Liquid Biopsies in the Early Diagnosis, Prognosis, and Tailored Treatment of Colorectal Cancer
Simple Summary
Abstract
1. Introduction
2. CRC Biology
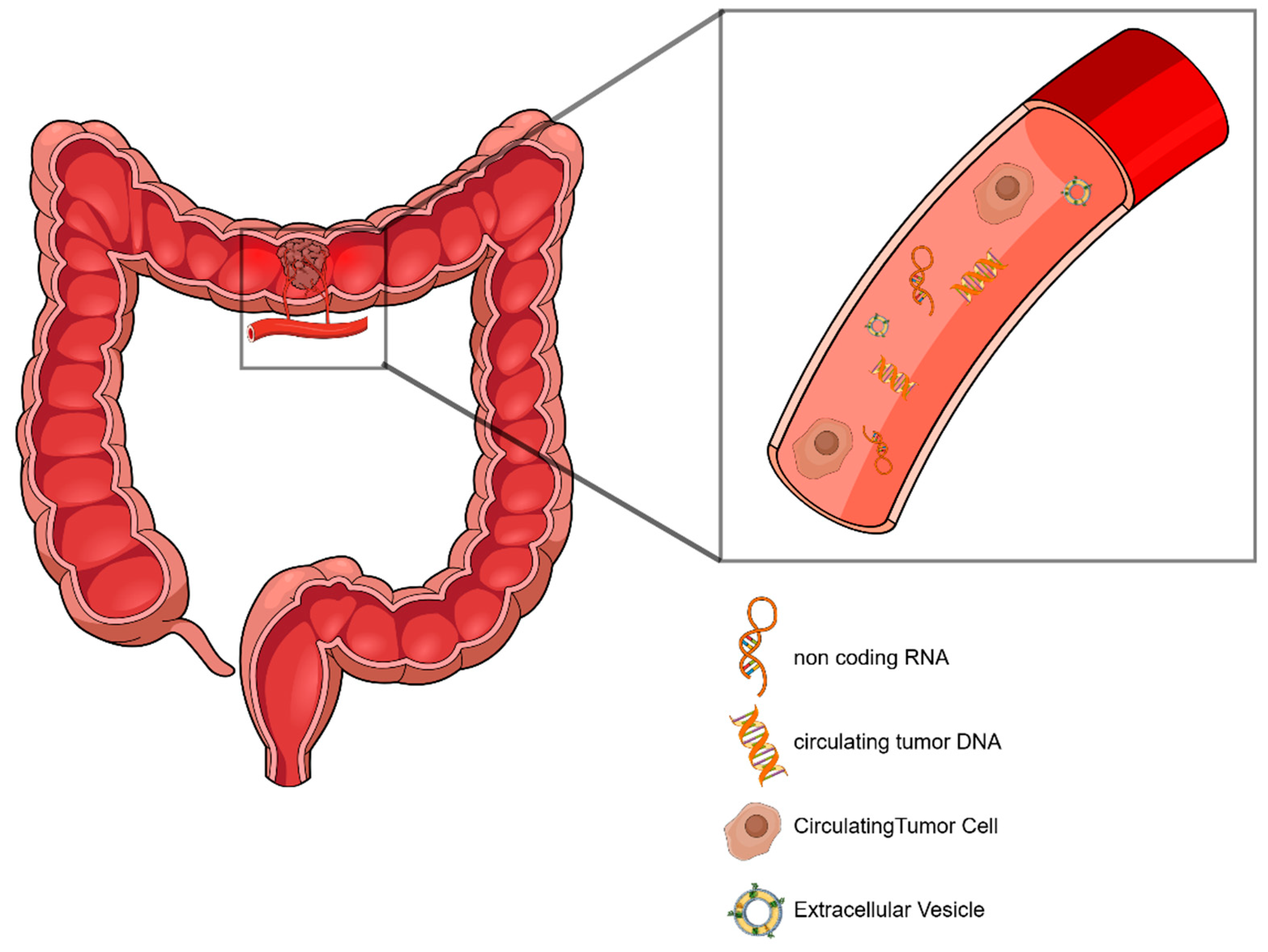
2.1. Liquid Biopsy–General Features
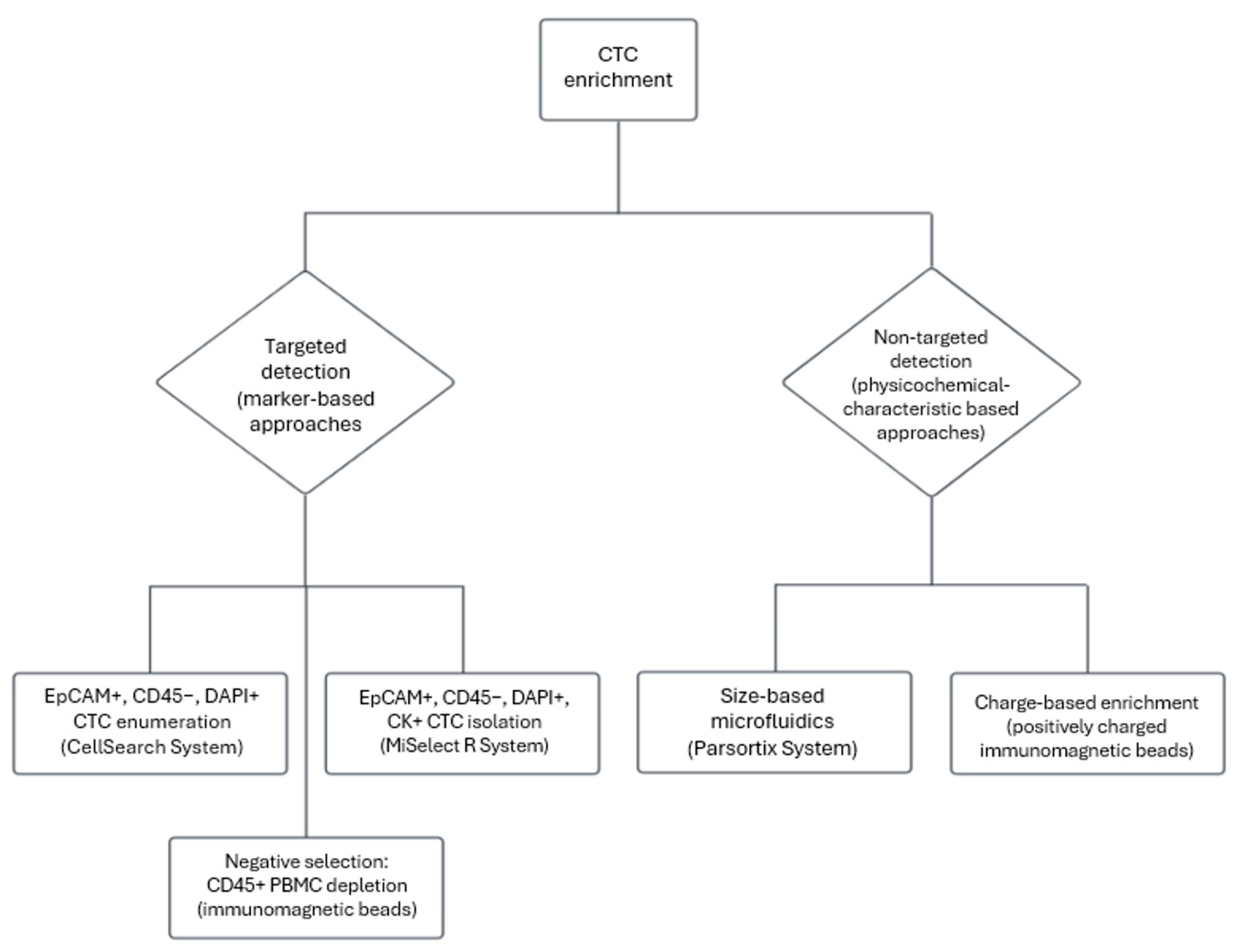
2.2. Circulating Tumor Cells–Biology and Clinical Utility in Early-Stage CRC
2.3. ctDNA Biology and Analysis in CRC
2.4. Non-Coding RNAs as Liquid Biopsy Biomarkers—The Role of EVs in ncRNA Analysis
2.5. Tumor-Educated Platelets—A Liquid Biopsy Perspective of the Tumor Microenvironment
2.6. Prognostic Utility of Liquid Biopsies
2.7. Prediction of Resistance to Treatment and Real-Time Monitoring of Response to Therapy
2.8. Comparison Between Liquid and Tissue Biopsies and Upcoming Limitations
3. Limitations and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Alinia, S.; Ahmadi, S.; Mohammadi, Z.; Shirvandeh, F.R.; Jafarabadi, M.A.; Mahmoudi, L.; Safari, M.; Roshanaei, G. Exploring the impact of stage and tumor site on colorectal cancer survival: Bayesian survival modeling. Sci. Rep. 2024, 14, 4270. [Google Scholar] [CrossRef]
- Galoș, D.; Gorzo, A.; Balacescu, O.; Sur, D. Clinical Applications of Liquid Biopsy in Colorectal Cancer Screening: Current Challenges and Future Perspectives. Cells 2022, 11, 3493. [Google Scholar] [CrossRef]
- Honoré, N.; Galot, R.; van Marcke, C.; Limaye, N.; Machiels, J.-P. Liquid Biopsy to Detect Minimal Residual Disease: Methodology and Impact. Cancers 2021, 13, 5364. [Google Scholar] [CrossRef] [PubMed]
- Oshima, M.; Oshima, H.; Kitagawa, K.; Kobayashi, M.; Itakura, C.; Taketo, M. Loss of Apc heterozygosity and abnormal tissue building in nascent intestinal polyps in mice carrying a truncated Apc gene. Proc. Natl. Acad. Sci. USA 1995, 92, 4482–4486. [Google Scholar] [CrossRef] [PubMed]
- Housini, M.; Dariya, B.; Ahmed, N.; Stevens, A.; Fiadjoe, H.; Nagaraju, G.P.; Basha, R. Colorectal cancer: Genetic alterations, novel biomarkers, current therapeutic strategies and clinical trials. Gene 2024, 892, 147857. [Google Scholar] [CrossRef]
- Mattox, A.K.; Bettegowda, C.; Zhou, S.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Applications of liquid biopsies for cancer. Sci. Transl. Med. 2019, 11, eaay1984. [Google Scholar] [CrossRef]
- Zhou, H.; Zhu, L.; Song, J.; Wang, G.; Li, P.; Li, W.; Luo, P.; Sun, X.; Wu, J.; Liu, Y.; et al. Liquid biopsy at the frontier of detection, prognosis and progression monitoring in colorectal cancer. Mol. Cancer 2022, 21, 86. [Google Scholar] [CrossRef]
- Mirza, S.; Bhadresha, K.; Mughal, M.J.; McCabe, M.; Shahbazi, R.; Ruff, P.; Penny, C. Liquid biopsy approaches and immunotherapy in colorectal cancer for precision medicine: Are we there yet? Front. Oncol. 2022, 12, 1023565. [Google Scholar] [CrossRef]
- Ladabaum, U.; Mannalithara, A.; Weng, Y.; Schoen, R.E.; Dominitz, J.A.; Desai, M.; Lieberman, D. Comparative Effectiveness and Cost-Effectiveness of Colorectal Cancer Screening with Blood-Based Biomarkers (Liquid Biopsy) vs. Fecal Tests or Colonoscopy. Gastroenterology 2024, 167, 378–391. [Google Scholar] [CrossRef]
- Ashworth, T. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust. Med. J. 1869, 14, 146. [Google Scholar]
- Yao, S.; Han, Y.; Yang, M.; Jin, K.; Lan, H. Integration of liquid biopsy and immunotherapy: Opening a new era in colorectal cancer treatment. Front. Immunol. 2023, 14, 1292861. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Wu, S.; Wang, Y.; Shi, D. Circulating tumor cell isolation for cancer diagnosis and prognosis. eBioMedicine 2022, 83, 104237. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Lu, J.; Kornmann, M.; Traub, B. Role of Epithelial to Mesenchymal Transition in Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 14815. [Google Scholar] [CrossRef]
