Electrocardiogram Signals Classification Using Deep-Learning-Based Incorporated Convolutional Neural Network and Long Short-Term Memory Framework †
Abstract
1. Introduction
2. Materials and Methods
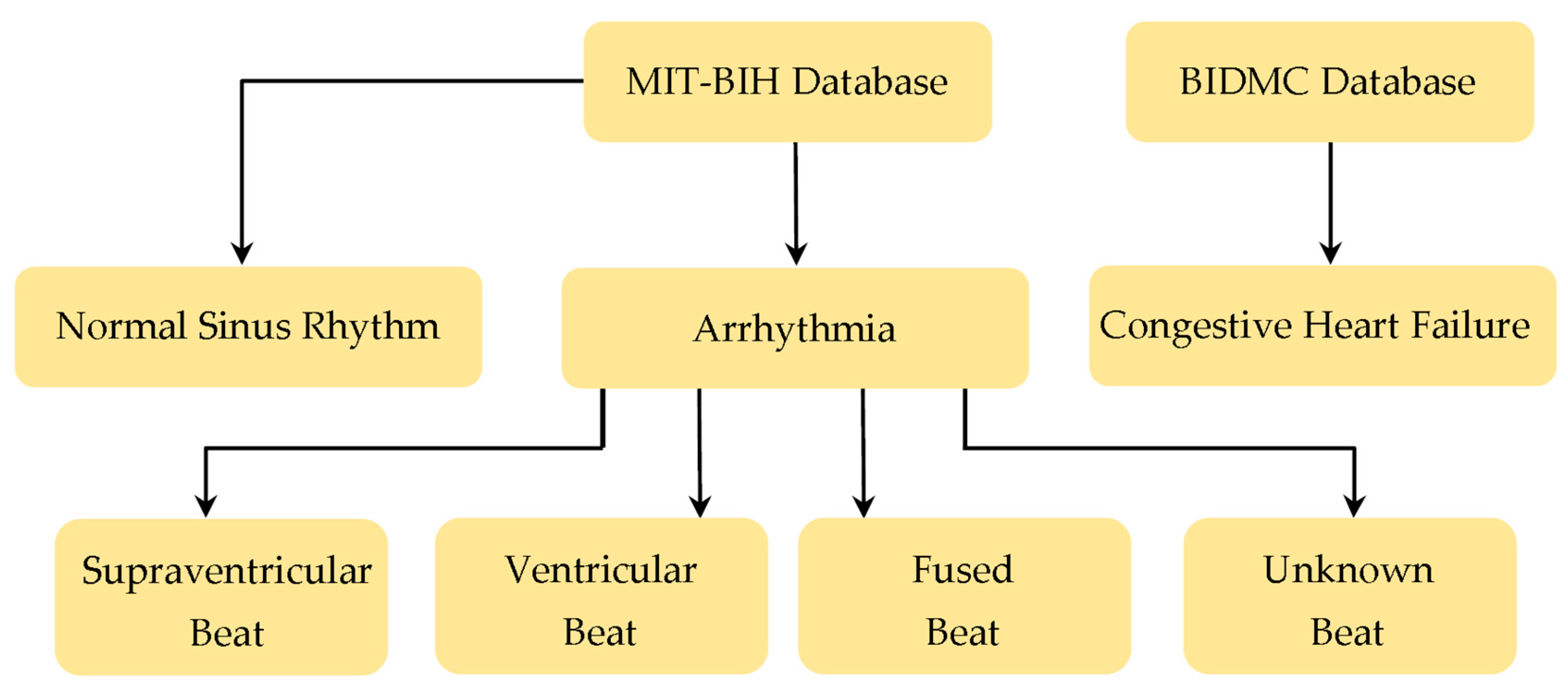
2.1. Database Preparation
2.2. Database Segmentation
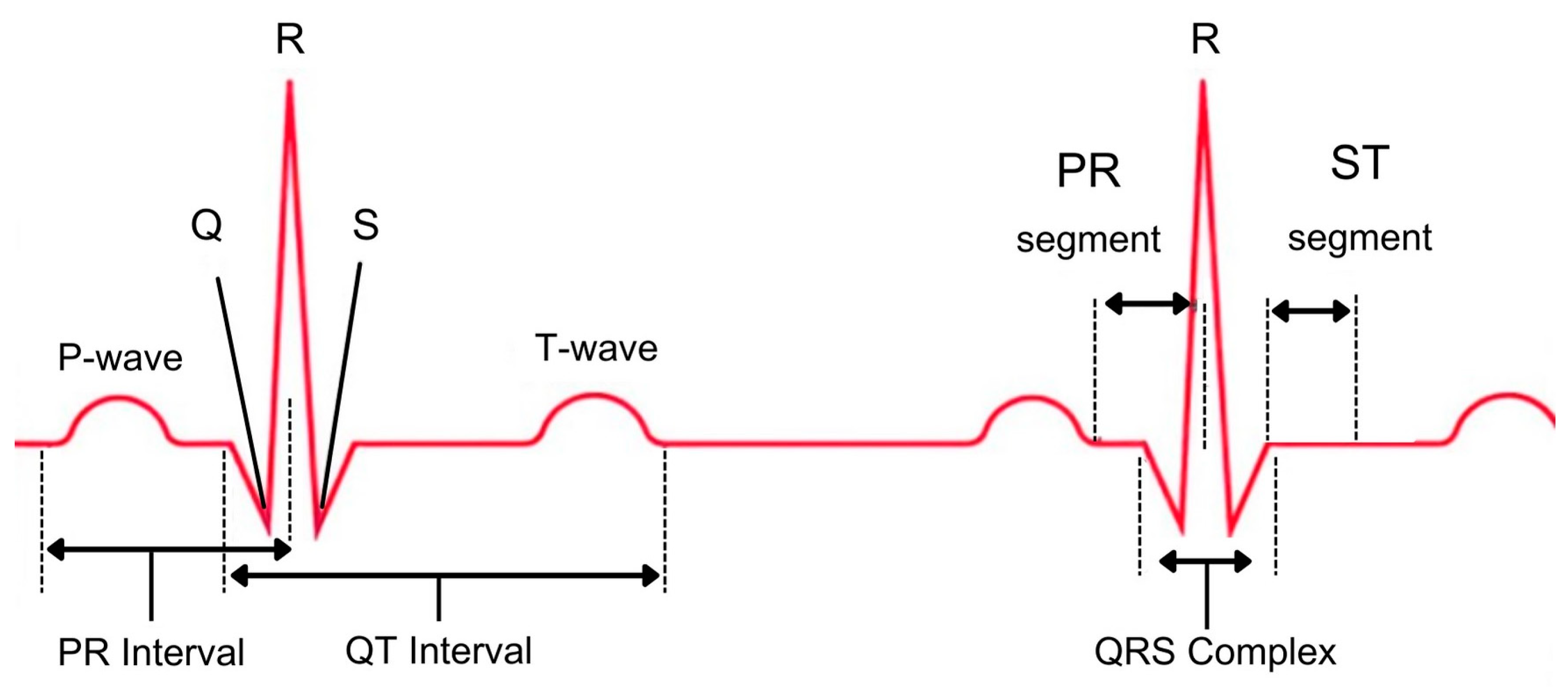
2.3. Feature Extraction
2.4. The Proposed Approach
3. Results and Discussion
3.1. Results of the First Scenario
3.2. Results of the Second Scenario
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Andresen, S.L.; McCarthy, J. Father of AI. IEEE Intell. Syst. 2002, 17, 84–85. [Google Scholar] [CrossRef]
- Rawat, W.; Wang, Z. Deep convolutional neural networks for image classification: A comprehensive review. Neural Comput. 2017, 29, 2352–2449. [Google Scholar] [CrossRef] [PubMed]
- Abdulrahman, M.; Gwadabe, T.R.; Abdu, F.J.; Eleyan, A. Gabor wavelet transform based facial expression recognition using PCA and LBP. In Proceedings of the 22nd Signal Processing and Communications Applications Conference (SIU), Trabzon, Turkey, 23–25 April 2014; pp. 2265–2268. [Google Scholar] [CrossRef]
- Zhang, Z.; Hamadi, H.A.; Damiani, E.; Yeun, C.Y.; Taher, F. Explainable artificial intelligence applications in cyber security: State-of-the-art in research. IEEE Access 2022, 10, 93104–93139. [Google Scholar] [CrossRef]
- Ren, X.; Chen, Y. How Can Artificial Intelligence Help with Space Missions—A Case Study: Computational Intelligence-Assisted Design of Space Tether for Payload Orbital Transfer Under Uncertainties. IEEE Access 2019, 7, 161449–161458. [Google Scholar] [CrossRef]
- Schrettenbrunnner, M.B. Artificial-Intelligence-Driven Management. IEEE Eng. Manag. Rev. 2020, 48, 15–19. [Google Scholar] [CrossRef]
- Lin, P.-H.; Wooders, A.; Wang, J.T.-Y.; Yuan, W.M. Artificial Intelligence, the Missing Piece of Online Education? IEEE Eng. Manag. Rev. 2018, 46, 25–28. [Google Scholar] [CrossRef]
- James, A.P. What and How of Artificial General Intelligence Chip Development. IEEE Trans. Cogn. Dev. Syst. 2022, 14, 333–347. [Google Scholar] [CrossRef]
- Bayram, F.; Eleyan, A. COVID-19 detection on chest radiographs using feature fusion based deep learning. Signal Image Video Process. 2022, 16, 1455–1462. [Google Scholar] [CrossRef]
- Ismael, A.M.; Sengür, A. Deep learning approaches for COVID19 detection based on chest X-ray images. Expert Syst. Appl. 2021, 164, 114054. [Google Scholar] [CrossRef]
