Loss of ING3 in the Prostate Leads to Activation of DNA Damage Repair Markers
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Care and Husbandry
2.2. Generation of the Ing3-LacZ Reporter and Conditional Ing3 Knockout Animals
2.3. PCR Genotyping
2.4. Digital PCR
2.5. SDS-PAGE and Western Blotting
2.6. Immunohistochemical Staining and Immunofluorescence
2.7. X-Gal Staining of Cryosections
2.8. Whole-Mount X-Gal Staining
3. Results
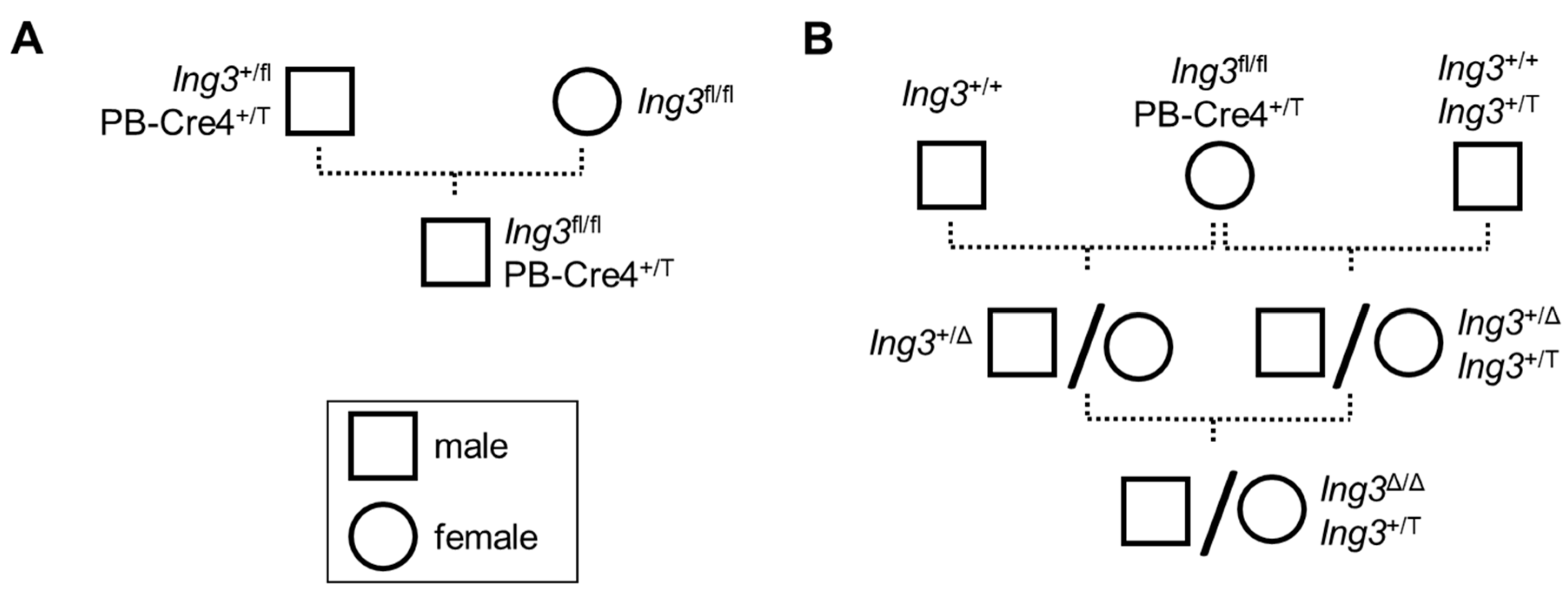
3.1. Animal Breeding
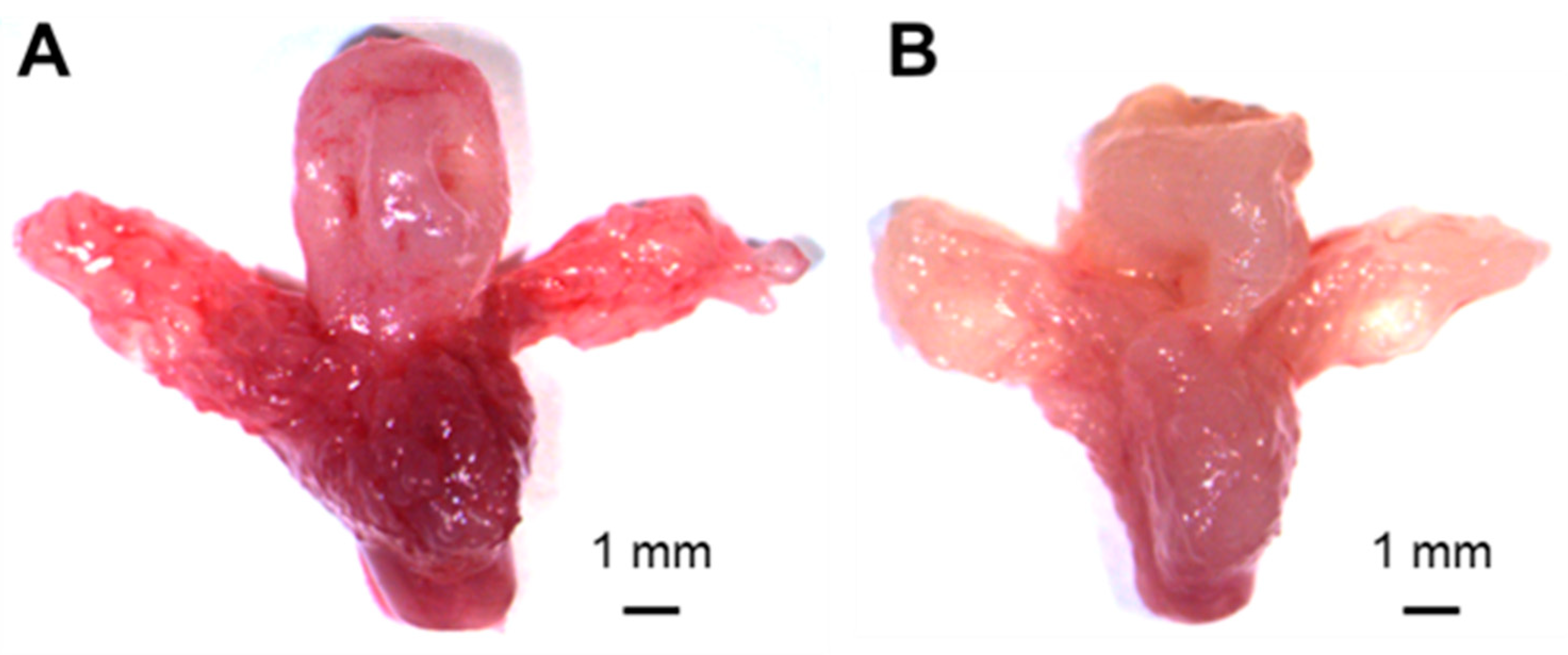
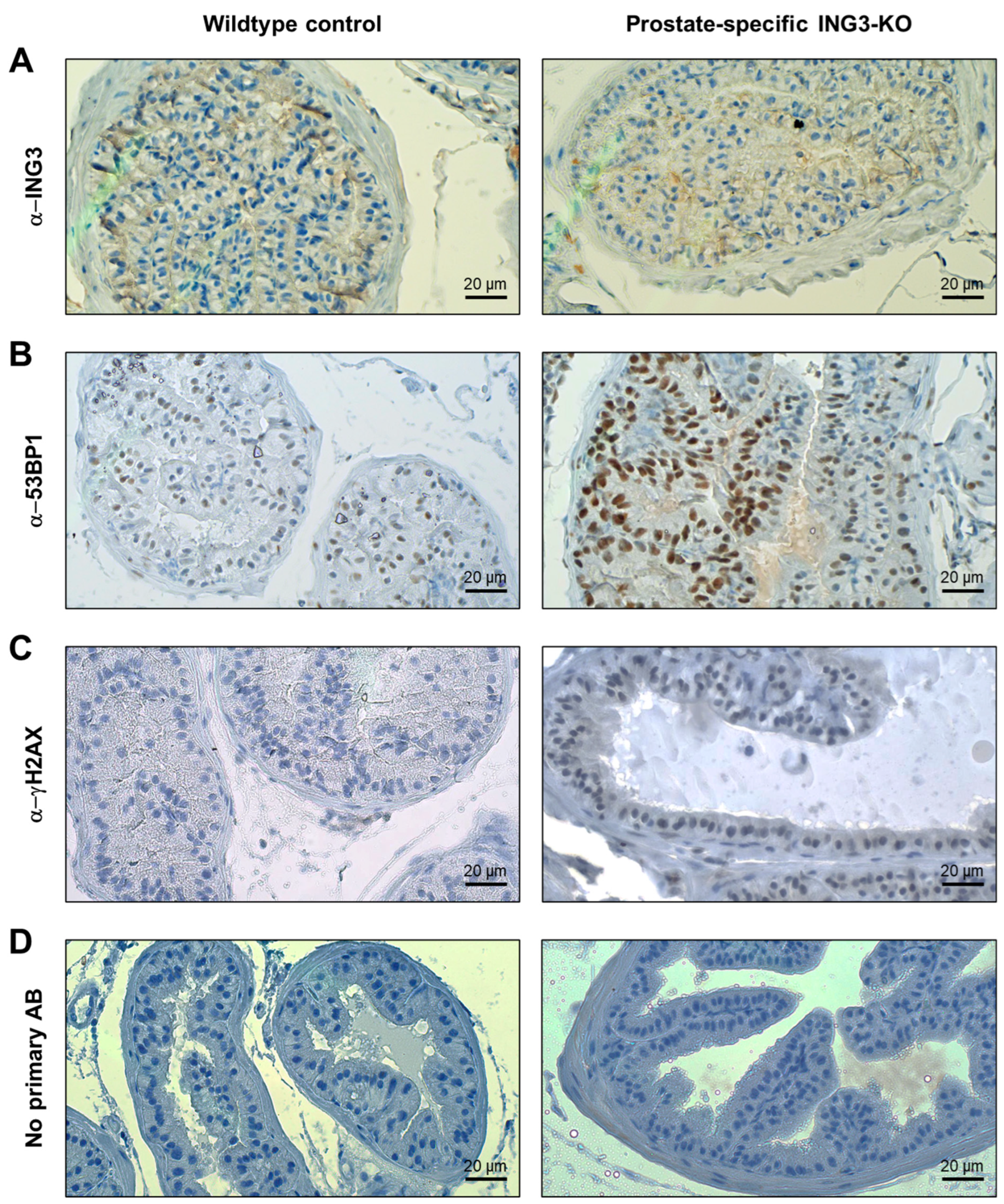
3.2. Immunohistochemistry of Prostate Tissues
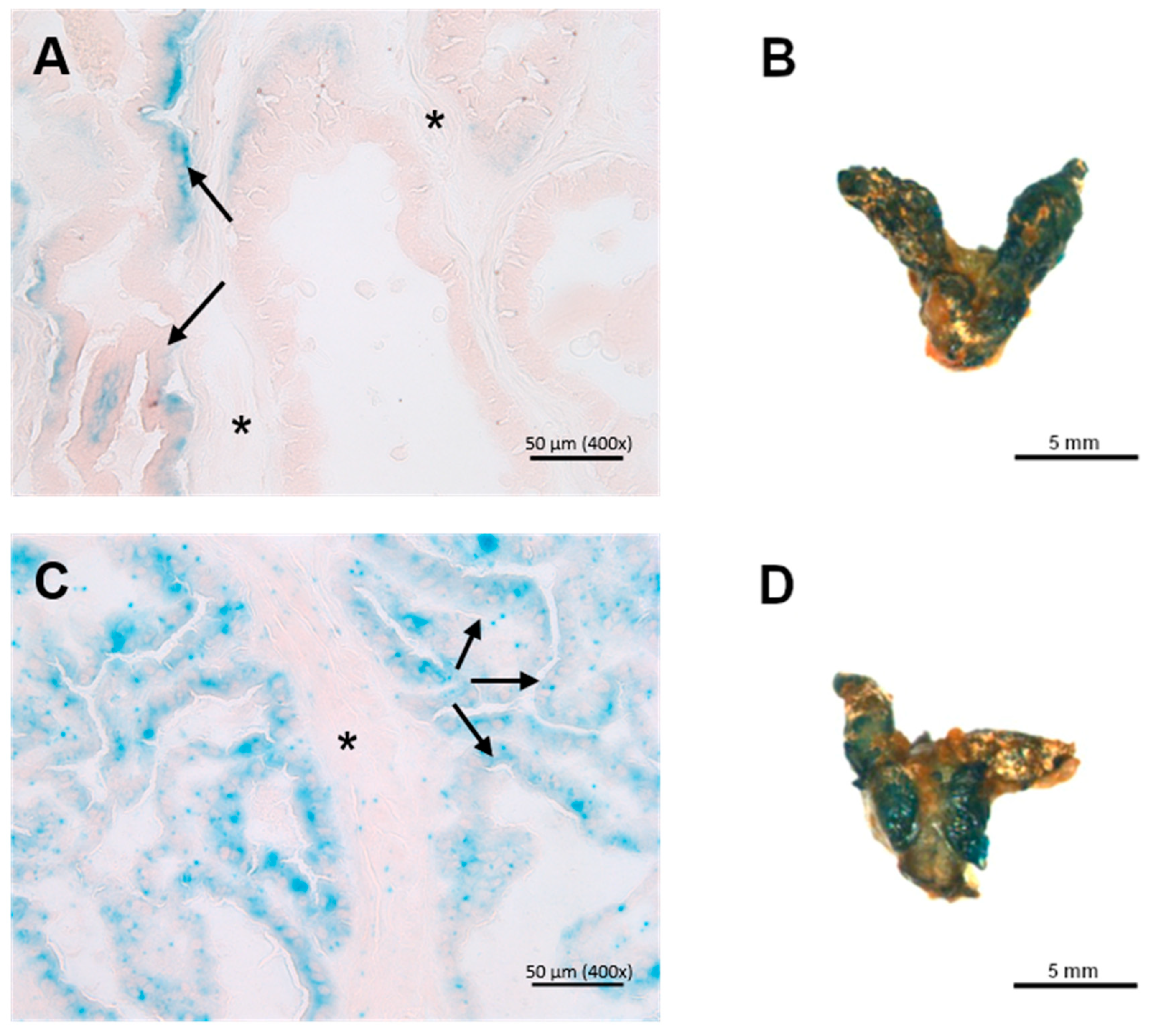
3.3. X-Gal Staining of Ing3-LacZ Reporter Mice
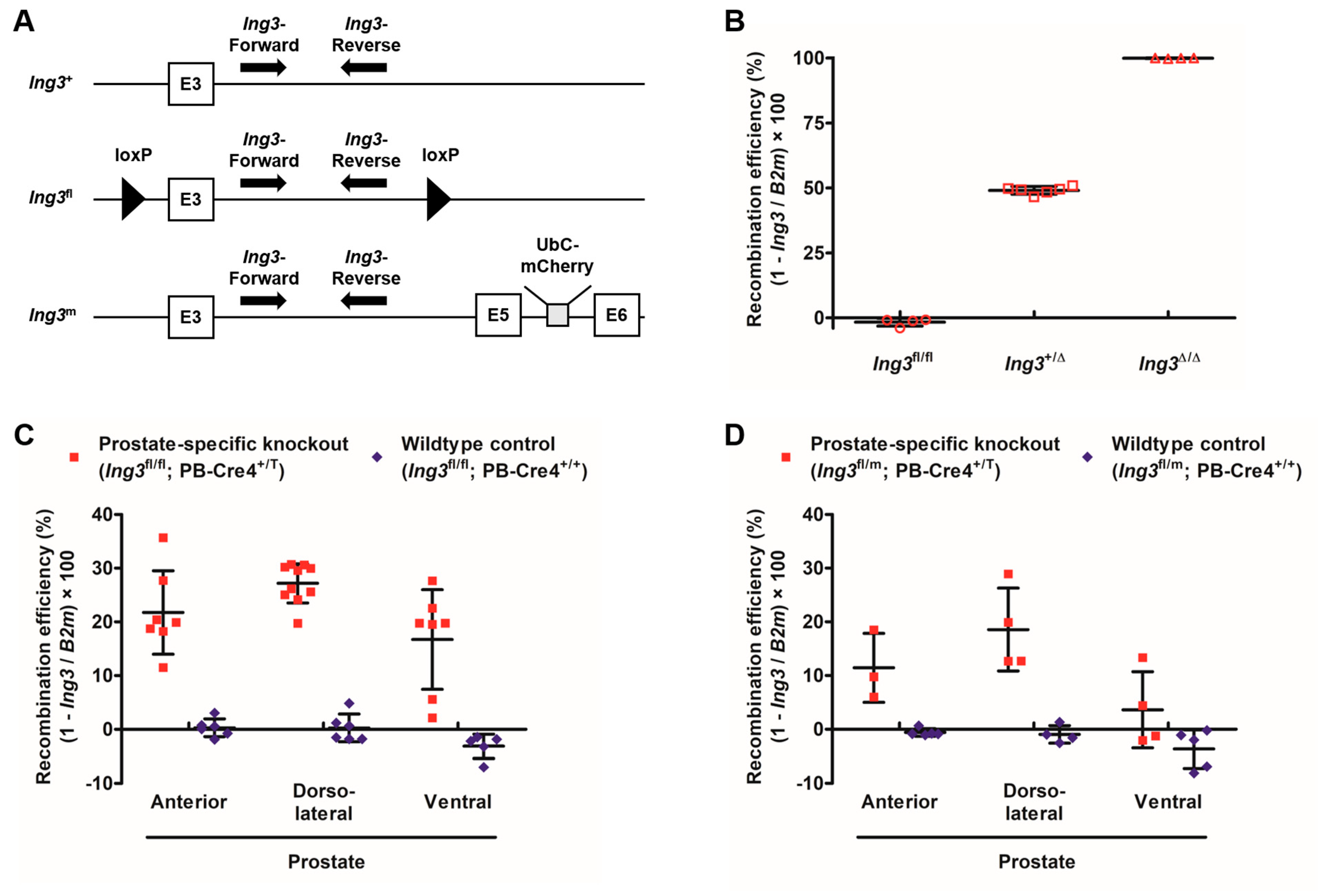
3.4. Digital PCR Calibration and Recombination Efficiency Assessment in the Prostate
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AP | Anterior prostate |
| AR | Androgen receptor |
| ATM | Ataxia-telangiectasia mutated |
| B2m | Beta-2-microglobulin |
| BSA | Bovine serum albumin |
| BSB | Basis staining buffer |
| Cdh1 | E-cadherin |
| CDKN1a | Cyclin-dependent kinase inhibitor 1A |
| Cre | Cyclization recombinase |
| DAB | 3,3′-Diaminobenzidine |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DLP | Dorsolateral prostate |
| DMSO | Dimethyl sulfoxide |
| dPCR | Digital polymerase chain reaction |
| EDTA | Ethylenediaminetetraacetic acid |
| EGTA | Ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid |
| EMT | Epithelial mesenchymal transition |
| FLP | Flippase |
| FRT | Flippase recognition target |
| HAT | Histone acetyltransferase |
| HRP | Horseradish peroxidase |
| IHC | Immunohistochemistry |
| ING3 | Inhibitor of growth 3 |
| lacZ | β-galactosidase gene |
| LoxP | Locus of X-over P1 |
| mRNA | Messenger RNA |
| NBS1 | Nibrin |
| NuA4 | Nucleosome acetyltransferase of histone H4 |
| PB | Probasin |
| PBS | Phosphate-buffered saline |
| PCR | Polymerase chain reaction |
