DICER1 Mutational Spectrum in Intracranial CNS-Neoplasias—A Review and a Report from the CNS-InterREST GPOH Study Center
Simple Summary
Abstract
1. Introduction
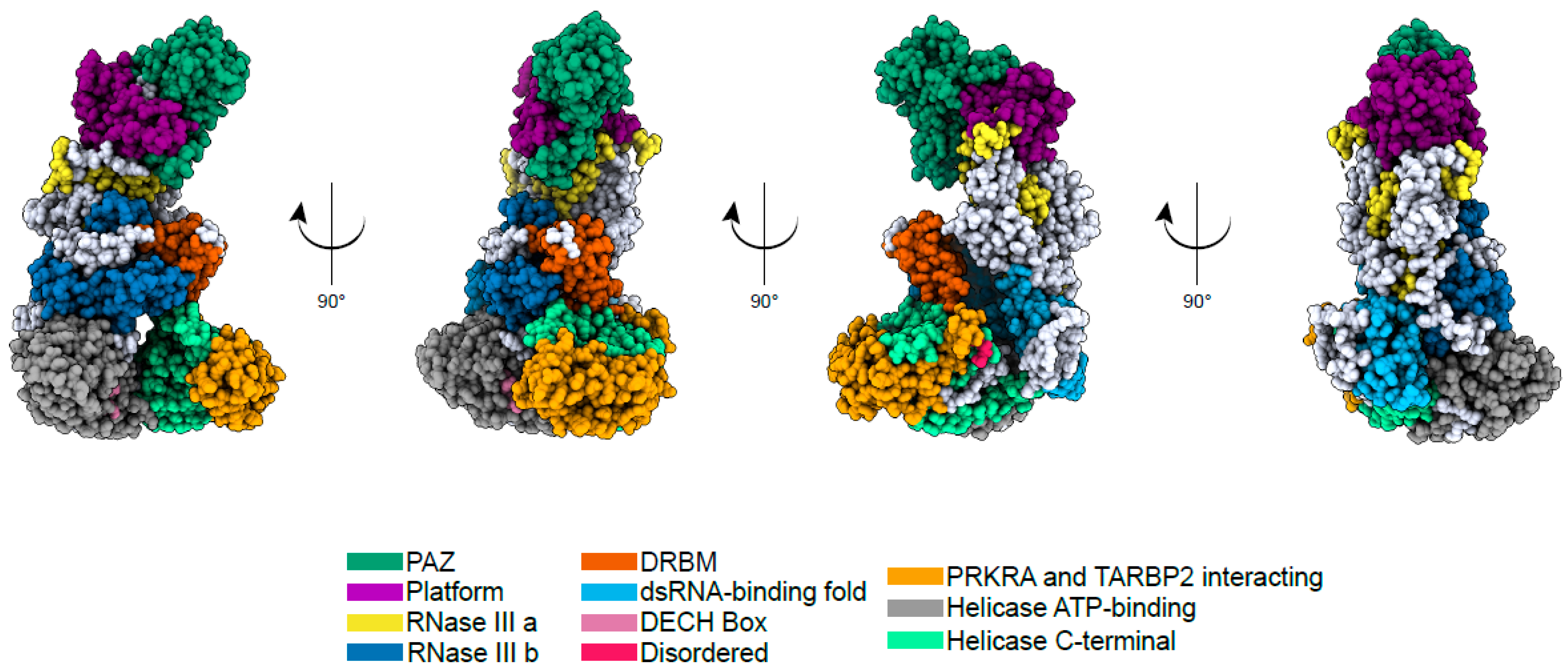
2. Biological Function and Structure of DICER1
3. Epidemiology of DICER1 Syndrome
4. DICER1—Mutations and Associated Cancer Types
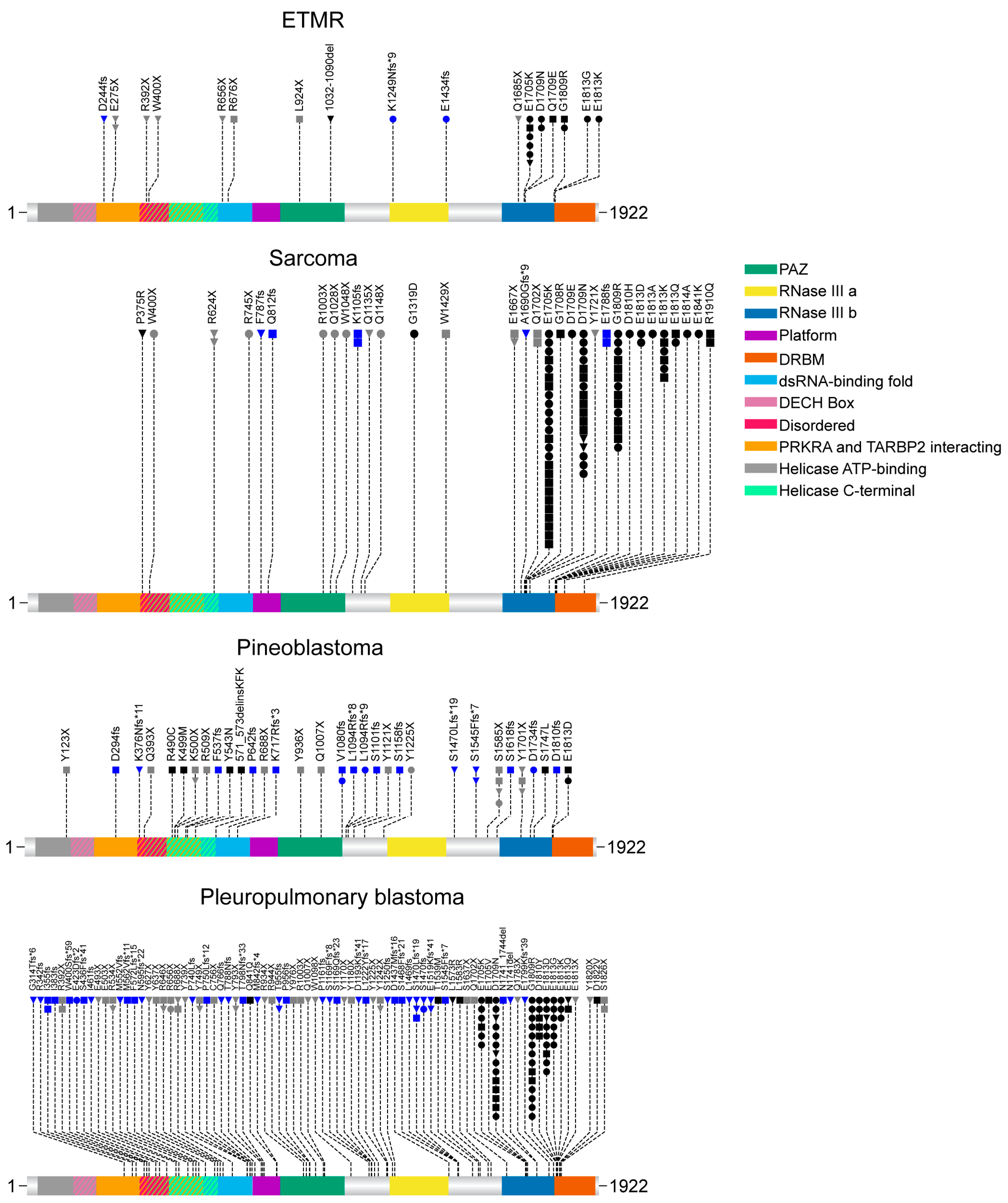
5. Embryonal Tumor with Multilayered Rosettes (ETMR)
6. Intracranial Sarcoma
7. Pineoblastoma (PinB)
8. Pleuropulmonary Blastoma (PPB)
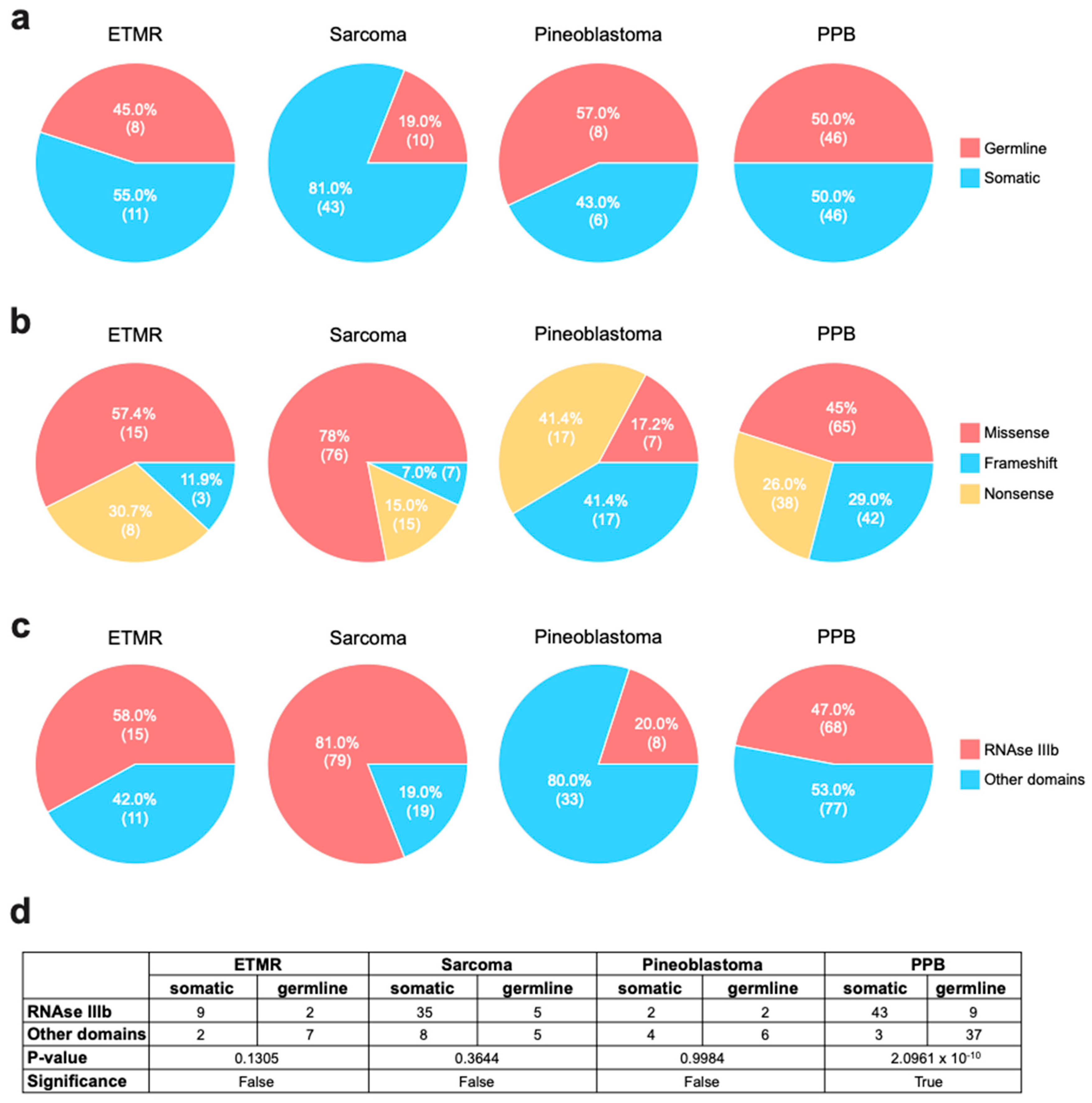
9. Type of Mutations in the Investigated Tumor Types
10. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mirshahi, U.L.; Kim, J.; Best, A.F.; Chen, Z.E.; Hu, Y.; Haley, J.S.; Golden, A.; Stahl, R.; Manickam, K.; Carr, A.G.; et al. A Genome-First Approach to Characterize DICER1 Pathogenic Variant Prevalence, Penetrance, and Phenotype. JAMA Netw. Open 2021, 4, e210112. [Google Scholar] [CrossRef] [PubMed]
- Cardona, A.F.; Chamorro Ortiz, D.F.; Ruíz-Patiño, A.; Gomez, D.; Muñoz, Á.; Ardila, D.V.; Garcia-Robledo, J.E.; Ordóñez-Reyes, C.; Sussmann, L.; Mosquera, A.; et al. DICER1-Associated Central Nervous System Sarcoma: A Comprehensive Clinical and Genomic Characterization of Case Series of Young Adult Patients. Neuro-Oncol. Pract. 2023, 10, 381–390. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Priest, J.R.; Duchaine, T.F. DICER1: Mutations, MicroRNAs and Mechanisms. Nat. Rev. Cancer 2014, 14, 662–672. [Google Scholar] [CrossRef]
- Lee, Y.Y.; Lee, H.; Kim, H.; Kim, V.N.; Roh, S.H. Structure of the Human DICER–Pre-MiRNA Complex in a Dicing State. Nature 2023, 615, 331–338. [Google Scholar] [CrossRef]
- Tian, Y.; Simanshu, D.K.; Ma, J.B.; Park, J.E.; Heo, I.; Kim, V.N.; Patel, D.J. A Phosphate-Binding Pocket within the Platform-PAZ-Connector Helix Cassette of Human Dicer. Mol. Cell 2014, 53, 606–616. [Google Scholar] [CrossRef]
- Lau, P.W.; Guiley, K.Z.; De, N.; Potter, C.S.; Carragher, B.; MacRae, I.J. The Molecular Architecture of Human Dicer. Nat. Struct. Mol. Biol. 2012, 19, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Zhou, K.; Smith, A.M.; Noland, C.L.; Doudna, J.A. Differential Roles of Human Dicer-Binding Proteins TRBP and PACT in Small RNA Processing. Nucleic Acids Res. 2013, 41, 6568–6576. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.C.; Jorcyk, C.L.; Oxford, J.T. DICER1 Syndrome: DICER1 Mutations in Rare Cancers. Cancers 2018, 10, 143. [Google Scholar] [CrossRef]
- Camino, L.P.; Dutta, A.; Barroso, S.; Sung, P.; Pé Rez-Calero, C.; Katz, J.N.; García-Rubio, M.; Gó Mez-Gonzá Lez, B.N.; Aguilera, A.S. DICER Ribonuclease Removes Harmful R-Loops. Mol. Cell 2023, 83, 3707–3719. [Google Scholar] [CrossRef]
- García-Muse, T.; Aguilera, A. R Loops: From Physiological to Pathological Roles. Cell 2019, 179, 604–618. [Google Scholar] [CrossRef]
