Serum-Based Proteomic Approach to Identify Clinical Biomarkers of Radiation Exposure
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Characteristics and Sample Collection
2.2. Sample Preparation
2.3. Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS) Analysis
2.4. Mass Spectrometry (MS) Data Analysis
2.5. Protein Identification and Quantification
2.6. Statistical and Bioinformatics Analysis
3. Results
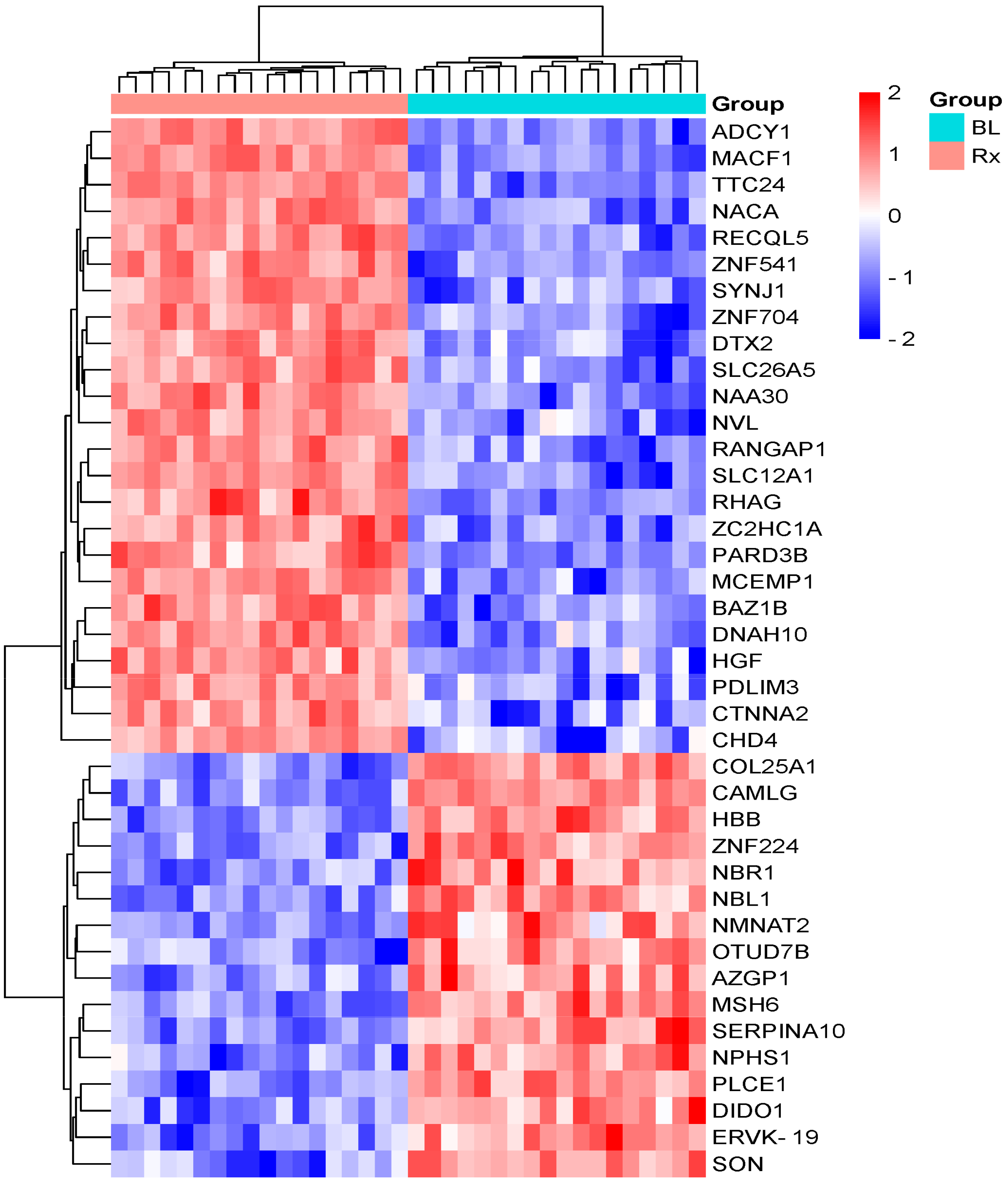
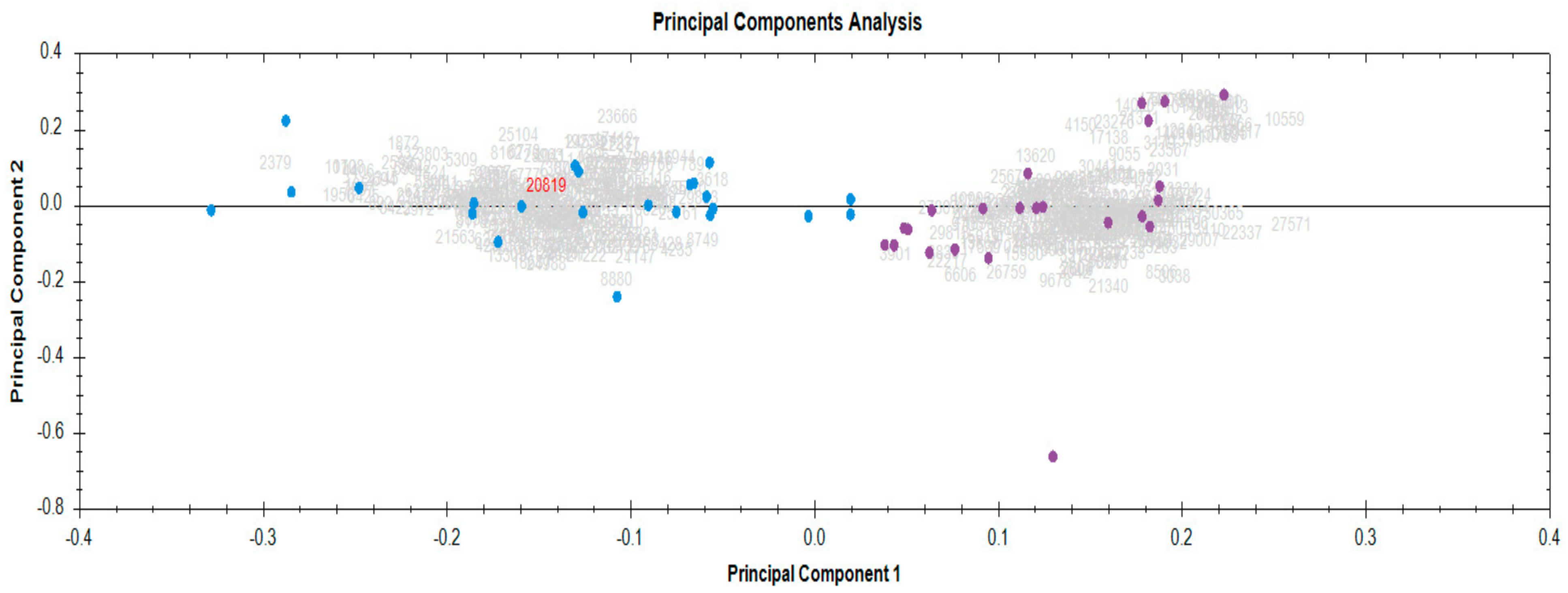
3.1. Identification and Quantification of Serum Proteome in Response to Radiotherapy
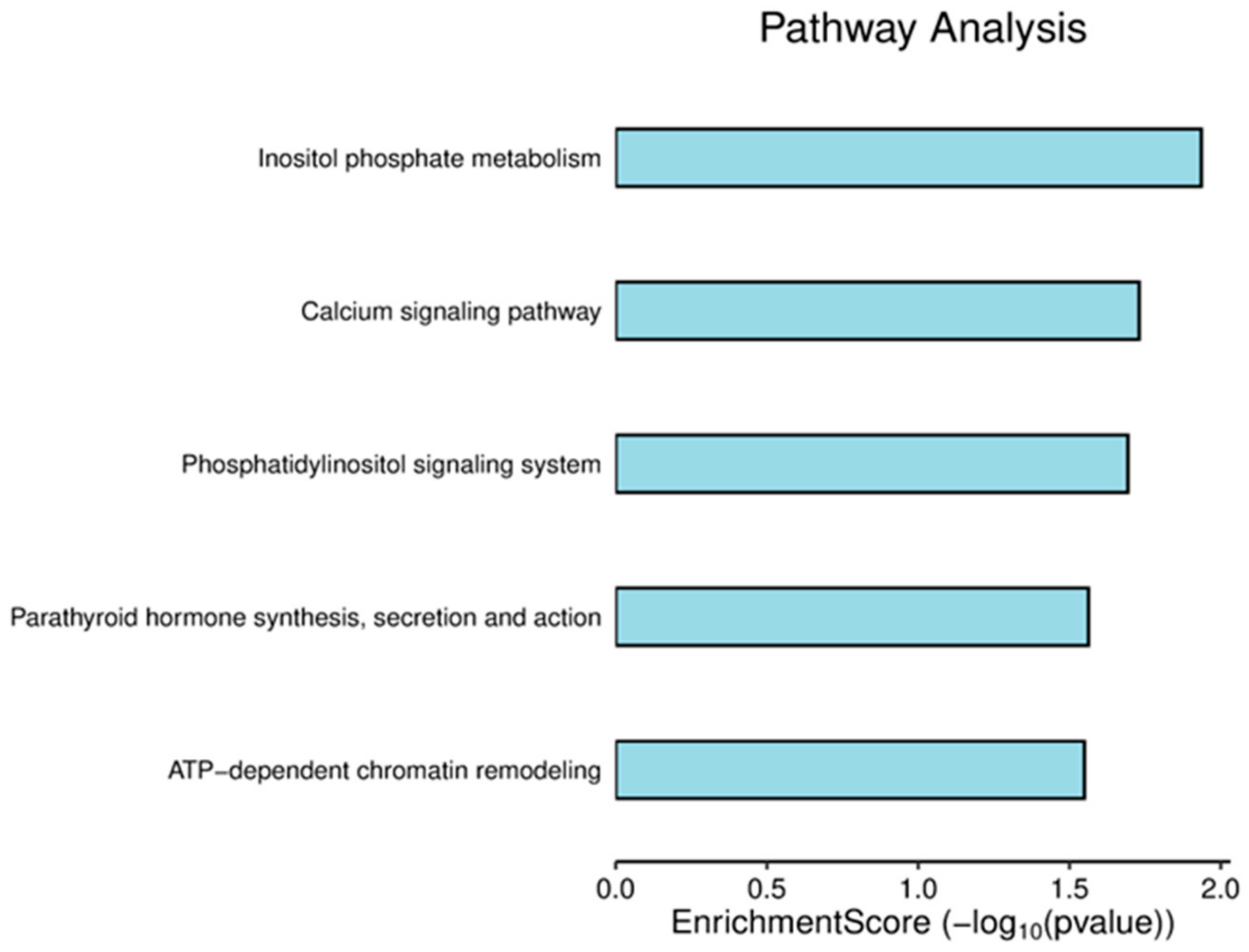
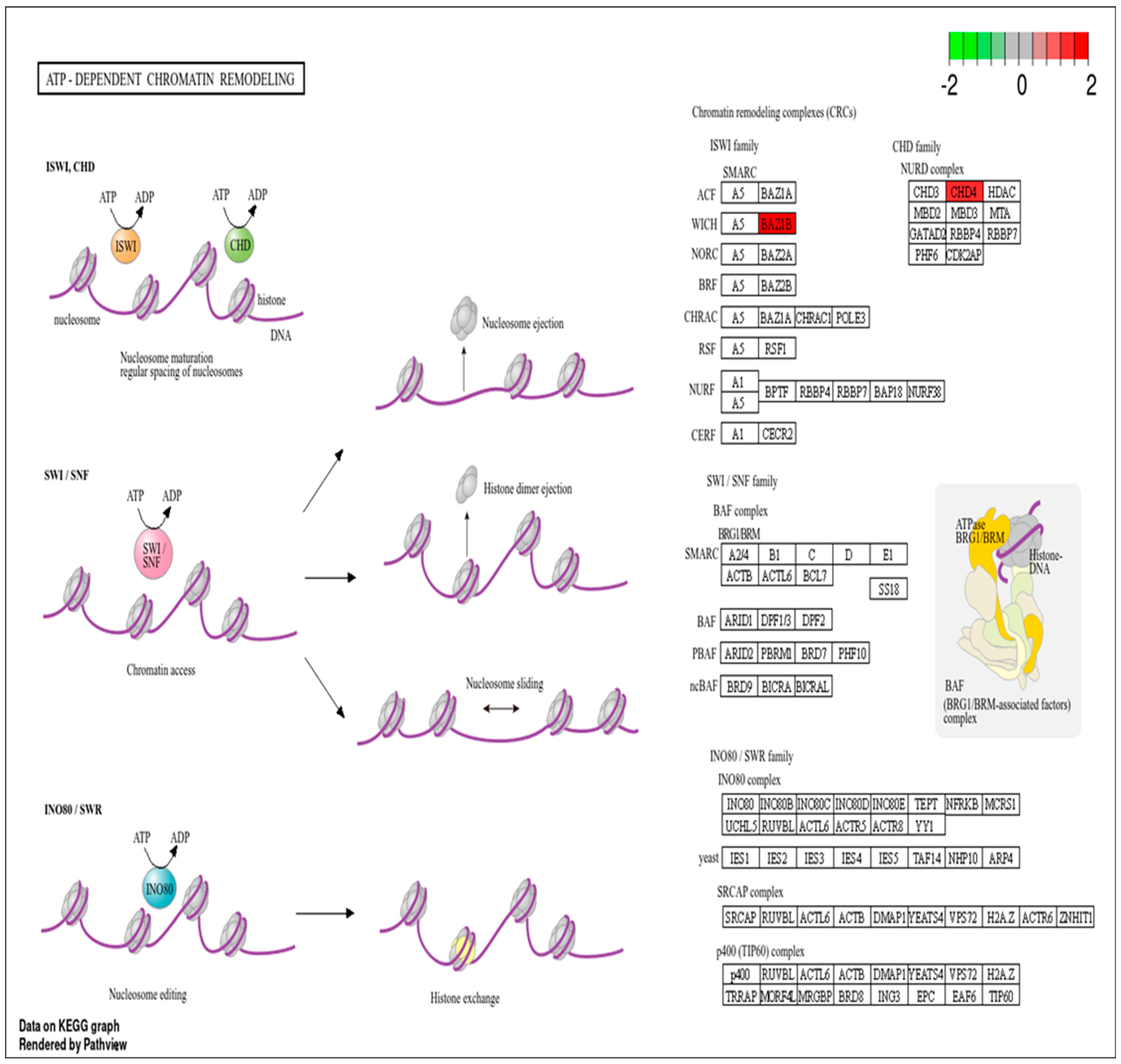
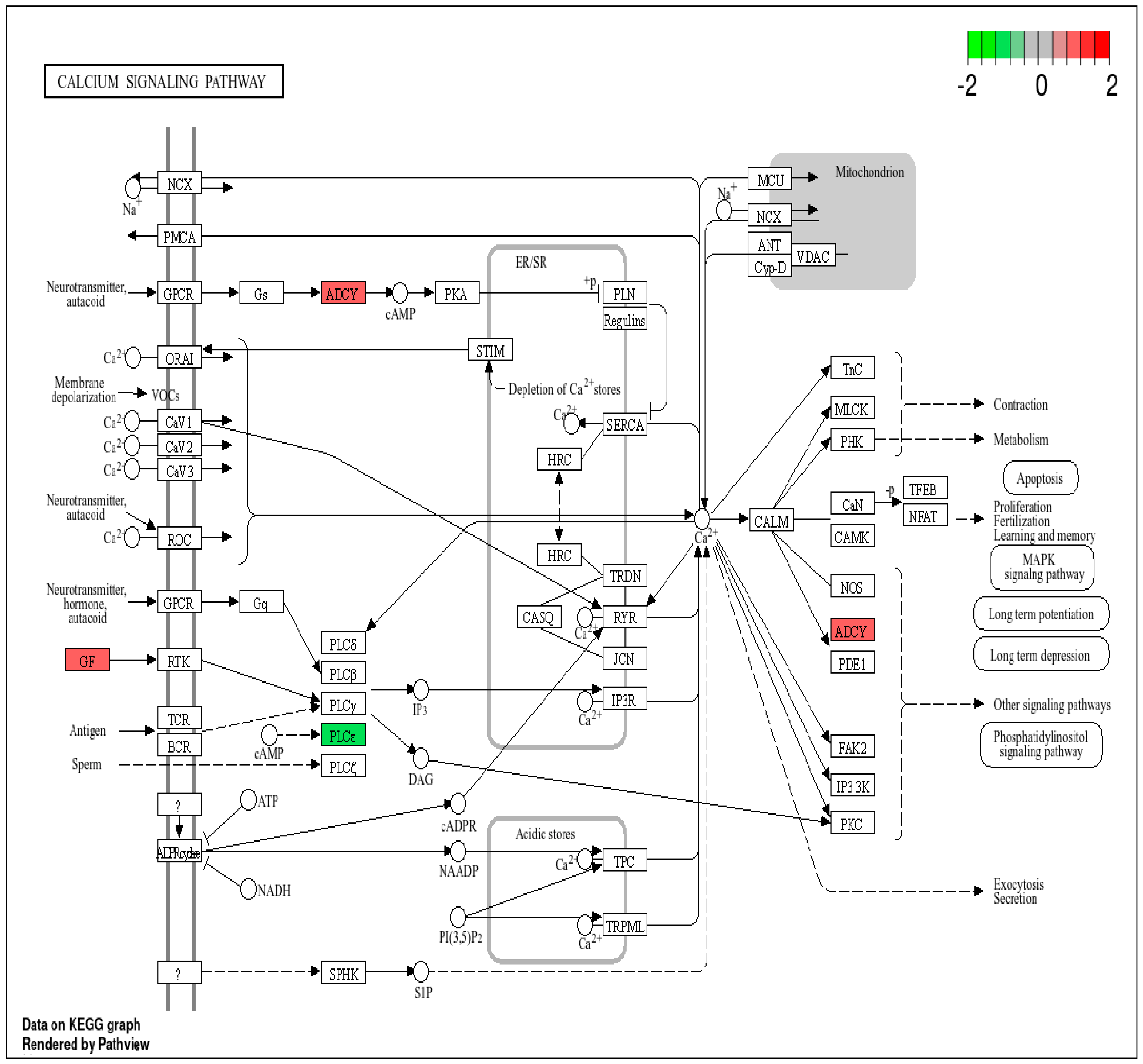
3.2. Pathway Analysis of DEPs
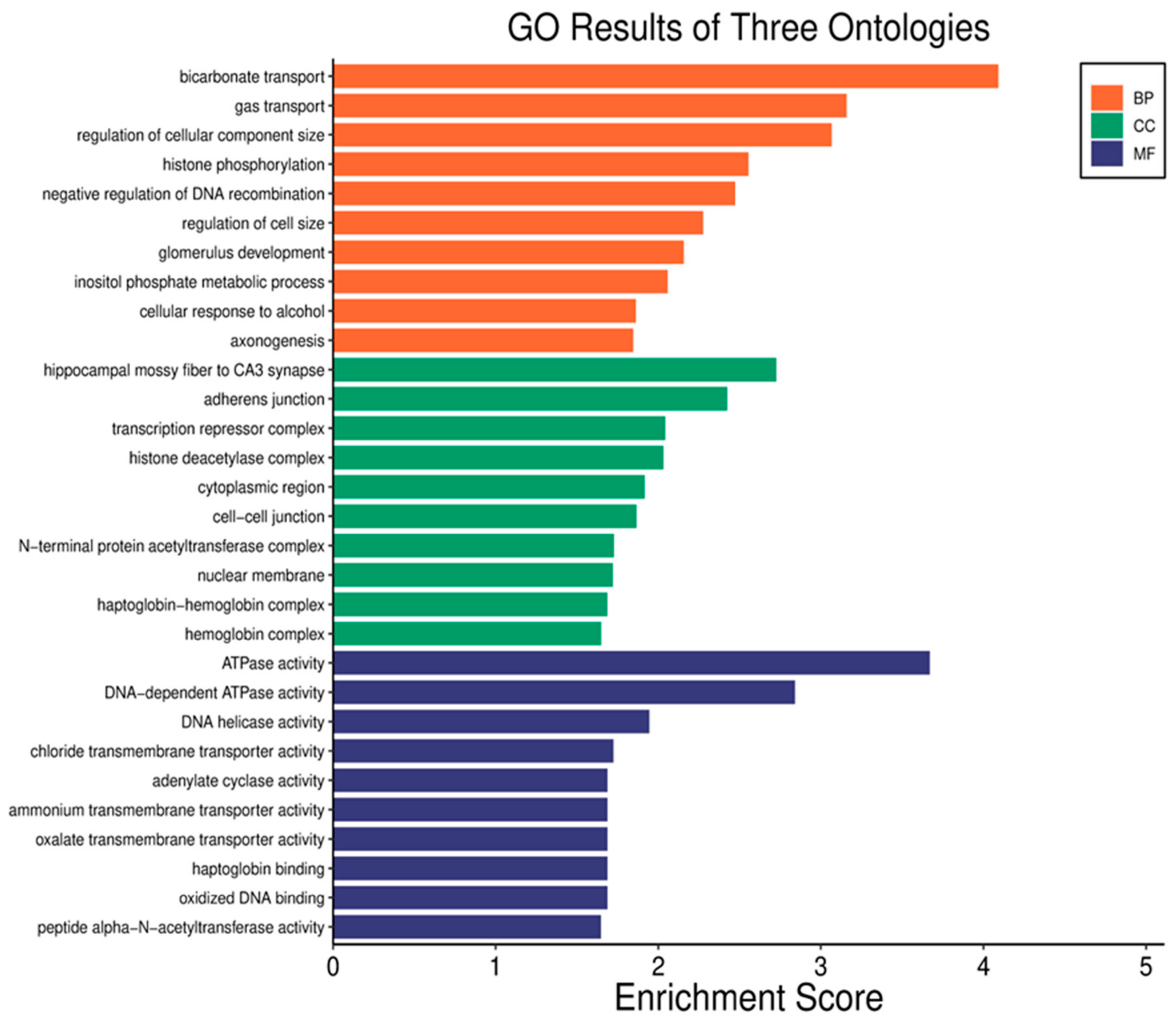
3.3. Gene Ontology (GO) Analysis
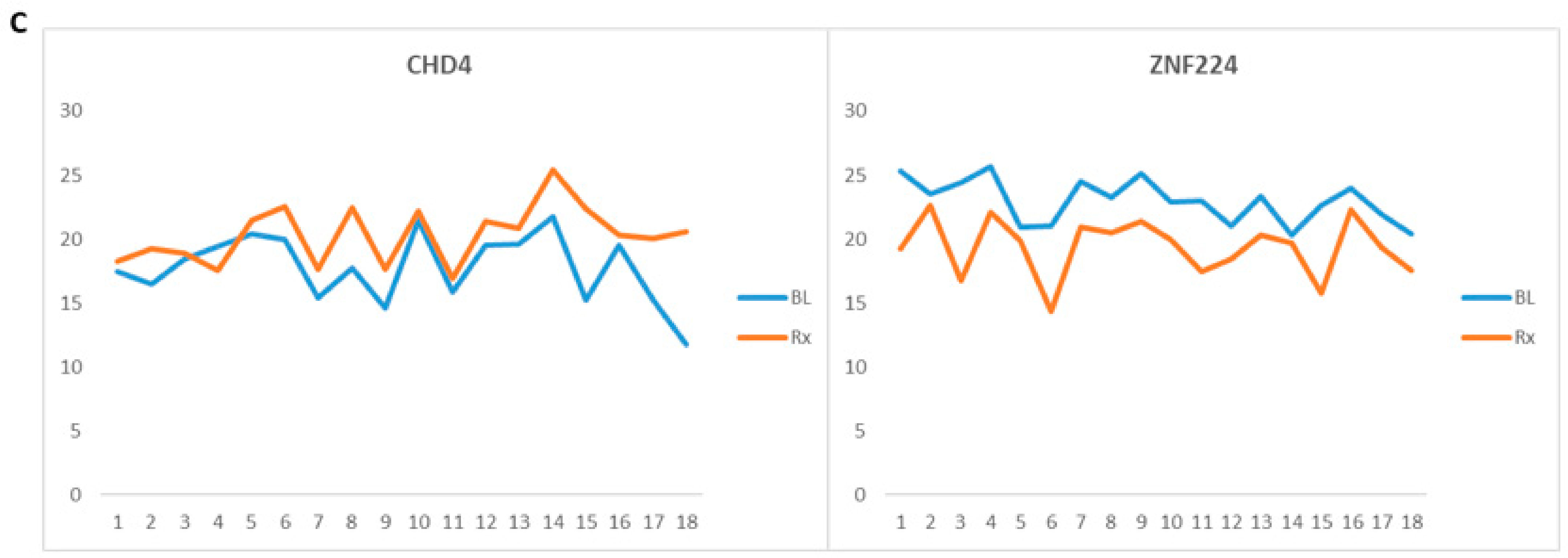
3.4. Ionizing Radiation Clinical Biomarkers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ARS | Acute radiation syndrome |
| BD-SST | Blood collection serum-separator |
| RT | Radiotherapy |
| BP | Biological process |
| CC | Cellular component |
| DNA | Deoxyribonucleic acid |
| DSBs | Double-strand breaks |
| FC | Fold change |
| GLOBOCAN | Global Cancer Observatory |
| GO | Gene ontology |
| GY | Gray |
| HNC | Head and neck cancer |
| HPLC | High performance liquid chromatography |
| IARC | International Agency for Cancer Research |
| IMRT | Intensity-modulated radiotherapy |
| IPA | Ingenuity pathway analysis |
| IR | Ionizing radiation |
| MF | Molecular function |
| MS | Mass spectrometer |
| PCA | Principal component analysis |
| PPI | Protein interaction |
| RMPs | Radiation-modulated proteins |
| RNA | Ribonucleic acid |
| TFA | Trifluoroacetic acid |
| TNM | Tumor, nodes, and metastasis |
References
- Zeegers, D.; Venkatesan, S.; Koh, S.W.; Kah, G.; Low, M.; Srivastava, P.; Sundaram, N.; Sethu, S.; Banerjee, B.; Jayapal, M.; et al. Biomarkers of Ionizing Radiation Exposure: A Multiparametric. Genome Integr. 2017, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Bai, H.; Feng, X.; Hua, J.; Long, K.; He, J.; Zhang, Y.; Ding, N.; Wang, J.; Zhou, H. Serum Proteins as New Biomarkers for Whole-Body Exposure to High- and Low-LET Ionizing Radiation. Dose-Response 2020, 18, 1559325820914172. [Google Scholar] [CrossRef] [PubMed]
- Subedi, P.; Gomolka, M.; Moertl, S.; Dietz, A. Ionizing radiation protein biomarkers in normal tissue and their correlation to radiosensitivity: A systematic review. J. Pers. Med. 2021, 11, 140. [Google Scholar] [CrossRef]
- Rosenblatt, E.; Izewska, J.; Anacak, Y.; Pynda, Y.; Scalliet, P.; Boniol, M.; Autier, P. Radiotherapy capacity in European countries: An analysis of the Directory of Radiotherapy Centres (DIRAC) database. Lancet Oncol. 2013, 14, e79–e86. [Google Scholar] [CrossRef]
