Assigning Transcriptomic Subtypes to Chronic Lymphocytic Leukemia Samples Using Nanopore RNA-Sequencing and Self-Organizing Maps
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. CLL Public Data Source
2.2. Transcriptome Portrayal Using Self-Organizing Maps (SOMs)
2.3. Phenotype Maps and Survival Analysis
2.4. Patients and Sample Collection
2.5. RNA Isolation
2.6. Sequencing Library Preparation
2.7. ONT Sequencing Data Preprocessing
2.8. Projection of ONT Gene Expression Data to SOMs Space
3. Results and Discussion
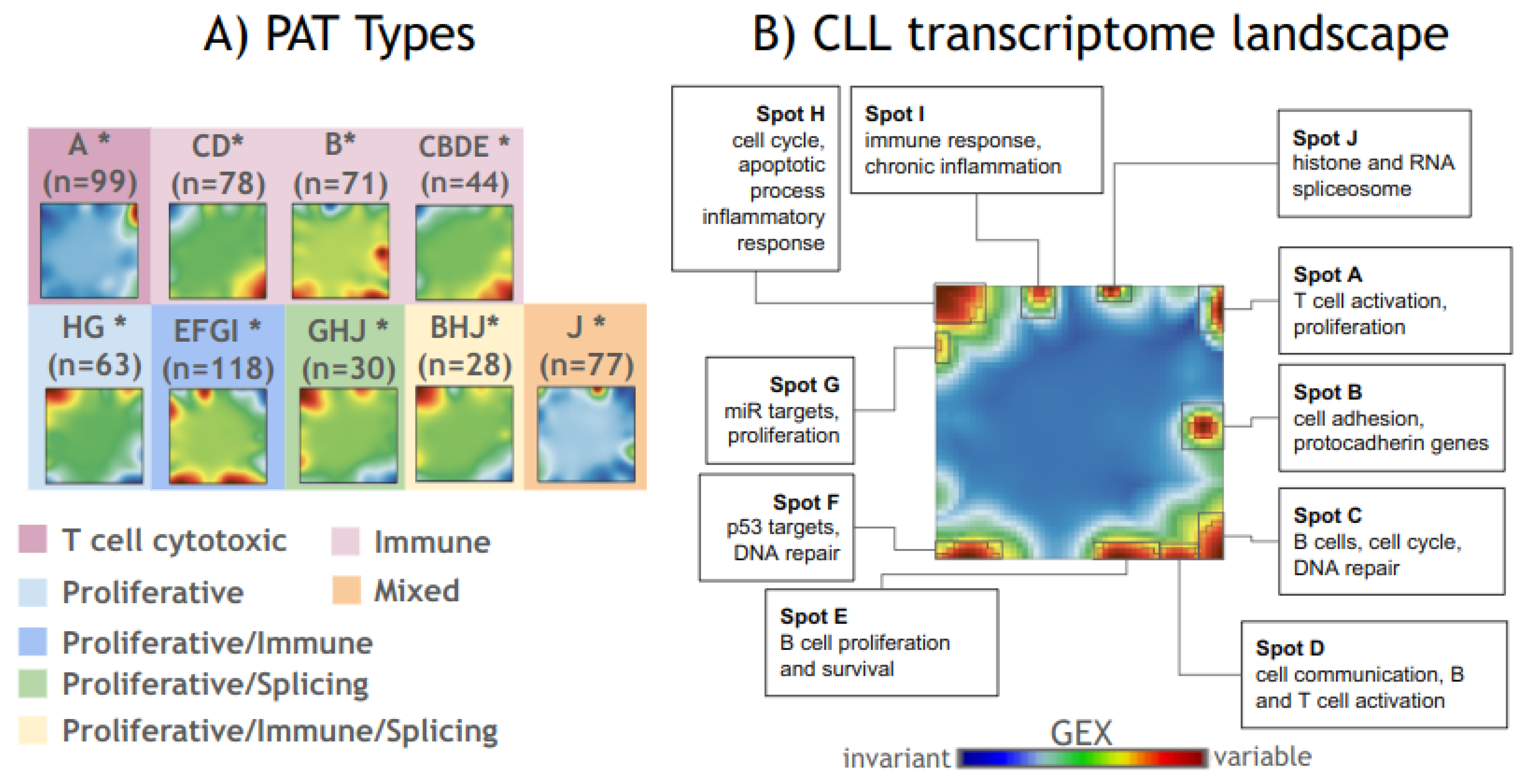
3.1. Transcriptome Portrayal of CLL
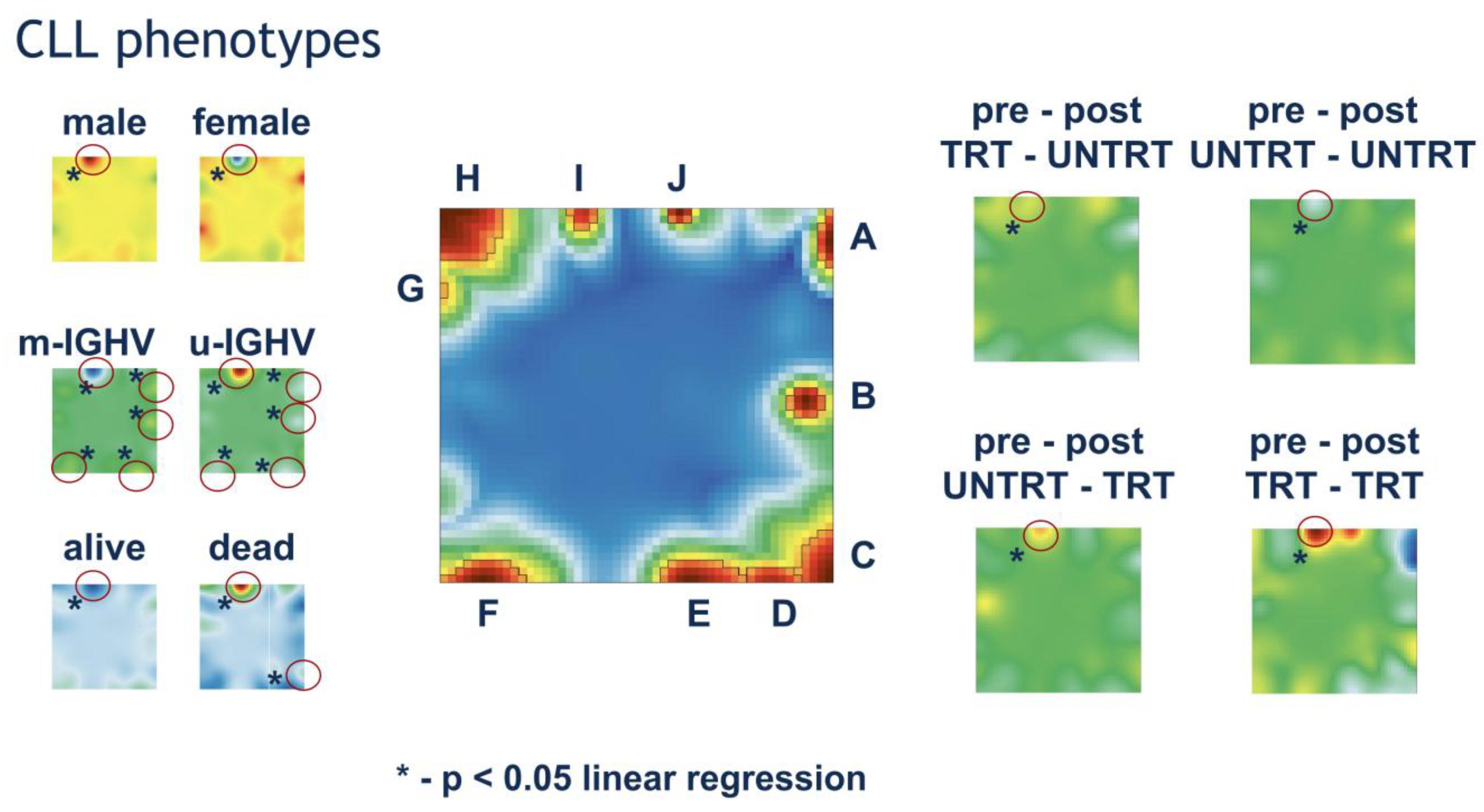
3.2. Phenotype Maps Associate Transcriptome Deregulations with Clinical Characteristics
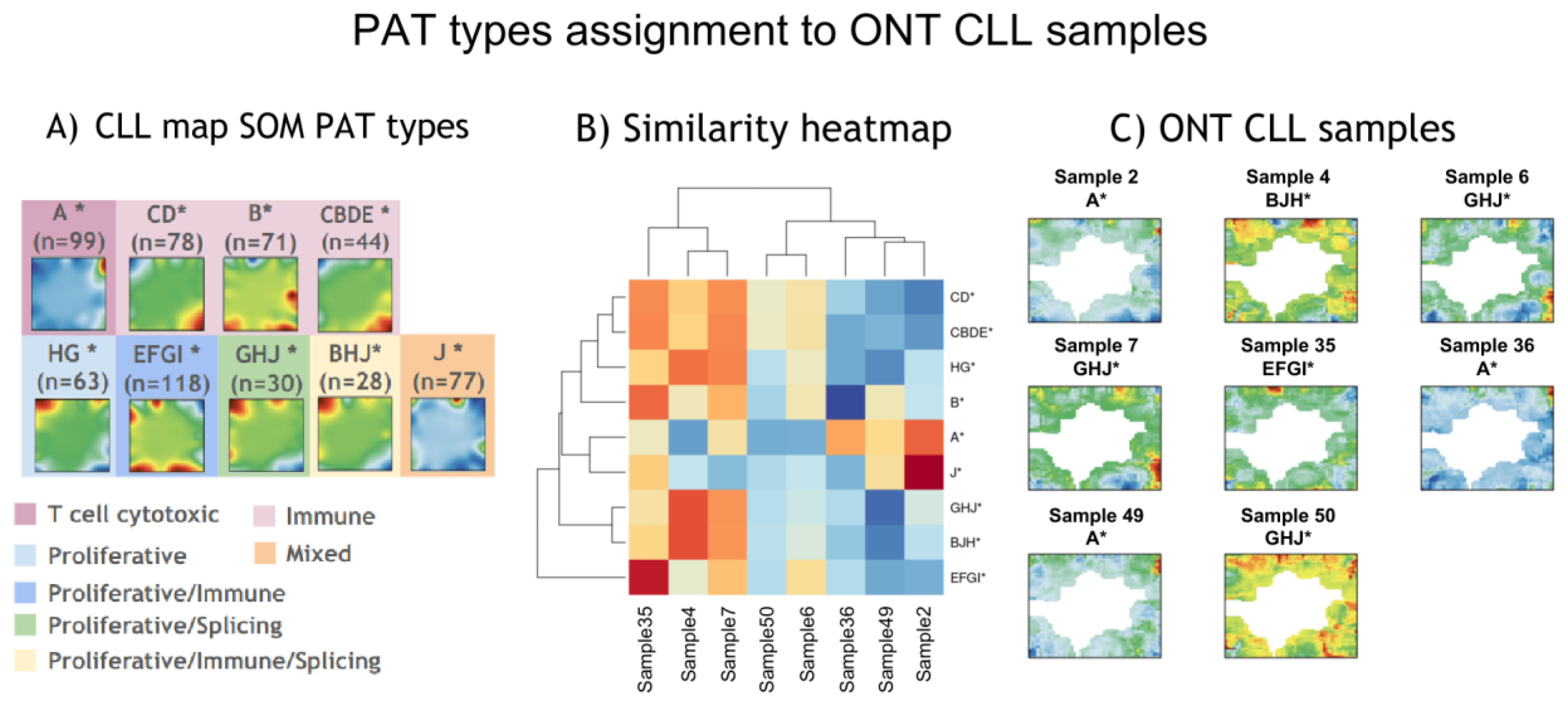
3.3. Projection of ONT CLL Data onto CLL SOMs Space
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hallek, M. Chronic Lymphocytic Leukemia: 2020 Update on Diagnosis, Risk Stratification and Treatment. Am. J. Hematol. 2019, 94, 1266–1287. [Google Scholar] [CrossRef] [PubMed]
- Sud, A.; Parry, E.M.; Wu, C.J. The Molecular Map of CLL and Richter’s Syndrome. Semin. Hematol. 2024, 61, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Turk, A.; Čeh, E.; Calin, G.A.; Kunej, T. Multiple Omics Levels of Chronic Lymphocytic Leukemia. Cell Death Discov. 2024, 10, 293. [Google Scholar] [CrossRef]
- Tsagiopoulou, M.; Gut, I.G. Machine Learning and Multi-Omics Data in Chronic Lymphocytic Leukemia: The Future of Precision Medicine? Front. Genet. 2023, 14, 1304661. [Google Scholar] [CrossRef]
- Ferreira, P.G.; Jares, P.; Rico, D.; Gómez-López, G.; Martínez-Trillos, A.; Villamor, N.; Ecker, S.; González-Pérez, A.; Knowles, D.G.; Monlong, J.; et al. Transcriptome Characterization by RNA Sequencing Identifies a Major Molecular and Clinical Subdivision in Chronic Lymphocytic Leukemia. Genome Res. 2014, 24, 212–226. [Google Scholar] [CrossRef]