- Schultz, K.A.P.; Rednam, S.P.; Kamihara, J.; Doros, L.; Achatz, M.I.; Wasserman, J.D.; Diller, L.R.; Brugières, L.; Druker, H.; Schneider, K.A.; et al. PTEN, DICER1, FH, and Their Associated Tumor Susceptibility Syndromes: Clinical Features, Genetics, and Surveillance Recommendations in Childhood. Clin. Cancer Res. 2017, 23, e76–e82. [Google Scholar] [CrossRef] [PubMed]
- Stewart, D.R.; Best, A.F.; Williams, G.M.; Harney, L.A.; Carr, A.G.; Harris, A.K.; Kratz, C.P.; Dehner, L.P.; Messinger, Y.H.; Rosenberg, P.S.; et al. Neoplasm Risk among Individuals with a Pathogenic Germline Variant in DICER1. J. Clin. Oncol. 2019, 37, 668–676. [Google Scholar] [CrossRef]
- Liu, K.X.; Shang, H.H.; Cacciotti, C.; Everdell, E.; Aizer, A.A.; Rahman, R.; Malinowski, S.; Meredith, D.M.; Kamihara, J.; Wen, P.Y.; et al. DICER1 Mutations in Primary Central Nervous System Tumors: New Insights into Histologies, Mutations, and Prognosis. J. Neurooncol. 2022, 157, 499–510. [Google Scholar] [CrossRef] [PubMed]
- de Kock, L.; Priest, J.R.; Foulkes, W.D.; Alexandrescu, S. An Update on the Central Nervous System Manifestations of DICER1 Syndrome. Acta Neuropathol. 2020, 139, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Lambo, S.; von Hoff, K.; Korshunov, A.; Pfister, S.M.; Kool, M. ETMR: A Tumor Entity in Its Infancy. Acta Neuropathol. 2020, 140, 249–266. [Google Scholar]
- Lambo, S.; Gröbner, S.N.; Rausch, T.; Waszak, S.M.; Schmidt, C.; Gorthi, A.; Romero, J.C.; Mauermann, M.; Brabetz, S.; Krausert, S.; et al. The Molecular Landscape of ETMR at Diagnosis and Relapse. Nature 2019, 576, 274–280. [Google Scholar] [CrossRef]
- Zhao, X.; He, X.; Han, X.; Yu, Y.; Ye, F.; Chen, Y.; Hoang, T.N.; Xu, X.; Mi, Q.S.; Xin, M.; et al. MicroRNA-Mediated Control of Oligodendrocyte Differentiation. Neuron 2010, 65, 612–626. [Google Scholar] [CrossRef]
- Khan, N.E.; Bauer, A.J.; Doros, L.; Schultz, K.A.P.; Decastro, R.M.; Harney, L.A.; Kase, R.G.; Carr, A.G.; Harris, A.K.; Williams, G.M.; et al. Macrocephaly Associated with the DICER1 Syndrome. Genet. Med. 2017, 19, 244–248. [Google Scholar] [CrossRef]
- Korshunov, A.; Sturm, D.; Ryzhova, M.; Hovestadt, V.; Gessi, M.; Jones, D.T.W.; Remke, M.; Northcott, P.; Perry, A.; Picard, D.; et al. Embryonal Tumor with Abundant Neuropil and True Rosettes (ETANTR), Ependymoblastoma, and Medulloepithelioma Share Molecular Similarity and Comprise a Single Clinicopathological Entity. Acta Neuropathol. 2014, 128, 279–289. [Google Scholar] [CrossRef]
- Edelbach, B.M.; Gospodarev, V.; Raghavan, R.; Dye, J. Primary Intracranial Sarcoma, DICER-1 Mutant, with Hemorrhagic Presentation: A Case Report. Surg. Neurol. Int. 2024, 15, 253. [Google Scholar] [CrossRef]
