A Novel Two-Stage Heart Arrhythmia Ensemble Classifier
Abstract
:1. Introduction
1.1. Cardiovascular Disease
1.2. Computer-Aided Diagnosis
1.3. Data Set
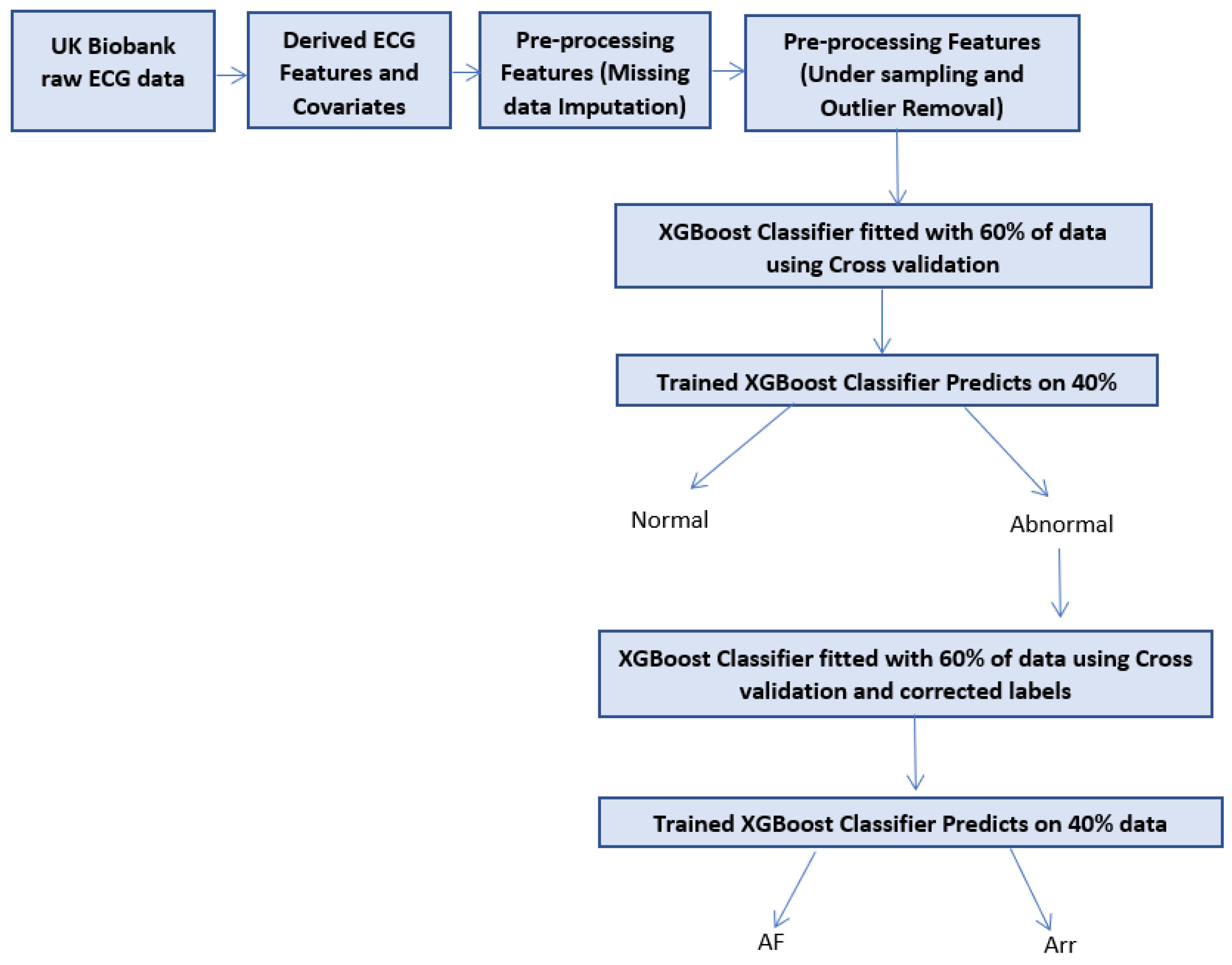
1.4. Two-Stage Concept
1.5. Paper Structure
2. Literature Background
2.1. Available Data Sets
2.2. Related Work
3. Proposed Two-Stage Method
3.1. Data Description
3.2. Pre-Processing
3.3. Missing Data
3.4. Undersampling and Outlier Removal
4. Classifier Description
4.1. Two-Stage Classifier
4.2. XGBoost Classifier
4.3. XGBoost Parameters
4.4. Cross Validation and Overfitting
5. Results and Discussion
5.1. Evaluation Metric Measures
5.2. Classifiers Implementation
5.3. Comparison with Other Methods
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Arr | Ventricular Arrhythmia |
| AF | Atrial Fibrillation |
| TP | True Positives |
| TN | True Negatives |
| FP | False Positives |
| FN | False Negatives |
References
- CVD statistics British Heart Foundation. Available online: https://www.bhf.org.uk (accessed on 22 April 2021).