- Griffen, T.L.; Dammer, E.B.; Dill, C.D.; Carey, K.M.; Young, C.D.; Nunez, S.K.; Ohandjo, A.Q.; Kornblau, S.M.; Lillard, J.W. Multivariate Transcriptome Analysis Identifies Networks and Key Drivers of Chronic Lymphocytic Leukemia Relapse Risk and Patient Survival. BMC Med. Genom. 2021, 14, 171. [Google Scholar] [CrossRef]
- Knisbacher, B.A.; Lin, Z.; Hahn, C.K.; Nadeu, F.; Duran-Ferrer, M.; Stevenson, K.E.; Tausch, E.; Delgado, J.; Barbera-Mourelle, A.; Taylor-Weiner, A.; et al. Molecular Map of Chronic Lymphocytic Leukemia and Its Impact on Outcome. Nat. Genet. 2022, 54, 1664–1674. [Google Scholar] [CrossRef]
- Ramkissoon, L.A.; Montgomery, N.D. Applications of Next-Generation Sequencing in Hematologic Malignancies. Hum. Immunol. 2021, 82, 859–870. [Google Scholar] [CrossRef]
- Helmy, M.; Awad, M.; Mosa, K.A. Limited Resources of Genome Sequencing in Developing Countries: Challenges and Solutions. Appl. Transl. Genom. 2016, 9, 15–19. [Google Scholar] [CrossRef]
- MacKenzie, M.; Argyropoulos, C. An Introduction to Nanopore Sequencing: Past, Present, and Future Considerations. Micromachines 2023, 14, 459. [Google Scholar] [CrossRef]
- Wang, J.; Bhakta, N.; Ayer Miller, V.; Revsine, M.; Litzow, M.R.; Paietta, E.; Fedoriw, Y.; Roberts, K.G.; Gu, Z.; Mullighan, C.G.; et al. Acute Leukemia Classification Using Transcriptional Profiles From Low-Cost Nanopore mRNA Sequencing. JCO Precis. Oncol. 2022, 6, e2100326. [Google Scholar] [CrossRef] [PubMed]
- Loeffler-Wirth, H.; Kalcher, M.; Binder, H. oposSOM: R-Package for High-Dimensional Portraying of Genome-Wide Expression Landscapes on Bioconductor. Bioinformatics 2015, 31, 3225–3227. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting Batch Effects in Microarray Expression Data Using Empirical Bayes Methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Wirth, H.; Löffler, M.; von Bergen, M.; Binder, H. Expression Cartography of Human Tissues Using Self Organizing Maps. BMC Bioinform. 2011, 12, 306. [Google Scholar] [CrossRef]
