generator (PRNG). Due to this increased randomness, an enhanced number of participants (e.g., physicians, scientists) and greater variation in the types of generated codes, SDNA produces unique ciphertexts generated in every session. In addition to maintaining the core benefits of NDNA (e.g., biologically accurate DNA sequences for each character), SDNA also greatly improves upon several security functions of NDNA (e.g., dynamic, encrypted ciphertext) and the level of simulation complexity. Therefore, the table’s main points show that while SDNA retains many of NDNA’s scientific advantages, it can address several of NDNA’s significant weaknesses.
Both SDNA and NDNA retain NDNA’s core biological transformation pipeline (DNA > mRNA > tRNA > Amino Acid) but differ in their implementation. In comparison, SDNA has made significant advances in three main areas: First, table generation shifted from static to dynamic methods. Second, instead of using intron design, introns are now generated using a PRNG (random number generator) to produce multi-parameter introns via an algorithm. Finally, the mapping pipeline has been upgraded to enable multi-block parallel processing, rather than being performed sequentially. All of these improvements provide a substantial increase in execution speed while maintaining the same security characteristics as NDNA.
4.8. Intron Generation Process
By using a cryptographically seeded pseudorandom number generator (PRNG), UC, LC, and S are randomly selected from the predetermined uppercase, lowercase, and special-character subsets, thereby maximizing unpredictability in intron generation. The hashes of time-stamped (timestamp) seeds create OD and ED and map them to alphabetic characters, thereby introducing additional random elements, rather than using fixed initials of the days of the week. Likewise, M is based on output generated via the PRNG rather than being the actual calendar month number, which adds to the randomness and uniqueness of the intron sequence for the session. UC, LC, S, OD, ED, M, sampled using cryptographically seeded PRNG, and intron_seq are not directly derived from plaintext or visible timestamp.
A hybrid seed comprising high-resolution system time (e.g., in microseconds or nanoseconds) and a confidential value that is exclusively owned by the DO serves as the basis for initializing the Pseudo-Random Number Generator (PRNG). The use of both seed types makes it impossible for an attacker to recreate the PRNG state. The value assigned to a data owner adds entropy that cannot be derived from system timestamps alone. An adversary may observe or estimate the timing of encryption, but without the ability to identify the unknown value specific to the Data Owner, the resulting intron sequence, Pseudo-Random Number Generator (PRNG) outputs, and total random mappings (TRMs) will all remain random and distinct from others during each session.
While the conceptualized description uses week and month characters, the intron sequence that is actually generated is produced by a PRNG seeded with cryptographic randomness. An attacker cannot reconstruct the intron from the approximate time the messages were encrypted, because the PRNG seed contains secret DO-specific entropy; thus, the intron patterns are not predictable from publicly available information, including dates or times.
5. Experimental Results
The proposed scheme, a secure DNA cryptosystem (SDNA) for ensuring data confidentiality in the cloud, was implemented in Ubuntu OpenStack Cloud, a private cloud setup, running on an Intel 4-core processor system with 16 GB of RAM. Sufficient clusters with nodes (data owners/users) are created in the proposed framework. The DO executes the secure DNA encryption algorithm, uploads the file to the CSS, and shares the key file. The DU receives the key file, downloads the cipher-text file from the cloud storage server, and executes the secure DNA decryption algorithm. With this private cloud setup, performance and security analyses were performed against the proposed framework. The results listed in this evaluation represent the mean of five identical execution runs conducted on the same cloud node under the same conditions to provide a fair comparison across all assessments. Averaging over five runs ensures that transient fluctuations due to other program executions (such as background processes running in parallel with this evaluation), temporary network latency, or CPU scheduling have little effect on the final results. Therefore, the values reported here represent a more stable and reliable estimation of the algorithm’s performance.
The NDNA (Normalized DNA Algorithm) [
6] is an earlier DNA-based encryption method that employs static encoding tables and fixed keys. It serves as the baseline for performance comparison with the SDNA.
5.1. Experimental Setup and Environment
The experiments described were conducted on an Ubuntu OpenStack private cloud with Ubuntu 20.04 LTS on an Intel® Quad-core processor (4 × 2.40 GHz), 16 GB of RAM, and all the SDNA algorithm prototype code (Python 3.8), developed without low-level optimizations (e.g., GPU). To evaluate the effectiveness of various data sources, the evaluation dataset comprised synthetically generated random plaintext file sizes based on ASCII text. As this is a character-based experiment, it can be assumed that the average word length is five characters, which follows the currently accepted methods of cryptographic benchmarking. Each reported time is an average of five independent runs conducted under the same conditions in the cloud.
The average word length in this experiment was assumed to be five characters, implying that 16,384 words equate to approximately 81,920 characters. The average times for the encryption and decryption processes (75 ms and 62 ms, respectively) are averaged based on five identical execution runs.
The results presented in this report reflect a lower performance bound due to the existing prototype rather than an optimally implemented version of the system. This shows the need for more efficient implementation using a C/C++ or GPU-based design, which may improve processing speed and increase overall throughput.
The fields shown in
Table 10 provide information about the types of information that compose the SDNA KEY file, and how each of these supports the accurate and secure decoding of your DNA. Hash_DOseq contains a digital hash of your DNA-related sequences (as your seeds), so that the receiver can check whether or not they successfully reconstructed your biological transformations. Seed_intron, seed_table(s) are the PRNG seeds used for regenerating the intron sequences and dynamic encoding tables, respectively, used in the encryption of your DNA, to ensure that the reproduction of the encoded DNA is deterministic without having to store any of the actual/real data. Table_refs provides compact (or abbreviated) identifiers for reconstructing the ELT/AT layout and allows for the elimination of the need to store large lookup tables, consequently reducing the chance of losing these sensitive data items.
Due to the regeneration of the intron sequence and encoding tables during each session using the secret pseudorandom number generator (PRNG) seeds saved exclusively in the encrypted key file, an adversary will not be able to compare ciphertexts across sessions or reconstruct the mapping unless they obtain access to the key file itself.
