Analysis of Genotype and Expression of FTO and ALKBH5 in a MENA-Region Renal Cell Carcinoma Cohort
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Specimens
2.2. DNA Extraction from FFPE Biopsies
2.3. Targeted DNA Sequencing
2.4. DNA Sequencing Data Analysis
2.5. Sanger Sequencing
2.6. Immunohistochemistry
2.7. RNA Extraction from FFPE Biopsies
2.8. RNA Sequencing
2.9. Gene-Specific cDNA Synthesis and RT-qPCR
2.10. Retrieval of FTO and ALKBH5 Targets
2.11. Statistical Analysis
3. Results
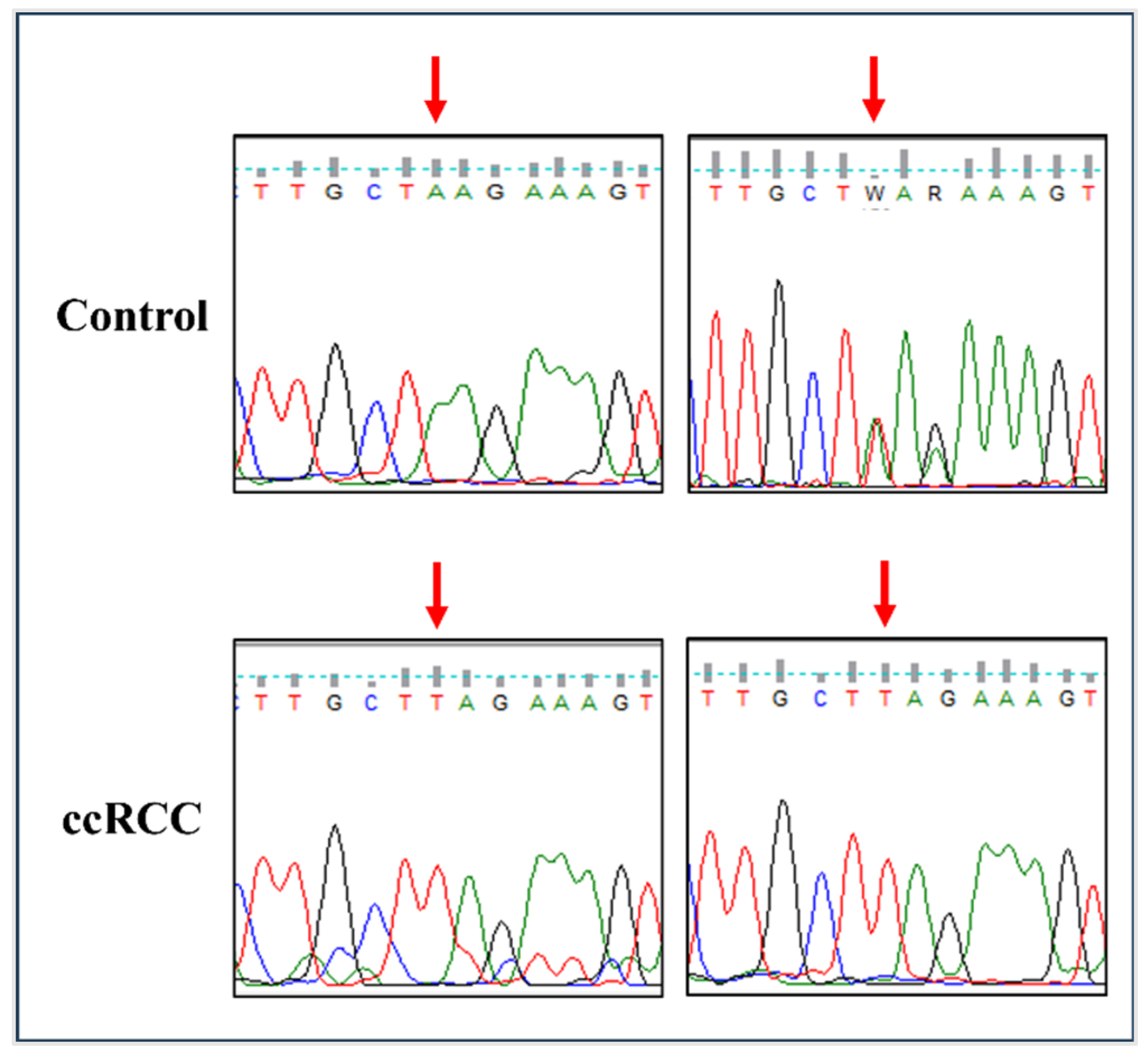
3.1. The rs11075995T Variant in FTO Is Associated with an Increased Risk of ccRCC
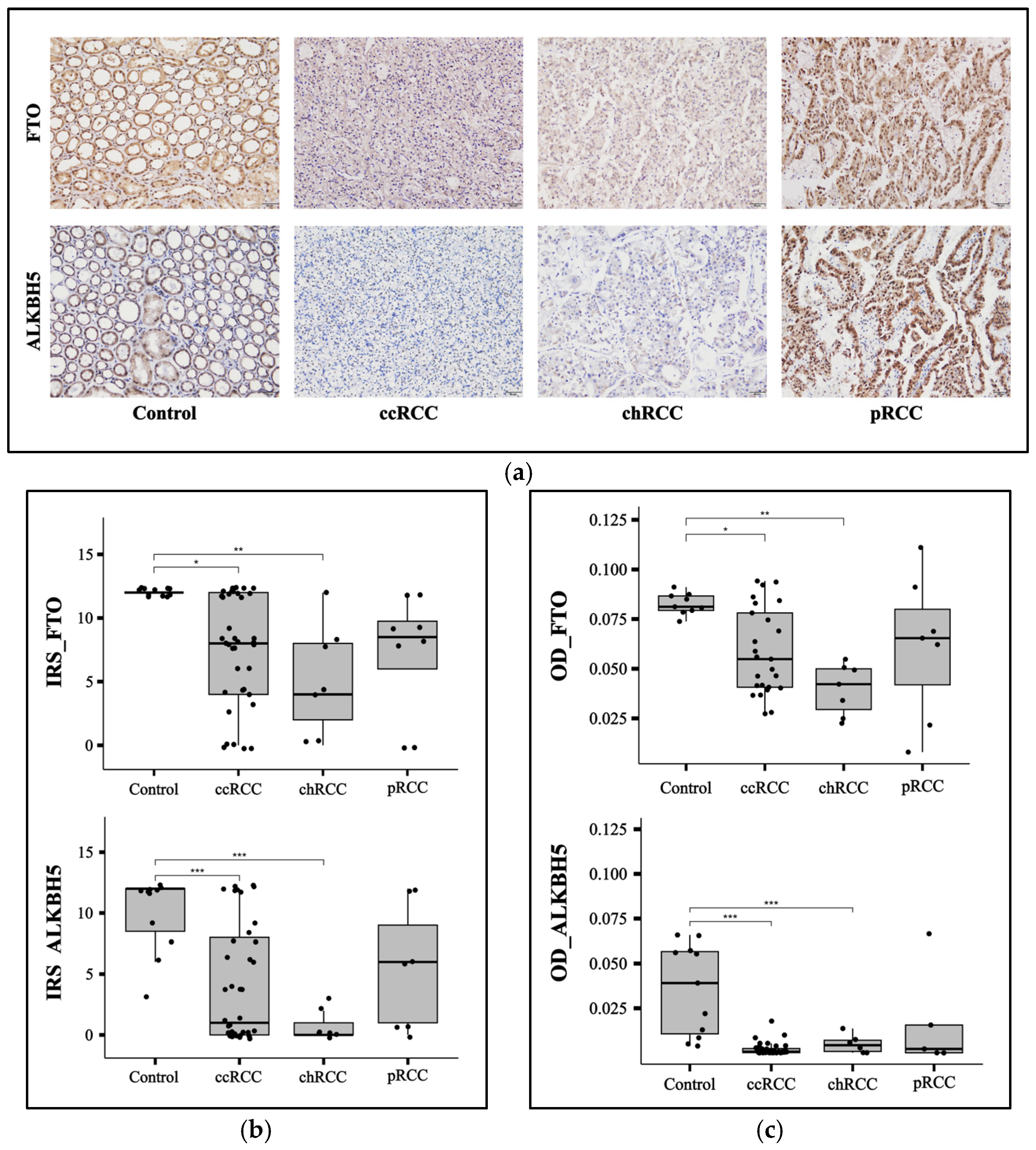