| PHD PIN | Plant homeodomain Prostatic intraepithelial neoplasia |
| Pten | Phosphatase and tensin homologue |
| SPB | Sodium phosphate buffer |
| TBST | Tris-buffered saline with Tween 20 |
| VP | Ventral prostate |
| X-gal | 5-Bromo-4-chloro-3-indolyl β-D-galactopyranoside |
References
- He, G.H.Y.; Helbing, C.C.; Wagner, M.J.; Sensen, C.W.; Riabowol, K. Phylogenetic Analysis of the ING Family of PHD Finger Proteins. Mol. Biol. Evol. 2005, 22, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Garkavtsev, I.; Kazarov, A.; Gudkov, A.; Riabowol, K. Suppression of the Novel Growth Inhibitor P33ING1 Promotes Neoplastic Transformation. Nat. Genet. 1996, 14, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Hara, Y.; Riabowol, K.T. Different HATS of the ING1 Gene Family. Trends Cell Biol. 2002, 12, 532–538. [Google Scholar] [CrossRef]
- Nagashima, M.; Shiseki, M.; Miura, K.; Hagiwara, K.; Linke, S.P.; Pedeux, R.; Wang, X.W.; Yokota, J.; Riabowol, K.; Harris, C.C. DNA Damage-Inducible Gene P33ING2 Negatively Regulates Cell Proliferation through Acetylation of P53. Proc. Natl. Acad. Sci. USA 2001, 98, 9671–9676. [Google Scholar] [CrossRef]
- Nagashima, M.; Shiseki, M.; Pedeux, R.M.; Okamura, S.; Kitahama-Shiseki, M.; Miura, K.; Yokota, J.; Harris, C.C. A Novel PHD-Finger Motif Protein, P47ING3, Modulates P53-Mediated Transcription, Cell Cycle Control, and Apoptosis. Oncogene 2003, 22, 343–350. [Google Scholar] [CrossRef]
- Shiseki, M.; Nagashima, M.; Pedeux, R.M.; Kitahama-Shiseki, M.; Miura, K.; Okamura, S.; Onogi, H.; Higashimoto, Y.; Appella, E.; Yokota, J.; et al. P29ING4 and P28ING5 Bind to P53 and P300, and Enhance P53 Activity. Cancer Res. 2003, 63, 2373–2378. [Google Scholar] [PubMed]
- Dantas, A.; Al Shueili, B.; Yang, Y.; Nabbi, A.; Fink, D.; Riabowol, K. Biological Functions of the ING Proteins. Cancers 2019, 11, 1817. [Google Scholar] [CrossRef]