- Fazel, R.; Gerber, T.C.; Balter, S.; Brenner, D.J.; Carr, J.J.; Cerqueira, M.D.; Chen, J.; Einstein, A.J.; Krumholz, H.M.; Mahesh, M.; et al. Approaches to enhancing radiation safety in cardiovascular imaging a scientific statement from the American Heart Association. Circulation 2014, 130, 1730–1748. [Google Scholar] [CrossRef]
- Donnelly, E.H.; Nemhauser, J.B.; Smith, J.M.; Kazzi, Z.N.; Farfán, E.B.; Chang, A.S.; Naeem, S.F. Acute radiation syndrome: Assessment and management. South Med. J. 2010, 103, 541–546. [Google Scholar] [CrossRef]
- Renner, R. Redrawing the Dose-Response Curve. Environ. Sci. Technol. 2004, 38, 90A–95A. [Google Scholar] [CrossRef]
- Fenech, M. Current status, new frontiers and challenges in radiation biodosimetry using cytogenetic, transcriptomic and proteomic technologies. Radiat. Meas. 2011, 46, 737–741. [Google Scholar] [CrossRef]
- Jia, M.; Wang, Z. MicroRNAs as Biomarkers for Ionizing Radiation Injury. Front. Cell Dev. Biol. 2022, 10, 861451. [Google Scholar] [CrossRef]
- Mzizi, Y.; Mbambara, S.; Moetlhoa, B.; Mahapane, J.; Mdanda, S.; Sathekge, M.; Kgatle, M. Ionising radiation exposure-induced regulation of selected biomarkers and their impact in cancer and treatment. Front. Nucl. Med. 2024, 4, 1469897. [Google Scholar] [CrossRef]
- Surace, L.; Angelika, N.; Gupta, A.; Broek, M. Van Den Radiotherapy supports tumor specific immunity by acute inflammation. Oncoimmunology 2016, 5, e1060391. [Google Scholar] [CrossRef] [PubMed]
- Budworth, H.; Snijders, A.M.; Marchetti, F.; Mannion, B.; Bhatnagar, S.; Kwoh, E.; Tan, Y.; Wang, S.X.; Blakely, W.F.; Coleman, M.; et al. DNA Repair and Cell Cycle Biomarkers of Radiation Exposure and Inflammation Stress in Human Blood. PLoS ONE 2012, 7, e48619. [Google Scholar] [CrossRef] [PubMed]
- Yakkioui, Y.; Temel, Y.; Chevet, E.; Negroni, L. Integrated and Quantitative Proteomics of Human Tumors, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2017; Volume 586. [Google Scholar]
- Winters, T.A.; Taliaferro, L.P.; Satyamitra, M.M. Development of Biomarkers for Radiation Biodosimetry and Medical Countermeasures Research: Current Status, Utility, and Regulatory Pathways. Radiat. Res. 2022, 197, 554–558. [Google Scholar] [CrossRef]
- Guipaud, O. Serum and plasma proteomics and its possible use as detector and predictor of radiation diseases. Adv. Exp. Med. Biol. 2013, 990, 61–86. [Google Scholar] [CrossRef]
- Gabriela, R.; Vera, V.; Pavel, R.; Helena, R.; Igor, S.; Marie, D.; Marketa, M.; Alena, M.F.; Ales, T. Discovering the Radiation Biomarkers in the Plasma of Total-Body Irradiated Leukemia Patients. Radiat. Res. 2024, 201, 418–428. [Google Scholar] [CrossRef]