- Muhammad Ashfaq Khan, Y.K. Cardiac Arrhythmia Disease Classification Using LSTM Deep Learning Approach. Comput. Mater. Contin. 2021, 67, 427–443. [Google Scholar] [CrossRef]
- Hizukuri, A.; Nakayama, R.; Nara, M.; Suzuki, M.; Namba, K. Computer-Aided Diagnosis Scheme for Distinguishing Between Benign and Malignant Masses on Breast DCE-MRI Images Using Deep Convolutional Neural Network with Bayesian Optimization. J. Digit. Imaging 2020, 34, 116–123. [Google Scholar] [CrossRef]
- UK Biobank ECG at Rest Repository. Available online: https://biobank.ndph.ox.ac.uk/ukb/field.cgi?id=20205 (accessed on 23 April 2021).
- Jha, C.K.; Kolekar, M.H. Cardiac arrhythmia classification using tunable Q-wavelet transform based features and support vector machine classifier. Biomed. Signal Process. Control 2020, 59, 101875. [Google Scholar] [CrossRef]
- Ramírez, E.; Melin, P.; Prado-Arechiga, G. Hybrid Model Based on Neural Networks and Fuzzy Logic for 2-Lead Cardiac Arrhythmia Classification. In Hybrid Intelligent Systems in Control, Pattern Recognition and Medicine Studies in Computational Intelligence; Springer: Berlin/Heidelberg, Germany, 2019; pp. 193–217. [Google Scholar] [CrossRef]
- Sahoo, S.; Subudhi, A.; Dash, M.; Sabut, S. Automatic Classification of Cardiac Arrhythmias Based on Hybrid Features and Decision Tree Algorithm. Int. J. Autom. Comput. 2020, 17, 551–561. [Google Scholar] [CrossRef]
- Swetha, R.; Ramakrishnan, S. k-means Clustering Optimized Fuzzy Logic Control Algorithm for Arrhythmia Classification. In Proceedings of the 2021 International Conference on Advances in Electrical, Computing, Communication and Sustainable Technologies (ICAECT), Bhilai, India, 19–20 February 2021; pp. 1–7. [Google Scholar] [CrossRef]
- Oster, J.; Hopewell, J.C.; Ziberna, K.; Wijesurendra, R.; Camm, C.F.; Casadei, B.; Tarassenko, L. Identification of patients with atrial fibrillation: A big data exploratory analysis of the UK Biobank. Physiol. Meas. 2020, 41, 025001. [Google Scholar] [CrossRef]
- Chen, C.; Hua, Z.; Zhang, R.; Liu, G.; Wen, W. Automated arrhythmia classification based on a combination network of CNN and LSTM. Biomed. Signal Process. Control 2020, 57, 101819. [Google Scholar] [CrossRef]
- Fujita, H.; Cimr, D. Computer Aided detection for fibrillations and flutters using deep convolutional neural network. Inf. Sci. 2019, 486, 231–239. [Google Scholar] [CrossRef]
- Nishio, M.; Sugiyama, O.; Yakami, M.; Ueno, S.; Kubo, T.; Kuroda, T.; Togashi, K. Computer-aided diagnosis of lung nodule classification between benign nodule, primary lung cancer, and metastatic lung cancer at different image size using deep convolutional neural network with transfer learning. PLoS ONE 2018, 13, e0200721. [Google Scholar] [CrossRef] [Green Version]
- Al-antari, M.A.; Han, S.M.; Kim, T.S. Evaluation of deep learning detection and classification towards computer-aided diagnosis of breast lesions in digital X-ray mammograms. Comput. Methods Programs Biomed. 2020, 196, 105584. [Google Scholar] [CrossRef] [PubMed]