- Han, X.; Feng, X.; Rattner, J.B.; Smith, H.; Bose, P.; Suzuki, K.; Soliman, M.A.; Scott, M.S.; Burke, B.E.; Riabowol, K. Tethering by Lamin A Stabilizes and Targets the ING1 Tumour Suppressor. Nat. Cell Biol. 2008, 10, 1333–1340. [Google Scholar] [CrossRef]
- Liang, G.; Lin, J.C.Y.; Wei, V.; Yoo, C.; Cheng, J.C.; Nguyen, C.T.; Weisenberger, D.J.; Egger, G.; Takai, D.; Gonzales, F.A.; et al. Distinct Localization of Histone H3 Acetylation and H3-K4 Methylation to the Transcription Start Sites in the Human Genome. Proc. Natl. Acad. Sci. USA 2004, 101, 7357–7362. [Google Scholar] [CrossRef]
- Shi, X.; Hong, T.; Walter, K.L.; Ewalt, M.; Michishita, E.; Hung, T.; Carney, D.; Pena, P.; Lan, F.; Kaadige, M.R.; et al. ING2 PHD Domain Links Histone H3 Lysine 4 Methylation to Active Gene Repression. Nature 2006, 442, 96–99. [Google Scholar] [CrossRef]
- Doyon, Y.; Selleck, W.; Lane, W.S.; Tan, S.; Cote, J. Structural and Functional Conservation of the NuA4 Histone Acetyltransferase Complex from Yeast to Humans. Mol. Cell Biol. 2004, 24, 1884–1896. [Google Scholar] [CrossRef]
- Berger, S.L. The Complex Language of Chromatin Regulation during Transcription. Nature 2007, 447, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, H.; Bonnefin, P.; Vieyra, D.; Feng, X.; Hara, Y.; Miura, Y.; Joh, T.; Nakabayashi, H.; Vaziri, H.; Harris, C.C.; et al. ING1 Represses Transcription by Direct DNA Binding and through Effects on P53. Cancer Res. 2003, 63, 5785–5792. [Google Scholar]
- Kitahara, J.; Chiba, N.; Sakamoto, H.; Nakagawa, Y. Alteration of Gene Expressions by the Overexpression of Mitochondrial Phospholipid Hydroperoxide Glutathione Peroxidase (MtPHGPx). Gene Expr. 2003, 11, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, G. ING3 Promotes UV-Induced Apoptosis via Fas/Caspase-8 Pathway in Melanoma Cells. J. Biol. Chem. 2006, 281, 11887–11893. [Google Scholar] [CrossRef]
- Gunduz, M.; Ouchida, M.; Fukushima, K.; Ito, S.; Jitsumori, Y.; Nakashima, T.; Nagai, N.; Nishizaki, K.; Shimizu, K. Allelic Loss and Reduced Expression of the ING3, a Candidate Tumor Suppressor Gene at 7q31, in Human Head and Neck Cancers. Oncogene 2002, 21, 4462–4470. [Google Scholar] [CrossRef]
