Supervised Machine Learning Models for Liver Disease Risk Prediction
Abstract
1. Introduction
- Data preprocessing is performed with the synthetic minority oversampling technique (SMOTE). In this way, the dataset’s instances are distributed in a balanced way allowing us to design efficient classification models and predict the occurrence of liver disease.
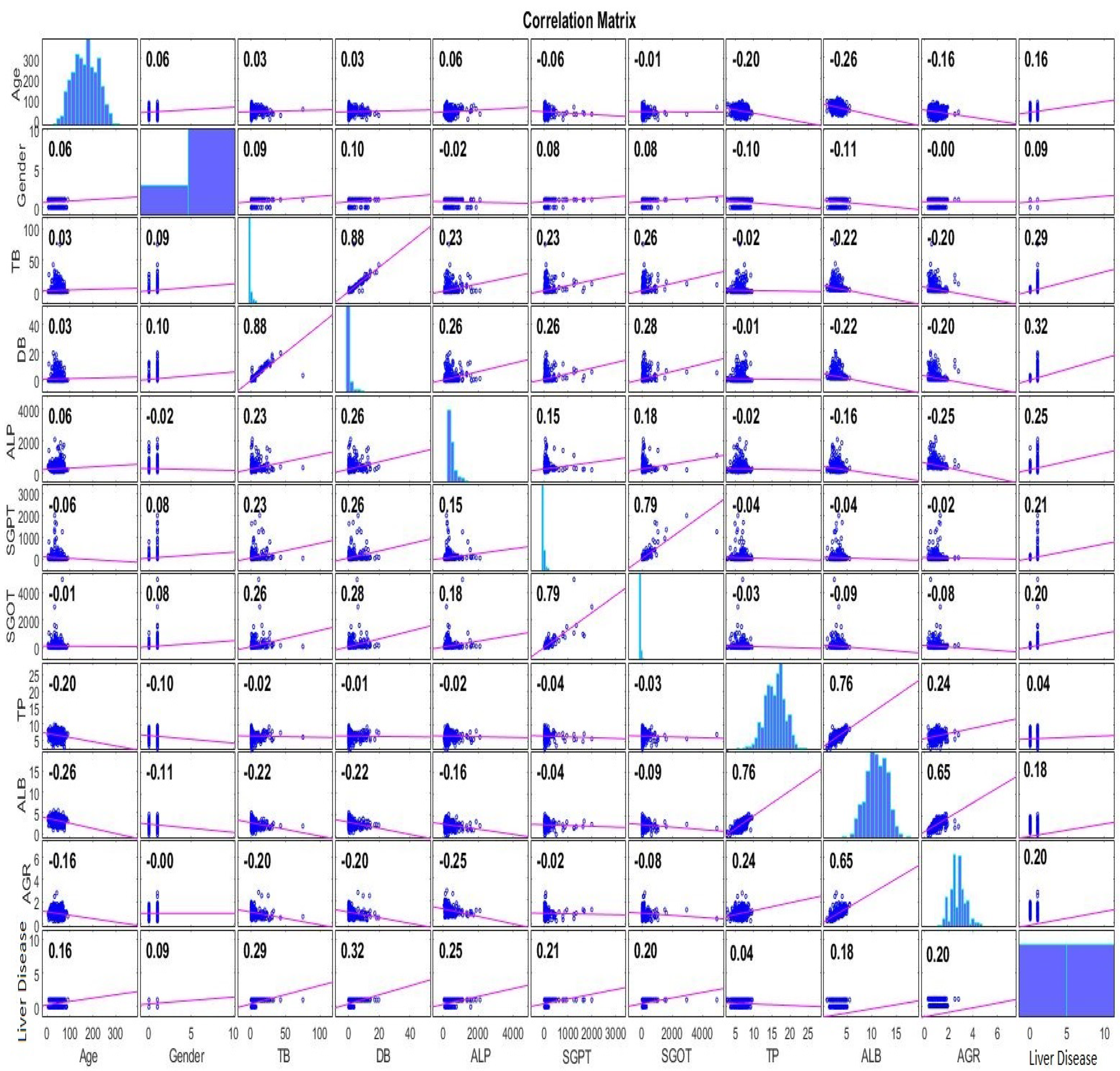
- Features’ importance evaluation based on the Pearson Correlation, Gain Ratio and Random Forest is carried out.
- A comparative evaluation of many ML models’ performance is illustrated considering well-known metrics, such as Precision, Recall, F-Measure, Accuracy and AUC. The experimental results indicated that the Voting classifier prevailed over the other models constituting the proposition of this research work.
- Considering published papers that were based on the same dataset with the same features we relied on, our main proposal (i.e., Voting classifier) outperformed in terms of accuracy.