- Wirth, H.; von Bergen, M.; Binder, H. Mining SOM Expression Portraits: Feature Selection and Integrating Concepts of Molecular Function. BioData Min. 2012, 5, 18. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. G:Profiler-Interoperable Web Service for Functional Enrichment Analysis and Gene Identifier Mapping (2023 Update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- Griffith, D.M.; Veech, J.A.; Marsh, C.J. Cooccur: Probabilistic Species Co-Occurrence Analysis in R. J. Stat. Softw. 2016, 69, 1–17. [Google Scholar] [CrossRef]
- Loeffler-Wirth, H.; Kreuz, M.; Hopp, L.; Arakelyan, A.; Haake, A.; Cogliatti, S.B.; Feller, A.C.; Hansmann, M.-L.; Lenze, D.; Möller, P.; et al. A Modular Transcriptome Map of Mature B Cell Lymphomas. Genome Med. 2019, 11, 27. [Google Scholar] [CrossRef]
- Arakelyan, A.; Melkonyan, A.; Hakobyan, S.; Boyarskih, U.; Simonyan, A.; Nersisyan, L.; Nikoghosyan, M.; Filipenko, M.; Binder, H. Transcriptome Patterns of BRCA1- and BRCA2- Mutated Breast and Ovarian Cancers. Int. J. Mol. Sci. 2021, 22, 1266. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R Package Rsubread Is Easier, Faster, Cheaper and Better for Alignment and Quantification of RNA Sequencing Reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Nikoghosyan, M.; Loeffler-Wirth, H.; Davidavyan, S.; Binder, H.; Arakelyan, A. Projection of High-Dimensional Genome-Wide Expression on SOM Transcriptome Landscapes. BioMedInformatics 2022, 2, 62–76. [Google Scholar] [CrossRef]
- Orchard, J.A.; Ibbotson, R.E.; Davis, Z.; Wiestner, A.; Rosenwald, A.; Thomas, P.W.; Hamblin, T.J.; Staudt, L.M.; Oscier, D.G. ZAP-70 Expression and Prognosis in Chronic Lymphocytic Leukaemia. Lancet 2004, 363, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Saiya-Cork, K.; Collins, R.; Parkin, B.; Ouillette, P.; Kuizon, E.; Kujawski, L.; Erba, H.; Campagnaro, E.; Shedden, K.; Kaminski, M.; et al. A Pathobiological Role of the Insulin Receptor in Chronic Lymphocytic Leukemia. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 2679–2692. [Google Scholar] [CrossRef]