- Lu, M.; Chen, F.; Wang, Q.; Wang, K.; Pan, Q.; Zhang, X. Downregulation of Inhibitor of Growth 3 Is Correlated with Tumorigenesis and Progression of Hepatocellular Carcinoma. Oncol. Lett. 2012, 4, 47–52. [Google Scholar] [CrossRef]
- Wang, Y.; Dai, D.L.; Martinka, M.; Li, G. Prognostic Significance of Nuclear ING3 Expression in Human Cutaneous Melanoma. Clin. Cancer Res. 2007, 13, 4111–4116. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Rotte, A.; Li, G.; Chen, X.; Chen, G.; Bhandaru, M. Nuclear Localization of ING3 Is Required to Suppress Melanoma Cell Migration, Invasion and Angiogenesis. Biochem. Biophys. Res. Commun. 2020, 527, 418–424. [Google Scholar] [CrossRef]
- Nabbi, A.; Almami, A.; Thakur, S.; Suzuki, K.; Boland, D.; Bismar, T.A.; Riabowol, K. ING3 Protein Expression Profiling in Normal Human Tissues Suggest Its Role in Cellular Growth and Self-Renewal. Eur. J. Cell Biol. 2015, 94, 214–222. [Google Scholar] [CrossRef]
- Almami, A.; Hegazy, S.A.; Nabbi, A.; Alshalalfa, M.; Salman, A.; Abou-Ouf, H.; Riabowol, K.; Bismar, T.A. ING3 Is Associated with Increased Cell Invasion and Lethal Outcome in ERG-Negative Prostate Cancer Patients. Tumor Biol. 2016, 37, 9731–9738. [Google Scholar] [CrossRef]
- McClurg, U.L.; Nabbi, A.; Ricordel, C.; Korolchuk, S.; McCracken, S.; Heer, R.; Wilson, L.; Butler, L.M.; Irving-Hooper, B.K.; Pedeux, R.; et al. Human Ex Vivo Prostate Tissue Model System Identifies ING3 as an Oncoprotein. Br. J. Cancer 2018, 118, 713–726. [Google Scholar] [CrossRef]
- Gaughan, L.; Logan, I.R.; Cook, S.; Neal, D.E.; Robson, C.N. Tip60 and Histone Deacetylase 1 Regulate Androgen Receptor Activity through Changes to the Acetylation Status of the Receptor. J. Biol. Chem. 2002, 277, 25904–25913. [Google Scholar] [CrossRef] [PubMed]