5.2. Performance Analysis
References [
11,
17] recognized that standardizing DNA-based cryptosystems with performance parameters is a research focus. The time required to generate the secure DNA encoding table appears to be the same across all cases, as it is independent of the plaintext. But the time taken to perform secure DNA encryption and decryption depends on the plaintext size. Similarly, a frequency analysis has been conducted between the plaintext and ciphertext in four different cases.
While the current experiment validated the encryption speed and efficiency of generating tables from text data alone, it also indicated that this new DNA-based approach may extend the encryption method to all types of rich data (e.g., images, audio). However, applying the technique to other rich data will require more pre-processing and larger tests than were conducted with text data, which can be performed in future research projects. This statement serves as a guide for researchers to visualize future opportunities to expand the implementation of rich data in the DNA-based system. The metric-based performance analysis is discussed below.
5.2.1. Comparison with Traditional Cryptographic Algorithms
Table 11 compares the proposed lightweight SDNA cryptosystem with other well-known cryptographic algorithms in terms of speed (execution time and throughput) and suitability for cloud environments. The traditional symmetric encryption algorithms, AES and DES, have an average encryption/decryption execution time of 2.4–3.1 s. RSA-2048 has low throughput when encrypting/decrypting bulk data; therefore, it is not practical for this type of application. The ECC + AES hybrid method yields a worse performance than SDNA, with execution times of 1.7–2.2 s. The SDNA offers fast encryption and decryption times of 75 ms and 62 ms, respectively, and high throughput when tested in an OpenStack-based cloud environment. Overall, SDNA outperforms conventional forms of cryptographic systems.
The improved performance obtained by SDNA compared to both NDNA and other traditional algorithms is the result of many design optimizations. The use of multi-block parallel encoding helps minimize data sequential dependencies. At the same time, the compact lookup table structure was applied to enable a relatively inexpensive way to convert codons into characters. In addition, SDNA can operate in both single-pass encoding and single-pass decoding modes, unlike AES or DES (which typically require several transformation rounds). Furthermore, reusable internally generated tables across multiple blocks within an individual session will contribute to SDNA’s overall efficiency.
5.2.2. Range of Characters
An increase in the length of characters in the plaintext, the execution times of the proposed system, SDNA, and the existing system, NDNA [
6], is shown in
Table 12. It compares the encryption and decryption times of the SDNA and NDNA [
6] DNA-based cryptographic algorithms as the input character count varies. This demonstrates that both encryption and decryption times increased with the number of characters, indicating scalability as the amount of data increased. The SDNA algorithm has faster encryption and decryption times than NDNA [
6], whose processing time significantly increases with larger inputs. Thus, NDNA [
6] is shown to provide strong security, albeit at the expense of reduced processing speed efficiency.
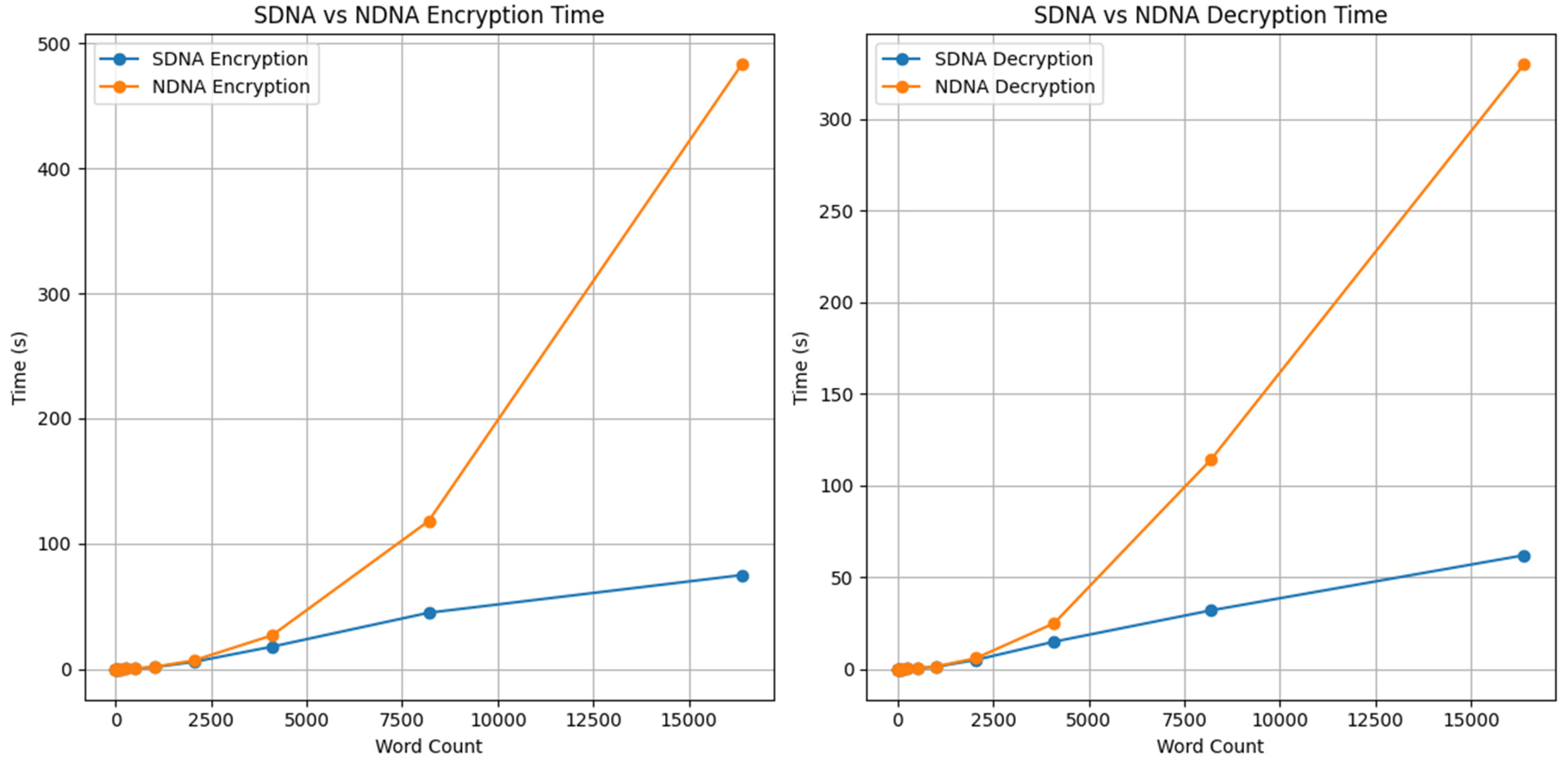
Figure 6 compares the encryption and decryption performance of SDNA and NDNA as the character count increases. In both plots, SDNA shows a gradual, consistent increase in processing time, demonstrating stable scalability. In contrast, NDNA exhibits a much steeper increase, especially for larger inputs, indicating significantly higher computational cost. Overall, the graphs clearly show that SDNA outperforms NDNA in both encryption and decryption, providing faster execution and better scalability as the data size grows.