- Zebene, E.D.; Lombardi, R.; Pucci, B.; Medhin, H.T.; Seife, E.; Di Gennaro, E.; Budillon, A.; Woldemichael, G.B. Proteomic Analysis of Biomarkers Predicting Treatment Response in Patients with Head and Neck Cancers. Int. J. Mol. Sci. 2024, 25, 12513. [Google Scholar] [CrossRef]
- Amundson, S.A. The transcriptomic revolution and radiation biology. Int. J. Radiat. Biol. 2022, 98, 428–438. [Google Scholar] [CrossRef]
- Pannkuk, E.L.; Fornace, A.J.; Laiakis, E.C. Metabolomic applications in radiation biodosimetry: Exploring radiation effects through small molecules. Int. J. Radiat. Biol. 2017, 93, 1151–1176. [Google Scholar] [CrossRef]
- Ghaderi, N.; Jung, J.; Brüningk, S.C.; Subramanian, A.; Nassour, L.; Peacock, J. A Century of Fractionated Radiotherapy: How Mathematical Oncology Can Break the Rules. Int. J. Mol. Sci. 2022, 23, 1316. [Google Scholar] [CrossRef]
- Winters, T.A.; Cassatt, D.R.; Harrison-Peters, J.R.; Hollingsworth, B.A.; Rios, C.I.; Satyamitra, M.M.; Taliaferro, L.P.; Dicarlo, A.L. Considerations of Medical Preparedness to Assess and Treat Various Populations during a Radiation Public Health Emergency. Radiat. Res. 2023, 199, 301–318. [Google Scholar] [CrossRef]
- Brien, G.O.; Cruz-garcia, L.; Majewski, M.; Grepl, J.; Abend, M.; Port, M.; Tichý, A.; Sirak, I.; Malkova, A.; Donovan, E.; et al. OPEN FDXR is a biomarker of radiation exposure in vivo. Sci. Rep. 2018, 8, 684. [Google Scholar] [CrossRef]
- Kanagaraj, K.; Phillippi, M.A.; Ober, E.H.; Shuryak, I.; Kleiman, N.J.; Olson, J.; Schaaf, G.; Cline, J.M.; Turner, H.C. BAX and DDB2 as biomarkers for acute radiation exposure in the human blood ex vivo and non-human primate models. Sci. Rep. 2024, 14, 19345. [Google Scholar] [CrossRef] [PubMed]
- Schaue, D.; Micewicz, E.D.; Ratikan, J.A.; Xie, M.W.; Cheng, G.; McBride, W.H. Radiation & Inflammation. Semin. Radiat. Oncol. 2015, 25, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kim, J.J.; Miller, K.M. Bromodomain proteins: Protectors against endogenous DNA damage and facilitators of genome integrity. Exp. Mol. Med. 2021, 53, 1268–1277. [Google Scholar] [CrossRef]
- Decrock, E.; Hoorelbeke, D.; Ramadan, R.; Delvaeye, T.; De Bock, M.; Wang, N.; Krysko, D.V.; Baatout, S.; Bultynck, G.; Aerts, A.; et al. Calcium, oxidative stress and connexin channels, a harmonious orchestra directing the response to radiotherapy treatment? Biochim. Biophys. Acta-Mol. Cell Res. 2017, 1864, 1099–1120. [Google Scholar] [CrossRef]
- McKelvey, K.J.; Hudson, A.L.; Back, M.; Eade, T.; Diakos, C.I. Radiation, inflammation and the immune response in cancer. Mamm. Genome 2018, 29, 843–865. [Google Scholar] [CrossRef]