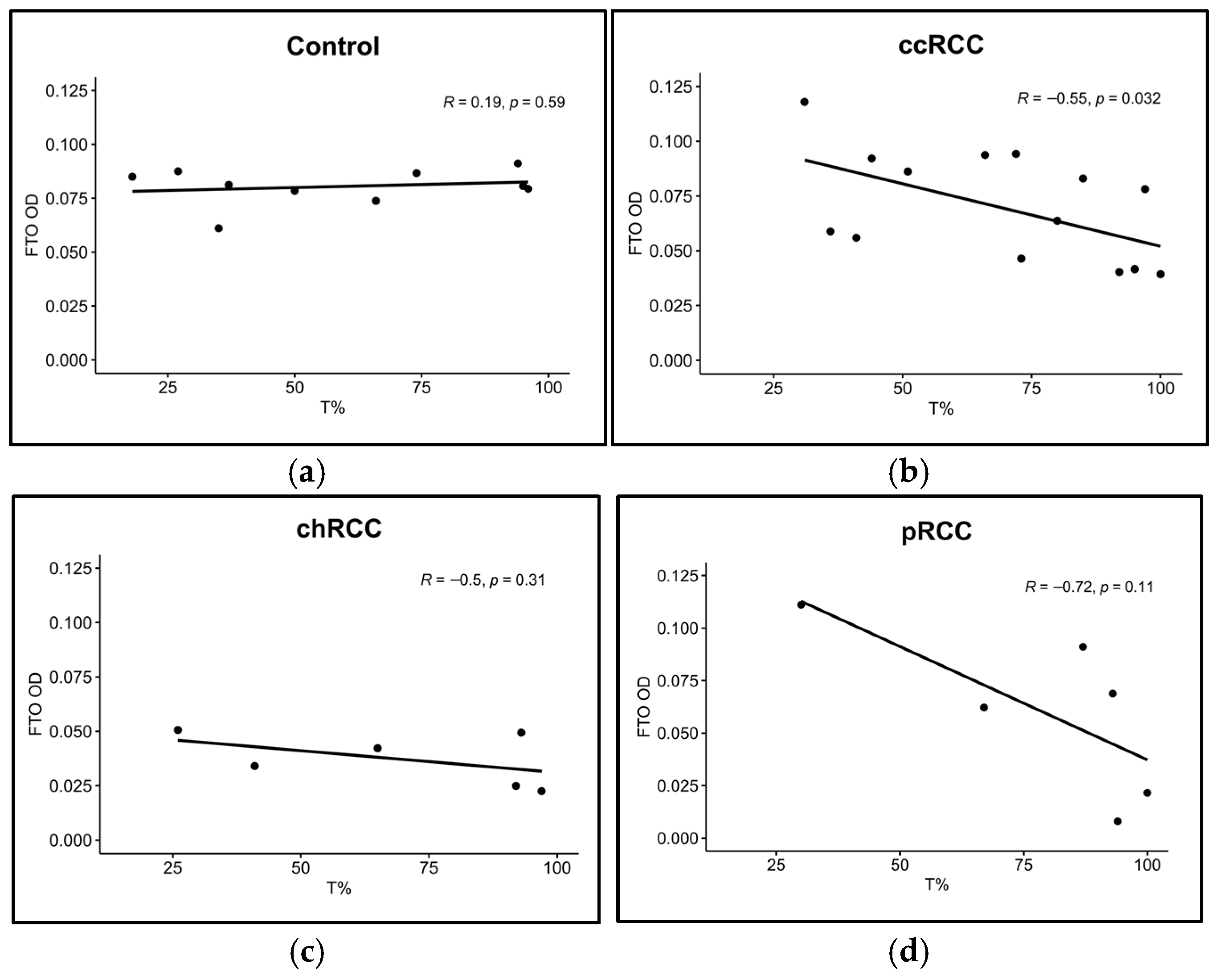
3.2. FTO and ALKBH5 Proteins Have Lower Expression in ccRCC and chRCC Patients
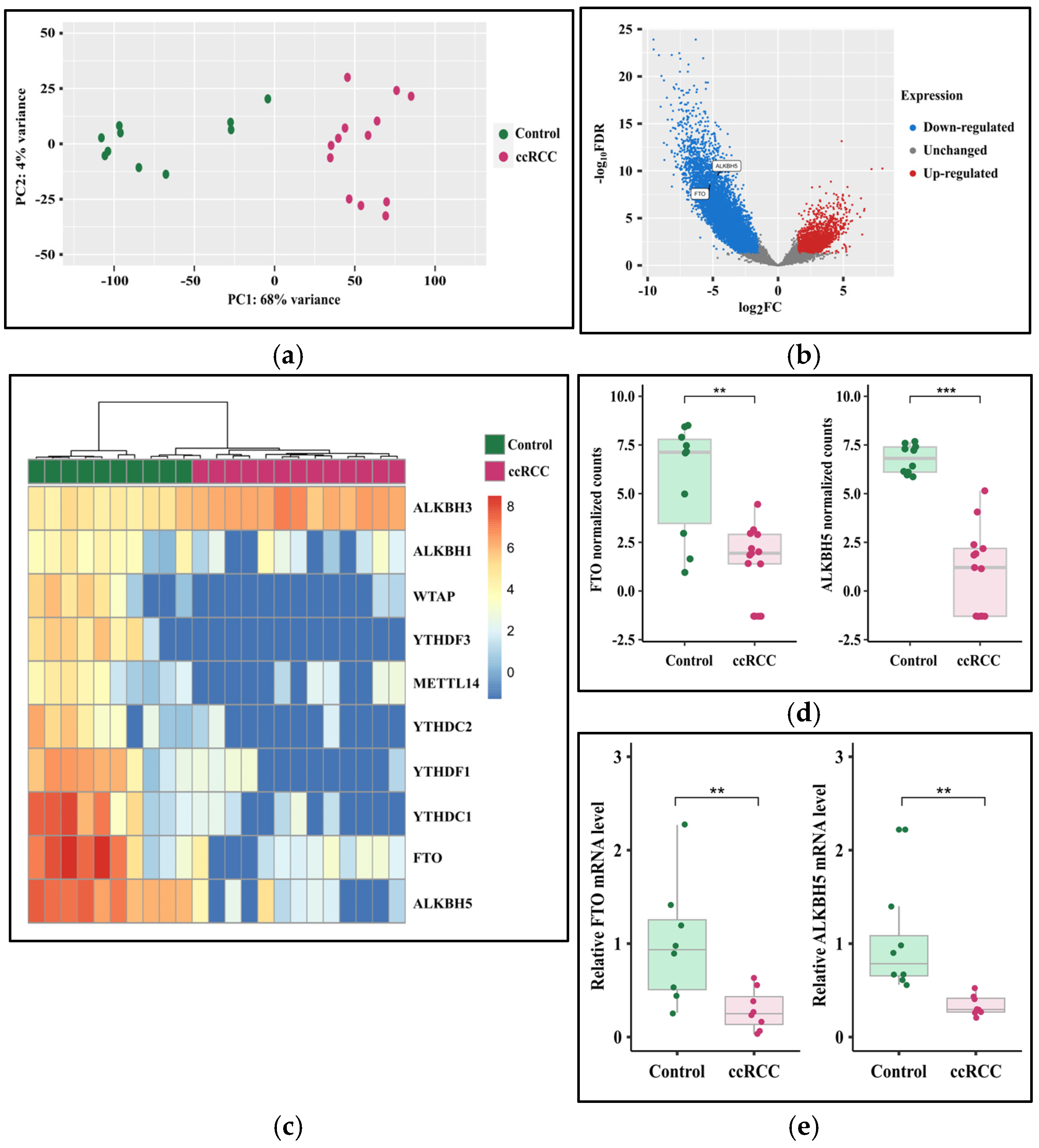
3.3. The Expression of FTO and ALKBH5 Genes Is Downregulated in ccRCC
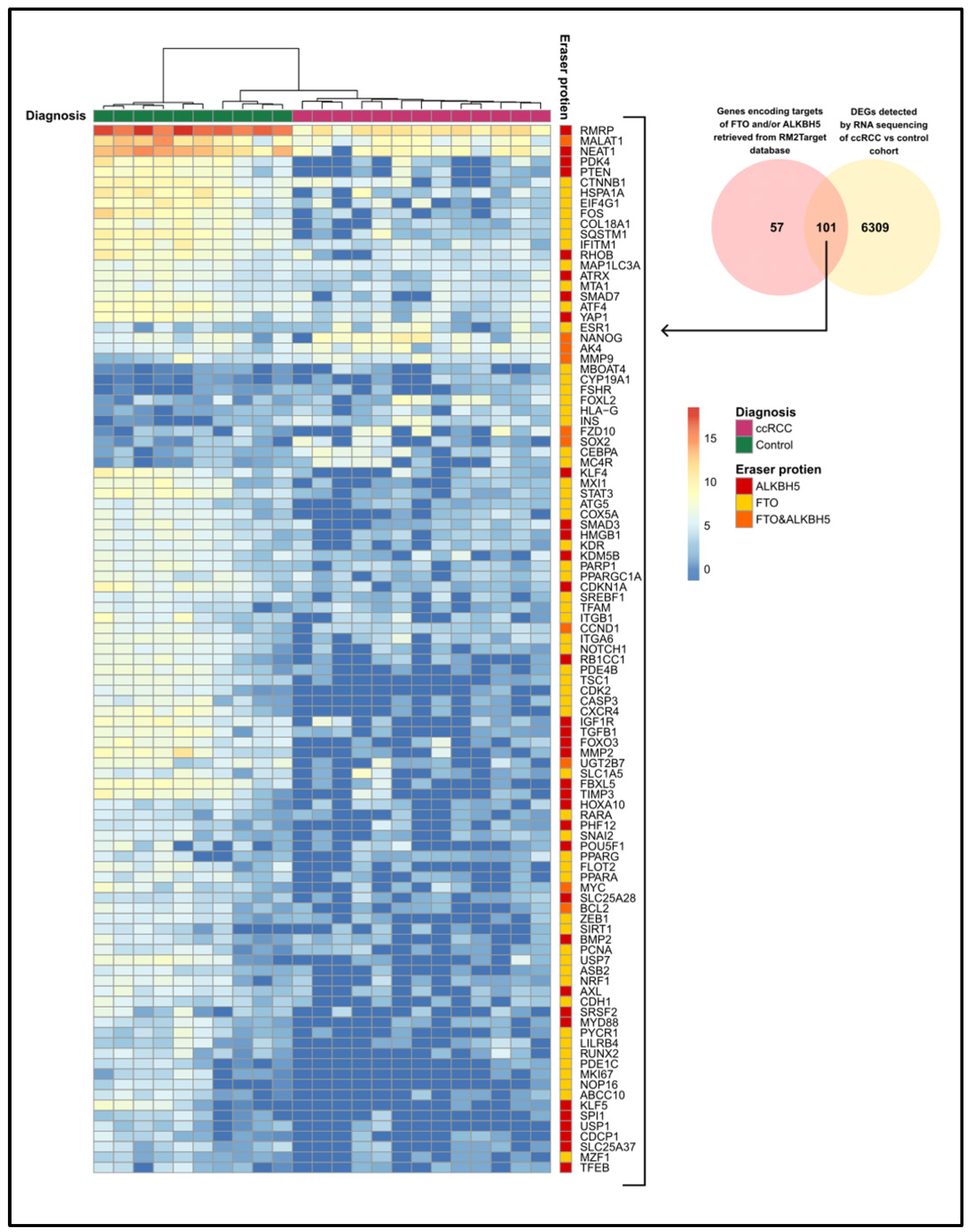
3.4. The Expression of FTO and ALKBH5 Target Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALKBH | AlkB homolog |
| ANOVA | Analysis of variance |
| BAM | Binary alignment map |
| ccRCC | Clear cell renal cell carcinoma |
| chRCC | Chromophobe renal cell carcinoma |
| CI | Confidence interval |
| CRC | Colorectal cancer |
| DAB | 3,3′-Diaminobenzidine |
| DEG | Differentially expressed gene |