- Zafeiriadou, A.; Kollias, I.; Londra, T.; Tsaroucha, E.; Georgoulias, V.; Kotsakis, A.; Lianidou, E.; Markou, A. Metabolism-Related Gene Expression in Circulating Tumor Cells from Patients with Early Stage Non-Small Cell Lung Cancer. Cancers 2022, 14, 3237. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, Y.; Kang, K.; Mao, Y.; Yu, Y.; Yi, Q.; Wu, Y. Controllable Environment Protein Corona-Disguised Immunomagnetic Beads for High-Performance Circulating Tumor Cell Enrichment. Anal. Chem. 2022, 94, 4650–4657. [Google Scholar] [CrossRef] [PubMed]
- Reduzzi, C.; Nicolo, E.; Singhal, S.; Venetis, K.; Ortega-Franco, A.; de Miguel-Perez, D.; Dipasquale, A.; Gouda, M.A.; Saldanha, E.F.; Kasi, P.M.; et al. Unveiling the impact of circulating tumor cells: Two decades of discovery and clinical advancements in solid tumors. Crit. Rev. Oncol./Hematol. 2024, 203, 104483. [Google Scholar] [CrossRef]
- Cohen, S.J.; Punt, C.J.A.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef]
- Köstler, C.; Polzer, B.; Alberter, B. Circulating Tumor Cell Enrichment and Single-Cell Isolation Combining the CellSearch® and DEPArrayTM Systems. In Single Cell Analysis: Methods and Protocols; Gužvić, M., Ed.; Springer: New York, NY, USA, 2024; pp. 11–42. [Google Scholar] [CrossRef]
- van Dalum, G.; Stam, G.J.; Scholten, L.F.A.; Mastboom, W.J.B.; Vermes, I.; Tibbe, A.G.J.; de Groot, M.R.; Terstappen, L.W.M.M. Importance of circulating tumor cells in newly diagnosed colorectal cancer. Int. J. Oncol. 2015, 46, 1361–1368. [Google Scholar] [CrossRef]
- Uen, Y.H.; Lin, S.R.; Wu, D.C.; Su, Y.C.; Wu, J.Y.; Cheng, T.L.; Chi, C.W.; Wang, J.W. Prognostic Significance of Multiple Molecular Markers for Patients with Stage II Colorectal Cancer Undergoing Curative Resection. Ann. Surg. 2007, 246, 1040–1046. [Google Scholar] [CrossRef]
- Yu, J.H.; Wang, D.; Jin, L.; Wang, J.; Zhao, X.M.; Wu, G.K.; Yao, H.W.; Yang, Y.C.; Zhang, Z.T. Utility of circulating tumor cells in stage II colorectal cancer patients undergoing curative resection. Transl. Cancer Res. 2020, 9, 1487–1494. [Google Scholar] [CrossRef]
- Lin, C.C.; Yang, C.Y.; Hung, T.C.; Wang, C.H.; Wei, S.W.; Schiro, P.; Tseng, J.Y.; Lin, C.H.; Jiang, J.K. MiSelect R System: The validation of a new detection system of CTCs and their correlation with prognosis in non-metastatic CRC patients. Sci. Rep. 2023, 13, 4773. [Google Scholar] [CrossRef] [PubMed]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the Serum of Cancer Patients and the Effect of Therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Andersen, L.; Kisistók, J.; Henriksen, T.V.; Bramsen, J.B.; Reinert, T.; Øgaard, N.; Mattesen, T.B.; Birkbak, N.J.; Andersen, C.L. Exploring the biology of ctDNA release in colorectal cancer. Eur. J. Cancer 2024, 207, 114186. [Google Scholar] [CrossRef] [PubMed]
- Torresan, S.; de Scordilli, M.; Bortolot, M.; Di Nardo, P.; Foltran, L.; Fumagalli, A.; Guardascione, M.; Ongaro, E.; Puglisi, F. Liquid biopsy in colorectal cancer: Onward and upward. Crit. Rev. Oncol./Hematol. 2024, 194, 104242. [Google Scholar] [CrossRef]
- Lianidou, E. Detection and relevance of epigenetic markers on ctDNA: Recent advances and future outlook. Mol. Oncol. 2021, 15, 1683–1700. [Google Scholar] [CrossRef]