2. Materials and Methods
2.1. Dataset Description
2.2. Liver Disease Risk Prediction
2.2.1. Data Preprocessing
2.2.2. Features Analysis
2.3. Machine Learning Models
2.4. Evaluation Metrics
- Accuracy: summarizes the performance of the classification task and measures the number of correctly predicted instances out of all the data instances.
- Recall: corresponds to the proportion of participants who were diagnosed with LD and were correctly classified as positive, concerning all positive participants.
- Precision: indicates how many of those who were identified as LD belong to this class.
- F-Measure: is the harmonic mean of the Precision and Recall and sums up the predictive performance of a model. The desired metrics will be calculated with the help of the Confusion matrix.In order to evaluate the distinguishability of a model, the AUC is exploited. It is a metric that varies in [0, 1]. The closer to one, the better the ML model performance is in distinguishing LD from Non-LD instances.
3. Results
3.1. Experimental Setup
3.2. Performance Evaluation
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Arias, I.M.; Alter, H.J.; Boyer, J.L.; Cohen, D.E.; Shafritz, D.A.; Thorgeirsson, S.S.; Wolkoff, A.W. The Liver: Biology and Pathobiology; John Wiley & Sons: Hoboken, NJ, USA, 2020. [Google Scholar]
- Singh, H.R.; Rabi, S. Study of morphological variations of liver in human. Transl. Res. Anat. 2019, 14, 1–5. [Google Scholar] [CrossRef]
- Razavi, H. Global epidemiology of viral hepatitis. Gastroenterol. Clin. 2020, 49, 179–189. [Google Scholar] [CrossRef]
- Ginès, P.; Krag, A.; Abraldes, J.G.; Solà, E.; Fabrellas, N.; Kamath, P.S. Liver cirrhosis. Lancet 2021, 398, 1359–1376. [Google Scholar] [CrossRef] [PubMed]
- Ringehan, M.; McKeating, J.A.; Protzer, U. Viral hepatitis and liver cancer. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160274. [Google Scholar] [CrossRef]
- Powell, E.E.; Wong, V.W.S.; Rinella, M. Non-alcoholic fatty liver disease. Lancet 2021, 397, 2212–2224. [Google Scholar] [CrossRef]
- Smith, A.; Baumgartner, K.; Bositis, C. Cirrhosis: Diagnosis and management. Am. Fam. Physician 2019, 100, 759–770. [Google Scholar] [PubMed]
- Rycroft, J.A.; Mullender, C.M.; Hopkins, M.; Cutino-Moguel, T. Improving the accuracy of clinical interpretation of serological testing for the diagnosis of acute hepatitis a infection. J. Clin. Virol. 2022, 155, 105239. [Google Scholar] [CrossRef]
- Thomas, D.L. Global elimination of chronic hepatitis. N. Engl. J. Med. 2019, 380, 2041–2050. [Google Scholar] [CrossRef]
- Rasche, A.; Sander, A.L.; Corman, V.M.; Drexler, J.F. Evolutionary biology of human hepatitis viruses. J. Hepatol. 2019, 70, 501–520. [Google Scholar] [CrossRef]
- Gust, I.D. Hepatitis A; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- Yuen, M.F.; Chen, D.S.; Dusheiko, G.M.; Janssen, H.L.; Lau, D.T.; Locarnini, S.A.; Peters, M.G.; Lai, C.L. Hepatitis B virus infection. Nat. Rev. Dis. Prim. 2018, 4, 1–20. [Google Scholar] [CrossRef]
- Manns, M.P.; Buti, M.; Gane, E.; Pawlotsky, J.M.; Razavi, H.; Terrault, N.; Younossi, Z. Hepatitis C virus infection. Nat. Rev. Dis. Prim. 2017, 3, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Mentha, N.; Clément, S.; Negro, F.; Alfaiate, D. A review on hepatitis D: From virology to new therapies. J. Adv. Res. 2019, 17, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Kamar, N.; Izopet, J.; Pavio, N.; Aggarwal, R.; Labrique, A.; Wedemeyer, H.; Dalton, H.R. Hepatitis E virus infection. Nat. Rev. Dis. Prim. 2017, 3, 1–16. [Google Scholar] [CrossRef]
- Marchesini, G.; Moscatiello, S.; Di Domizio, S.; Forlani, G. Obesity-associated liver disease. J. Clin. Endocrinol. Metab. 2008, 93, s74–s80. [Google Scholar] [CrossRef]
- Seitz, H.K.; Bataller, R.; Cortez-Pinto, H.; Gao, B.; Gual, A.; Lackner, C.; Mathurin, P.; Mueller, S.; Szabo, G.; Tsukamoto, H. Alcoholic liver disease. Nat. Rev. Dis. Prim. 2018, 4, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Åberg, F.; Färkkilä, M. Drinking and obesity: Alcoholic liver disease/nonalcoholic fatty liver disease interactions. In Seminars in Liver Disease; Thieme Medical Publishers: New York, NY, USA, 2020; Volume 40, pp. 154–162. [Google Scholar]