- Nabbi, A.; McClurg, U.L.; Thalappilly, S.; Almami, A.; Mobahat, M.; Bismar, T.A.; Binda, O.; Riabowol, K.T. ING3 Promotes Prostate Cancer Growth by Activating the Androgen Receptor. BMC Med. 2017, 15, 103. [Google Scholar] [CrossRef]
- Fink, D.; Yau, T.; Nabbi, A.; Wagner, B.; Wagner, C.; Hu, S.M.; Lang, V.; Handschuh, S.; Riabowol, K.; Rulicke, T. Loss of Ing3 Expression Results in Growth Retardation and Embryonic Death. Cancers 2019, 12, 80. [Google Scholar] [CrossRef] [PubMed]
- Ryder, E.; Gleeson, D.; Sethi, D.; Vyas, S.; Miklejewska, E.; Dalvi, P.; Habib, B.; Cook, R.; Hardy, M.; Jhaveri, K.; et al. Molecular Characterization of Mutant Mouse Strains Generated from the EUCOMM/KOMP-CSD ES Cell Resource. Mamm. Genome 2013, 24, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Skarnes, W.C.; Rosen, B.; West, A.P.; Koutsourakis, M.; Bushell, W.; Iyer, V.; Mujica, A.O.; Thomas, M.; Harrow, J.; Cox, T.; et al. A Conditional Knockout Resource for the Genome-Wide Study of Mouse Gene Function. Nature 2011, 474, 337–342. [Google Scholar] [CrossRef]
- Mahler Convenor, M.; Berard, M.; Feinstein, R.; Gallagher, A.; Illgen-Wilcke, B.; Pritchett-Corning, K.; Raspa, M. FELASA Recommendations for the Health Monitoring of Mouse, Rat, Hamster, Guinea Pig and Rabbit Colonies in Breeding and Experimental Units. Lab. Anim. 2014, 48, 178–192. [Google Scholar] [CrossRef]
- Wu, X.; Wu, J.; Huang, J.; Powell, W.C.; Zhang, J.; Matusik, R.J.; Sangiorgi, F.O.; Maxson, R.E.; Sucov, H.M.; Roy-Burman, P. Generation of a Prostate Epithelial Cell-Specific Cre Transgenic Mouse Model for Tissue-Specific Gene Ablation. Mech. Dev. 2001, 101, 61–69. [Google Scholar] [CrossRef]
- Zuker, M. Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Huggett, J.F.; Foy, C.A.; Benes, V.; Emslie, K.; Garson, J.A.; Haynes, R.; Hellemans, J.; Kubista, M.; Mueller, R.D.; Nolan, T.; et al. The Digital MIQE Guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clin. Chem. 2013, 59, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Gou, W.-F.; Yang, X.-F.; Shen, D.-F.; Zhao, S.; Sun, H.-Z.; Luo, J.