| EMT | Epithelial-mesenchymal transition |
| FDR | False discovery rate |
| FFPE | Formalin-fixed paraffin-embedded |
| HWE | Hardy-Weinberg equilibrium |
| IFC | Integrated fluidic circuit |
| IGV | Integrative Genomics Viewer |
| IHC | Immunohistochemistry |
| IQR | Interquartile range |
| IRS | Immunoreactive score |
| MENA | Middle East and Northern Africa |
| NGS | Next generation sequencing |
| NSCLC | Non-small-cell lung cancer |
| OD | Optical density |
| OR | Odds ratio |
| PC | Principal component |
| PCA | Principal component analysis |
| PD-L1 | Programmed death-ligand 1 |
| pRCC | Papillary renal cell carcinoma |
| RCC | Renal cell carcinoma |
| RMP | RNA-modifying protein |
| RNA-seq | RNA sequencing |
| RT-qPCR | Quantitative real-time PCR |
| SNP | Single-nucleotide polymorphism |
| TMAP | Torrent Mapping Alignment Program |
| UTR | Untranslated region |
References
- Cancer Today. Available online: https://gco.iarc.fr/today/en/dataviz/tables?mode=population&cancers=29&types=1 (accessed on 29 November 2024).
- Cancer Today. Available online: https://gco.iarc.fr/today/en/dataviz/bars?mode=cancer&types=0_1&sort_by=value1&populations=12_275_368_376_400_414_422_434_48_504_512_634_682_729_760_784_788_818_887&group_populations=1&key=total (accessed on 29 November 2024).