- Papageorgiou, E.P.; Boser, B.E.; Anwar, M. Chip-Scale Angle-Selective Imager for In Vivo Microscopic Cancer Detection. IEEE Trans. Biomed. Circuits Syst. 2020, 14, 91–103. [Google Scholar] [CrossRef]
- Eleyan, A.; Alboghbaish, E. Multi-Classifier Deep Learning based System for ECG Classification Using Fourier Transform. In Proceedings of the 5th International Conference on Bioengineering for Smart Technologies (BioSMART), Paris, France, 7–9 June 2023; pp. 1–4. [Google Scholar] [CrossRef]
- Mironovova, M.; Bíla, J. Fast Fourier transform for feature extraction and neural network for classification of electrocardiogram signals. In Proceedings of the Fourth International Conference on Future Generation Communication Technology (FGCT), Luton, UK, 29–31 July 2015; pp. 1–6. [Google Scholar] [CrossRef]
- Sahoo, S.; Kanungo, B.; Behera, S.; Sabut, S. Multiresolution wavelet transform based feature extraction and ECG classification to detect cardiac abnormalities. Measurement 2017, 108, 55–66. [Google Scholar] [CrossRef]
- Aravind, S.; Sanjay, M. ECG Classification and Arrhythmia Detection Using Wavelet Transform and Convolutional Neural Network. In Proceedings of the International Conference on Communication, Control and Information Sciences (ICCISc), Idukki, India, 16–18 June 2021; pp. 1–5. [Google Scholar] [CrossRef]
- Nahak, S.; Saha, G. A Fusion Based Classification of Normal, Arrhythmia and Congestive Heart Failure in ECG. In Proceedings of the National Conference on Communications (NCC), Kharagpur, India, 21–23 February 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Rahuja, N.; Valluru, S.K. A Deep Neural Network Approach to Automatic Multi-Class Classification of Electrocardiogram Signals. In Proceedings of the International Conference on Intelligent Technologies (CONIT), Hubli, India, 25–27 June 2021; pp. 1–4. [Google Scholar] [CrossRef]
- Rahuja, N.; Valluru, S.K. A Comparative Analysis of Deep Neural Network Models using Transfer Learning for Electrocardiogram Signal Classification. In Proceedings of the International Conference on Recent Trends on Electronics, Information, Communication & Technology (RTEICT), Bangalore, India, 27–28 August 2021; pp. 285–290. [Google Scholar] [CrossRef]
- Ebrahimi, Z.; Loni, M.; Daneshtalab, M.; Gharehbaghi, A. A review on deep learning methods for ECG arrhythmia classification. Expert Syst. Appl. X 2020, 7, 100033. [Google Scholar] [CrossRef]
- Ying, Z.; Zhang, G.; Pan, Z.; Chu, C.; Liu, X. FedECG: A federated semi-supervised learning framework for electrocardiogram abnormalities prediction. J. King Saud Univ.—Comput. Inf. Sci. 2023, 35, 101568. [Google Scholar] [CrossRef]