-S.; Zheng, H.-C. Immunohistochemical Profile of ING3 Protein in Normal and Cancerous Tissues. Oncol. Lett. 2017, 13, 1631–1636. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Willet, S.G.; Bankaitis, E.D.; Xu, Y.; Wright, C.V.E.; Gu, G. Non-Parallel Recombination Limits Cre-LoxP-Based Reporters as Precise Indicators of Conditional Genetic Manipulation. Genesis 2013, 51, 436–442. [Google Scholar] [CrossRef]
- Powell, W.; Cardiff, R.; Cohen, M.; Miller, G.; Roy-Burman, P. Mouse Strains for Prostate Tumorigenesis Based on Genes Altered in Human Prostate Cancer. Curr. Drug Targets 2003, 4, 263–279. [Google Scholar] [CrossRef]
- Wang, S.; Gao, J.; Lei, Q.; Rozengurt, N.; Pritchard, C.; Jiao, J.; Thomas, G.V.; Li, G.; Roy-Burman, P.; Nelson, P.S.; et al. Prostate-Specific Deletion of the Murine Pten Tumor Suppressor Gene Leads to Metastatic Prostate Cancer. Cancer Cell 2003, 4, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Olson, A.; Le, V.; Aldahl, J.; Yu, E.J.; Hooker, E.; He, Y.; Lee, D.H.; Kim, W.K.; Cardiff, R.D.; Geradts, J.; et al. The Comprehensive Role of E-Cadherin in Maintaining Prostatic Epithelial Integrity during Oncogenic Transformation and Tumor Progression. PLoS Genet. 2019, 15, e1008451. [Google Scholar] [CrossRef]
- Melekhova, A.; Leeder, M.; Pungsrinont, T.; Schmäche, T.; Kallenbach, J.; Ehsani, M.; Mirzakhani, K.; Rasa, S.M.M.; Neri, F.; Baniahmad, A. A Novel Splice Variant of the Inhibitor of Growth 3 Lacks the Plant Homeodomain and Regulates Epithelial-Mesenchymal Transition in Prostate Cancer Cells. Biomolecules 2021, 11, 1152. [Google Scholar] [CrossRef]
- Kim, S.; Natesan, S.; Cornilescu, G.; Carlson, S.; Tonelli, M.; McClurg, U.L.; Binda, O.; Robson, C.N.; Markley, J.L.; Balaz, S.; et al. Mechanism of Histone H3K4me3 Recognition by the Plant Homeodomain of Inhibitor of Growth 3. J. Biol. Chem. 2016, 291, 18326–18341. [Google Scholar] [CrossRef]