- Bae, M.; Park, Y.K.; Lee, J.Y. Food components with antifibrotic activity and implications in prevention of liver disease. J. Nutr. Biochem. 2018, 55, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Zhang, X.J.; Li, H. Progress and challenges in the prevention and control of nonalcoholic fatty liver disease. Med. Res. Rev. 2019, 39, 328–348. [Google Scholar] [CrossRef] [PubMed]
- Fazakis, N.; Kocsis, O.; Dritsas, E.; Alexiou, S.; Fakotakis, N.; Moustakas, K. Machine learning tools for long-term type 2 diabetes risk prediction. IEEE Access 2021, 9, 103737–103757. [Google Scholar] [CrossRef]
- Dritsas, E.; Trigka, M. Data-Driven Machine-Learning Methods for Diabetes Risk Prediction. Sensors 2022, 22, 5304. [Google Scholar] [CrossRef]
- Alexiou, S.; Dritsas, E.; Kocsis, O.; Moustakas, K.; Fakotakis, N. An approach for Personalized Continuous Glucose Prediction with Regression Trees. In Proceedings of the 2021 6th South-East Europe Design Automation, Computer Engineering, Computer Networks and Social Media Conference (SEEDA-CECNSM), Preveza, Greece, 24–26 September 2021; pp. 1–6. [Google Scholar]
- Dritsas, E.; Alexiou, S.; Konstantoulas, I.; Moustakas, K. Short-term Glucose Prediction based on Oral Glucose Tolerance Test Values. In Proceedings of the International Joint Conference on Biomedical Engineering Systems and Technologies—HEALTHINF, Lisbon, Portugal, 9–11 February 2022; Volume 5, pp. 249–255. [Google Scholar]
- Fazakis, N.; Dritsas, E.; Kocsis, O.; Fakotakis, N.; Moustakas, K. Long-Term Cholesterol Risk Prediction with Machine Learning Techniques in ELSA Database. In Proceedings of the 13th International Joint Conference on Computational Intelligence (IJCCI), Online, 24–26 October 2021; pp. 445–450. [Google Scholar]
- Dritsas, E.; Fazakis, N.; Kocsis, O.; Fakotakis, N.; Moustakas, K. Long-Term Hypertension Risk Prediction with ML Techniques in ELSA Database. In Proceedings of the International Conference on Learning and Intelligent Optimization, Athens, Greece, 20–25 June 2021; pp. 113–120. [Google Scholar]
- Dritsas, E.; Alexiou, S.; Moustakas, K. Efficient Data-driven Machine Learning Models for Hypertension Risk Prediction. In Proceedings of the 2022 International Conference on INnovations in Intelligent SysTems and Applications (INISTA), Biarritz, France, 8–10 August 2022; pp. 1–6. [Google Scholar]
- Dritsas, E.; Trigka, M. Machine Learning Methods for Hypercholesterolemia Long-Term Risk Prediction. Sensors 2022, 22, 5365. [Google Scholar] [CrossRef]
- Dritsas, E.; Alexiou, S.; Moustakas, K. COPD Severity Prediction in Elderly with ML Techniques. In Proceedings of the 15th International Conference on PErvasive Technologies Related to Assistive Environments, Corfu Island, Greece, 29 June–1 July 2022; pp. 185–189. [Google Scholar]
- Dritsas, E.; Trigka, M. Supervised Machine Learning Models to Identify Early-Stage Symptoms of SARS-CoV-2. Sensors 2023, 23, 40. [Google Scholar] [CrossRef]
- Dritsas, E.; Trigka, M. Stroke Risk Prediction with Machine Learning Techniques. Sensors 2022, 22, 4670. [Google Scholar] [CrossRef]
- Dritsas, E.; Trigka, M. Machine Learning Techniques for Chronic Kidney Disease Risk Prediction. Big Data Cogn. Comput. 2022, 6, 98. [Google Scholar] [CrossRef]
- Dritsas, E.; Trigka, M. Lung Cancer Risk Prediction with Machine Learning Models. Big Data Cogn. Comput. 2022, 6, 139. [Google Scholar] [CrossRef]
- Konstantoulas, I.; Kocsis, O.; Dritsas, E.; Fakotakis, N.; Moustakas, K. Sleep Quality Monitoring with Human Assisted Corrections. In Proceedings of the International Joint Conference on Computational Intelligence (IJCCI), Online, 24–26 October 2021; pp. 435–444. [Google Scholar]
- Konstantoulas, I.; Dritsas, E.; Moustakas, K. Sleep Quality Evaluation in Rich Information Data. In Proceedings of the 2022 13th International Conference on Information, Intelligence, Systems & Applications (IISA), Corfu, Greece, 18–20 July 2022; pp. 1–4. [Google Scholar]
- Dritsas, E.; Alexiou, S.; Moustakas, K. Cardiovascular Disease Risk Prediction with Supervised Machine Learning Techniques. In Proceedings of the ICT4AWE, Online, 23–25 April 2022; pp. 315–321. [Google Scholar]
- Indian Liver Patient Records. Available online: https://www.kaggle.com/datasets/uciml/indian-liver-patient-records (accessed on 14 November 2022).