- Kosteniuk, S.E.; Michaiel, G.; Dunham, C. A Case of Primary Intracranial Sarcoma, DICER1-Mutant, in a Child with a Germline DICER1 Mutation. Brain Sci. 2023, 13, 1040. [Google Scholar] [CrossRef] [PubMed]
- Kamihara, J.; Paulson, V.; Breen, M.A.; Laetsch, T.W.; Rakheja, D.; Shulman, D.S.; Schoettler, M.L.; Clinton, C.M.; Ward, A.; Reidy, D.; et al. DICER1-Associated Central Nervous System Sarcoma in Children: Comprehensive Clinicopathologic and Genetic Analysis of a Newly Described Rare Tumor. Mod. Pathol. 2020, 33, 1910–1921. [Google Scholar] [CrossRef]
- Vuong, H.G.; Le, M.K.; Dunn, I.F. A Systematic Review of the Clinicopathological Features and Prognostic Outcomes of DICER1-Mutant Malignant Brain Neoplasms. J. Neurosurg. Pediatr. 2022, 30, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Villanueva-Meyer, J.E.; Ferris, S.P.; Sloan, E.A.; Hofmann, J.W.; Hattab, E.M.; Williams, B.J.; Guo, H.; Torkildson, J.; Florez, A.; et al. Primary Intracranial Sarcomas with DICER1 Mutation Often Contain Prominent Eosinophilic Cytoplasmic Globules and Can Occur in the Setting of Neurofibromatosis Type 1. Acta Neuropathol. 2019, 137, 521–525. [Google Scholar] [CrossRef]
- Diaz Coronado, R.Y.; Mynarek, M.; Koelsche, C.; Mora Alferez, P.; Casavilca Zambrano, S.; Wachtel Aptowitzer, A.; Sahm, F.; von Deimling, A.; Schüller, U.; Spohn, M.; et al. Primary Central Nervous System Sarcoma with DICER1 Mutation—Treatment Results of a Novel Molecular Entity in Pediatric Peruvian Patients. Cancer 2022, 128, 697–707. [Google Scholar] [CrossRef]
- de Kock, L.; Sabbaghian, N.; Druker, H.; Weber, E.; Hamel, N.; Miller, S.; Choong, C.S.; Gottardo, N.G.; Kees, U.R.; Rednam, S.P.; et al. Germ-Line and Somatic DICER1 Mutations in Pineoblastoma. Acta Neuropathol. 2014, 128, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Abdelbaki, M.S.; Abu-Arja, M.H.; Davidson, T.B.; Fangusaro, J.R.; Stanek, J.R.; Dunkel, I.J.; Dhall, G.; Gardner, S.L.; Finlay, J.L. Pineoblastoma in Children Less than Six Years of Age: The Head Start I, II, and III Experience. Pediatr. Blood Cancer 2020, 67, e28252. [Google Scholar] [CrossRef]
- Pfaff, E.; Aichmüller, C.; Sill, M.; Stichel, D.; Snuderl, M.; Karajannis, M.A.; Schuhmann, M.U.; Schittenhelm, J.; Hasselblatt, M.; Thomas, C.; et al. Molecular Subgrouping of Primary Pineal Parenchymal Tumors Reveals Distinct Subtypes Correlated with Clinical Parameters and Genetic Alterations. Acta Neuropathol. 2020, 139, 243–257. [Google Scholar] [CrossRef]
- Hill, D.A.; Ivanovich, J.; Priest, J.R.; Gurnett, C.A.; Dehner, L.P.; Desruisseau, D.; Jarzembowski, J.A.; Wikenheiser-Brokamp, K.A.; Suarez, B.K.; Whelan, A.J.; et al. DICER1 Mutations in Familial Pleuropulmonary Blastoma. Science 2009, 325, 965. [Google Scholar] [CrossRef]
- Masarweh, K.; Mordechai, O.; Gur, M.; Bar-Yoseph, R.; Bentur, L.; Ilivitzki, A. Challenges in DICER1-Associated Lung Disease. J. Clin. Med. 2023, 12, 1918. [Google Scholar] [CrossRef]