- Markou, A.; Tzanikou, E.; Lianidou, E. The potential of liquid biopsy in the management of cancer patients. Semin. Cancer Biol. 2022, 84, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Kasprzak, A.; Adamek, A. The Neuropeptide System and Colorectal Cancer Liver Metastases: Mechanisms and Management. Int. J. Mol. Sci. 2020, 21, 3494. [Google Scholar] [CrossRef]
- Mishra, M.; Ahmed, R.; Das, D.K.; Pramanik, D.D.; Dash, S.K.; Pramanik, A. Recent Advancements in the Application of Circulating Tumor DNA as Biomarkers for Early Detection of Cancers. ACS Biomater. Sci. Eng. 2024, 10, 4740–4756. [Google Scholar] [CrossRef]
- Raunkilde, L.; Hansen, T.F.; Andersen, R.F.; Mayland Havelund, B.; Brenner Thomsen, C.; Jensen, L.H. NPY Gene Methylation in Circulating Tumor DNA as an Early Biomarker for Treatment Effect in Metastatic Colorectal Cancer. Cancers 2022, 14, 4459. [Google Scholar] [CrossRef] [PubMed]
- Zandonadi, F.S.; Santa Cruz, E.C.; Korvala, J. New SDC function prediction based on protein-protein interaction using bioinformatics tools. Comput. Biol. Chem. 2019, 83, 107087. [Google Scholar] [CrossRef]
- Sun, J.; Zheng, M.-Y.; Li, Y.-W.; Zhang, S.-W. Structure and function of Septin 9 and its role in human malignant tumors. World J. Gastrointest. Oncol. 2020, 12, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Y.; Zhang, D.; Xiong, X.; Hao, T.; Zhong, L.; Zhao, Y. Diagnostic accuracy of DNA-based SDC2 methylation test in colorectal cancer screening: A meta-analysis. BMC Gastroenterol. 2022, 22, 314. [Google Scholar] [CrossRef]
- Zhang, J.; Li, C.; An, Y.; Wang, B.; Liang, G. Comparative analysis of SDC2 and SEPT9 methylation tests in the early detection of colorectal cancer: A systematic review and meta-analysis. Front. Med. 2024, 11, 1460233. [Google Scholar] [CrossRef]
- Chan, H.T.; Nagayama, S.; Otaki, M.; Chin, Y.M.; Fukunaga, Y.; Ueno, M.; Nakamura, Y.; Low, S.K. Tumor-informed or tumor-agnostic circulating tumor DNA as a biomarker for risk of recurrence in resected colorectal cancer patients. Front. Oncol. 2023, 12, 1055968. [Google Scholar] [CrossRef]
- Nakamura, Y.; Watanabe, J.; Akazawa, N.; Hirata, K.; Kataoka, K.; Yokota, M.; Kato, K.; Kotaka, M.; Kagawa, Y.; Yeh, K.H.; et al. ctDNA-based molecular residual disease and survival in resectable colorectal cancer. Nat. Med. 2024, 30, 3272–3283. [Google Scholar] [CrossRef] [PubMed]
- Kasi, P.M.; Aushev, V.N.; Ensor, J.; Langer, N.; Wang, C.G.; Cannon, T.L.; Berim, L.D.; Feinstein, T.; Grothey, A.; McCollom, J.W.; et al. Circulating tumor DNA (ctDNA) for informing adjuvant chemotherapy (ACT) in stage II/III colorectal cancer (CRC): Interim analysis of BESPOKE CRC study. J. Clin. Oncol. 2024, 42, 9. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Lahouel, K.; Lo, S.N.; Wang, Y.; Kosmider, S.; Wong, R.; Shapiro, J.; Lee, M.; Harris, S.; et al. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N. Engl. J. Med. 2022, 386, 2261–2272. [Google Scholar] [CrossRef]
- Jácome, A.A.; Johnson, B. Minimal Residual Disease in Colorectal Cancer: Are We Finding the Needle in a Haystack? Cells 2023, 12, 1068. [Google Scholar] [CrossRef]
- Lin, Y.; Zhao, W.; Pu, R.; Lv, Z.; Xie, H.; Li, Y.; Zhang, Z. Long non-coding RNAs as diagnostic and prognostic biomarkers for colorectal cancer. Oncol. Lett. 2024, 28, 486. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Shan, Z.; Duan, S. Harnessing extracellular vesicles using liquid biopsy for cancer diagnosis and monitoring: Highlights from AACR Annual Meeting 2024. J. Hematol. Oncol. 2024, 17, 55. [Google Scholar] [CrossRef] [PubMed]