- Mouche, A.; Archambeau, J.; Ricordel, C.; Chaillot, L.; Bigot, N.; Guillaudeux, T.; Grenon, M.; Pedeux, R. ING3 Is Required for ATM Signaling and DNA Repair in Response to DNA Double Strand Breaks. Cell Death Differ. 2019, 26, 2344–2357. [Google Scholar] [CrossRef]
- Song, Y.; Hou, G.; Diep, J.; Ooi, Y.S.; Akopyants, N.S.; Beverley, S.M.; Carette, J.E.; Greenberg, H.B.; Ding, S. Inhibitor of Growth Protein 3 Epigenetically Silences Endogenous Retroviral Elements and Prevents Innate Immune Activation. Nucleic Acids Res. 2021, 49, 12706–12715. [Google Scholar] [CrossRef] [PubMed]
- Couto, S.S.; Cao, M.; Duarte, P.C.; Banach-Petrosky, W.; Wang, S.; Romanienko, P.; Wu, H.; Cardiff, R.D.; Abate-Shen, C.; Cunha, G.R. Simultaneous Haploinsufficiency of Pten and Trp53 Tumor Suppressor Genes Accelerates Tumorigenesis in a Mouse Model of Prostate Cancer. Differentiation 2008, 77, 103. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.; Liu, Y.-N.; Pierce, R.; Abou-Kheir, W.; Casey, O.; Seng, V.; Camacho, D.; Simpson, R.M.; Kelly, K. Prostate Epithelial Pten/TP53 Loss Leads to Transformation of Multipotential Progenitors and Epithelial to Mesenchymal Transition. Am. J. Pathol. 2011, 179, 422–435. [Google Scholar] [CrossRef] [PubMed]
| Allele Designation | Description | Reference |
|---|---|---|
| Ing3+ | Wild-type allele | NCBI Gene: 71777 |
| Ing3-LacZ | Targeted knockout-first; reporter-tagged insertion with conditional potential (Ing3tm1a(EUCOMM)Wtsi/Biat) | MGI: 4432585 This work |
| Ing3fl | Derived from Ing3-LacZ through FLP-mediated recombination; conditional allele (Ing3-tm1c) | This work |
| Ing3Δ | Derived from Ing3fl through Cre-mediated recombination; null allele (Ing3-tm1d) | This work |
| Ing3m | Insertional mutant; disruption of Ing3 expression by mCherry reporter cassette (Tg(UBC-mCherry)1Phbs) | MGI: 5296812 [25] |
| Ing3T | Transgenic; ectopic overexpression of Ing3 (TgTn(sb-CAG-Ing3-P2A-eGFP)774.1Biat) | [25] |
| PB-Cre4 | Transgenic; Cre expression in prostate epithelial cells (Tg(Pbsn-cre)4Prb) | MGI: 2385927 [29] |