- Sproull, M.; Kramp, T.; Tandle, A.; Shankavaram, U.; Camphausen, K. Serum Amyloid A as a Biomarker for Radiation Exposure. Radiat. Res. 2015, 184, 14–23. [Google Scholar] [CrossRef]
- Widlak, P.; Jelonek, K.; Wojakowska, A.; Pietrowska, M.; Polanska, J. Serum Proteome Signature of Radiation Response: Upregulation of Inflammation-Related Factors and Downregulation of Apolipoproteins and Coagulation Factors in Cancer Patients Treated With Radiation Therapy A Pilot Study. Int. J. Radiat. Oncol. Biol. Phys. 2015, 92, 1108–1115. [Google Scholar] [CrossRef]
- Tavares, L.P.; Negreiros-Lima, G.L.; Lima, K.M.; Silva, P.M.R.E.; Pinho, V.; Teixeira, M.M.; Sousa, L.P. Blame the signaling: Role of cAMP for the resolution of inflammation. Pharmacol. Res. 2020, 159, 105030. [Google Scholar] [CrossRef]
- Marchetti, F.; Coleman, M.A.; Jones, I.M.; Wyrobek, A.J. Candidate protein biodosimeters of human exposure to ionizing radiation. Int. J. Radiat. Biol. 2006, 82, 605–639. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, S.; Zhou, Y.; Feng, F.; Wang, Q.; Zhu, X.; Ai, H.; Huang, X.; Zhang, X. Hepatocyte growth factor gene-modified adipose-derived mesenchymal stem cells ameliorate radiation induced liver damage in a rat model. PLoS ONE 2014, 9, e114670. [Google Scholar] [CrossRef]
- Dong, L.H.; Jiang, Y.Y.; Liu, Y.J.; Cui, S.; Xia, C.C.; Qu, C.; Jiang, X.; Qu, Y.Q.; Chang, P.Y.; Liu, F. The anti-fibrotic effects of mesenchymal stem cells on irradiated lungs via stimulating endogenous secretion of HGF and PGE2. Sci. Rep. 2015, 5, 8713. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Lu, Y.; Xiao, F.; Cheng, X.; Hu, C.; Zhu, X.; Wang, X.; Duan, H.; Du, L.; Zhang, Q. A recombinant plasmid encoding human hepatocyte growth factor promotes healing of combined radiation-trauma skin injury involved in regulating Nrf2 pathway in mice. J. Radiat. Res. 2024, 65, 279–290. [Google Scholar] [CrossRef]
- Landy, R.E.; Stross, W.C.; May, J.M.; Kaleem, T.A.; Malouff, T.D.; Waddle, M.R.; Vallow, L.A. Idiopathic mast cell activation syndrome and radiation therapy: A case study, literature review, and discussion of mast cell disorders and radiotherapy. Radiat. Oncol. 2019, 14, 222. [Google Scholar] [CrossRef]
- Smith, J.; Tan, J.K.H.; Short, C.; O’Neill, H.; Moro, C. The effect of myeloablative radiation on urinary bladder mast cells. Sci. Rep. 2024, 14, 6219. [Google Scholar] [CrossRef]
- Dworak, N.; Makosa, D.; Chatterjee, M.; Jividen, K.; Yang, C.S.; Snow, C.; Simke, W.C.; Johnson, I.G.; Kelley, J.B.; Paschal, B.M. A nuclear lamina-chromatin-Ran GTPase axis modulates nuclear import and DNA damage signaling. Aging Cell 2019, 18, e12851. [Google Scholar] [CrossRef]