- Najafi, S.; Majidpoor, J.; Mortezaee, K. Liquid biopsy in colorectal cancer. Clin. Chim. Acta 2024, 553, 117674. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Xi, N.; Han, Z.; Luo, W.; Shen, J.; Wang, S.; Li, J.; Guo, Z.; Cheng, H. The Role of Liquid Biopsy Analytes in Diagnosis, Treatment and Prognosis of Colorectal Cancer. Front. Endocrinol. 2022, 13, 875442. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Bakhsh, T.; Alhazmi, S.; Farsi, A.; Yusuf, A.S.; Alharthi, A.; Qahl, S.H.; Alghamdi, A.; Alzahrani, F.A.; Elgaddar, O.H.; Ibrahim, M.A.; et al. Molecular detection of exosomal miRNAs of blood serum for prognosis of colorectal cancer. Sci. Rep. 2024, 14, 8902. [Google Scholar] [CrossRef]
- Shakerian, N.; Darzi-Eslam, E.; Afsharnoori, F.; Bana, N.; Ghahroodi, F.N.; Tarin, M.; Mard-soltani, M.; Khalesi, B.; Hashemi, Z.S.; Khalili, S. Therapeutic and diagnostic applications of exosomes in colorectal cancer. Med. Oncol. 2024, 41, 203. [Google Scholar] [CrossRef]
- Wu, Y.; Xiao, Y.; Ding, Y.; Ran, R.; Wei, K.; Tao, S.; Mao, H.; Wang, J.; Pang, S.; Shi, J.; et al. Colorectal cancer cell-derived exosomal miRNA-372-5p induces immune escape from colorectal cancer via PTEN/AKT/NF-κB/PD-L1 pathway. Int. Immunopharmacol. 2024, 143, 113261. [Google Scholar] [CrossRef]
- Pan, B.; Qin, J.; Liu, X.; He, B.; Wang, X.; Pan, Y.; Sun, H.; Xu, T.; Xu, M.; Chen, X.; et al. Identification of Serum Exosomal hsa-circ-0004771 as a Novel Diagnostic Biomarker of Colorectal Cancer. Front. Genet. 2019, 10, 1096. [Google Scholar] [CrossRef]
- Jia, Z.; An, J.; Liu, Z.; Zhang, F. Non-Coding RNAs in Colorectal Cancer: Their Functions and Mechanisms. Front. Oncol. 2022, 12, 783079. [Google Scholar] [CrossRef]
- Glass, S.E.; Coffey, R.J. Recent Advances in the Study of Extracellular Vesicles in Colorectal Cancer. Gastroenterology 2022, 163, 1188–1197. [Google Scholar] [CrossRef] [PubMed]
- Heck, K.A.; Lindholm, H.T.; Niederdorfer, B.; Tsirvouli, E.; Kuiper, M.; Flobak, A.; Lægreid, A.; Thommesen, L. Characterisation of Colorectal Cancer Cell Lines through Proteomic Profiling of Their Extracellular Vesicles. Proteomes 2023, 11, 3. [Google Scholar] [CrossRef]
- Zheng, R.; Zhang, K.; Tan, S.; Gao, F.; Zhang, Y.; Xu, W.; Wang, H.; Gu, D.; Zhu, L.; Li, S.; et al. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3–eIF3h interaction. Mol. Cancer 2022, 21, 49. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Mi, Y.; Zheng, B.; Wei, P.; Gu, Y.; Zhang, Z.; Xu, Y.; Cai, S.; Li, X.; Li, D. Highly-metastatic colorectal cancer cell released miR-181a-5p-rich extracellular vesicles promote liver metastasis by activating hepatic stellate cells and remodelling the tumour microenvironment. J. Extracell. Vesicles 2022, 11, e12186. [Google Scholar] [CrossRef] [PubMed]
- Najafi, S.; Asemani, Y.; Majidpoor, J.; Mahmoudi, R.; Aghaei-Zarch, S.M.; Mortezaee, K. Tumor-educated platelets. Clin. Chim. Acta 2024, 552, 117690. [Google Scholar] [CrossRef]
- Plantureux, L.; Mège, D.; Crescence, L.; Carminita, E.; Robert, S.; Cointe, S.; Brouilly, N.; Ezzedine, W.; Dignat-George, F.; Dubois, C. The Interaction of Platelets with Colorectal Cancer Cells Inhibits Tumor Growth but Promotes Metastasis. Cancer Res. 2020, 80, 291–303. [Google Scholar] [CrossRef]