The NDNA [
6] serves as the baseline for comparison, proposing the first DNA-based cryptosystem that employs fixed encoding and static key generation without dynamic table construction. The concrete results in the comparison tables reveal the advantages of dynamic encoding and session key variation, as the SDNA achieves faster encryption and decryption times with equivalent security to the static NDNA.
5.2.3. Range of Words
When the length of words in the plaintext increases, the execution time of the proposed system, SDNA, and the existing system, NDNA [
6], are shown in
Table 13. The experimental results clearly show that the Data User can perform fast data retrieval operations in the cloud.
Figure 7 shows the performance characteristics (encryption and decryption) of SDNA and NDNA across smaller and larger word counts, as well as the scaling of each algorithm’s execution time with data size. For example, both graphs indicate that SDNA’s execution time increases slowly and steadily as data size grows. Conversely, NDNA’s execution time increases rapidly with large input sizes, resulting in higher computational costs. Thus, the difference between SDNA and NDNA shows that SDNA has a significant advantage over NDNA for both encryption and decryption due to its greater efficiency in handling large amounts of data and a lower workload.
A comparison of the encrypted and decrypted times for the SDNA and NDNA [
6] under randomization is shown in
Table 13 for different word counts. As the number of words grows, the time required to process them increases for both encryption and decryption. However, the SDNA encryption and decryption were consistently more efficient than the NDNA [
6] encryption and decryption, which take considerably longer, especially as data sizes grow. Thus, the SDNA displays greater efficiency and scalability than NDNA [
6]. Still, it is worth noting that the NDNA [
6] multiplex scheme provides an additional layer of security at the expense of speed.
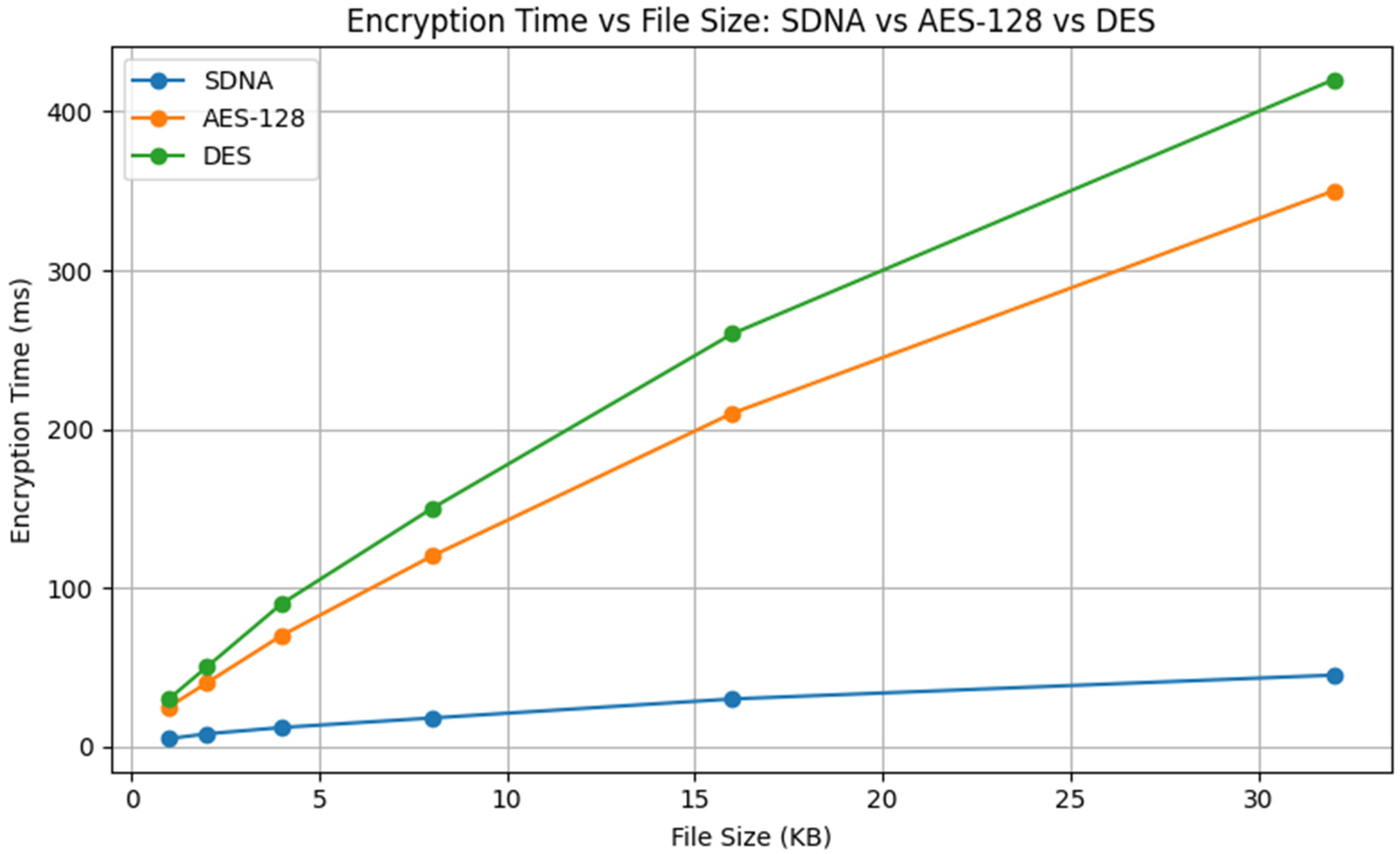
A comparison of the encrypted time (time to encrypt) for SDNA, AES-128, and DES shows that, across the range of file sizes (from 1 KB to 32 KB), SDNA always has the shortest encryption time. In contrast, AES-128 and DES both incur considerable encryption delays, and these delays increase sharply as file sizes grow. DES takes the longest to encrypt overall, followed by AES-128, with SDNA exhibiting the most efficient (linear) and least dramatic growth in encrypted time. Thus, it is clear that SDNA provides a substantial performance increase and slower growth compared to traditional symmetric algorithms, as shown in
Figure 8.