- Ali, Y.F.; Cucinotta, F.A.; Ning-Ang, L.; Zhou, G. Cancer Risk of Low Dose Ionizing Radiation. Front. Phys. 2020, 8, 234. [Google Scholar] [CrossRef]
- Noel, C.K.; Bruce, E.D.; Ryan, B.J. Suit Up: A Systematic Review of the Personal Protective Equipment (PPE) Recommended and Utilized by Various Classes of Responders to Nuclear Radiological Disasters at Nuclear Power Plants. Prehosp. Disaster Med. 2024, 39, 85–93. [Google Scholar] [CrossRef]
- Qiu, S.; Wu, Q.; Wang, H.; Liu, D.; Chen, C.; Zhu, Z.; Zheng, H.; Yang, G.; Li, L.; Yang, M. AZGP1 in POMC neurons modulates energy homeostasis and metabolism through leptin-mediated STAT3 phosphorylation. Nat. Commun. 2024, 15, 3377. [Google Scholar] [CrossRef]
- Fang, Y.Y.; Huang, J.M.; Wen, J.Y.; Li, J.D.; Shen, J.H.; Zeng, D.T.; Pan, Y.F.; Huang, H.Q.; Huang, Z.G.; Liu, L.M.; et al. AZGP1 Up-Regulation is a Potential Target for Andrographolide Reversing Radioresistance of Colorectal Cancer. Pharmgenom. Pers. Med. 2022, 15, 999–1017. [Google Scholar] [CrossRef]
- Varland, S.; Silva, R.D.; Kjosås, I.; Faustino, A.; Bogaert, A.; Billmann, M.; Boukhatmi, H.; Kellen, B.; Costanzo, M.; Drazic, A.; et al. N-terminal acetylation shields proteins from degradation and promotes age-dependent motility and longevity. Nat. Commun. 2023, 14, 6774. [Google Scholar] [CrossRef]
- Van Damme, P.; Kalvik, T.V.; Starheim, K.K.; Jonckheere, V.; Myklebust, L.M.; Menschaert, G.; Varhaug, J.E.; Gevaert, K.; Arnesen, T. A role for human N-alpha acetyltransferase 30 (Naa30) in maintaining mitochondrial integrity. Mol. Cell. Proteom. 2016, 15, 3361–3372. [Google Scholar] [CrossRef]
- Garcia-Montojo, M.; Doucet-O’Hare, T.; Henderson, L.; Nath, A. Human endogenous retrovirus-K (HML-2): A comprehensive review. Crit. Rev. Microbiol. 2018, 44, 715–738. [Google Scholar] [CrossRef]
- Suntsova, M.; Garazha, A.; Ivanova, A.; Kaminsky, D.; Zhavoronkov, A.; Buzdin, A. Molecular functions of human endogenous retroviruses in health and disease. Cell. Mol. Life Sci. 2015, 72, 3653–3675. [Google Scholar] [CrossRef]
- Kelly-robinson, G.A.; Reihill, J.A.; Lundy, F.T.; McGarvey, L.P.; Lockhart, J.C.; Litherland, G.J.; Thornbury, K.D.; Martin, S.L. The serpin superfamily and their role in the regulation and dysfunction of serine protease activity in copd and other chronic lung diseases. Int. J. Mol. Sci. 2021, 22, 6351. [Google Scholar] [CrossRef]
- Lans, H.; Marteijn, J.A.; Vermeulen, W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin 2012, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Larsen, D.H.; Poinsignon, C.; Gudjonsson, T.; Dinant, C.; Payne, M.R.; Hari, F.J.; Danielsen, J.M.R.; Menard, P.; Sand, J.C.; Stucki, M.; et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J. Cell Biol. 2010, 190, 731–740. [Google Scholar] [CrossRef]