- Gonzalez, D.; Else, M.; Wren, D.; Usai, M.; Buhl, A.M.; Parker, A.; Oscier, D.; Morgan, G.; Catovsky, D. CLLU1 Expression Has Prognostic Value in Chronic Lymphocytic Leukemia after First-Line Therapy in Younger Patients and in Those with Mutated IGHV Genes. Haematologica 2013, 98, 274–278. [Google Scholar] [CrossRef]
- Eisele, L.; Prinz, R.; Klein-Hitpass, L.; Nückel, H.; Lowinski, K.; Thomale, J.; Moeller, L.C.; Dührsen, U.; Dürig, J. Combined PER2 and CRY1 Expression Predicts Outcome in Chronic Lymphocytic Leukemia. Eur. J. Haematol. 2009, 83, 320–327. [Google Scholar] [CrossRef]
- Massarweh, S.; Osborne, C.K.; Creighton, C.J.; Qin, L.; Tsimelzon, A.; Huang, S.; Weiss, H.; Rimawi, M.; Schiff, R. Tamoxifen Resistance in Breast Tumors Is Driven by Growth Factor Receptor Signaling with Repression of Classic Estrogen Receptor Genomic Function. Cancer Res. 2008, 68, 826–833. [Google Scholar] [CrossRef]
- Creighton, C.J.; Massarweh, S.; Huang, S.; Tsimelzon, A.; Hilsenbeck, S.G.; Osborne, C.K.; Shou, J.; Malorni, L.; Schiff, R. Development of Resistance to Targeted Therapies Transforms the Clinically Associated Molecular Profile Subtype of Breast Tumor Xenografts. Cancer Res. 2008, 68, 7493–7501. [Google Scholar] [CrossRef]
- Zhu, Y.; Gan, X.; Qin, R.; Lin, Z. Identification of Six Diagnostic Biomarkers for Chronic Lymphocytic Leukemia Based on Machine Learning Algorithms. J. Oncol. 2022, 2022, 3652107. [Google Scholar] [CrossRef]
- Campo, E.; Cymbalista, F.; Ghia, P.; Jäger, U.; Pospisilova, S.; Rosenquist, R.; Schuh, A.; Stilgenbauer, S. TP53 Aberrations in Chronic Lymphocytic Leukemia: An Overview of the Clinical Implications of Improved Diagnostics. Haematologica 2018, 103, 1956–1968. [Google Scholar] [CrossRef]