- Mauvais-Jarvis, F.; Merz, N.B.; Barnes, P.J.; Brinton, R.D.; Carrero, J.J.; DeMeo, D.L.; De Vries, G.J.; Epperson, C.N.; Govindan, R.; Klein, S.L.; et al. Sex and gender: Modifiers of health, disease, and medicine. Lancet 2020, 396, 565–582. [Google Scholar] [CrossRef]
- Lin, H.; Yip, T.C.F.; Zhang, X.; Li, G.; Tse, Y.K.; Hui, V.W.K.; Liang, L.Y.; Lai, J.C.T.; Chan, S.L.; Chan, H.L.Y.; et al. Age and the relative importance of liver-related deaths in nonalcoholic fatty liver disease. Hepatology 2022. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, A.R.G.; Crespo, J.; Martínez, R.M.L.; Iruzubieta, P.; Mercadal, G.C.; Garcés, M.L.; Lavin, B.; Ruiz, M.M. Measurement and clinical usefulness of bilirubin in liver disease. Adv. Lab. Med. Med. Lab. 2021, 2, 352–361. [Google Scholar]
- Liu, Y.; Cavallaro, P.M.; Kim, B.M.; Liu, T.; Wang, H.; Kühn, F.; Adiliaghdam, F.; Liu, E.; Vasan, R.; Samarbafzadeh, E.; et al. A role for intestinal alkaline phosphatase in preventing liver fibrosis. Theranostics 2021, 11, 14. [Google Scholar] [CrossRef]
- Goodarzi, R.; Sabzian, K.; Shishehbor, F.; Mansoori, A. Does turmeric/curcumin supplementation improve serum alanine aminotransferase and aspartate aminotransferase levels in patients with nonalcoholic fatty liver disease? A systematic review and meta-analysis of randomized controlled trials. Phytother. Res. 2019, 33, 561–570. [Google Scholar] [CrossRef]
- He, B.; Shi, J.; Wang, X.; Jiang, H.; Zhu, H.J. Genome-wide pQTL analysis of protein expression regulatory networks in the human liver. BMC Biol. 2020, 18, 1–16. [Google Scholar] [CrossRef]
- Carvalho, J.R.; Machado, M.V. New insights about albumin and liver disease. Ann. Hepatol. 2018, 17, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Chen, W.; Gu, M.; Xian, G.; Pan, B.; Zheng, L.; Zhang, Z.; Sheng, P. Serum globulin and albumin to globulin ratio as potential diagnostic biomarkers for periprosthetic joint infection: A retrospective review. J. Orthop. Surg. Res. 2020, 15, 1–7. [Google Scholar] [CrossRef]
- Maldonado, S.; López, J.; Vairetti, C. An alternative SMOTE oversampling strategy for high-dimensional datasets. Appl. Soft Comput. 2019, 76, 380–389. [Google Scholar] [CrossRef]
- Dritsas, E.; Fazakis, N.; Kocsis, O.; Moustakas, K.; Fakotakis, N. Optimal Team Pairing of Elder Office Employees with Machine Learning on Synthetic Data. In Proceedings of the 2021 12th International Conference on Information, Intelligence, Systems & Applications (IISA), Chania Crete, Greece, 12–14 July 2021; pp. 1–4. [Google Scholar]
- Jain, D.; Singh, V. Feature selection and classification systems for chronic disease prediction: A review. Egypt. Inform. J. 2018, 19, 179–189. [Google Scholar] [CrossRef]
- Liu, Y.; Mu, Y.; Chen, K.; Li, Y.; Guo, J. Daily activity feature selection in smart homes based on Pearson correlation coefficient. Neural Process. Lett. 2020, 51, 1771–1787. [Google Scholar] [CrossRef]
- Gnanambal, S.; Thangaraj, M.; Meenatchi, V.; Gayathri, V. Classification algorithms with attribute selection: An evaluation study using WEKA. Int. J. Adv. Netw. Appl. 2018, 9, 3640–3644. [Google Scholar]
- Aldrich, C. Process variable importance analysis by use of random forests in a shapley regression framework. Minerals 2020, 10, 420. [Google Scholar] [CrossRef]
- Berrar, D. Bayes’ theorem and naive Bayes classifier. Encycl. Bioinform. Comput. Biol. ABC Bioinform. 2018, 1, 403. [Google Scholar]
- Nusinovici, S.; Tham, Y.C.; Yan, M.Y.C.; Ting, D.S.W.; Li, J.; Sabanayagam, C.; Wong, T.Y.; Cheng, C.Y. Logistic regression was as good as machine learning for predicting major chronic diseases. J. Clin. Epidemiol. 2020, 122, 56–69. [Google Scholar] [CrossRef]