- Braga, E.A.; Fridman, M.V.; Loginov, V.I.; Dmitriev, A.A.; Morozov, S.G. Molecular Mechanisms in Clear Cell Renal Cell Carcinoma: Role of miRNAs and Hypermethylated miRNA Genes in Crucial Oncogenic Pathways and Processes. Front. Genet. 2019, 10, 320. [Google Scholar] [CrossRef] [PubMed]
- Padala, S.A.; Barsouk, A.; Thandra, K.C.; Saginala, K.; Mohammed, A.; Vakiti, A.; Rawla, P.; Barsouk, A. Epidemiology of renal cell carcinoma. World J. Oncol. 2020, 11, 79. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, S.; Arzalluz-Luque, A.; Conesa, A. Undisclosed, unmet and neglected challenges in multi-omics studies. Nat. Comput. Sci. 2021, 1, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Long, Z.; Sun, C.; Tang, M.; Wang, Y.; Ma, J.; Yu, J.; Wei, J.; Ma, J.; Wang, B.; Xie, Q. Single-cell multiomics analysis reveals regulatory programs in clear cell renal cell carcinoma. Cell Discov. 2022, 8, 68. [Google Scholar] [CrossRef]
- Koch, E.; Finne, K.; Eikrem, Ø.; Landolt, L.; Beisland, C.; Leh, S.; Marti, H.P. Transcriptome-proteome integration of archival human renal cell carcinoma biopsies enables identification of molecular mechanisms. Am. J. Physiol. Renal Physiol. 2019, 316, F1053–F1067. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Patel, H.; Chen, J.; Wang, J.; Chen, Z.-S.; Wang, H. Epigenetic modification of m6A regulator proteins in cancer. Mol. Cancer 2023, 22, 102. [Google Scholar] [CrossRef]
- Tang, Q.; Li, L.; Wang, Y.; Wu, P.; Hou, X.; Ouyang, J.; Fan, C.; Li, Z.; Wang, F.; Guo, C. RNA modifications in cancer. Br. J. Cancer 2023, 129, 204–221. [Google Scholar] [CrossRef]
- Tu, W.; Huang, X.; Liu, S.; Zhan, Y.; Cai, X.; Shao, L. The m6A demethylase fat mass and obesity-associated protein mitigates pyroptosis and inflammation in doxorubicin-induced heart failure via the toll-like receptor 4/NF-κB pathway. Cardiovasc. Diagn. Ther. 2024, 14, 158. [Google Scholar] [CrossRef]
- Qu, M.; Zuo, L.; Zhang, M.; Cheng, P.; Guo, Z.; Yang, J.; Li, C.; Wu, J. High glucose induces tau hyperphosphorylation in hippocampal neurons via inhibition of ALKBH5-mediated Dgkh m6A demethylation: A potential mechanism for diabetic cognitive dysfunction. Cell Death Dis. 2023, 14, 385. [Google Scholar] [CrossRef]
- Li, R.; Kuang, Y.; Niu, Y.; Zhang, S.; Chen, S.; Su, F.; Wang, J.; Lin, S.; Liu, D.; Shen, C. FTO-mediated RNA m6A methylation regulates synovial aggression and inflammation in rheumatoid arthritis. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2024, 1870, 167341. [Google Scholar] [CrossRef]
- Perry, G.S.; Das, M.; Woon, E.C.Y. Inhibition of AlkB Nucleic Acid Demethylases: Promising New Epigenetic Targets. J. Med. Chem. 2021, 64, 16974–17003. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hu, X.; Yu, H.; Sun, H.; Zhang, L.; Shao, C. The FTO mediated N6-methyladenosine modification of DDIT4 regulation with tumorigenesis and metastasis in prostate cancer. Research 2024, 7, 0313. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Wan, A.H.; Sun, L.; Liang, H.; Niu, Y.; Deng, Y.; Yan, S.; Wang, Q.-P.; Bu, X.; Zhang, X. N6-methyladenosine demethylase FTO enhances chemo-resistance in colorectal cancer through SIVA1-mediated apoptosis. Mol. Ther. 2023, 31, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Xu, Q.; Wu, R.; Sun, W.; Gu, Y.; Zhu, S.; Liu, X.; Lv, T.; Song, Y. ALKBH5 promotes non-small cell lung cancer progression and susceptibility to anti-PD-L1 therapy by modulating interactions between tumor and macrophages. J. Exp. Clin. Cancer Res. 2024, 43, 164. [Google Scholar] [CrossRef]
- Guo, X.; Li, K.; Jiang, W.; Hu, Y.; Xiao, W.; Huang, Y.; Feng, Y.; Pan, Q.; Wan, R. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol. Cancer 2020, 19, 91. [Google Scholar] [CrossRef]
- Alhammadi, M.A.; Bajbouj, K.; Talaat, I.M.; Hamoudi, R. The role of RNA-modifying proteins in renal cell carcinoma. Cell Death Dis. 2024, 15, 227. [Google Scholar] [CrossRef]
- Hu, W.; Klümper, N.; Schmidt, D.; Ritter, M.; Ellinger, J.; Hauser, S. Depletion of the m6A demethylases FTO and ALKBH5 impairs growth and metastatic capacity through EMT phenotype change in clear cell renal cell carcinoma. Am. J. Transl. Res. 2023, 15, 1744. [Google Scholar]
- Xiao, Y.; Thakkar, K.N.; Zhao, H.; Broughton, J.; Li, Y.; Seoane, J.A.; Diep, A.N.; Metzner, T.J.; von Eyben, R.; Dill, D.L.; et al. The m6A RNA demethylase FTO is a HIF-independent synthetic lethal partner with the VHL tumor suppressor. Proc. Natl. Acad. Sci. USA 2020, 117, 21441–21449. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, L.; Lou, W.; Su, J.; Huang, J.; Liu, A.; Xu, Y.; He, H.; Gao, Y.; Xu, D. Aberrant activation of m6A demethylase FTO renders HIF2αlow/− clear cell renal cell carcinoma sensitive to BRD9 inhibitors. Sci. Transl. Med. 2021, 13, eabf6045. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, F.; Wang, Z.; Yang, X.; Yu, H.; Si, S.; Lu, J.; Zhou, Z.; Lu, Q.; Wang, Z.; et al. ALKBH5 promotes the proliferation of renal cell carcinoma by regulating AURKB expression in an m6A-dependent manner. Ann. Transl. Med. 2020, 8, 646. [Google Scholar] [CrossRef]