- Lütge, A.; Lu, J.; Hüllein, J.; Walther, T.; Sellner, L.; Wu, B.; Rosenquist, R.; Oakes, C.C.; Dietrich, S.; Huber, W.; et al. Subgroup-Specific Gene Expression Profiles and Mixed Epistasis in Chronic Lymphocytic Leukemia. Haematologica 2023, 108, 2664–2676. [Google Scholar] [CrossRef] [PubMed]
- Vlachonikola, E.; Stamatopoulos, K.; Chatzidimitriou, A. T Cells in Chronic Lymphocytic Leukemia: A Two-Edged Sword. Front. Immunol. 2021, 11, 612244. [Google Scholar] [CrossRef] [PubMed]
- Roessner, P.M.; Seiffert, M. T-Cells in Chronic Lymphocytic Leukemia: Guardians or Drivers of Disease? Leukemia 2020, 34, 2012–2024. [Google Scholar] [CrossRef]
- Plander, M.; Seegers, S.; Ugocsai, P.; Diermeier-Daucher, S.; Iványi, J.; Schmitz, G.; Hofstädter, F.; Schwarz, S.; Orsó, E.; Knüchel, R.; et al. Different Proliferative and Survival Capacity of CLL-Cells in a Newly Established in Vitro Model for Pseudofollicles. Leukemia 2009, 23, 2118–2128. [Google Scholar] [CrossRef][Green Version]
- Catovsky, D.; Wade, R.; Else, M. The Clinical Significance of Patients’ Sex in Chronic Lymphocytic Leukemia. Haematologica 2014, 99, 1088–1094. [Google Scholar] [CrossRef]
- Schmidt, J.; Berghaus, S.; Blessing, F.; Herbeck, H.; Blessing, J.; Schierack, P.; Rödiger, S.; Roggenbuck, D.; Wenzel, F. Genotyping of Familial Mediterranean Fever Gene (MEFV)-Single Nucleotide Polymorphism-Comparison of Nanopore with Conventional Sanger Sequencing. PLoS ONE 2022, 17, e0265622. [Google Scholar] [CrossRef]
- Sun, X.; Song, J.; Leng, X.; Li, F.; Wang, H.; He, J.; Zhai, W.; Wang, Z.; Wu, Q.; Li, Z.; et al. A Preliminary Evaluation of Targeted Nanopore Sequencing Technology for the Detection of Mycobacterium Tuberculosis in Bronchoalveolar Lavage Fluid Specimens. Front. Cell. Infect. Microbiol. 2023, 13, 1107990. [Google Scholar] [CrossRef]
- Avetyan, D.; Hakobyan, S.; Nikoghosyan, M.; Ghukasyan, L.; Khachatryan, G.; Sirunyan, T.; Muradyan, N.; Zakharyan, R.; Chavushyan, A.; Hayrapetyan, V.; et al. Molecular Analysis of SARS-CoV-2 Lineages in Armenia. Viruses 2022, 14, 1074. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An Immeasurable Source of Knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef]