- Ghosh, S.; Dasgupta, A.; Swetapadma, A. A study on support vector machine based linear and non-linear pattern classification. In Proceedings of the 2019 International Conference on Intelligent Sustainable Systems (ICISS), Palladam, India, 21–22 February 2019; pp. 24–28. [Google Scholar]
- Emon, S.U.; Trishna, T.I.; Ema, R.R.; Sajal, G.I.H.; Kundu, S.; Islam, T. Detection of hepatitis viruses based on J48, KStar and Naïve Bayes Classifier. In Proceedings of the 2019 10th International Conference on Computing, Communication and Networking Technologies (ICCCNT), Kanpur, India, 6–8 July 2019; pp. 1–7. [Google Scholar]
- Joloudari, J.H.; Hassannataj Joloudari, E.; Saadatfar, H.; Ghasemigol, M.; Razavi, S.M.; Mosavi, A.; Nabipour, N.; Shamshirband, S.; Nadai, L. Coronary artery disease diagnosis; ranking the significant features using a random trees model. Int. J. Environ. Res. Public Health 2020, 17, 731. [Google Scholar] [CrossRef]
- Catherine, O. Lower Respiratory Tract Infection Clinical Diagnostic System Driven by Reduced Error Pruning Tree (REP Tree). Am. J. Compt. Sci. Inf. Technol. 2020, 8, 53. [Google Scholar]
- Dong, X.; Yu, Z.; Cao, W.; Shi, Y.; Ma, Q. A survey on ensemble learning. Front. Comput. Sci. 2020, 14, 241–258. [Google Scholar] [CrossRef]
- González, S.; García, S.; Del Ser, J.; Rokach, L.; Herrera, F. A practical tutorial on bagging and boosting based ensembles for machine learning: Algorithms, software tools, performance study, practical perspectives and opportunities. Inf. Fusion 2020, 64, 205–237. [Google Scholar] [CrossRef]
- Palimkar, P.; Shaw, R.N.; Ghosh, A. Machine learning technique to prognosis diabetes disease: Random forest classifier approach. In Advanced Computing and Intelligent Technologies; Springer: Berlin/Heidelberg, Germany, 2022; pp. 219–244. [Google Scholar]
- Ani, R.; Jose, J.; Wilson, M.; Deepa, O. Modified rotation forest ensemble classifier for medical diagnosis in decision support systems. In Progress in Advanced Computing and Intelligent Engineering; Springer: Berlin/Heidelberg, Germany, 2018; pp. 137–146. [Google Scholar]
- Polat, K.; Sentürk, U. A novel ML approach to prediction of breast cancer: Combining of mad normalization, KMC based feature weighting and AdaBoostM1 classifier. In Proceedings of the 2018 2nd International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), Kanpur, India, 19–21 October 2018; pp. 1–4. [Google Scholar]
- Kumari, S.; Kumar, D.; Mittal, M. An ensemble approach for classification and prediction of diabetes mellitus using soft voting classifier. Int. J. Cogn. Comput. Eng. 2021, 2, 40–46. [Google Scholar] [CrossRef]
- Pavlyshenko, B. Using stacking approaches for machine learning models. In Proceedings of the 2018 IEEE Second International Conference on Data Stream Mining & Processing (DSMP), Lviv, Ukraine, 21–25 August 2018; pp. 255–258. [Google Scholar]
- Masih, N.; Naz, H.; Ahuja, S. Multilayer perceptron based deep neural network for early detection of coronary heart disease. Health Technol. 2021, 11, 127–138. [Google Scholar] [CrossRef]
- Cunningham, P.; Delany, S.J. k-Nearest neighbour classifiers-A Tutorial. ACM Comput. Surv. (CSUR) 2021, 54, 1–25. [Google Scholar] [CrossRef]
- Handelman, G.S.; Kok, H.K.; Chandra, R.V.; Razavi, A.H.; Huang, S.; Brooks, M.; Lee, M.J.; Asadi, H. Peering into the black box of artificial intelligence: Evaluation metrics of machine learning methods. Am. J. Roentgenol. 2019, 212, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Gandomi, A.H.; Chen, F.; Holzinger, A. Evaluating the quality of machine learning explanations: A survey on methods and metrics. Electronics 2021, 10, 593. [Google Scholar] [CrossRef]
- Weka. Available online: https://www.weka.io/ (accessed on 14 November 2022).