- Zhuang, C.; Zhuang, C.; Luo, X.; Huang, X.; Yao, L.; Li, J.; Li, Y.; Xiong, T.; Ye, J.; Zhang, F.; et al. N6-methyladenosine demethylase FTO suppresses clear cell renal cell carcinoma through a novel FTO-PGC-1α signalling axis. J. Cell Mol. Med. 2019, 23, 2163–2173. [Google Scholar] [CrossRef] [PubMed]
- Strick, A.; Hagen, F.; Gundert, L.; Klümper, N.; Tolkach, Y.; Schmidt, D.; Kristiansen, G.; Toma, M.; Ritter, M.; Ellinger, J. The N6-methyladenosine (m6A) erasers alkylation repair homologue 5 (ALKBH5) and fat mass and obesity-associated protein (FTO) are prognostic biomarkers in patients with clear cell renal carcinoma. BJU Int. 2020, 125, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, R.; Martignoni, G.; Hartmann, A.; Caliò, A.; Segala, D.; Stöhr, C.; Wach, S.; Erlmeier, F.; Weichert, W.; Autenrieth, M. Multi-institutional re-evaluation of prognostic factors in chromophobe renal cell carcinoma: Proposal of a novel two-tiered grading scheme. Virchows Arch. 2020, 476, 409–418. [Google Scholar] [CrossRef]
- Gouda, H.R.; Talaat, I.M.; Bouzid, A.; El-Assi, H.; Nabil, A.; Venkatachalam, T.; Manasa Bhamidimarri, P.; Wohlers, I.; Mahdami, A.; El-Gendi, S. Genetic analysis of CFH and MCP in Egyptian patients with immune-complex proliferative glomerulonephritis. Front. Immunol. 2022, 13, 960068. [Google Scholar] [CrossRef]
- Jalaleddine, N.; Bouzid, A.; Hachim, M.; Sharif-Askari, N.S.; Mahboub, B.; Senok, A.; Halwani, R.; Hamoudi, R.A.; Al Heialy, S. ACE2 polymorphisms impact COVID-19 severity in obese patients. Sci. Rep. 2022, 12, 21491. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Kraft, I.L.; Godley, L.A. Identifying potential germline variants from sequencing hematopoietic malignancies. Hematol. Am. Soc. Hematol. Educ. Program. 2020, 2020, 219–227. [Google Scholar] [CrossRef]
- Mahfood, M.; Chouchen, J.; Mohamed, W.K.E.A.; Al Mutery, A.; Harati, R.; Tlili, A. Whole exome sequencing, in silico and functional studies confirm the association of the GJB2 mutation p. Cys169Tyr with deafness and suggest a role for the TMEM59 gene in the hearing process. Saudi J. Biol. Sci. 2021, 28, 4421–4429. [Google Scholar] [CrossRef]
- Remmele, W.; Stegner, H. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Der Pathol. 1987, 8, 138–140. [Google Scholar]
- Liu, L.; Hou, Y.; Deng, C.; Tao, Z.; Chen, Z.; Hu, J.; Chen, K. Single cell sequencing reveals that CD39 inhibition mediates changes to the tumor microenvironment. Nat. Commun. 2022, 13, 6740. [Google Scholar] [CrossRef]
- Shu, J.; Qiu, G.; Mohammad, I. A semi-automatic image analysis tool for biomarker detection in immunohistochemistry analysis. In Proceedings of the 2013 Seventh International Conference on Image and Graphics, Qingdao, China, 26–28 July 2013; pp. 937–942. [Google Scholar]
- Abdul Razzaq, E.A.; Bajbouj, K.; Bouzid, A.; Alkhayyal, N.; Hamoudi, R.; Bendardaf, R. Transcriptomic changes associated with ERBB2 overexpression in colorectal cancer implicate a potential role of the wnt signaling pathway in tumorigenesis. Cancers 2022, 15, 130. [Google Scholar] [CrossRef] [PubMed]
- Rabjerg, M. Identification and validation of novel prognostic markers in renal cell carcinoma. Dan. Med. J. 2017, 64, B5339. [Google Scholar] [PubMed]
- Bao, X.; Zhang, Y.; Li, H.; Teng, Y.; Ma, L.; Chen, Z.; Luo, X.; Zheng, J.; Zhao, A.; Ren, J. RM2Target: A comprehensive database for targets of writers, erasers and readers of RNA modifications. Nucleic Acids Res. 2023, 51, D269–D279. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Zhang, H.; Zhu, K.; Li, X.; Ye, Y.; Li, R.; Liu, X.; Lin, D.; Zuo, Z.; Zheng, J. M6A2Target: A comprehensive database for targets of m 6 A writers, erasers and readers. Brief. Bioinform. 2021, 22, bbaa055. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots, 0.5.0; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Kuhn, M.; Jackson, S.; Cimentada, J. corrr: Correlations in R, 0.4.4; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Kolde, R. pheatmap: Pretty Heatmaps, 1.0.12; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Kassambara, A. rstatix: Pipe-Friendly Framework for Basic Statistical Tests, 0.7.1; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Chen, H.; Liu, H.; Zhang, C.; Xiao, N.; Li, Y.; Zhao, X.; Zhang, R.; Gu, H.; Kang, Q.; Wan, J. RNA methylation-related inhibitors: Biological basis and therapeutic potential for cancer therapy. Clin. Transl. Med. 2024, 14, e1644. [Google Scholar] [CrossRef]