- Gangaraju, V.K.; Bartholomew, B. Mechanisms of ATP dependent chromatin remodeling. Mutat. Res.-Fundam. Mol. Mech. Mutagen. 2007, 618, 3–17. [Google Scholar] [CrossRef]
- Borrego-Soto, G.; Ortiz-López, R.; Rojas-Martínez, A. Ionizing radiation-induced DNA injury and damage detection in patients with breast cancer. Genet. Mol. Biol. 2015, 38, 420–432. [Google Scholar] [CrossRef]
- Datasets. 2024. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ZNF541 (accessed on 11 November 2024).
- Snyder, N.A.; Silva, G.M. Deubiquitinating enzymes (DUBs): Regulation, homeostasis, and oxidative stress response. J. Biol. Chem. 2021, 297, 101077. [Google Scholar] [CrossRef]
- Pizzagalli, M.D.; Bensimon, A.; Superti-Furga, G. A guide to plasma membrane solute carrier proteins. FEBS J. 2021, 288, 2784–2835. [Google Scholar] [CrossRef]
- Fasano, D.; Parisi, S.; Pierantoni, G.M.; De Rosa, A.; Picillo, M.; Amodio, G.; Pellecchia, M.T.; Barone, P.; Moltedo, O.; Bonifati, V.; et al. Alteration of endosomal trafficking is associated with early-onset parkinsonism caused by SYNJ1 mutations. Cell Death Dis. 2018, 9, 385. [Google Scholar] [CrossRef]
- Juarez-Navarro, K.; Ayala-Garcia, V.M.; Ruiz-Baca, E.; Meneses-Morales, I.; Rios-Banuelos, J.L.; Lopez-Rodriguez, A. Assistance for folding of disease-causing plasma membrane proteins. Biomolecules 2020, 10, 728. [Google Scholar] [CrossRef]
- Guo, B.; Huang, J.; Wu, W.; Feng, D.; Wang, X.; Chen, Y.; Zhang, H. The nascent polypeptide-associated complex is essential for autophagic flux. Autophagy 2014, 10, 1738–1748. [Google Scholar] [CrossRef]
- Wahba, A.; Lehman, S.L.; Tofilon, P.J. Radiation-induced translational control of gene expression. Translation 2017, 5, e1265703. [Google Scholar] [CrossRef]
- Butler, P.; Pascheto, I.; Lizzi, M.; St-Onge, R.; Lanner, C.; Guo, B.; Masilamani, T.; Pritzker, L.B.; Kovala, A.T.; Parissenti, A.M. RNA disruption is a widespread phenomenon associated with stress-induced cell death in tumour cells. Sci. Rep. 2023, 13, 1711. [Google Scholar] [CrossRef]
- Wurtmann, E.J.; Wolin, S.L. RNA under attack: Cellular handling of RNA damage RNA under attack: Cellular handling of RNA damage. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 34–49. [Google Scholar] [CrossRef]
- Ladeira, C.; Smajdova, L. The use of genotoxicity biomarkers in molecular epidemiology: Applications in environmental, occupational and dietary studies. AIMS Genet. 2017, 4, 166–191. [Google Scholar] [CrossRef]