- Dhamodharan, S. Liver Disease Prediction Using Bayesian Classification. 2016. Available online: https://www.ijact.in/index.php/ijact/article/viewFile/443/378 (accessed on 14 November 2022).
- Gajendran, G.; Varadharajan, R. Classification of Indian liver patients data set using MAMFFN. In Proceedings of the AIP Conference Proceedings, Coimbatore, India, 17–18 July 2020; Volume 2277, p. 120001. [Google Scholar]
- Geetha, C.; Arunachalam, A. Evaluation based Approaches for Liver Disease Prediction using Machine Learning Algorithms. In Proceedings of the 2021 International Conference on Computer Communication and Informatics (ICCCI), Coimbatore, India, 27–29 January 2021; pp. 1–4. [Google Scholar]
- Rahman, A.S.; Shamrat, F.J.M.; Tasnim, Z.; Roy, J.; Hossain, S.A. A comparative study on liver disease prediction using supervised machine learning algorithms. Int. J. Sci. Technol. Res. 2019, 8, 419–422. [Google Scholar]
- Srivastava, A.; Kumar, V.V.; Mahesh, T.; Vivek, V. Automated Prediction of Liver Disease using Machine Learning (ML) Algorithms. In Proceedings of the 2022 Second International Conference on Advances in Electrical, Computing, Communication and Sustainable Technologies (ICAECT), Bhilai, India, 21–22 April 2022; pp. 1–4. [Google Scholar]
- Singh, A.S.; Irfan, M.; Chowdhury, A. Prediction of liver disease using classification algorithms. In Proceedings of the 2018 4th International Conference on Computing Communication and Automation (ICCCA), Greater Noida, India, 14–15 December 2018; pp. 1–3. [Google Scholar]
- Choudhary, R.; Gopalakrishnan, T.; Ruby, D.; Gayathri, A.; Murthy, V.S.; Shekhar, R. An Efficient Model for Predicting Liver Disease Using Machine Learning. In Data Analytics in Bioinformatics: A Machine Learning Perspective; Wiley Online Library: Hoboken, NJ, USA, 2021; pp. 443–457. [Google Scholar]
- Bahramirad, S.; Mustapha, A.; Eshraghi, M. Classification of liver disease diagnosis: A comparative study. In Proceedings of the 2013 Second International Conference on Informatics & Applications (ICIA), Lodz, Poland, 23–25 September 2013; pp. 42–46. [Google Scholar]
- Kumar, P.; Thakur, R.S. Early detection of the liver disorder from imbalance liver function test datasets. Int. J. Innov. Technol. Explor. Eng. 2019, 8, 179–186. [Google Scholar]
- Idris, K.; Bhoite, S. Applications of machine learning for prediction of liver disease. Int. J. Comput. Appl. Technol. Res 2019, 8, 394–396. [Google Scholar] [CrossRef]
- Muthuselvan, S.; Rajapraksh, S.; Somasundaram, K.; Karthik, K. Classification of liver patient dataset using machine learning algorithms. Int. J. Eng. Technol. 2018, 7, 323. [Google Scholar] [CrossRef]
- Azam, M.S.; Rahman, A.; Iqbal, S.H.S.; Ahmed, M.T. Prediction of liver diseases by using few machine learning based approaches. Aust. J. Eng. Innov. Technol. 2020, 2, 85–90. [Google Scholar]
- Sontakke, S.; Lohokare, J.; Dani, R. Diagnosis of liver diseases using machine learning. In Proceedings of the 2017 International Conference on Emerging Trends & Innovation in ICT (ICEI), Pune, India, 3–5 February 2017; pp. 129–133. [Google Scholar]
- Sokoliuk, A.; Kondratenko, G.; Sidenko, I.; Kondratenko, Y.; Khomchenko, A.; Atamanyuk, I. Machine learning algorithms for binary classification of liver disease. In Proceedings of the 2020 IEEE International Conference on Problems of Infocommunications. Science and Technology (PIC S&T), Kharkiv, Ukraine, 6–9 October 2020; pp. 417–421. [Google Scholar]