- Antunes-Ferreira, M.; D’Ambrosi, S.; Arkani, M.; Post, E.; In ‘t Veld, S.G.J.G.; Ramaker, J.; Zwaan, K.; Kucukguzel, E.D.; Wedekind, L.E.; Griffioen, A.W. Tumor-educated platelet blood tests for Non-Small Cell Lung Cancer detection and management. Sci. Rep. 2023, 13, 9359. [Google Scholar] [CrossRef] [PubMed]
- Giannakeas, V.; Kotsopoulos, J.; Cheung, M.C.; Rosella, L.; Brooks, J.D.; Lipscombe, L.; Akbari, M.R.; Austin, P.C.; Narod, S.A. Analysis of Platelet Count and New Cancer Diagnosis Over a 10-Year Period. JAMA Netw. Open 2022, 5, e2141633. [Google Scholar] [CrossRef]
- Huang, J.; Swieringa, F.; Solari, F.A.; Provenzale, I.; Grassi, L.; De Simone, I.; Baaten, C.C.F.M.J.; Cavill, R.; Sickmann, A.; Frontini, M.; et al. Assessment of a complete and classified platelet proteome from genome-wide transcripts of human platelets and megakaryocytes covering platelet functions. Sci. Rep. 2021, 11, 12358. [Google Scholar] [CrossRef]
- Tao, X.-Y.; Li, Q.-Q.; Zeng, Y. Clinical application of liquid biopsy in colorectal cancer: Detection, prediction, and treatment monitoring. Mol. Cancer 2024, 23, 145. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Chen, X.; Song, Y.; Huang, X.; Chen, Q.; Lv, X.; Gao, P.; Wang, Z. The platelet to lymphocyte ratio is a potential inflammatory marker predicting the effects of adjuvant chemotherapy in patients with stage II colorectal cancer. BMC Cancer 2021, 21, 792. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zheng, X.; Wu, M.; Zhang, F.; Xu, S.; Wang, X.; Song, M.; You, C.; Zhang, T.; Jiang, M.; et al. Development and validation of postoperative and preoperative platelets ratio (PPR) to predict the prognosis of patients undergoing surgery for colorectal cancer: A dual-center retrospective cohort study. Cancer Med. 2022, 12, 111–121. [Google Scholar] [CrossRef]
- Xu, L.; Li, X.; Li, X.; Wang, X.; Ma, Q.; She, D.; Lu, X.; Zhang, J.; Yang, Q.; Lei, S.; et al. RNA profiling of blood platelets noninvasively differentiates colorectal cancer from healthy donors and noncancerous intestinal diseases: A retrospective cohort study. Genome Med. 2022, 14, 26. [Google Scholar] [CrossRef]
- Raza, A.; Khan, A.Q.; Inchakalody, V.P.; Mestiri, S.; Yoosuf, Z.S.K.M.; Bedhiafi, T.; El-Ella, D.M.A.; Taib, N.; Hydrose, S.; Akbar, S.; et al. Dynamic liquid biopsy components as predictive and prognostic biomarkers in colorectal cancer. J. Exp. Clin. Cancer Res. 2022, 41, 99. [Google Scholar] [CrossRef]
- Tsilimigras, D.I.; Ntanasis-Stathopoulos, I.; Pawlik, T.M. Liquid Biopsies in Colorectal Liver Metastases: Towards the Era of Precision Oncologic Surgery. Cancers 2022, 14, 4237. [Google Scholar] [CrossRef]
- Tol, J.; Koopman, M.; Miller, M.C.; Tibbe, A.; Cats, A.; Creemers, G.J.; Vos, A.H.; Nagtegaal, I.D.; Terstappen, L.W.; Punt, C.J. Circulating tumour cells early predict progression-free and overall survival in advanced colorectal cancer patients treated with chemotherapy and targeted agents. Ann. Oncol. 2010, 21, 1006–1012. [Google Scholar] [CrossRef]
- Tan, Y.; Wu, H. The significant prognostic value of circulating tumor cells in colorectal cancer: A systematic review and meta-analysis. Curr. Probl. Cancer 2018, 42, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Reinert, T.; Schøler, L.V.; Thomsen, R.; Tobiasen, H.; Vang, S.; Nordentoft, I.; Lamy, P.; Kannerup, A.S.; Mortensen, F.V.; Stribolt, K.; et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016, 65, 625–634. [Google Scholar] [CrossRef]