- Messinger, Y.H.; Stewart, D.R.; Priest, J.R.; Williams, G.M.; Harris, A.K.; Schultz, K.A.P.; PhD, J.Y.; Doros, L.; Rosenberg, P.S.; Ashley Hill, D.; et al. Pleuropulmonary Blastoma: A Report on 350 Central Pathology-Confirmed Pleuropulmonary Blastoma Cases by the International Pleuropulmonary Blastoma Registry. Cancer 2015, 121, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, A.; Ozeki, M.; Kawasaki, R.; Nakama, M.; Iwata, H.; Yamamoto, T.; Fukao, T. Identification of Homozygous Somatic DICER1 Mutation in Pleuropulmonary Blastoma. J. Pediatr. Hematol. Oncol. 2020, 42, 307–309. [Google Scholar] [CrossRef]
- Ricarte-Filho, J.C.; Casado-Medrano, V.; Reichenberger, E.; Spangler, Z.; Scheerer, M.; Isaza, A.; Baran, J.; Patel, T.; MacFarland, S.P.; Brodeur, G.M.; et al. DICER1 RNase IIIb Domain Mutations Trigger Widespread MiRNA Dysregulation and MAPK Activation in Pediatric Thyroid Cancer. Front. Endocrinol. 2023, 14, 1083382. [Google Scholar] [CrossRef]
- Abbo, O.; Pinnagoda, K.; Brouchet, L.; Leobon, B.; Savagner, F.; Oliver, I.; Galinier, P.; Castex, M.P.; Pasquet, M. Wilms tumor, pleuropulmonary blastoma, and DICER1: Case report and literature review. World J. Surg. Oncol. 2018, 16, 164. [Google Scholar] [CrossRef]
- Alexandrescu, S.; Meredith, D.M.; Lidov, H.G.; Alaggio, R.; Novello, M.; Ligon, K.L.; Vargas, S.O. Loss of histone H3 trimethylation on lysine 27 and nuclear expression of transducin-like enhancer 1 in primary intracranial sarcoma, DICER1-mutant. Histopathology 2021, 78, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Bahubeshi, A.; Tischkowitz, M.; Foulkes, W.D. miRNA processing and human cancer: DICER1 cuts the mustard. Sci. Transl. Med. 2011, 3, 111ps46. [Google Scholar] [CrossRef]
- Apellaniz-Ruiz, M.; Segni, M.; Kettwig, M.; Glüer, S.; Pelletier, D.; Nguyen, V.-H.; Wagener, R.; López, C.; Muchantef, K.; Bouron-Dal Soglio, D.; et al. Mesenchymal Hamartoma of the Liver and DICER1 Syndrome. N. Engl. J. Med. 2019, 380, 1834–1842. [Google Scholar] [CrossRef]
- Brenneman, M.; Field, A.; Yang, J.; Williams, G.; Doros, L.; Rossi, C.; Schultz, K.A.; Rosenberg, A.; Ivanovich, J.; Turner, J.; et al. Temporal order of RNase IIIb and loss-of-function mutations during development determines phenotype in DICER1 syndrome: A unique variant of the two-hit tumor suppression model. F1000Research 2015, 4, 214. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Wang, X.; Zhao, W.; Fu, L.; Ma, X.; Peng, X. DICER1 mutations in twelve Chinese patients with pleuropulmonary blastoma. Sci. China Life Sci. 2017, 60, 714–720. [Google Scholar] [CrossRef]