5.2.4. Block Size IMPACT
Ciphertext length depends on the plaintext bits’ length. For experimental purposes, plaintext length was fixed at 512 bits, as shown in
Table 14. Thus, the change in block size enables the generation of ciphertexts of varying lengths, thereby improving the cryptosystem’s security.
Table 14 compares plaintext length, block size, and resulting ciphertext size for both SDNA and NDNA [
6] encryption techniques. As shown in the table, when the block size increases from 16 to 1024 bits, the ciphertext length also steadily increases; however, NDNA [
6] has a slightly larger ciphertext than SDNA, indicating that NDNA [
6] exhibits somewhat higher data expansion due to longer encoding.
5.2.5. Impact of File Size
The structure of the key file is constant because the ciphertext file size increases as the plaintext size increases, as shown in
Table 15.
Compared to NDNA [
6], the SDNA requires less computational time. Thus, the experimental results demonstrate that, without compromising the security of the proposed framework, the time and space complexities are balanced. The inability to perform cryptanalysis reflects the security of the proposed system, as this makes it hard to correlate plaintext with ciphertext to infer the algorithm and the key file.
5.3. Frequency Analysis
The frequency of plaintext and of different ciphertexts must be distinct. Here, the frequency distributions of characters in both plaintext and ciphertext are analyzed across different encoding tables, intron sequences, and input sequences.
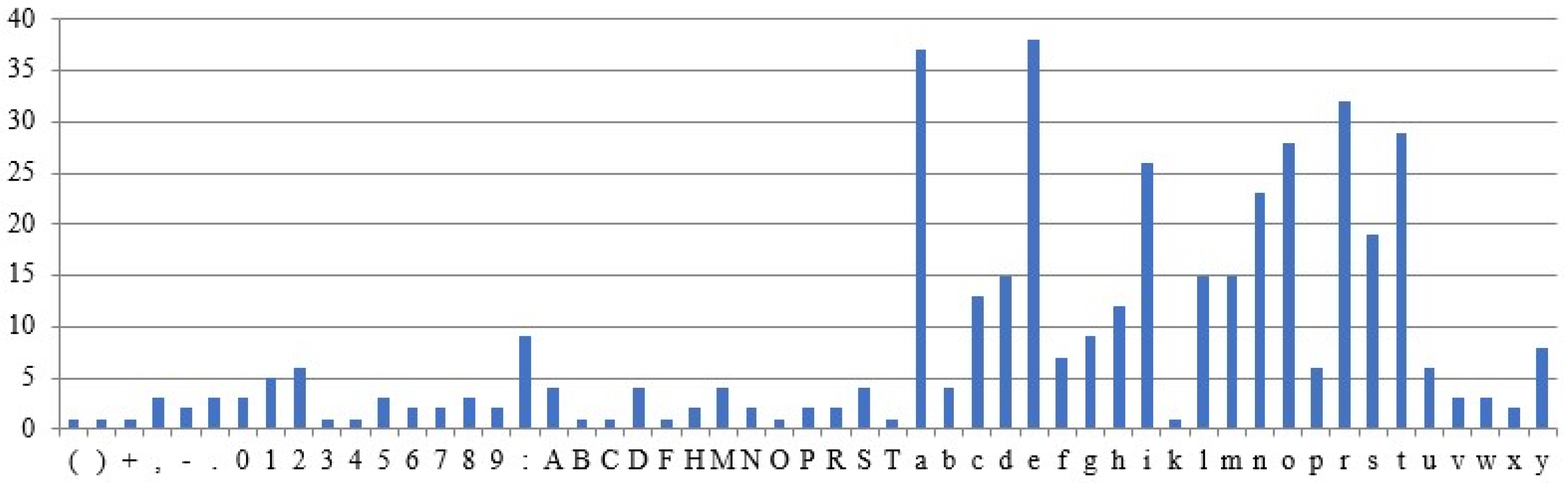
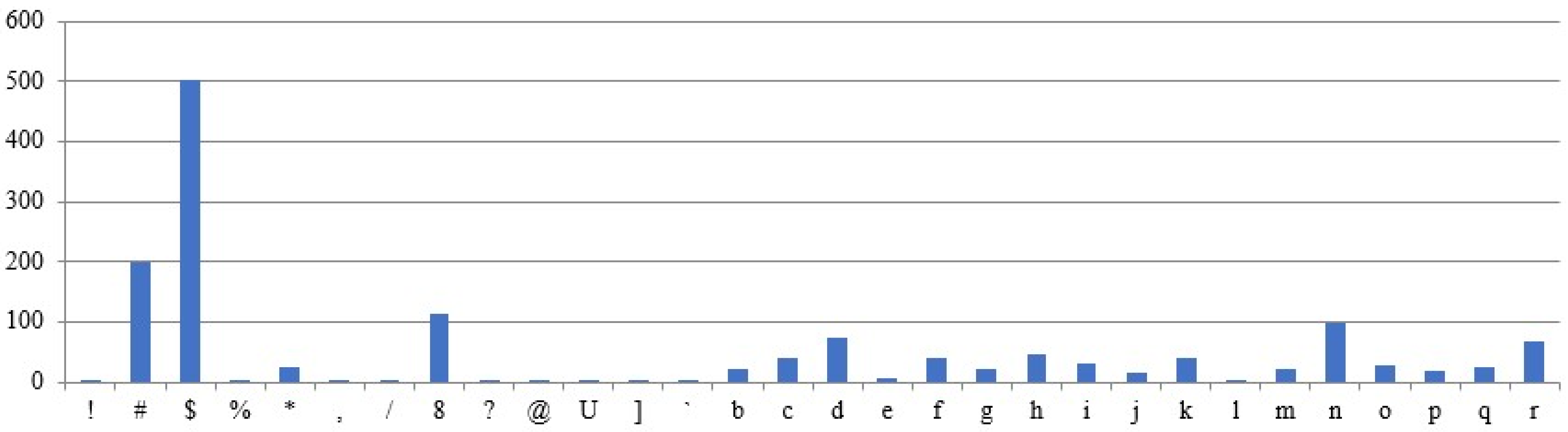
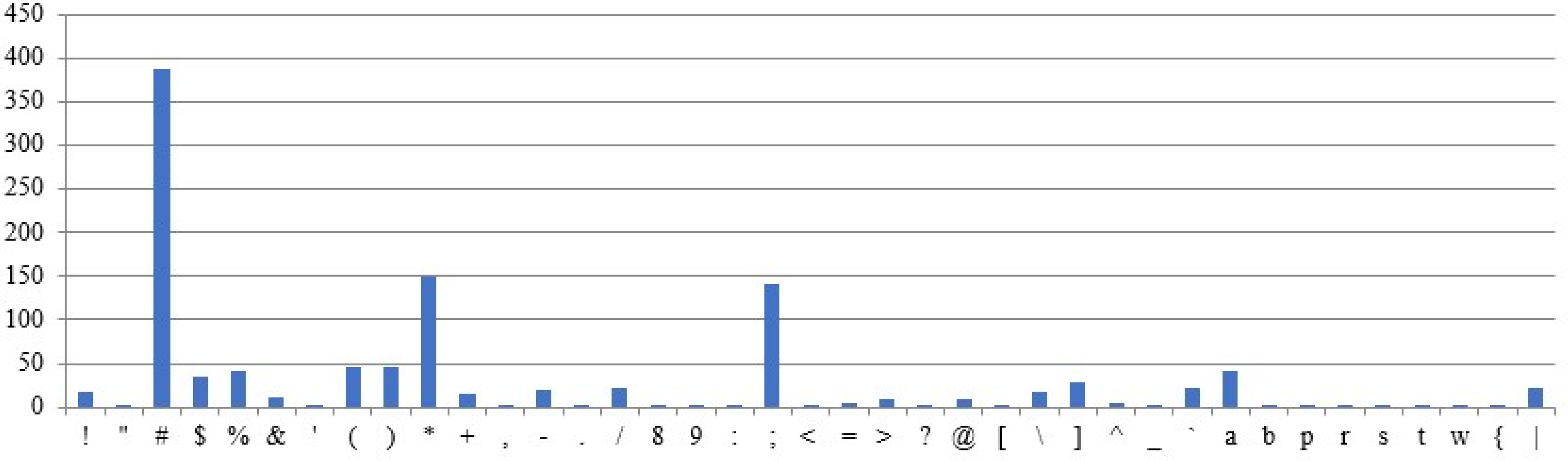
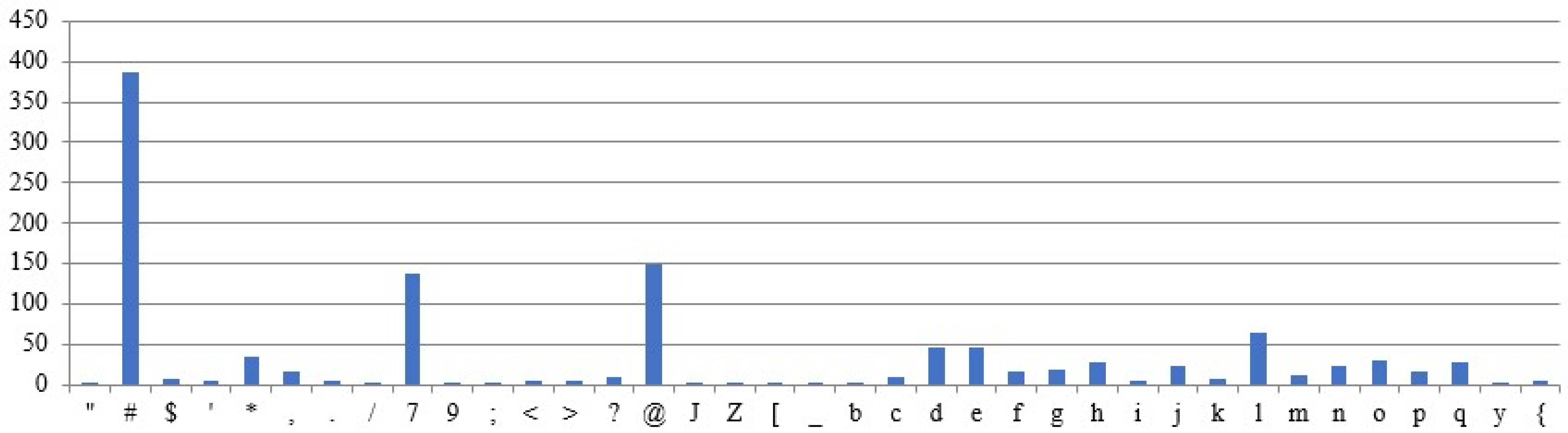
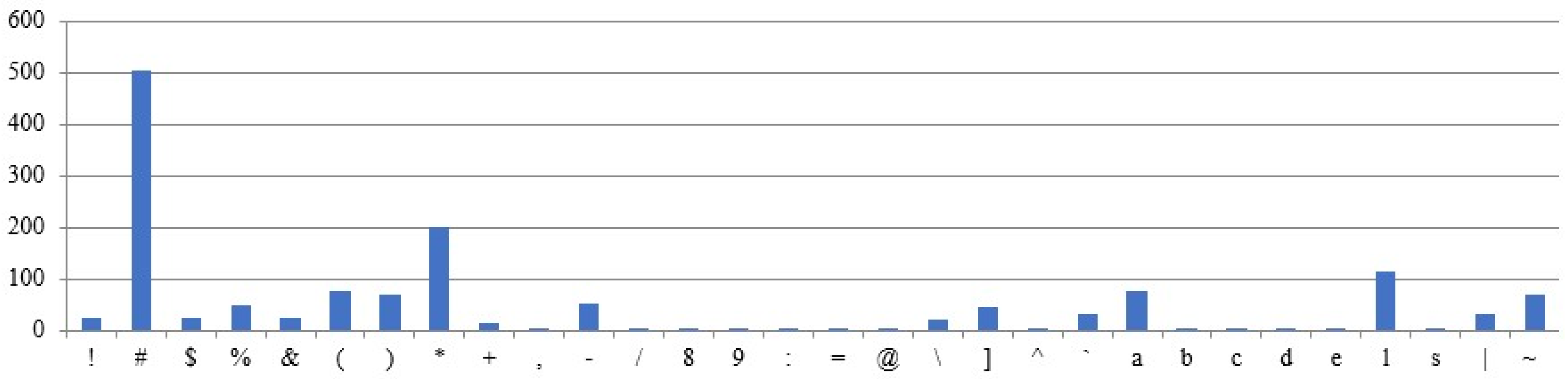
Figure 9 shows the occurrence of the characters in the sample plaintext. The plaintext is assumed to be fixed across all test cases, while the remaining parameters are treated as variables in the experimental results. The above assumptions are designed to examine the correlation between ciphertexts and their corresponding plaintexts. The graphs use the X-axis for characters and the Y-axis for frequency counts.