- Cook, R.; Brown, N.; Rihtman, B.; Michniewski, S.; Redgwell, T.; Clokie, M.; Stekel, D.J.; Chen, Y.; Scanlan, D.J.; Hobman, J.L.; et al. The Long and Short of It: Benchmarking Viromics Using Illumina, Nanopore and PacBio Sequencing Technologies. Microb. Genom. 2024, 10, 001198. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arakelyan, A.; Sirunyan, T.; Khachatryan, G.; Hakobyan, S.; Minasyan, A.; Nikoghosyan, M.; Hakobyan, M.; Chavushyan, A.; Martirosyan, G.; Hakobyan, Y.; et al. Assigning Transcriptomic Subtypes to Chronic Lymphocytic Leukemia Samples Using Nanopore RNA-Sequencing and Self-Organizing Maps. Cancers 2025, 17, 964. https://doi.org/10.3390/cancers17060964
Arakelyan A, Sirunyan T, Khachatryan G, Hakobyan S, Minasyan A, Nikoghosyan M, Hakobyan M, Chavushyan A, Martirosyan G, Hakobyan Y, et al. Assigning Transcriptomic Subtypes to Chronic Lymphocytic Leukemia Samples Using Nanopore RNA-Sequencing and Self-Organizing Maps. Cancers. 2025; 17(6):964. https://doi.org/10.3390/cancers17060964
Chicago/Turabian StyleArakelyan, Arsen, Tamara Sirunyan, Gisane Khachatryan, Siras Hakobyan, Arpine Minasyan, Maria Nikoghosyan, Meline Hakobyan, Andranik Chavushyan, Gevorg Martirosyan, Yervand Hakobyan, and et al. 2025. "Assigning Transcriptomic Subtypes to Chronic Lymphocytic Leukemia Samples Using Nanopore RNA-Sequencing and Self-Organizing Maps" Cancers 17, no. 6: 964. https://doi.org/10.3390/cancers17060964
APA StyleArakelyan, A., Sirunyan, T., Khachatryan, G., Hakobyan, S., Minasyan, A., Nikoghosyan, M., Hakobyan, M., Chavushyan, A., Martirosyan, G., Hakobyan, Y., & Binder, H. (2025). Assigning Transcriptomic Subtypes to Chronic Lymphocytic Leukemia Samples Using Nanopore RNA-Sequencing and Self-Organizing Maps. Cancers, 17(6), 964. https://doi.org/10.3390/cancers17060964