- Komeda, Y.; Handa, H.; Matsui, R.; Kashida, H.; Watanabe, T.; Sakurai, T.; Kudo, M. Computer-Aided Diagnosis (Cad) Based On Convolutional Neural Network (Cnn) System Using Artificial Intelligence (Ai) For Colorectal Polyp Classification. ESGE Days 2019 2019. [Google Scholar] [CrossRef]
- Acharya, U.R.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M.; Gertych, A.; Tan, R.S. A deep convolutional neural network model to classify heartbeats. Comput. Biol. Med. 2017, 89, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Kutlu, Y.; Kuntalp, D. A multi-stage automatic arrhythmia recognition and classification system. Comput. Biol. Med. 2011, 41, 37–45. [Google Scholar] [CrossRef]
- Manju, B.R.; Nair, A.R. Classification of Cardiac Arrhythmia of 12 Lead ECG Using Combination of SMOTEENN, XGBoost and Machine Learning Algorithms. In Proceedings of the 2019 9th International Symposium on Embedded Computing and System Design (ISED), Kollam, India, 13–14 December 2019; pp. 1–7. [Google Scholar] [CrossRef]
- Zheng, J.; Chu, H.; Struppa, D.; Zhang, J.; Yacoub, S.M.; El-Askary, H.; Chang, A.; Ehwerhemuepha, L.; Abudayyeh, I.; Barrett, A.; et al. Optimal Multi-Stage Arrhythmia Classification Approach. Sci. Rep. 2020, 10, 2898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambhia, J.; Naik, A.; Nagori, N. Arrhythmia Detection and Classification by Neural Network Using ECG Features. SSRN Electron. J. 2019. [Google Scholar] [CrossRef]
- Alonso, E.; Irusta, U.; Aramendi, E. A machine learning approach for detecting ventricular fibrillation during out-of-hospital cardiac arrest. Resuscitation 2018, 130, e53–e54. [Google Scholar] [CrossRef]
- PÅ‚awiak, P. Novel genetic ensembles of classifiers applied to myocardium dysfunction recognition based on ECG signals. Swarm Evol. Comput. 2018, 39, 192–208. [Google Scholar] [CrossRef]
- Trapeznikov, K.; Saligrama, V.; Castanon, D. Multi-Stage Classifier Design. Mach. Learn. 2012, 92. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Si, Y.; Wang, D. LSTM Neural Network for Beat Classification in ECG Identity Recognition. Intell. Autom. Soft Comput. 2019. [Google Scholar] [CrossRef]
- Jan, A.; Khurshid, N.; Khattak, M.I. Developing Resource Efficient Heart Arrhythmia Classifier. Int. J. Comput. Appl. 2015, 109, 35–39. [Google Scholar] [CrossRef]
- Eoghan Keany, What makes “XGBoost” so Extreme? 2020. Available online: https://https://medium.com/analytics-vidhya/what-makes-xgboost-so-extreme-e1544a4433bb (accessed on 23 April 2021).
- Fordham, S. How to Use the SimpleImputer Class in Machine Learning with Python. 2020. Available online: https://towardsdatascience.com/how-to-use-the-simpleimputer-class-in-machine-learning-with-python-10b321c94861 (accessed on 23 April 2021).
- Kim, H. Isolation Forest Step by Step. 2021. Available online: https://hyunsukim-9320.medium.com/isolation-forest-step-by-step-341b82923168 (accessed on 23 April 2021).
- Morde, V. XGBoost Algorithm: Long May She Reign! 2019. Available online: https://towardsdatascience.com/https-medium-com-vishalmorde-XGBoost-algorithm-long-she-may-rein-edd9f99be63d (accessed on 23 April 2021).