5.3.1. Different Intron Sequences Are Used for Ciphertext Generation
Different ciphertexts are generated for the same plaintext and DNA sequences, depending on the intron sequence, as shown by the results (
Figure 10 and
Figure 11). The character frequencies differ significantly between the two ciphertexts.
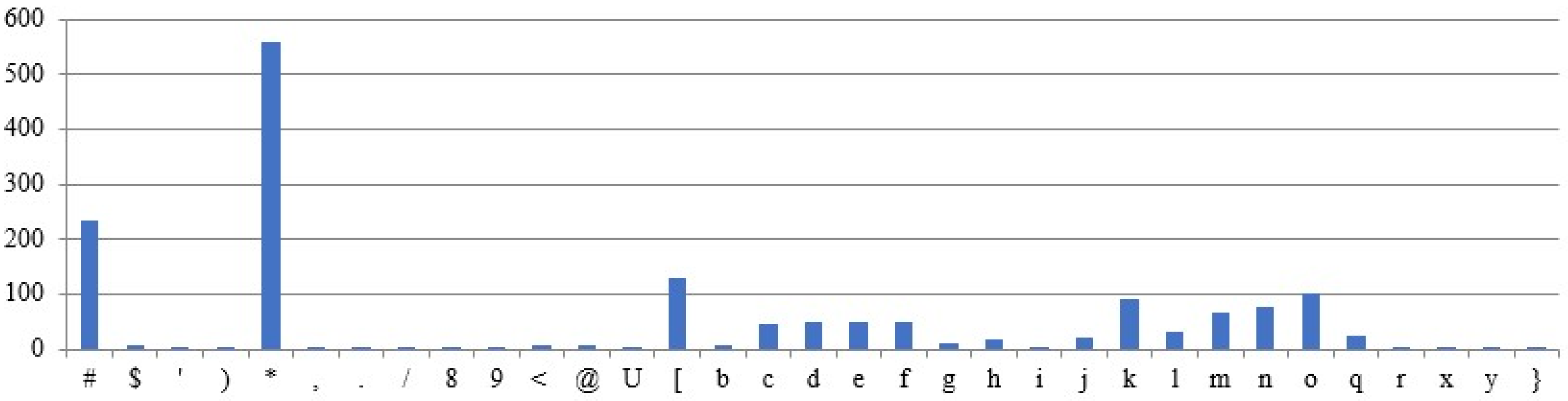
5.3.2. Different DNA Sequences Are Used for Ciphertext Generation
Ciphertexts are generated for the same plaintext and intron sequences, but different DNA sequences, as shown in
Figure 12 and
Figure 13. The obtained ciphertexts are entirely different.
5.3.3. The Same Plaintext, but Different Input Sequences Are Used for the Ciphertext Generation
Ciphertexts are generated for the same plaintext, but with varying sequences of intron, as shown in
Figure 14 and
Figure 15. Both ciphertexts are correlated.
5.3.4. Different Plaintexts and Different Input Sequences Were Utilized for the Ciphertext Generated
For the given different plaintext, intron, and DNA sequences (
Figure 9 and
Figure 16), Different ciphertext frequencies are generated (
Figure 17 and
Figure 18). Thus, the analysis reveals no correlation between the generated ciphertexts and the given plaintexts.
Established DNA-based cryptosystems, such as NDNA and hybrid DNA–AES approaches, have demonstrated strong security, although at increased computational cost and slower performance. In comparison, the proposed SDNA algorithm delivers faster performance while maintaining security through repeatable encoding across multiple data files, dynamic key mapping, and a compact lookup structure. As a result, SDNA offers an innovative, computationally simple DNA encryption/decryption tool that is faster, equally secure, and scalable—making it ideally suited for modern lightweight applications compared to prior DNA-based cryptosystems.
5.4. Security Validation
The proposed SDNA needs to be evaluated against security threats to enable real-time implementation in the cloud. A security analysis of the proposed SDNA was conducted against password guessing, collisions, and internal attacks, as well as the properties of DNA cryptography.