- Das, A.; Roy, P.; Modi, S.K.; Achari, R.B.; Sen, S.; Singh, A.; Sukumaran, R.; Bhattacharyya, A. Germline DICER1-mutant intracranial sarcoma with dual chondroid and spindle cell morphology and pulmonary metastases treated with multimodal therapy. Pediatr. Blood Cancer 2019, 66, e27744. [Google Scholar] [CrossRef]
- de Kock, L.; Geoffrion, D.; Rivera, B.; Wagener, R.; Sabbaghian, N.; Bens, S.; Ellezam, B.; Bouron-Dal Soglio, D.; Ordóñez, J.; Sacharow, S.; et al. Multiple DICER1-related tumors in a child with a large interstitial 14q32 deletion. Genes Chromosomes Cancer 2018, 57, 223–230. [Google Scholar] [CrossRef]
- de Kock, L.; Plourde, F.; Carter, M.T.; Hamel, N.; Srivastava, A.; Meyn, M.S.; Arseneau, J.; Soglio, D.B.D.; Foulkes, W.D. Germ-line and somatic DICER1 mutations in a pleuropulmonary blastoma. Pediatr. Blood Cancer 2013, 60, 2091–2092. [Google Scholar] [CrossRef] [PubMed]
- Doros, L.; Yang, J.; Dehner, L.; Rossi, C.T.; Skiver, K.; Jarzembowski, J.A.; Messinger, Y.; Schultz, K.A.; Williams, G.; André, N.; et al. DICER1 Mutations in embryonal rhabdomyosarcomas from children with and without familial PPB-tumor predisposition syndrome. Pediatr. Blood Cancer 2012, 59, 558–560. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Martínez, L.; Villegas, J.A.; Santamaría, Í.; Pitiot, A.S.; Alvarado, M.G.; Fernández, S.; Torres, H.; Paredes, Á.; Blay, P.; Balbín, M. Identification of somatic and germ-line DICER1 mutations in pleuropulmonary blastoma, cystic nephroma and rhabdomyosarcoma tumors within a DICER1 syndrome pedigree. BMC Cancer 2017, 17, 146. [Google Scholar] [CrossRef] [PubMed]
- Foulkes, W.D.; Bahubeshi, A.; Hamel, N.; Pasini, B.; Asioli, S.; Baynam, G.; Choong, C.S.; Charles, A.; Frieder, R.P.; Dishop, M.K.; et al. Extending the phenotypes associated with DICER1 mutations. Hum. Mutat. 2011, 32, 1381–1384. [Google Scholar] [CrossRef]
- Hiemcke-Jiwa, L.S.; van Belle, S.; Eijkelenboom, A.; Merks, J.H.M.; van Noesel, M.M.; Kaal, S.E.J.; Pijnenborg, J.M.A.; Bulten, J.; Tops, B.B.J.; van de Ven, C.P.; et al. Pleuropulmonary blastoma (PPB) and other DICER1-associated high-grade malignancies are morphologically, genetically and epigenetically related—A comparative study of 4 PPBs and 6 sarcomas. Ann. Diagn. Pathol. 2022, 60, 152002. [Google Scholar] [CrossRef]
- Hurdogan, O.; Yilmaz, I.; Bay, S.B.; Vural, S.; Tugcu, D.; Kebudi, R.; Gun, F.; Ozkan, B.; Bilgic, B.; Firat, P.; et al. DICER1 Hotspot Mutations in Pleuropulmonary Blastoma: A Case Series From a Tertiary Center. Pediatr. Dev. Pathol. 2020, 23, 204–209. [Google Scholar] [CrossRef]
- Kim, E.E.; Lee, K.; Phi, J.H.; Kim, M.S.; Kang, H.J.; Yun, H.; Park, S.H. Methylation-based Subclassifications of Embryonal Tumor with Multilayered Rosettes in Not Just Pediatric Brains. Exp. Neurobiol. 2023, 32, 354–361. [Google Scholar] [CrossRef]