- Vidal, J.; Casadevall, D.; Bellosillo, B.; Pericay, C.; Garcia-Carbonero, R.; Losa, F.; Layos, L.; Alonso, V.; Capdevila, J.; Gallego, J.; et al. Clinical Impact of Presurgery Circulating Tumor DNA after Total Neoadjuvant Treatment in Locally Advanced Rectal Cancer: A Biomarker Study from the GEMCAD 1402 Trial. Clin. Cancer Res. 2021, 27, 2890–2898. [Google Scholar] [CrossRef]
- Mason, M.C.; Tzeng, C.D.; Tran Cao, H.S.; Aloia, T.A.; Newhook, T.E.; Overman, M.J.; Kopetz, S.E.; Vauthey, J.N.; Chun, Y.S. Preliminary Analysis of Liquid Biopsy After Hepatectomy for Colorectal Liver Metastases. J. Am. Coll. Surg. 2021, 233, 82–89.e1. [Google Scholar] [CrossRef] [PubMed]
- Barták, B.K.; Fodor, T.; Kalmár, A.; Nagy, Z.B.; Zsigrai, S.; Szigeti, K.A.; Valcz, G.; Igaz, P.; Dank, M.; Takács, I.; et al. A Liquid Biopsy-Based Approach for Monitoring Treatment Response in Post-Operative Colorectal Cancer Patients. Int. J. Mol. Sci. 2022, 23, 3774. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, G.; Burotto, M.; Marcelain, K.; González-Montero, J. Liquid biopsy to detect resistance mutations against anti-epidermal growth factor receptor therapy in metastatic colorectal cancer. World J. Gastrointest. Oncol. 2022, 14, 1654–1664. [Google Scholar] [CrossRef]
- Mauri, G.; Vitiello, P.P.; Sogari, A.; Crisafulli, G.; Sartore-Bianchi, A.; Marsoni, S.; Siena, S.; Bardelli, A. Liquid biopsies to monitor and direct cancer treatment in colorectal cancer. Br. J. Cancer 2022, 127, 394–407. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, E.; Martini, G.; Famiglietti, V.; Troiani, T.; Napolitano, S.; Pietrantonio, F.; Ciardiello, D.; Terminiello, M.; Borrelli, C.; Vitiello, P.P.; et al. Cetuximab Rechallenge Plus Avelumab in Pretreated Patients With RAS Wild-type Metastatic Colorectal Cancer: The Phase 2 Single-Arm Clinical CAVE Trial. JAMA Oncol. 2021, 7, 1529–1535. [Google Scholar] [CrossRef]
- Sartore-Bianchi, A.; Pietrantonio, F.; Lonardi, S.; Mussolin, B.; Rua, F.; Crisafulli, G.; Bartolini, A.; Fenocchio, E.; Amatu, A.; Manca, P.; et al. Circulating tumor DNA to guide rechallenge with panitumumab in metastatic colorectal cancer: The phase 2 CHRONOS trial. Nat. Med. 2022, 28, 1612–1618. [Google Scholar] [CrossRef]
- Kotsiliti, E. Liquid biopsy guides anti-EGFR rechallenge in metastatic CRC. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 624. [Google Scholar] [CrossRef]
- Szász, I.; Kiss, T.; Mokánszki, A.; Koroknai, V.; Deák, J.; Patel, V.; Jámbor, K.; Ádány, R.; Balázs, M. Identification of liquid biopsy-based mutations in colorectal cancer by targeted sequencing assays. Mol. Cell. Probes 2023, 67, 101888. [Google Scholar] [CrossRef]
- Bachet, J.B.; Bouché, O.; Taieb, J.; Dubreuil, O.; Garcia, M.L.; Meurisse, A.; Normand, C.; Gornet, J.M.; Artru, P.; Louafi, S.; et al. RAS mutation analysis in circulating tumor DNA from patients with metastatic colorectal cancer: The AGEO RASANC prospective multicenter study. Ann. Oncol. 2018, 29, 1211–1219. [Google Scholar] [CrossRef]
- Siena, S.; Sartore-Bianchi, A.; Garcia-Carbonero, R.; Karthaus, M.; Smith, D.; Tabernero, J.; Van Cutsem, E.; Guan, X.; Boedigheimer, M.; Ang, A.; et al. Dynamic molecular analysis and clinical correlates of tumor evolution within a phase II trial of panitumumab-based therapy in metastatic colorectal cancer. Ann. Oncol. 2018, 29, 119–126. [Google Scholar] [CrossRef]