The experimental results are strengthened by the addition of a simple security validation, called an Avalanche Test. This test will determine how a one-bit change to the input plaintext affects multiple bits throughout the encryption cycle. By showing that the ciphertext is highly sensitive to slight differences in the plaintext and that these differences will continue to spread throughout the entire ciphertext, it can be demonstrated that the ciphertext will be highly diffused as well. Applying random-number tests to assess the amount of structural information, such as Shannon entropy or chi-square tests, will demonstrate that the ciphertext shows no regularity. The first evaluation showed an average Shannon entropy of 7.98, indicating that the ciphertext exhibits a nearly uniform distribution and is highly resistant to statistical attacks. These initial results suggest that the DNA-based method has beneficial security properties, and more extensive security testing will be conducted.
The security assessment indicates the same levels of randomness and diffusion in the ciphertext created with SDNA as with NDNA. The entropy of the SDNA ciphertext is 7.98, which is almost equal to the ideal value of 8.0, thereby demonstrating that the randomness of SDNA is as strong or stronger than that provided by NDNA. In addition, the avalanche testing performed on SDNA indicates that a one-bit change in the plaintext will create approximately a 50% change in the corresponding SDNA ciphertext, thereby indicating that the SDNA ciphertext preserves the diffusion properties of an NDNA ciphertext. The results of this study support the claim that SDNA provides an equal level of security and improved performance compared to NDNA.
Capabilities of Adversaries: There are two types of adversaries: active and passive. A passive adversary can inspect ciphertext stored in the CSS and also measure the DO communication and the DU. If a passive adversary manages to collect an intercepted ciphertext through electronic means, they can also use this ciphertext for offline analysis. An active adversary, on the other hand, can alter the ciphertext and can also replay previously received messages or compromise access to the CSS (the place that saves the ciphertext) to gain access to the data. An insider adversary is a malicious employee, potentially a Do/DU/CSS employee. They can also attempt to infer key materials or internal parameters of the SDNA system.
Types of attacks that are evaluated with SDNA: SDNA is evaluated using standard cryptology models, including the following:
Ciphertext-Only Attack (COA)—A hacker will only see the ciphertext.
Known Plaintext Attack (KPA)—A hacker will know a pair of clear text and ciphertext.
Chosen Plaintext Attack (CPA)—A hacker may provide a clear text and see the resulting ciphertext.
Limited Chosen Ciphertext Attack (CCA-Lite)—A hacker can attempt to replay or slightly modify the attempted ciphertext.
Through the use of session-based introns randomization and dynamic encoding tables, SDNA provides unique intron randomization and dynamic encoding tables that mitigate the ability to correlate across multiple CPA Attempts, thereby creating an IND-CPA confidentiality value.
Assumptions About Security: The asymmetric key exchange (RSA/ElGamal) can be considered secure based on established hardness assumptions (or computational security), such as, for example, an adversary’s inability to solve either the integer factoring or discrete logarithms problem. The hash functions in the key file are thought to be collision-resistant and preimage-resistant, producing unique hashes for each random sequence of characters (e.g., DNA sequences, introns, and encoded tables). Randomness for DNA sequence generation, intron sequence generation, and the creation of encoded tables is generated by a PRNG with a cryptographically secure seed, so that its seeds cannot be predicted even if the adversary makes its time stamps public.
Computational Security Justification: The adversary model provides a basis for the evidence of computational security provided by the SDNA through the following mechanisms:
Combined adequate key space of about 4256 ≈ 10154. Hard to attack through brute force enumeration.
Shannon entropy of 7.98 for ciphertext randomness. This indicates nearly uniform output distribution.
Avalanche effect of approximately 50%. This shows that, through brute-force enumeration, strong diffusion exists.
Dynamic ELT and intron generation. This ensures that no identical plaintexts will ever yield identical ciphertexts during session processing.
All the above evidence supports the claims made about the computational security of the SDNA. In particular, confidentiality claims, comparable to symmetric schemes with IND-CPA security, can be supported by SDNA.
5.4.1. Password Guessing Attack
DO encrypts the input file using the secure DNA encoding table. A secure DNA-encoding table is generated with real-time parameters that depend on the DO. It is not easy to guess or determine the secret data of DO that is required to create the secure DNA encoding table for an attacker or malicious users. Therefore, the proposed cryptosystem is resistant to password-guessing attacks.
5.4.2. Collision Attack
In the proposed framework, the DNA sequence is obtained by encrypting a large amount of data. A DNA sequence is generated based on uppercase and lowercase characters, months, odd–even days, and even weekdays. If there is a greedy user, unauthorized access is not possible because all the values are the same as the DO to generate a DNA sequence with an encoding table; guessing the real-time value is not easy. The greedy user cannot create the intron sequence and the encoding table. This scheme does not reveal sensitive information. Therefore, the proposed cryptosystem is resistant to collision attacks.
5.4.3. Internal Attack
The proposed scheme is not easy to cryptanalyze against internal attacks. An internal attack may originate from a CSP, a DO, a DU, or a third party. Insiders may attempt to access the cloud data, but this is not easy due to the dynamic encoding table. Every character of the plain text of a DNA sequence is encoded with a unique sequence, and biological process simulation concepts are implemented in the proposed framework. Therefore, the proposed cryptosystem is resistant to internal attacks.
The suggested adversarial model encompasses both passive and active adversaries, each of which can intercept ciphertexts, brute-force the cipher, or try to compute the encryption key. As stated in the above equations, mercy/covenant arises from randomness in “intron” selection (gate function or exit function), and the table is dynamically generated for “DNA” encoding. The research estimates the key space for an alphabet of four bases, yielding an encoding table of size 256, with ~4256 (≈10154 possibilities), exceeding the complexity of AES-128. The ample key space provides resistance to brute-force attacks. The dynamic encoding table enhances resilience against chosen-plaintext attacks; similarly, session-based key variations protect against replay attacks. Further formal analysis may be needed to evaluate resistance against further adversaries using more advanced adaptive methods.
Table 16 provides a comparative summary of the SDNA system’s strength against various cryptographic attacks and adversaries’ capabilities. The SDNA system’s security mechanisms, compared with the adversary’s ability to use a given method to attack it, show that brute-force attacks are impractical due to the huge key space (approximately 10
154 keys). The use of ciphertext-only attacks is mitigated by the high entropy and very variable encoding table. Known-plaintext and/or chosen-plaintext attacks are countered by changing all mappings in each session, and because a PRNG generates introns, no correlation or reuse between sessions can occur. Replay attacks will not work due to the session-based variation, and because compact key elements do not store the mapping tables, the number of internal attack vectors is limited.