| Clinical and Demographic Features | HNC = 18 | p-Value |
|---|---|---|
| Median age (95% CI range) | 39.5 (32.3–42.9) | |
| Radiation dose (95% CI) | 70 (60.8–69.1) | |
| Gender | ||
| Male | 9 (50%) | 0.00 |
| Female | 9 (50%) | |
| T1–T2 (T1 = 3, T2 = 7) | 10 (55.5) | 0.00 |
| T3 = 8 | 8 (44.4%) | |
| N0 | 9 (50%) | 0.00 |
| N+ | 9 (50%) | |
| Clinical and demographic features | Rectal = 5 | |
| Median age (95% CI range) | 26 (18.0–42.7) | |
| Radiation dose (95% CI) | 45 (43–49) | |
| Gender | ||
| Male | 0 (0.0%) | |
| Female | 5 (100%) | |
| TNM | ||
| T1–T2 (T1 = 2, T2 = 2) | 4 (80%) | 0.082 |
| T3 = 8 (T3 = 1) | 1 (20%) | |
| N0 | 5 (100%) |
| Protein Description | Gene Name | Acc. UniProtKB | Fold Change (Tx/CTR) | p-Value | Regulation |
|---|---|---|---|---|---|
| Tyrosine protein kinase BAZ1B | BAZ1B | Q9UIG0 | 10.5 | 0.02 | Upregulated |
| ATP-dependent DNA helicase Q5 | RECQL5 | O94762 | 5.2 | 0.04 | Upregulated |
| Zinc finger C2HC domain-containing protein 1A | ZC2HC1A | Q96GY0 | 5.2 | 0.04 | Upregulated |
| Zinc finger protein 541 | ZNF541 | Q9H0D2 | 3.7 | 0.02 | Upregulated |
| Prestin | SLC26A5 | P58743 | 3.2 | 0.02 | Upregulated |
| Ran GTPase-activating protein 1 | RANGAP1 | P46060 | 3.2 | 0.01 | Upregulated |
| Solute carrier family 12 member 1 | SLC12A1 | Q13621 | 3.1 | 0.04 | Upregulated |
| Dynein heavy chain 10, axonemal | DNAH10 | Q8IVF4 | 3.0 | 0.03 | Upregulated |
| Catenin alpha-2 | CTNNA2 | P26232 | 2.9 | 0.02 | Upregulated * |
| *Chromodomain-helicase-DNA-binding protein 4 | CHD4 | Q14839 | 2.6 | 0.03 | Upregulated * |
| Synaptojanin-1 | SYNJ1 | O43426 | 2.3 | 0.03 | Upregulated |
| Ammonium transporter Rh type A | RHAG | Q02094 | 2.3 | 0.04 | Upregulated |
| Hepatocyte growth factor-like protein | HGF | P14210 | 2.2 | 0.04 | Upregulated |
| Adenylate cyclase type 1 | ADCY1 | Q08828 | 2.1 | 0.01 | Upregulated |
| PDZ and LIM domain protein 3 | PDLIM3 | Q53GG5 | 2.1 | 0.03 | Upregulated |
| Mast cell-expressed membrane protein 1 | MCEMP1 | Q8IX19 | 1.8 | 0.03 | Upregulated |
| N-alpha-acetyltransferase 30 | NAA30 | Q147X3 | 1.8 | 0.03 | Upregulated * |
| Zinc finger protein 704 | ZNF704 | Q6ZNC4 | 1.7 | 0.04 | Upregulated |
| Tetratricopeptide repeat protein 24 | TTC24 | A2A3L6 | 1.6 | 0.01 | Upregulated |
| Nascent polypeptide-associated complex subunit alpha, muscle-specific form | NACA | E9PAV3 | 1.6 | 0.03 | Upregulated |
| Probable E3 ubiquitin protein ligase DTX2 | DTX2 | Q86UW9 | 1.6 | 0.02 | Upregulated |
| Partitioning defective 3 homolog B | PARD3B | Q8TEW8 | 1.5 | 0.03 | Upregulated |
| Microtubule–actin cross-linking factor 1, isoforms 1/2/3/5 | MACF1 | O94854 | 1.5 | 0.02 | Upregulated * |