- Swapna, K.; Prasad Babu, M. Critical analysis of Indian liver patients dataset using ANOVA method. Int. J. Eng. Technol 2017, 7, 19–33. [Google Scholar]
- Gulia, A.; Vohra, R.; Rani, P. Liver patient classification using intelligent techniques. Int. J. Comput. Sci. Inf. Technol. 2014, 5, 5110–5115. [Google Scholar]
- Khan, B.; Naseem, R.; Ali, M.; Arshad, M.; Jan, N. Machine learning approaches for liver disease diagnosing. Int. J. Data Sci. Adv. Anal. (ISSN 2563-4429) 2019, 1, 27–31. [Google Scholar]
- Jin, H.; Kim, S.; Kim, J. Decision factors on effective liver patient data prediction. Int. J. Bio-Sci. Bio-Technol. 2014, 6, 167–178. [Google Scholar] [CrossRef]
- Ramana, B.V.; Boddu, R.S.K. Performance comparison of classification algorithms on medical datasets. In Proceedings of the 2019 IEEE 9th Annual Computing and Communication Workshop and Conference (CCWC), Las Vegas, NV, USA, 7–9 January 2019; pp. 140–145. [Google Scholar]

| Feature | Type | Description |
|---|---|---|
| Gender [38] | nominal | This feature illustrates the participant’s gender. |
| Age (years) [39] | numeric | The age range of the participants is 4–90 years. |
| Total Bilirubin—TB (mg/dL) [40] | numeric | This feature captures the participant’s total bilirubin. |
| Direct Bilirubin—DB (mg/dL) [40] | numeric | This feature captures the participant’s direct bilirubin. |
| Alkaline Phosphatase—ALP (IU/L) [41] | numeric | This feature captures the participant’s alkaline phosphatase. |
| Alanine Aminotransferase—SGPT (U/L) [42] | numeric | This feature captures the participant’s alanine aminotransferase. |
| Aspartate Aminotransferase—SGOT (U/L) [42] | numeric | This feature captures the participant’s aspartate aminotransferase. |
| Total Protein—TP (g/L) [43] | numeric | This feature captures the participant’s total protein. |
| Albumin—ALB (g/dL) [44] | numeric | This feature captures the participant’s albumin. |
| Albumin and Globulin Ratio—AGR [45] | numeric | This feature captures the participant’s albumin and globulin Ratio. |
| Liver Disease | nominal | This feature stands for whether the participant has been diagnosed with liver disease or not. |
| Min | Max | Mean ± Stdv | |
|---|---|---|---|
| Age | 4 | 90 | 43.55 ± 16.28 |
| TB | 0.4 | 75 | 2.65 ± 5.32 |
| DB | 0.1 | 19.7 | 1.16 ± 2.42 |
| ALP | 63 | 2110 | 267.26 ± 212.62 |
| SGPT | 10 | 2000 | 66.78 ± 155.16 |
| SGOT | 10 | 4929 | 88.78 ± 245.07 |
| TP | 2.7 | 9.6 | 6.50 ± 1.02 |
| ALB | 0.9 | 5.5 | 3.19 ± 0.76 |
| GR | 0.3 | 2.8 | 0.98 ± 0.30 |
| Feature | Pearson Rank | Feature | Gain Ratio | Feature | Random Forest |
|---|---|---|---|---|---|
| DB | 0.3205 | DB | 0.1421 | DB | 0.2895 |
| TB | 0.2874 | TB | 0.1373 | ALB | 0.2883 |
| ALP | 0.246 | SGOT | 0.1005 | TB | 0.2848 |
| SGPT | 0.2141 | ALP | 0.0867 | Age | 0.2625 |
| AGR | 0.2046 | AGR | 0.0822 | SGPT | 0.2613 |
| SGOT | 0.2017 | SGPT | 0.0701 | AGR | 0.2599 |
| ALB | 0.1836 | ALB | 0.0408 | SGOT | 0.2393 |
| Age | 0.1596 | Age | 0.0372 | TP | 0.2161 |
| Gender | 0.0857 | Gender | 0.0065 | ALP | 0.1936 |
| TP | 0.0443 | TP | 0 | Gender | 0.0255 |
| Model | Parameters |
|---|---|
| NB | useKernelEstimator: False useSupervisedDiscretization: True |
| SVM | eps = 0.001 gamma = 0.0 kernel type: linear loss = 0.1 |
| LR | ridge = useConjugateGradientDescent: True |
| ANN | hidden layers: ‘a’ learning rate = 0.1 momentum = 0.2 training time = 200 |
| J48 | reducedErrorPruning: True savelnstanceData: True useMDLCorrection: True subtreeRaising: True binarysplits = True collapseTree = True |