- van Engelen, J.E.; Hoos, H.H. A survey on semi-supervised learning. Mach. Learn. 2020, 109, 373–440. [Google Scholar] [CrossRef]
- Kachuee, M.; Fazeli, S.; Sarrafzadeh, M. ECG Heartbeat Classification: A Deep Transferable Representation. In Proceedings of the IEEE International Conference on Healthcare Informatics (ICHI), New York, NY, USA, 4–7 June 2018; pp. 443–444. [Google Scholar] [CrossRef]
- Safdar, M.F.; Pałka, P.; Nowak, R.M.; AlFaresi, A. A novel data augmentation approach for enhancement of ECG signal classification. Biomed. Signal Process. Control 2023, 86, 105114. [Google Scholar] [CrossRef]
- Chen, C.; Hua, Z.; Zhang, R.; Liu, G.; Wen, W. Automated arrhythmia classification based on a combination network of CNN and LSTM. Biomed. Signal Process. Control 2020, 57, 101819. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, A.; Gao, M.; Chen, X.; Zhang, X.; Chen, X. ECG-based multi-class arrhythmia detection using spatio-temporal attention-based convolutional recurrent neural network. Artif. Intell. Med. 2020, 106, 101856. [Google Scholar] [CrossRef]
- Arhin, J.R.; Zhang, X.; Coker, K.; Agyemang, I.O.; Attipoe, W.K.; Sam, F.; Adjei-Mensah, I.; Agyei, E. ADCGNet: Attention-based dual channel Gabor network towards efficient detection and classification of electrocardiogram images. J. King Saud Univ.—Comput. Inf. Sci. 2023, 35, 101763. [Google Scholar] [CrossRef]
- Cheng, J.; Zou, Q.; Zhao, Y. ECG signal classification based on deep CNN and BiLSTM. BMC Med. Inform. Decis. Mak. 2021, 21, 365. [Google Scholar] [CrossRef]
- Madan, P.; Singh, V.; Singh, D.P.; Diwakar, M.; Pant, B.; Kishor, A. A Hybrid Deep Learning Approach for ECG-Based Arrhythmia Classification. Bioengineering 2022, 9, 152. [Google Scholar] [CrossRef]
- Khan, S.D.; Alarabi, L.; Basalamah, S. Toward Smart Lockdown: A Novel Approach for COVID-19 Hotspots Prediction Using a Deep Hybrid Neural Network. Computers 2020, 9, 99. [Google Scholar] [CrossRef]
- Nassif, A.B.; Shahin, I.; Bader, M.; Hassan, A.; Werghi, N. COVID-19 Detection Systems Using Deep-Learning Algorithms Based on Speech and Image Data. Mathematics 2022, 10, 564. [Google Scholar] [CrossRef]
- Zhu, C.; Sun, Y.; Pan, C. Speech Enhancement with Fractional Fourier Transform. In Proceedings of the International Symposium on Communications and Information Technologies (ISCIT), Xi’an, China, 27–30 September 2022; pp. 296–302. [Google Scholar] [CrossRef]