- Bao, J. Multi-features Based Arrhythmia Diagnosis Algorithm Using Xgboost. In Proceedings of the 2020 International Conference on Computing and Data Science (CDS), Stanford, CA, USA, 1–2 August 2020. [Google Scholar] [CrossRef]
- Brownlee, J. Hyperparameter Optimization With Random Search and Grid Search. 2020. Available online: https://machinelearningmastery.com/hyperparameter-optimization-with-random-search-and-grid-search/ (accessed on 22 April 2021).
- Brownlee, J. A Gentle Introduction to k-Fold Cross-Validation. 2020. Available online: https://machinelearningmastery.com/k-fold-cross-validation/ (accessed on 22 April 2021).
- Singh, V.; Tewary, S.; Sardana, V.; Sardana, H.K. Arrhythmia Detection—A Machine Learning based Comparative Analysis with MIT-BIH ECG Data. In Proceedings of the 2019 IEEE 5th International Conference for Convergence in Technology (I2CT), Bombay, India, 29–31 March 2019. [Google Scholar] [CrossRef]
- Apandi, Z.F.M.; Ikeura, R.; Hayakawa, S. Arrhythmia Detection Using MIT-BIH Dataset: A Review. In Proceedings of the 2018 International Conference on Computational Approach in Smart Systems Design and Applications (ICASSDA), Kuching, Malaysia, 15–17 August 2018. [Google Scholar] [CrossRef]
- Bulbul, H.I.; Usta, N.; Yildiz, M. Classification of ECG Arrhythmia with Machine Learning Techniques. In Proceedings of the 2017 16th IEEE International Conference on Machine Learning and Applications (ICMLA), Cancun, Mexico, 18–21 December 2017. [Google Scholar] [CrossRef]
- Ma, F.; Zhang, J.; Liang, W.; Xue, J. Automated Classification of Atrial Fibrillation Using Artificial Neural Network for Wearable Devices. Math. Probl. Eng. 2020, 2020, 9159158. [Google Scholar] [CrossRef] [Green Version]
- Islam, S.; Ammour, N.; Alajlan, N. Atrial fibrillation detection with multiparametric RR interval feature and machine learning technique. In Proceedings of the 2017 International Conference on Informatics, Health & Technology (ICIHT), Riyadh, Saudi Arabia, 21–23 February 2017. [Google Scholar] [CrossRef]
- Mitra, M.; Samanta, R. Cardiac Arrhythmia Classification Using Neural Networks with Selected Features. Procedia Technol. 2013, 10, 76–84. [Google Scholar] [CrossRef] [Green Version]
- Rajagopal, R. Automated arrhythmia classification for monitoring cardiac patients using machine learning techniques. In Classification Techniques for Medical Image Analysis and Computer Aided Diagnosis; Academic Press: Cambridge, MA, USA, 2019; pp. 153–177. [Google Scholar] [CrossRef]
- Mustaqeem, A.; Anwar, S.M.; Majid, M. Multiclass Classification of Cardiac Arrhythmia Using Improved Feature Selection and SVM Invariants. Comput. Math. Methods Med. 2018, 2018, 7310496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khare, S.; Bhandari, A.; Singh, S.; Arora, A. ECG Arrhythmia Classification Using Spearman Rank Correlation and Support Vector Machine. In Proceedings of the International Conference on Soft Computing for Problem Solving (SocProS 2011) December 20–22, 2011; Deep, K., Nagar, A., Pant, M., Bansal, J.C., Eds.; Springer: New Delhi, India, 2012; pp. 591–598. [Google Scholar]
- Alarsan, F.I.; Younes, M. Analysis and classification of heart diseases using heartbeat features and machine learning algorithms. J. Big Data 2019, 6. [Google Scholar] [CrossRef] [Green Version]
- Dua, D.; Karra Taniskidou, E. UCI Machine Learning Repository. 2017. Available online: http://archive.ics.uci.edu/ml (accessed on 21 April 2021).