- Gutierrez, R.; O’Connor, T.R. DNA direct reversal repair and alkylating agent drug resistance. Cancer Drug Resist. 2021, 4, 414–423. [Google Scholar] [CrossRef]
- Shen, D.; Ding, L.; Lu, Z.; Wang, R.; Yu, C.; Wang, H.; Zheng, Q.; Wang, X.; Xu, W.; Yu, H.; et al. METTL14-mediated Lnc-LSG1 m6A modification inhibits clear cell renal cell carcinoma metastasis via regulating ESRP2 ubiquitination. Mol. Ther. Nucleic Acids 2022, 27, 547–561. [Google Scholar] [CrossRef]
- Cuykendall, T.N.; Rubin, M.A.; Khurana, E. Non-coding genetic variation in cancer. Curr. Opin. Syst. Biol. 2017, 1, 9–15. [Google Scholar] [CrossRef]
- Robles-Espinoza, C.D.; Mohammadi, P.; Bonilla, X.; Gutierrez-Arcelus, M. Allele-specific expression: Applications in cancer and technical considerations. Curr. Opin. Genet. Dev. 2021, 66, 10–19. [Google Scholar] [CrossRef]
- Wang, M.; Sunkel, B.D.; Ray, W.C.; Stanton, B.Z. Chromatin structure in cancer. BMC Mol. Cell Biol. 2022, 23, 35. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Weidhaas, J.B. Functional micro RNA binding site variants. Mol. Oncol. 2019, 13, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Caballero, M.E.; Sierra-Ramírez, J.A. Single nucleotide polymorphisms of the FTO gene and cancer risk: An overview. Mol. Biol. Rep. 2015, 42, 699–704. [Google Scholar] [CrossRef] [PubMed]
- Kaklamani, V.; Yi, N.; Sadim, M.; Siziopikou, K.; Zhang, K.; Xu, Y.; Tofilon, S.; Agarwal, S.; Pasche, B.; Mantzoros, C. The role of the fat mass and obesity associated gene (FTO) in breast cancer risk. BMC Med. Genet. 2011, 12, 1–10. [Google Scholar] [CrossRef]
- Garcia-Closas, M.; Couch, F.J.; Lindstrom, S.; Michailidou, K.; Schmidt, M.K.; Brook, M.N.; Orr, N.; Rhie, S.K.; Riboli, E.; Feigelson, H.S. Genome-wide association studies identify four ER negative–specific breast cancer risk loci. Nat. Genet. 2013, 45, 392–398. [Google Scholar] [CrossRef]
- Wiggins, G.A.; Black, M.A.; Dunbier, A.; Merriman, T.R.; Pearson, J.F.; Walker, L.C. Variable expression quantitative trait loci analysis of breast cancer risk variants. Sci. Rep. 2021, 11, 7192. [Google Scholar] [CrossRef]
- Zhang, B.; Li, Y.; Li, L.; Chen, M.; Zhang, C.; Zuo, X.-b.; Zhou, F.-s.; Liang, B.; Zhu, J.; Li, P. Association study of susceptibility loci with specific breast cancer subtypes in Chinese women. Breast Cancer Res. Treat. 2014, 146, 503–514. [Google Scholar] [CrossRef]
- Kang, Y.; Liu, F.; Liu, Y. Is FTO gene variant related to cancer risk independently of adiposity? An updated meta-analysis of 129,467 cases and 290,633 controls. Oncotarget 2017, 8, 50987. [Google Scholar] [CrossRef]
- Da Cunha, P.A.; de Carlos Back, L.K.; Sereia, A.F.R.; Kubelka, C.; Ribeiro, M.C.M.; Fernandes, B.L.; de Souza, I.R. Interaction between obesity-related genes, FTO and MC4R, associated to an increase of breast cancer risk. Mol. Biol. Rep. 2013, 40, 6657–6664. [Google Scholar] [CrossRef]
- Nock, N.L.; Plummer, S.J.; Thompson, C.L.; Casey, G.; Li, L. FTO polymorphisms are associated with adult body mass index (BMI) and colorectal adenomas in African-Americans. Carcinogenesis 2011, 32, 748–756. [Google Scholar] [CrossRef]
- Hubacek, J.A.; Dlouha, D.; Bobak, M.; Jiraskova, A.; Vitek, L. The risk of sporadic colorectal cancer development is not influenced by fat mass and obesity related gene polymorphism in Slavs. Eur. J. Intern. Med. 2012, 23, e175–e176. [Google Scholar] [CrossRef]
- Tarabra, E.; Actis, G.; Fadda, M.; De Paolis, P.; Comandone, A.; Coda, R.; Rosina, F. The obesity gene and colorectal cancer risk: A population study in Northern Italy. Eur. J. Intern. Med. 2012, 23, 65–69. [Google Scholar] [CrossRef]
- Guan, Q.; Lin, H.; Hua, W.; Lin, L.; Liu, J.; Deng, L.; Zhang, J.; Cheng, J.; Yang, Z.; Li, Y. Variant rs8400 enhances ALKBH5 expression through disrupting miR-186 binding and promotes neuroblastoma progression. Chin. J. Cancer Res. 2023, 35, 140. [Google Scholar] [CrossRef]
- Hua, R.X.; Liu, J.; Fu, W.; Zhu, J.; Zhang, J.; Cheng, J.; Li, S.; Zhou, H.; Xia, H.; He, J. ALKBH5 gene polymorphisms and Wilms tumor risk in Chinese children: A five-center case-control study. J. Clin. Lab. Anal. 2020, 34, e23251. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Lu, Y. Dexmedetomidine suppressed the biological behavior of HK-2 cells treated with LPS by down-regulating ALKBH5. Inflammation 2020, 43, 2256–2263. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-T.; Hu, X.-W.; Chen, H.-Y.; Yang, Q.; Li, H.-D.; Dong, Y.-H.; Zhang, Y.; Wang, J.-N.; Jin, J.; Wu, Y.-G. DNA methylation of FTO promotes renal inflammation by enhancing m6A of PPAR-α in alcohol-induced kidney injury. Pharmacol. Res. 2021, 163, 105286. [Google Scholar] [CrossRef] [PubMed]