- Shutko, V.; Tereshchenko, L.; Shutko, M.; Silantieva, I.; Kolganova, O. Application of Spline-Fourier Transform for Radar Signal Processing. In Proceedings of the IEEE 15th International Conference on the Experience of Designing and Application of CAD Systems (CADSM), Polyana, Ukraine, 26 February–2 March 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Wang, B.; Chen, G.; Rong, L.; Liu, Y.; Yu, A.; He, X.; Wen, T.; Zhang, Y.; Hu, B. Arrhythmia Disease Diagnosis Based on ECG Time–Frequency Domain Fusion and Convolutional Neural Network. IEEE J. Transl. Eng. Health Med. 2023, 11, 116–125. [Google Scholar] [CrossRef]
- Thalluri, L.N.; Koripalli, H.; Nukala, P.K.N.; Mandava, V.N.S.R.; Gudapati, G.; Yaswanth, V.V.N. ECG Signal Classification using Deep Neural Networks with Ensemble Techniques. In Proceedings of the 7th International Conference on Communication and Electronics Systems (ICCES), Coimbatore, India, 22–24 June 2022; pp. 273–280. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Vyas, P.; Liu, J.; El-Gayar, O. Fake New Detection on the Web: An LSTM-based Approach. In Proceedings of the AMCIS 2021 Proceedings, Virtual, 9–13 August 2021; p. 5. [Google Scholar]
- Yang, S.; Zhang, Y.; Cho, S.-Y.; Correia, R.; Morgan, S.P. Non-invasive cuff-less blood pressure estimation using a hybrid deep learning model. Opt. Quantum Electron. 2021, 53, 1–20. [Google Scholar] [CrossRef]
- Ji, X.; Dong, Z.; Han, Y.; Lai, C.S.; Zhou, G.; Qi, D. EMSN: An Energy-Efficient Memristive Sequencer Network for Human Emotion Classification in Mental Health Monitoring. IEEE Trans. Consum. Electron. 2023. [Google Scholar] [CrossRef]
- Turk, M.; Pentland, A. Eigenfaces for Recognition. J. Cogn. Neurosci. 1991, 3, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Eleyan, A.; Demirel, H. Face Recognition System Based on PCA and Feedforward Neural Networks. In Computational Intelligence and Bioinspired Systems; IWANN. Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2005; Volume 3512. [Google Scholar] [CrossRef]
- Bartlett, M.S.; Movellan, J.R.; Sejnowski, T.J. Face recognition by independent component analysis. IEEE Trans. Neural Netw. 2002, 13, 1450–1464. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Hu, Z.; Zhang, L.; Li, L.; Liang, Z.; Zhang, Z. Removal of eye-blinking artifacts by ICA in cross-modal long-term EEG recording. In Proceedings of the 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 217–220. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Majumder, S.; Debnath, P.; Chanda, M. Arrhythmic Heartbeat Classification Using Ensemble of Random Forest and Support Vector Machine Algorithm. IEEE Trans. Artif. Intell. 2021, 2, 260–268. [Google Scholar] [CrossRef]