- Hill, N.R.; Ayoubkhani, D.; Mcewan, P.; Sugrue, D.M.; Farooqui, U.; Lister, S.; Lumley, M.; Bakhai, A.; Cohen, A.T.; O’Neill, M.; et al. Predicting atrial fibrillation in primary care using machine learning. PLoS ONE 2019, 14, e0224582. [Google Scholar] [CrossRef] [PubMed]
| Column Name | Missing Count |
|---|---|
| Tperest | 145 |
| QTc | 145 |
| QRS rest | 707 |
| BMI | 1 |
| Diab | 222 |
| smoke | 253 |
| DBP | 133 |
| SBP | 134 |
| PR | 1788 |
| Parameter | Classifier 1 Value | Classifier 2 Value |
|---|---|---|
| Objective | binary logistic | binary logistic |
| Subsample | 0.75 | 0.75 |
| No estimators | 50 | 100 |
| Colsample by tree | 0.7 | 0.7 |
| Learning rate | 0.49 | 0.99 |
| Max depth | 10 | 20 |
| Gamma | 5 | 5 |
| Alpha | 1 | 1 |
| Seed | 134 | 134 |
| Label | Sensitivity | Specificity | F1Score |
|---|---|---|---|
| Normal | 0.990 | 0.960 | 0.975 |
| AF | 0.390 | 0.720 | 0.506 |
| Arr | 0.020 | 0.120 | 0.034 |
| Label | Sensitivity | Specificity | F1Score |
|---|---|---|---|
| First-Stage | 0.785 | 0.810 | 0.797 |
| Second-Stage | 0.986 | 0.909 | 0.946 |
| Method | Dataset | Sensitivity | Specificity | Accuracy | F1Score |
|---|---|---|---|---|---|
| KNN Classifier [16] | MIT-BIH | 85.59 | 95.46 | - | - |
| XGBoost and SMOTENN [17] | UCI | - | - | 97.48 | - |
| Deep learning and SVM [9] | UK Biobank | - | - | - | 84.8 |
| Random Forest [41] | MIT-BIH | - | - | 97.98 | - |
| Gradient Boosted Trees [41] | MIT-BIH | - | - | 96.75 | - |
| Our Method | UK Biobank | 98.6 | 90.9 | 99 | 94.6 |
| Class Code | Class | No of Instances |
|---|---|---|
| 01 | Normal | 245 |
| 02 | Ischemic changes (Coronary Artery Disease) | 44 |
| 03 | Old Anterior Myocardial Infarction | 15 |
| 04 | Old Inferior Myocardial Infarction | 15 |
| 05 | Sinus tachycardy | 13 |
| 06 | Sinus bradycardy | 25 |
| 07 | Ventricular Premature Contraction (PVC) | 3 |
| 08 | Superventricular Premature Contraction) | 2 |
| 09 | Left bundle branch block | 9 |
| 10 | Right bundle branch block | 50 |
| 11 | 1. degree Atrioventricular block | 0 |
| 12 | 2. degree AV block | 0 |
| 13 | 3. degree AV block | 0 |
| 14 | Left ventricle hypertrophy | 4 |
| 15 | Atrial Fibrillation or Flutter | 5 |
| 16 | Other | 22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rezaei, M.J.; Woodward, J.R.; Ramírez, J.; Munroe, P. A Novel Two-Stage Heart Arrhythmia Ensemble Classifier. Computers 2021, 10, 60. https://doi.org/10.3390/computers10050060
Rezaei MJ, Woodward JR, Ramírez J, Munroe P. A Novel Two-Stage Heart Arrhythmia Ensemble Classifier. Computers. 2021; 10(5):60. https://doi.org/10.3390/computers10050060
Chicago/Turabian StyleRezaei, Mercedeh J., John R. Woodward, Julia Ramírez, and Patricia Munroe. 2021. "A Novel Two-Stage Heart Arrhythmia Ensemble Classifier" Computers 10, no. 5: 60. https://doi.org/10.3390/computers10050060
APA StyleRezaei, M. J., Woodward, J. R., Ramírez, J., & Munroe, P. (2021). A Novel Two-Stage Heart Arrhythmia Ensemble Classifier. Computers, 10(5), 60. https://doi.org/10.3390/computers10050060