- Normanno, N.; Esposito Abate, R.; Lambiase, M.; Forgione, L.; Cardone, C.; Iannaccone, A.; Sacco, A.; Rachiglio, A.M.; Martinelli, E.; Rizzi, D.; et al. RAS testing of liquid biopsy correlates with the outcome of metastatic colorectal cancer patients treated with first-line FOLFIRI plus cetuximab in the CAPRI-GOIM trial. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018, 29, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Verschoor, N.; Bos, M.K.; Oomen-de Hoop, E.; Martens, J.W.M.; Sleijfer, S.; Jager, A.; Beije, N. A review of trials investigating ctDNA-guided adjuvant treatment of solid tumors: The importance of trial design. Eur. J. Cancer 2024, 207, 114159. [Google Scholar] [CrossRef] [PubMed]
- Kotani, D.; Oki, E.; Nakamura, Y.; Yukami, H.; Mishima, S.; Bando, H.; Shirasu, H.; Yamazaki, K.; Watanabe, J.; Kotaka, M.; et al. Molecular residual disease and efficacy of adjuvant chemotherapy in patients with colorectal cancer. Nat. Med. 2023, 29, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Heitzer, E. ctDNA-guided adjuvant chemotherapy for colorectal cancer-ready for prime time? Cancer Cell 2022, 40, 911–913. [Google Scholar] [CrossRef]
- Vacante, M.; Ciuni, R.; Basile, F.; Biondi, A. The Liquid Biopsy in the Management of Colorectal Cancer: An Overview. Biomedicines 2020, 8, 308. [Google Scholar] [CrossRef]
| Clinical Trial ID | Population | Clinical Outcome | Liquid Biopsy Method | Status | Estimated Number of Participants | Sponsor |
|---|---|---|---|---|---|---|
| NCT04852653 (RECC-EV) | Adenocarcinoma of the rectum | Response to neoadjuvant treatment by chemotherapy alone or by radiochemotherapy | Onco-exosomes and/or exo-DNA as a response to neoadjuvant treatment after 6 months follow up | Recruiting | 40 | University Hospital, Bordeaux, France |
| NCT06076811 (DANISH.MRD) | Stages I–III adenocarcinoma receiving intensive surgery | 3-year disease-free survival | ctDNA | Recruiting | 1600 | University of Aarhus, Denmark |
| NCT06111105 (GUIDE.MRD-01-CRC) | Stage III adenocarcinoma with received curative intention surgery | The 3-year recurrence-free survival | ctDNA | Recruiting | 440 | Guardant Health, Inc. |
| NCT05935384 (SIBYL) | Stages III–IV colorectal, non-small-cell lung, and breast cancer | Disease progression, RECIST, PFS, Lead Time | ctDNA (Guardant 360) | Recruiting | 440 | Guardant Health, Inc. |
|
NCT04776655
(LIBImAb) | RASmut at liquid biopsy and RASwt on tissue | Progression-free survival | ctDNA | Unknown Status | 280 | Azienda Unità Sanitaria Locale Reggio Emilia, Italy |
|
NCT05031975
(ERASE-TMZ) | Stage III or T4N0 stage II colon cancer | The activity in terms of seroreversion of TEMIRI consolidation regimen administered to patients, 2 years disease free | ctDNA | Unknown Status | 35 | Fondazione IRCCS Istituto Nazionale dei Tumori, Milano, Italy |
| NCT04259944 (PEGASUS) | Stage III and T4N0 stage II colon cancer | Number of post-surgery and post-adjuvant false negative cases | ctDNA | Active, not recruiting | 140 | IFOM ETS-The AIRC Institute of Molecular Oncology, Milan, Italy |
| NCT03401957 | Metastatic with cetuximab-based regimen as first-line setting | Time duration between the start of cetuximab treatment and detection of RAS mutation | ctDNA | Unknown status | 120 | National Health Research Institutes, Taiwan |
| NCT04258137 (COPE) | Locally advanced/unresectable and/or metastatic solid tumor | Overall survival | ctDNA | Recruiting | 332 | Institut Bergonié, Bordeuax, France |
| NCT04425239 (Phase II) (IMPROVE) | Metastatic colorectal, IMPROVE trial | Progression-free survival up to one year after randomization | cfDNA | Completed | 151 | National Cancer Institute, Naples, Italy |