- Zou, C.; Muller, A.; Wolfgang, U.; Ruckert, D.; Muller, P.; Becker, M.; Steger, A.; Martens, E. Heartbeat Classification by Random Forest with a Novel Context Feature: A Segment Label. IEEE J. Transl. Eng. Health Med. 2022, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chumachenko, D.; Butkevych, M.; Lode, D.; Frohme, M.; Schmailzl, K.J.G.; Nechyporenko, A. Machine Learning Methods in Predicting Patients with Suspected Myocardial Infarction Based on Short-Time HRV Data. Sensors 2022, 22, 7033. [Google Scholar] [CrossRef] [PubMed]
- Yakut, Ö.; Bolat, E.; Hatice, E.F.E. K-Means Clustering Algorithm Based Arrhythmic Heartbeat Detection in ECG Signal. Balk. J. Electr. Comput. Eng. 2021, 9, 53–58. [Google Scholar] [CrossRef]
- Rahman Khan, M.M.; Bakr Siddique, M.A.; Sakib, S.; Aziz, A.; Tanzeem, A.K.; Hossain, Z. Electrocardiogram Heartbeat Classification Using Convolutional Neural Networks for the Detection of Cardiac Arrhythmia. In Proceedings of the 2020 Fourth International Conference on I-SMAC (IoT in Social, Mobile, Analytics and Cloud) (I-SMAC), Palladam, India, 7–9 October 2020; pp. 915–920. [Google Scholar]
- Wang, T.; Lu, C.; Ju, W.; Liu, C. Imbalanced heartbeat classification using Easy Ensemble technique and global heartbeat information. Biomed. Signal Process. Control 2022, 71, 103–105. [Google Scholar] [CrossRef]
- Kumari, C.U.; Ankita, R.; Pavani, T.; Vignesh, N.A.; Varma, N.T.; Manzar, M.A.; Reethika, A. Heart Rhythm Abnormality Detection and Classification using Machine Learning Technique. In Proceedings of the 4th International Conference on Trends in Electronics and Informatics (ICOEI) (48184), Tirunelveli, India, 15–17 June 2020; pp. 580–584. [Google Scholar] [CrossRef]
- Phillips, P.; Hahn, C.; Fontana, P.; Yates, A.; Greene, K.; Broniatowski, D.; Przybocki, M. Four Principles of Explainable Artificial Intelligence; National Institute of Standards and Technology (NIST) Report; NIST: Gaithersburg, MD, USA, 2021. [CrossRef]
- Adamczyk, M.; Malatras, A.; Agrafiotis, I. Cybersecurity and Privacy in AI—Medical Imaging Diagnosis; European Union Agency for Cybersecurity (ENISA) Report; ENISA: Athens, Greece, 2023. [Google Scholar]






| Proposed Approach | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| RFSb (ResNet) [44] | 96.6 | 96.6 | 96.6 | 96.6 |