Table 16 shows that the SDNA system provides high resiliency to multiple attack vectors.
5.4.4. Key Space-Combat and Effective Bit Length
The key space generated with SDNA derives from three sources, as indicated by the following components:
Two random DNA sequences (DO1 and DO2): each of length n, providing 4n combinations.
The dynamic intron sequence is generated from six PRNG-driven parameters (UC, LC, OD, ED, S, M); these parameters provide approximately 248–260 combinations, depending on the character pools.
The dynamic encoding table (ELT) has 256 entries arising from the permutations of cs256 on D64. This results in approximately 256! ≈ 21684 different mappings.
Combining all components leads to K_total, which is dominated by the ELT permutations. Therefore, K_total ≈ (4256) × (260) × (256!), which offers approximately ≈23000 + or ≈10900, giving us an adequate key strength of roughly 3000 bits. In comparison, AES-256 only has 2256 key strength; hence, the key strength provided by SDNA is much greater than AES-256 and is impractical to search exhaustively.
The total number of possible keys in an asymmetric cryptographic key space is on the order of 10900. Therefore, even if an attacker can check (i.e., brute force) 1012 keys every second, it would take over 10888 years to search through that entire key space. This is many orders of magnitude longer than the estimated age of the universe (1010 years). Therefore, exhaustive key-space searches are computationally infeasible under any realistic threat model, further crediting the practicality and strength of the key-space protection found in asymmetric cryptography.
5.4.5. Resistance to Standard Cryptographic Attacks
- (a)
COA: The dynamic ELT combined with the dynamic intron sequence method ensures that all identical plaintext messages will produce different ciphertext messages every time the encryption is used. As seen in
Figure 9,
Figure 10,
Figure 11,
Figure 12,
Figure 13,
Figure 14,
Figure 15,
Figure 16 and
Figure 17, all of the frequency distributions are different (for a one-sentence reference, see the included references), as is evident from the lack of any plaintext–ciphertext correlation (supported by the entropy measurement of 7.98).
- (b)
KPA: ELT and intron values are dynamic and based upon the user session ID, and thus do not allow for any derivation of the table mappings, even with the knowledge of the plaintext–ciphertext pairs.
- (c)
CPA: The randomness associated with the per-session nature results in selected plaintext users generating many unrelated ciphertexts when the same plaintext is selected over many different sessions. This is similar to IND-CPA-type behavior, as the mappings are not deterministic at any point across sessions.
- (d)
CCA (limited): The attempt to manipulate the ciphertext renders itself ineffective unless the intron and ELT are reconstructed, as they are unknown to both the CSS and the attacker. If the intron is incorrectly reconstructed, it will not produce a valid mRNA-to-tRNA conversion and thus will ultimately render any CCA attempts futile.
5.4.6. Empirical Randomness Evaluation
The randomness of our ciphertext was statistically evaluated using the following tests:
Shannon Entropy: The average entropy of our ciphertext, =7.98, was close to the maximum theoretical value (8) and indicates a relatively uniform distribution.
Avalanche Effect: Flipping one bit of our input (message) created a change of approximately 50% of our ciphertext’s bits and provided evidence of a good level of diffusion.
Correlation Coefficient: The correlation between plaintext and ciphertext averaged approximately 0.002–0.01, which supports no statistical correlation.
The SDNA cryptosystem was evaluated for its security characteristics using a combination of measurements and supporting evidence, as shown in
Table 17. The SDNA cryptosystem provides an enormous key space of approximately 10
900 possible combinations, has a high ciphertext entropy of 7.98, shows sound diffusion, as evidenced by the approximately 50% bit flips in the avalanche tests, and has correlation values close to zero. This means that there is minimal statistical similarity between the plaintext and ciphertext, and the SDNA cryptosystem has shown strong resistance to both chosen-plaintext and known-plaintext attacks due to the use of dynamic ELT and intron generation. Additionally, replay attacks were reduced by incorporating session-based randomness, and brute-force attacks will take an estimated 10
888 years to succeed, even when performed at very high rates.
7. Conclusions and Future Works
In this paper, an SDNA was proposed to preserve the privacy of sensitive material during storage and retrieval among the Data Owner, Data User, and Cloud Storage Server. The proposed system employs DNA encoding table generation, which operates in constant time for variable DNA and collating sequences. Plaintext data is securely hidden using DNA nucleobases, while the incorporation of intron-sequence implementation and dynamic encoding-table generation significantly increases the complexity of cryptanalysis for potential attackers. By integrating theoretical concepts of DNA-based biological processes, the cryptosystem introduces randomness and dynamism into the encryption process, thereby minimizing the likelihood of successful cryptanalysis. The encryption and decryption algorithms are computationally efficient, as demonstrated through the experimental results. For a character count of 16,384, encryption and decryption times were 852 ms and 822 ms, respectively, while for a word count of 16,384, the times were significantly lower, at 75 ms and 62 ms, respectively. These findings confirm that the proposed cryptosystem achieves high performance with low computational overhead. Furthermore, the SDNA effectively resists collisions, password guessing, and internal attacks. In the proposed cloud framework, data transmission is protected using a cryptographic system. At the same time, key management and sharing are handled efficiently using existing asymmetric cryptographic techniques such as RSA and El-Gamal, resulting in a hybrid cloud security model that enhances overall system robustness. SDNA provides greater efficiency and scalability than existing cryptosystems, such as AES and RSA, notably for lightweight encryption in IoT environments. The biological encoding layer augments existing standards; it does not disrupt or replace existing standards.
In future work, the efficiency and scalability of the SDNA can be further improved to support real-time large-scale applications, including IoT-based cloud systems, edge computing, and distributed blockchain environments. Additionally, integrating machine learning-based optimization for dynamic key generation and adaptive encoding strategies may further strengthen security while reducing computational complexity.