| NCT05524012 (PRIMO) | Locally advanced rectal cancer stages II–III with neoadjuvant chemoradiotherapy (CRT) | Tumor regression grading (TRG) | CTC | Recruiting | 50 | Jena University Hospital, Germany |
| NCT04554836 (Phase II) (MoLiMoR) | MoLiMoR, stage IV adenocarcinoma with FOLFIRI-based first-line therapy with or without cetuximab | Progression-free survival | ctDNA | Active, not recruiting | 144 | TheraOp, Germany |
| NCT06509126 (Phase III) (IMPROVE-2) | Metastatic colorectal cancer panitumumab plus Folfiri (IMPROVE-2) | Time to treatment failure | ctDNA | Recruiting | 500 | National Cancer Institute, Naples, Italy |
| NCT03699410 (observational) (LIBReCa) | Locally advanced rectal cancer (T3 or T4 and/or N+) requiring long-course nCRT | Negative prognostic value | ctDNA | Terminated | 25 | Dimitri Christoforidis, Ente Ospedaliero Cantonale, Bellinzona, Italy |
| NCT03751176 (Phase II) (BEYOND) | Second-line treatment after first-line panitumumab and FOLFOX in wild-type mCRC | Progression-free survival (PFS), conversion rate of RAS/BRAF status after second-line treatment | ctDNA | Unknown Status | 31 | Grupo Espanol Multidisciplinario del Cancer Digestivo, Spain |
| NCT06287814 (observational) (FRENCH.MRD.CRC) | Stages I–III colon and rectal cancer, curative intent resection surgery | MRD assessment | ctDNA | Not yet recruiting | 70 | University Hospital, Montpellier, France |
| NCT06287723 (observational) (FRENCH.MRD.CRLM) | Stage IV liver metastasis | Three-year disease-free survival | ctDNA | Not yet recruiting | 30 | University Hospital, Montpellier, France |
| NCT04735900 (Interventional) (FOLICOLOR) | Metastatic colorectal cancer patients receiving first-line FOLFOX/FOLFIRI and panitumumab | Optimization of the cut-off value for NPY methylation in liquid biopsies | ctDNA | Unknown status | 60 | University Hospital, Antwerp, Belgium |
| NCT04232891 (observational) (REDCLOUD) | Adenocarcinoma of the colon or rectum with synchronous metastatic disease (localized in liver or in liver and lung) eligible for surgery immediately or after neo-adjuvant treatment | Relapse rate in MRD+ patients | cfDNA | Unknown status | 141 | Fondazione del Piemonte per l’Oncologia, Italy |
| NCT04776837 | Metastatic colorectal, pancreatobiliary, or esophagogastric cancer | Treatment response at first scan | ctDNA | Completed | 200 | Massachusetts General Hospital, USA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liatsou, E.; Kollias, I.; Trapali, M.; Tsilimigras, D.I.; Gavriatopoulou, M.; Ntanasis-Stathopoulos, I. Liquid Biopsies in the Early Diagnosis, Prognosis, and Tailored Treatment of Colorectal Cancer. Cancers 2025, 17, 927. https://doi.org/10.3390/cancers17060927
Liatsou E, Kollias I, Trapali M, Tsilimigras DI, Gavriatopoulou M, Ntanasis-Stathopoulos I. Liquid Biopsies in the Early Diagnosis, Prognosis, and Tailored Treatment of Colorectal Cancer. Cancers. 2025; 17(6):927. https://doi.org/10.3390/cancers17060927
Chicago/Turabian StyleLiatsou, Efstathia, Ioannis Kollias, Maria Trapali, Diamantis I. Tsilimigras, Maria Gavriatopoulou, and Ioannis Ntanasis-Stathopoulos. 2025. "Liquid Biopsies in the Early Diagnosis, Prognosis, and Tailored Treatment of Colorectal Cancer" Cancers 17, no. 6: 927. https://doi.org/10.3390/cancers17060927
APA StyleLiatsou, E., Kollias, I., Trapali, M., Tsilimigras, D. I., Gavriatopoulou, M., & Ntanasis-Stathopoulos, I. (2025). Liquid Biopsies in the Early Diagnosis, Prognosis, and Tailored Treatment of Colorectal Cancer. Cancers, 17(6), 927. https://doi.org/10.3390/cancers17060927