- Kline, C.N.; Joseph, N.M.; Grenert, J.P.; van Ziffle, J.; Talevich, E.; Onodera, C.; Aboian, M.; Cha, S.; Raleigh, D.R.; Braunstein, S.; et al. Targeted next-generation sequencing of pediatric neuro-oncology patients improves diagnosis, identifies pathogenic germline mutations, and directs targeted therapy. Neuro-Oncol. 2017, 19, 699–709. [Google Scholar] [CrossRef]
- Koelsche, C.; Mynarek, M.; Schrimpf, D.; Bertero, L.; Serrano, J.; Sahm, F.; Reuss, D.E.; Hou, Y.; Baumhoer, D.; Vokuhl, C.; et al. Primary intracranial spindle cell sarcoma with rhabdomyosarcoma-like features share a highly distinct methylation profile and DICER1 mutations. Acta Neuropathol. 2018, 136, 327–337. [Google Scholar] [CrossRef]
- Kommoss, F.K.F.; Chong, A.S.; Chong, A.L.; Pfaff, E.; Jones, D.T.W.; Hiemcke-Jiwa, L.S.; Kester, L.A.; Flucke, U.; Gessler, M.; Schrimpf, D.; et al. Genomic characterization of DICER1-associated neoplasms uncovers molecular. Nat. Commun. 2023, 14, 1677. [Google Scholar] [CrossRef] [PubMed]
- Kuhlen, M.; Hönscheid, A.; Schemme, J.; Merz, H.; Mauz-Körholz, C.; Borkhardt, A.; Troeger, A. Hodgkin lymphoma as a novel presentation of familial DICER1 syndrome. Eur. J. Pediatr. 2016, 175, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Leanne de Kock, W.D.F. Sarcoma and germ-line DICER1 mutations. Lancet Oncol. 2016, 17, e470. [Google Scholar] [CrossRef]
- Lee, J.C.; Mazor, T.; Lao, R.; Wan, E.; Diallo, A.B.; Hill, N.S.; Thangaraj, N.; Wendelsdorf, K.; Samuel, D.; Kline, C.N.; et al. Recurrent KBTBD4 small in-frame insertions and absence of DROSHA deletion or DICER1 mutation differentiate pineal parenchymal tumor of intermediate differentiation (PPTID) from pineoblastoma. Acta Neuropathol. 2019, 137, 851–854. [Google Scholar] [CrossRef] [PubMed]
- Leelatian, N.; Goss, J.; Pastakia, D.; Dewan, M.C.; Snuderl, M.; Mobley, B.C. Primary Intracranial Sarcoma, DICER1-Mutant Presenting as a Pineal Region Tumor Mimicking Pineoblastoma: Case Report and Review of the Literature. J. Neuropathol. Exp. Neurol. 2022, 81, 762–764. [Google Scholar] [CrossRef]
- Li, B.K.; Vasiljevic, A.; Dufour, C.; Yao, F.; Ho, B.L.B.; Lu, M.; Hwang, E.I.; Gururangan, S.; Hansford, J.R.; Fouladi, M.; et al. Pineoblastoma segregates into molecular sub-groups with distinct clinico-pathologic features: A Rare Brain Tumor Consortium registry study. Acta Neuropathol. 2020, 139, 223–241. [Google Scholar] [CrossRef]
- Liu, A.P.Y.; Gudenas, B.; Lin, T.; Orr, B.A.; Klimo, P.; Kumar, R.; Bouffet, E.; Gururangan, S.; Crawford, J.R.; Kellie, S.J.; et al. Risk-adapted therapy and biological heterogeneity in pineoblastoma: Integrated clinico-pathological analysis from the prospective, multi-center SJMB03 and SJYC07 trials. Acta Neuropathol. 2020, 139, 259–271. [Google Scholar] [CrossRef]