- Dina, C.; Meyre, D.; Gallina, S.; Durand, E.; Körner, A.; Jacobson, P.; Carlsson, L.M.; Kiess, W.; Vatin, V.; Lecoeur, C. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 2007, 39, 724–726. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | ccRCC, n = 39 | chRCC, n = 7 | pRCC, n = 8 |
|---|---|---|---|
| Age | 55 (49, 62) | 57 (51, 60) | 62 (58, 70) |
| Unknown | 1 | 0 | 0 |
| Sex | |||
| Female | 12 (31%) | 4 (57%) | 3 (38%) |
| Male | 26 (67%) | 3 (43%) | 5 (62%) |
| Unknown | 1 (2.6%) | 0 | 0 |
| Obesity | |||
| Underweight | 4 (10%) | 0 (0%) | 0 (0%) |
| Normal weight | 21 (54%) | 6 (86%) | 6 (75%) |
| Overweight | 10 (26%) | 1 (14%) | 1 (12%) |
| Obese | 4 (10%) | 0 (0%) | 1 (12%) |
| Diabetes | 11 (28%) | 3 (43%) | 4 (50%) |
| Nuclear grade | |||
| 1 | 3 (7.7%) | 0 | 0 |
| 2 | 13 (33%) | 0 | 3 (38%) |
| 3 | 15 (38%) | 0 | 4 (50%) |
| 4 | 8 (21%) | 0 | 1 (12%) |
| Not applicable 1 | 0 | 7 | 0 |
| Capsular invasion | |||
| Negative | 28 (72%) | 3 (43%) | 7 (88%) |
| Positive | 11 (28%) | 4 (57%) | 1 (12%) |
| Renal sinus invasion | |||
| Negative | 31 (79%) | 5 (71%) | 5 (62%) |
| Positive | 8 (21%) | 2 (29%) | 3 (38%) |
| Extent | |||
| Localized | 27 (69%) | 7 (100%) | 7 (88%) |
| Metastatic | 12 (31%) | 0 | 1 (12%) |
| Tumor stage | |||
| I | 12 (31%) | 2 (29%) | 3 (38%) |
| II | 10 (26%) | 1 (14%) | 2 (25%) |
| III | 8 (21%) | 4 (57%) | 2 (25%) |
| IV | 9 (23%) | 0 (0%) | 1 (12%) |
| Systemic treatment | 11 (28%) | 0 | 3 (38%) |
| Status | |||
| Alive with disease | 5 (13%) | 0 | 0 |
| Died of disease | 9 (23%) | 0 | 1 (12%) |
| Cured | 25 (64%) | 7 (100%) | 7 (88%) |
| Characteristic | n = 11 |
|---|---|
| Age | 50 (42, 60) |
| Unknown | 3 |
| Sex | |
| Female | 6 (86%) |
| Male | 1 (14%) |
| Unknown | 4 |
| Diagnosis | |
| Hydronephrosis | 1 (9.1%) |
| Mild lymphocytic infiltration | 1 (9.1%) |
| Normal | 2 (18%) |
| Pyelonephritis | 7 (64%) |
| Nationality | |
| Egypt | 8 (80%) |
| Iraq | 1 (10%) |
| United Arab Emirates | 1 (10%) |
| Unknown | 1 |
| Gene | SNP | SNP Position * | Controls (HWE p-Value) | Subtype (Cases) | OR | 95% CI | p-Value *** |
|---|---|---|---|---|---|---|---|
| FTO | rs1121980 (G>A) | 16:53809247 Intron | n = 11 (0.001) | ccRCC (n = 22) | 1.00 | (0.36, 2.78) | 1.00 |
| pRCC (n = 7) | 1.00 | (0.26, 3.82) | 1.00 | ||||
| chRCC (n = 6) | 1.00 | (0.25, 4.08) | 1.00 | ||||
| rs17817449 (T>G) | 16:53813367 Intron | n = 10 (0.43) | ccRCC (n = 19) | 0.875 | (0.288, 2.658) | 1.00 | |
| pRCC (n = 6) | 0.3 | (0.052, 1.747) | 0.248 | ||||
| chRCC (n = 5) | 0.167 | (0.018, 1.583) | 0.204 | ||||
| rs9939609 (T>A) | 16:53820527 Intron | n = 11 (0.87) | ccRCC (n = 20) | 1.11 | (0.095, 12.92) | 1.00 | |
| pRCC (n= 6) | 1.74 | (0.07, 46.15) ** | 1.00 | ||||
| chRCC (n = 6) | 1.74 | (0.07, 46.15) ** | 1.00 | ||||
| rs8050136 (C>A) | 16:53816275 Intron | n = 11 (0.058) | ccRCC (n = 20) | 0.942 | (0.32, 2.79) | 1.00 | |
| pRCC (n = 7) | 0.7 | (0.164, 2.981) | 0.73 | ||||
| chRCC (n = 6) | 0.875 | (0.199, 3.85) | 1.00 | ||||
| rs11075995 (A>T) | 16:53855291 Intron | n = 11 (0.38) | ccRCC (n = 22) | 3.24 | (1.06, 9.87) | 0.046 | |
| pRCC (n= 7) | 2.08 | (0.5, 8.7) | 0.49 | ||||
| chRCC (n = 6) | 1.67 | (0.39, 7.2) | 0.72 | ||||
| ALKBH5 | rs8400 (G>A) | 17:18112845 3′ UTR | n = 9 (2.56) | ccRCC (n=15) | 2.68 | (0.71, 10.07) | 0.214 |
| pRCC (n= 4) | 5.83 | (0.95, 35.72) | 0.078 | ||||
| chRCC (n = 4) | 5.83 | (0.95, 35.72) | 0.078 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhammadi, M.A.; Ilce, B.Y.; Bhamidimarri, P.M.; Bouzid, A.; Ali, N.; Alhamidi, R.S.; Hamad, A.M.; Mahfood, M.; Tlili, A.; Talaat, I.M.; et al. Analysis of Genotype and Expression of FTO and ALKBH5 in a MENA-Region Renal Cell Carcinoma Cohort. Cancers 2025, 17, 1395. https://doi.org/10.3390/cancers17091395
Alhammadi MA, Ilce BY, Bhamidimarri PM, Bouzid A, Ali N, Alhamidi RS, Hamad AM, Mahfood M, Tlili A, Talaat IM, et al. Analysis of Genotype and Expression of FTO and ALKBH5 in a MENA-Region Renal Cell Carcinoma Cohort. Cancers. 2025; 17(9):1395. https://doi.org/10.3390/cancers17091395
Chicago/Turabian StyleAlhammadi, Muna Abdalla, Burcu Yener Ilce, Poorna Manasa Bhamidimarri, Amal Bouzid, Nival Ali, Reem Sami Alhamidi, Alaa Mohamed Hamad, Mona Mahfood, Abdelaziz Tlili, Iman M. Talaat, and et al. 2025. "Analysis of Genotype and Expression of FTO and ALKBH5 in a MENA-Region Renal Cell Carcinoma Cohort" Cancers 17, no. 9: 1395. https://doi.org/10.3390/cancers17091395
APA StyleAlhammadi, M. A., Ilce, B. Y., Bhamidimarri, P. M., Bouzid, A., Ali, N., Alhamidi, R. S., Hamad, A. M., Mahfood, M., Tlili, A., Talaat, I. M., & Hamoudi, R. (2025). Analysis of Genotype and Expression of FTO and ALKBH5 in a MENA-Region Renal Cell Carcinoma Cohort. Cancers, 17(9), 1395. https://doi.org/10.3390/cancers17091395







_Talaat.jpg)