| RFSc (CNN) [44] | 96.8 | 96.7 | 96.7 | 96.7 |
| CNN [47] | 95.2 | 95.2 | 95.4 | 95.3 |
| ADA-Boost [48] | 95.6 | 66.2 | 80.1 | 66.2 |
| PCA-ICA+RF | 96.9 | 96.8 | 96.8 | 96.8 |
| FFT+CNN | 97.3 | 97.2 | 97.3 | 97.2 |
| FFT+CNN–LSTM | 97.4 | 97.3 | 97.4 | 97.3 |
| Actual | ||||||
|---|---|---|---|---|---|---|
| N | S | V | F | Q | ||
| Predicted | N | 17,996 | 31 | 63 | 10 | 18 |
| S | 188 | 352 | 15 | 0 | 1 | |
| V | 97 | 2 | 1334 | 10 | 5 | |
| F | 37 | 0 | 20 | 105 | 0 | |
| Q | 26 | 0 | 20 | 0 | 1532 | |
| Approach | # Used Recordings (ARR/CHF/NSR) | Feature Vector Length | Train/Test | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|---|---|---|
| Fusion+SVM [16] | 30/30/30 | - | - | 93.33 | - | - | |
| Fusion+RF [16] | 30/30/30 | - | - | 92.75 | - | - | |
| CWT+SVM [49] | 96/30/36 | 190 | 70/30 | 95.92 | 96.11 | 92.59 | 93.82 |
| CWT+AlexNet [17] | 96/30/36 | 500 | 80/20 | 97.3 | 97.3 | 96.6 | 96.9 |
| CWT+SqueezeNet [18] | 30/30/30 | 500 | 80/20 | 97.22 | 97.3 | 97.2 | 97.3 |
| CWT+GoogLeNet [18] | 30/30/30 | 500 | 80/20 | 97.78 | 97.8 | 97.7 | 97.7 |
| CWT+AlexNet [18] | 30/30/30 | 500 | 80/20 | 97.8 | 97.7 | 97.8 | 97.7 |
| CWT+CNN–LSTM [28] | 30/30/30 | 500 | 75/25 | 98.9% | 98% | 98% | 97.3% |
| FFT+CNN–LSTM | 30/30/30 | 500 | 80/20 | 99.2 | 99.2 | 99.2 | 99.2 |
| Actual | ||||
|---|---|---|---|---|
| ARR | CHF | NSR | ||
| Predicted | ARR | 761 | 3 | 4 |
| CHF | 0 | 769 | 1 | |
| NSR | 6 | 4 | 810 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eleyan, A.; Alboghbaish, E. Electrocardiogram Signals Classification Using Deep-Learning-Based Incorporated Convolutional Neural Network and Long Short-Term Memory Framework. Computers 2024, 13, 55. https://doi.org/10.3390/computers13020055
Eleyan A, Alboghbaish E. Electrocardiogram Signals Classification Using Deep-Learning-Based Incorporated Convolutional Neural Network and Long Short-Term Memory Framework. Computers. 2024; 13(2):55. https://doi.org/10.3390/computers13020055
Chicago/Turabian StyleEleyan, Alaa, and Ebrahim Alboghbaish. 2024. "Electrocardiogram Signals Classification Using Deep-Learning-Based Incorporated Convolutional Neural Network and Long Short-Term Memory Framework" Computers 13, no. 2: 55. https://doi.org/10.3390/computers13020055
APA StyleEleyan, A., & Alboghbaish, E. (2024). Electrocardiogram Signals Classification Using Deep-Learning-Based Incorporated Convolutional Neural Network and Long Short-Term Memory Framework. Computers, 13(2), 55. https://doi.org/10.3390/computers13020055