| Target Gene | NCBI Gene ID | Primer/Probe Designation | Primer Sequence (5′-3′) | Amplicon Length (bp) |
|---|---|---|---|---|
| Ing3 | 71777 | Ing3-Forward | CATTGGGACCCTCTAGGAGAGAT | 82 |
| Ing3-Reverse | GCCCCCAAGTCCCTCATAA | |||
| Ing3-Probe | /56-FAM/TTACGTAGA/ZEN/TACCT GGATATGGAGTGAGGGCA/3IABkFQ/ | |||
| B2m | 12010 | B2m-Forward | CTCAGAAACCCCTCAAATTCAAGTA | 96 |
| B2m-Reverse | GGCGGGTGGAACTGTGTTAC | |||
| B2m-Probe | /PentaYellow/CTCACGCCACCCAC CGGAGAATG/BHQ-1/ |
| Component | Volume Per Reaction (µL) | Stock Concentration | Final Concentration |
|---|---|---|---|
| QuantStudio 3D Digital PCR Master Mix v2 | 9.00 | 2 × | 1 × |
| Ing3-Forward | 0.36 | 10 µM | 0.2 µM |
| Ing3-Reverse | 0.36 | 10 µM | 0.2 µM |
| Ing3-Probe | 0.36 | 10 µM | 0.2 µM |
| B2m-Forward | 0.36 | 10 µM | 0.2 µM |
| B2m-Reverse | 0.36 | 10 µM | 0.2 µM |
| B2m-Probe | 0.36 | 10 µM | 0.2 µM |
| H2O | 1.84 | - | - |
| Template DNA | 5.00 | - | - |
| Total volume | 18.00 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lang, V.; Barones, L.; Hu, S.M.; Hashemi, F.; Blote, K.; Riabowol, K.; Fink, D. Loss of ING3 in the Prostate Leads to Activation of DNA Damage Repair Markers. Cancers 2025, 17, 1037. https://doi.org/10.3390/cancers17061037
Lang V, Barones L, Hu SM, Hashemi F, Blote K, Riabowol K, Fink D. Loss of ING3 in the Prostate Leads to Activation of DNA Damage Repair Markers. Cancers. 2025; 17(6):1037. https://doi.org/10.3390/cancers17061037
Chicago/Turabian StyleLang, Viktor, Lisa Barones, ShiTing Misaki Hu, Fatemeh Hashemi, Karen Blote, Karl Riabowol, and Dieter Fink. 2025. "Loss of ING3 in the Prostate Leads to Activation of DNA Damage Repair Markers" Cancers 17, no. 6: 1037. https://doi.org/10.3390/cancers17061037
APA StyleLang, V., Barones, L., Hu, S. M., Hashemi, F., Blote, K., Riabowol, K., & Fink, D. (2025). Loss of ING3 in the Prostate Leads to Activation of DNA Damage Repair Markers. Cancers, 17(6), 1037. https://doi.org/10.3390/cancers17061037