- Lyle, A.N.J.; Ohlsen, T.J.D.; Miller, D.E.; Brown, G.; Waligorski, N.; Stark, R.; Taylor, M.R.; Puia-Dumitrescu, M. Congenital pleuropulmonary blastoma in a newborn with a variant of uncertain significance in DICER1 evaluated by RNA-sequencing. Matern. Health Neonatol. Perinatol. 2023, 9, 4. [Google Scholar] [CrossRef]
- Raleigh, D.R.; Solomon, D.A.; Lloyd, S.A.; Lazar, A.; Garcia, M.A.; Sneed, P.K.; Clarke, J.L.; McDermott, M.W.; Berger, M.S.; Tihan, T.; et al. Histopathologic review of pineal parenchymal tumors identifies novel morphologic subtypes and prognostic factors for outcome. Neuro-Oncol. 2017, 19, 78–88. [Google Scholar] [CrossRef]
- Sabbaghian, N.; Hamel, N.; Srivastava, A.; Albrecht, S.; Priest, J.R.; Foulkes, W.D. Germline DICER1 mutation and associated loss of heterozygosity in a pineoblastoma. J. Med. Genet. 2012, 49, 417–419. [Google Scholar] [CrossRef]
- Sakaguchi, M.; Nakano, Y.; Honda-Kitahara, M.; Kinoshita, M.; Tanaka, S.; Oishi, M.; Noguchi, K.; Fukuda, M.; Maeba, H.; Watanabe, T.; et al. Two cases of primary supratentorial intracranial rhabdomyosarcoma with DICER1 mutation which may belong to a “spindle cell sarcoma with rhabdomyosarcoma-like feature, DICER1 mutant”. Brain Tumor Pathol. 2019, 36, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Yoshida, K.; Shiraishi, Y.; Shimamura, T.; Sato, Y.; Nishimura, R.; Okuno, Y.; Chiba, K.; Tanaka, H.; Kato, K.; et al. Biallelic DICER1 mutations in sporadic pleuropulmonary blastoma. Cancer Res. 2014, 74, 2742–2749. [Google Scholar] [CrossRef]
- Rath, S.R.; Bartley, A.; Charles, A.; Powers, N.; Baynam, G.; Jones, T.; Priest, J.R.; Foulkes, W.D.; Choong, C.S.Y. Multinodular Goiter in Children: An Important Pointer to a Germline DICER1 Mutation. J. Clin. Endocrinol. Metab. 2014, 99, 1947–1948. [Google Scholar] [CrossRef] [PubMed]
- Slade, I.; Bacchelli, C.; Davies, H.; Murray, A.; Abbaszadeh, F.; Hanks, S.; Barfoot, R.; Burke, A.; Chisholm, J.; Hewitt, M.; et al. DICER1 syndrome: Clarifying the diagnosis, clinical features and management implications of a pleiotropic tumour predisposition syndrome. J. Med. Genet. 2011, 48, 273–278. [Google Scholar] [CrossRef]
- Stewart, D.R.; Messinger, Y.; Williams, G.M.; Yang, J.; Field, A.; Schultz, K.A.P.; Harney, L.A.; Doros, L.A.; Dehner, L.P.; Hill, D.A. Nasal chondromesenchymal hamartomas arise secondary to germline and somatic mutations of DICER1 in the pleuropulmonary blastoma tumor predisposition disorder. Hum. Genet. 2014, 133, 1443–1450. [Google Scholar] [CrossRef] [PubMed]
- Thorner, P.S.; Chong, A.S.; Nadaf, J.; Benlimame, N.; Marrano, P.; Chami, R.; Fu, L.; Foulkes, W.D. PRAME protein expression in DICER1-related tumours. J. Pathol. Clin. Res. 2022, 8, 294–304. [Google Scholar] [CrossRef]
- Uro-Coste, E.; Masliah-Planchon, J.; Siegfried, A.; Blanluet, M.; Lambo, S.; Kool, M.; Roujeau, T.; Boetto, S.; Palenzuela, G.; Bertozzi, A.I.; et al. ETMR-like infantile cerebellar embryonal tumors in the extended morphologic spectrum of DICER1-related tumors. Acta Neuropathol. 2019, 137, 175–177. [Google Scholar] [CrossRef]