| Nuclear valosin-containing protein-like | NVL | O15381 | 1.5 | 0.02 | Upregulated * |
| DNA mismatch repair protein Msh6 | MSH6 | P52701 | 0.5 | 0.04 | Downregulated * |
| Collagen alpha-1(XXV) chain | COL25A1 | Q9BXS0 | 0.5 | 0.04 | Downregulated |
| Protein SON OS = Homo sapiens | SON | P18583 | 0.5 | 0.02 | Downregulated |
| Protein Z-dependent protease inhibitor | SERPINA10 | Q9UK55 | 0.5 | 0.01 | Downregulated |
| 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase epsilon-1 | PLCE1 | Q9P212 | 0.49 | 0.04 | Downregulated |
| Death-inducer obliterator 1 | DIDO1 | Q9BTC0 | 0.49 | 0.007 | Downregulated |
| Endogenous retrovirus group K member 19 Pro protein | ERVK-19 | O71037 | 0.48 | 0.02 | Downregulated |
| Zinc-alpha-2-glycoprotein | AZGP1 | P25311 | 0.47 | 0.003 | Downregulated |
| Nephrin OS = Homo sapiens | NPHS1 | O60500 | 0.47 | 0.02 | Downregulated |
| Next to BRCA1 gene 1 protein | NBR1 | Q14596 | 0.39 | 0.02 | downregulated |
| Calcium signal-modulating cyclophilin ligand | CAMLG | P49069 | 0.38 | 0.02 | Downregulated |
| Nicotinamide/nicotinic acid mononucleotide adenylyltransferase 2 | NMNAT2 | Q9BZQ4 | 0.36 | 0.04 | Downregulated |
| Neuroblastoma suppressor of tumorigenicity 1 | NBL1 | P41271 | 0.32 | 0.04 | Downregulated |
| Hemoglobin subunit beta | HBB | P68871 | 0.32 | 0.03 | Downregulated |
| Zinc finger protein 224 | ZNF224 | Q9NZL3 | 0.19 | 0.01 | Downregulated * |
| OTU domain-containing protein 7B | OTUD7B | Q6GQQ9 | 0.15 | 0.03 | Downregulated |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zebene, E.D.; Pucci, B.; Lombardi, R.; Medhin, H.T.; Seife, E.; Di Gennaro, E.; Budillon, A.; Woldemichael, G.B. Serum-Based Proteomic Approach to Identify Clinical Biomarkers of Radiation Exposure. Cancers 2025, 17, 1010. https://doi.org/10.3390/cancers17061010
Zebene ED, Pucci B, Lombardi R, Medhin HT, Seife E, Di Gennaro E, Budillon A, Woldemichael GB. Serum-Based Proteomic Approach to Identify Clinical Biomarkers of Radiation Exposure. Cancers. 2025; 17(6):1010. https://doi.org/10.3390/cancers17061010
Chicago/Turabian StyleZebene, Emeshaw Damtew, Biagio Pucci, Rita Lombardi, Hagos Tesfay Medhin, Edom Seife, Elena Di Gennaro, Alfredo Budillon, and Gurja Belay Woldemichael. 2025. "Serum-Based Proteomic Approach to Identify Clinical Biomarkers of Radiation Exposure" Cancers 17, no. 6: 1010. https://doi.org/10.3390/cancers17061010
APA StyleZebene, E. D., Pucci, B., Lombardi, R., Medhin, H. T., Seife, E., Di Gennaro, E., Budillon, A., & Woldemichael, G. B. (2025). Serum-Based Proteomic Approach to Identify Clinical Biomarkers of Radiation Exposure. Cancers, 17(6), 1010. https://doi.org/10.3390/cancers17061010