| RT | maxDepth = 0 minNum = 1.0 minVarianceProp = 0.001 |
| RepTree | maxDepth = 1 minNum = 2.0 minVarianceProp = 0.001 |
| RF | breakTiesRandomly:True numIterations = 100 numFeatures = 0 |
| RotF | classifier: Random Forest numberOfGroups: False projectionFilter: PrincipalComponents |
| kNN | k = 1 Search Algorithm: LinearNNSearch with Euclidean cross-validate = True |
| AdaBoostM1 | classifier: Random Forest resume: True useResampling: True |
| Stacking | classifiers: Random Forest and AdaBoostM1 metaClassifier: Logistic Regression numFolds = 10 |
| Voting | classifiers: Random Forest and AdaBoostM1 combinationRule: average of probabilities |
| Bagging | classifiers: Random Forest printClassifiers: True storeOutOfBagPredictions: True |
| Class “No” | Precision | Recall | ||
|---|---|---|---|---|
| No SMOTE | SMOTE | No SMOTE | SMOTE | |
| NB | 0.429 | 0.671 | 0.679 | 0.831 |
| SVM | 0.000 | 0.648 | 0.000 | 0.908 |
| LR | 0.544 | 0.659 | 0.261 | 0.826 |
| MLP | 0.415 | 0.653 | 0.297 | 0.787 |
| 1-NN | 0.405 | 0.670 | 0.467 | 0.744 |
| J48 | 0.391 | 0.647 | 0.055 | 0.792 |
| RF | 0.495 | 0.763 | 0.291 | 0.848 |
| RT | 0.475 | 0.717 | 0.509 | 0.717 |
| RepTree | 0.358 | 0.687 | 0.206 | 0.763 |
| RotF | 0.564 | 0.726 | 0.321 | 0.884 |
| AdaBoostM1 | 0.529 | 0.756 | 0.442 | 0.853 |
| Stacking | 0.494 | 0.770 | 0.255 | 0.831 |
| Bagging | 0.494 | 0.749 | 0.267 | 0.853 |
| Voting | 0.515 | 0.791 | 0.309 | 0.875 |
| Accuracy | Precision | Recall | F-Measure | AUC | |
|---|---|---|---|---|---|
| NB | 0.711 | 0.724 | 0.711 | 0.707 | 0.771 |
| SVM | 0.708 | 0.748 | 0.708 | 0.708 | 0.708 |
| LR | 0.700 | 0.713 | 0.700 | 0.700 | 0.754 |
| MLP | 0.690 | 0.701 | 0.690 | 0.690 | 0.742 |
| 1-NN | 0.688 | 0.691 | 0.688 | 0.687 | 0.693 |
| J48 | 0.732 | 0.733 | 0.732 | 0.732 | 0.735 |
| RF | 0.794 | 0.798 | 0.793 | 0.793 | 0.877 |
| RT | 0.718 | 0.717 | 0.717 | 0.717 | 0.717 |
| RepTree | 0.708 | 0.710 | 0.708 | 0.707 | 0.761 |
| RotF | 0.775 | 0.789 | 0.775 | 0.773 | 0.869 |
| AdaBoostM1 | 0.795 | 0.797 | 0.795 | 0.795 | 0.879 |
| Stacking | 0.794 | 0.795 | 0.793 | 0.793 | 0.881 |
| Bagging | 0.785 | 0.791 | 0.785 | 0.784 | 0.872 |
| Voting | 0.801 | 0.804 | 0.801 | 0.801 | 0.884 |
| Research Work | Proposed Model | Accuracy |
|---|---|---|
| Present Work | Voting | 80.10% |
| [70] | NB | 75.54% |
| [71] | MAMFFN | 75.30% |
| [72] | SVM | 75.04% |
| [73,74,75,76,77] | LR | 75% |
| [78] | kNN | 74.67% |
| [79] | AdaBoostM1 | 74.36% |
| [80] | RT | 74.20% |
| [81] | KNNWFST | 74% |
| [82] | Back Propagation | 73.20% |
| [83] | Gradient Tree Boosting | 72% |
| [84,85] | RF | 71.87% |
| [86] | CHIRP | 71.30% |
| [87] | DT | 69.40% |
| [88] | Bagging | 69.30% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dritsas, E.; Trigka, M. Supervised Machine Learning Models for Liver Disease Risk Prediction. Computers 2023, 12, 19. https://doi.org/10.3390/computers12010019
Dritsas E, Trigka M. Supervised Machine Learning Models for Liver Disease Risk Prediction. Computers. 2023; 12(1):19. https://doi.org/10.3390/computers12010019
Chicago/Turabian StyleDritsas, Elias, and Maria Trigka. 2023. "Supervised Machine Learning Models for Liver Disease Risk Prediction" Computers 12, no. 1: 19. https://doi.org/10.3390/computers12010019
APA StyleDritsas, E., & Trigka, M. (2023). Supervised Machine Learning Models for Liver Disease Risk Prediction. Computers, 12(1), 19. https://doi.org/10.3390/computers12010019