- van Engelen, K.; Villani, A.; Wasserman, J.D.; Aronoff, L.; Greer, M.L.C.; Tijerin Bueno, M.; Gallinger, B.; Kim, R.H.; Grant, R.; Meyn, M.S.; et al. DICER1 syndrome: Approach to testing and management at a large pediatric tertiary care center. Pediatr. Blood Cancer 2018, 65, e26720. [Google Scholar] [CrossRef]
- Wang, L.; Lu, D.; Piao, Y. A 2-year-old girl with posterior fossa mass. Brain Pathol. 2022, 32, e13026. [Google Scholar] [CrossRef]
- Warren, M.; Hiemenz, M.C.; Schmidt, R.; Shows, J.; Cotter, J.; Toll, S.; Parham, D.M.; Biegel, J.A.; Mascarenhas, L.; Shah, R. Expanding the spectrum of dicer1-associated sarcomas. Mod. Pathol. 2020, 33, 164–174. [Google Scholar] [CrossRef]
| Variable | Overall | ETMR | Sarcoma | PinB | PPB |
|---|---|---|---|---|---|
| Age, y | |||||
| Median | 6.8 | 3.7 | 11.9 | 8.3 | 3.7 |
| Range | 0.1–76 | 1–30 | 0.1–76 | 1–30 | 0.1–27 |
| Gender | |||||
| Male | 60 | 6 | 19 | 14 | 21 |
| Female | 79 | 8 | 26 | 8 | 47 |
| Not Available | 108 | 2 | 25 | 21 | 60 |
| Total | 247 | 16 | 70 | 43 | 118 |
| Mutation type | |||||
| Missense | 163 | 15 | 76 | 65 | |
| Nonsense | 78 | 8 | 15 | 17 | 38 |
| Frameshift | 69 | 3 | 7 | 17 | 42 |
| Total | 310 | 26 | 98 | 41 | 145 |
| Germline/Somatic | |||||
| Germline | 73 | 9 | 10 | 8 | 46 |
| Somatic | 106 | 11 | 43 | 6 | 46 |
| Not Available | 131 | 6 | 45 | 27 | 53 |
| Total | 310 | 26 | 98 | 41 | 145 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manea, S.; Fincke, V.E.; Frühwald, M.C.; Sturm, D.; von Zezschwitz, B.; Johann, P.D.; Mucha, M. DICER1 Mutational Spectrum in Intracranial CNS-Neoplasias—A Review and a Report from the CNS-InterREST GPOH Study Center. Cancers 2025, 17, 1513. https://doi.org/10.3390/cancers17091513
Manea S, Fincke VE, Frühwald MC, Sturm D, von Zezschwitz B, Johann PD, Mucha M. DICER1 Mutational Spectrum in Intracranial CNS-Neoplasias—A Review and a Report from the CNS-InterREST GPOH Study Center. Cancers. 2025; 17(9):1513. https://doi.org/10.3390/cancers17091513
Chicago/Turabian StyleManea, Selma, Victoria E. Fincke, Michael C. Frühwald, Dominik Sturm, Barbara von Zezschwitz, Pascal D. Johann, and Marlena Mucha. 2025. "DICER1 Mutational Spectrum in Intracranial CNS-Neoplasias—A Review and a Report from the CNS-InterREST GPOH Study Center" Cancers 17, no. 9: 1513. https://doi.org/10.3390/cancers17091513
APA StyleManea, S., Fincke, V. E., Frühwald, M. C., Sturm, D., von Zezschwitz, B., Johann, P. D., & Mucha, M. (2025). DICER1 Mutational Spectrum in Intracranial CNS-Neoplasias—A Review and a Report from the CNS-InterREST GPOH Study Center. Cancers, 17(9), 1513. https://doi.org/10.3390/cancers17091513





