The Cx43-Mediated Autophagy Mechanism Influences Triple-Negative Breast Cancer Through the Regulation of Rab31
Simple Summary
Abstract
1. Background
2. Materials and Methods
2.1. Clinical Breast Cancer Tissue Samples
2.2. Cell Lines and Culture Conditions
2.3. siRNA Transfection, Lentivirus Infection, and Overexpression Constructs
2.4. qRT-PCR
2.5. Western Blot (WB)
2.6. Transwell
2.7. Wound Healing
2.8. CCK-8
2.9. Immunofluorescence
2.10. Co-IP
2.11. Immunohistochemistry (IHC)
2.12. In Vivo Tumorigenesis Assay
2.13. Statistical Analysis
2.14. Bioinformatics and Database Analyses
3. Results
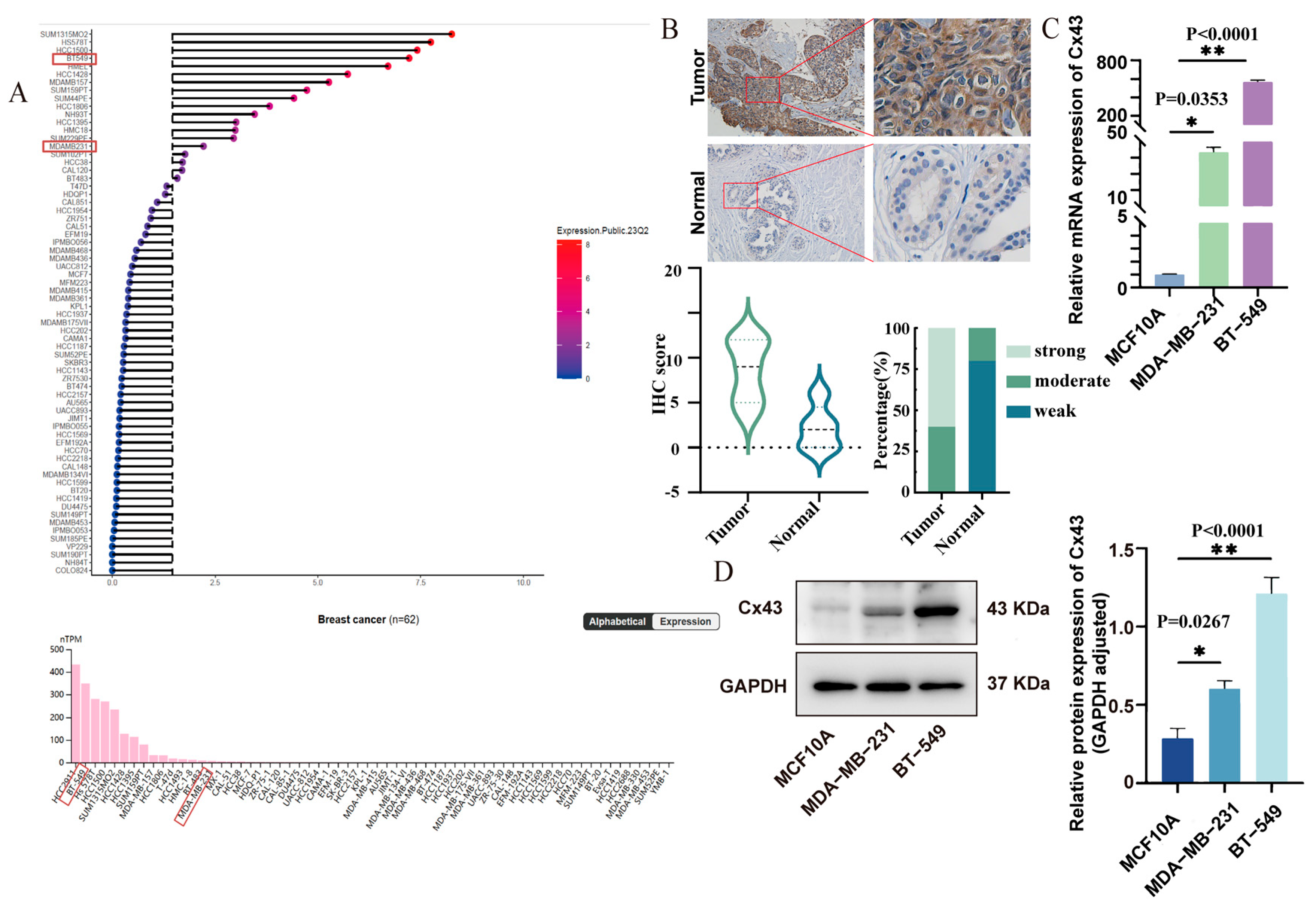
3.1. Cx43 Was Upregulated in Triple-Negative Breast Cancer Tissues and Cell Lines
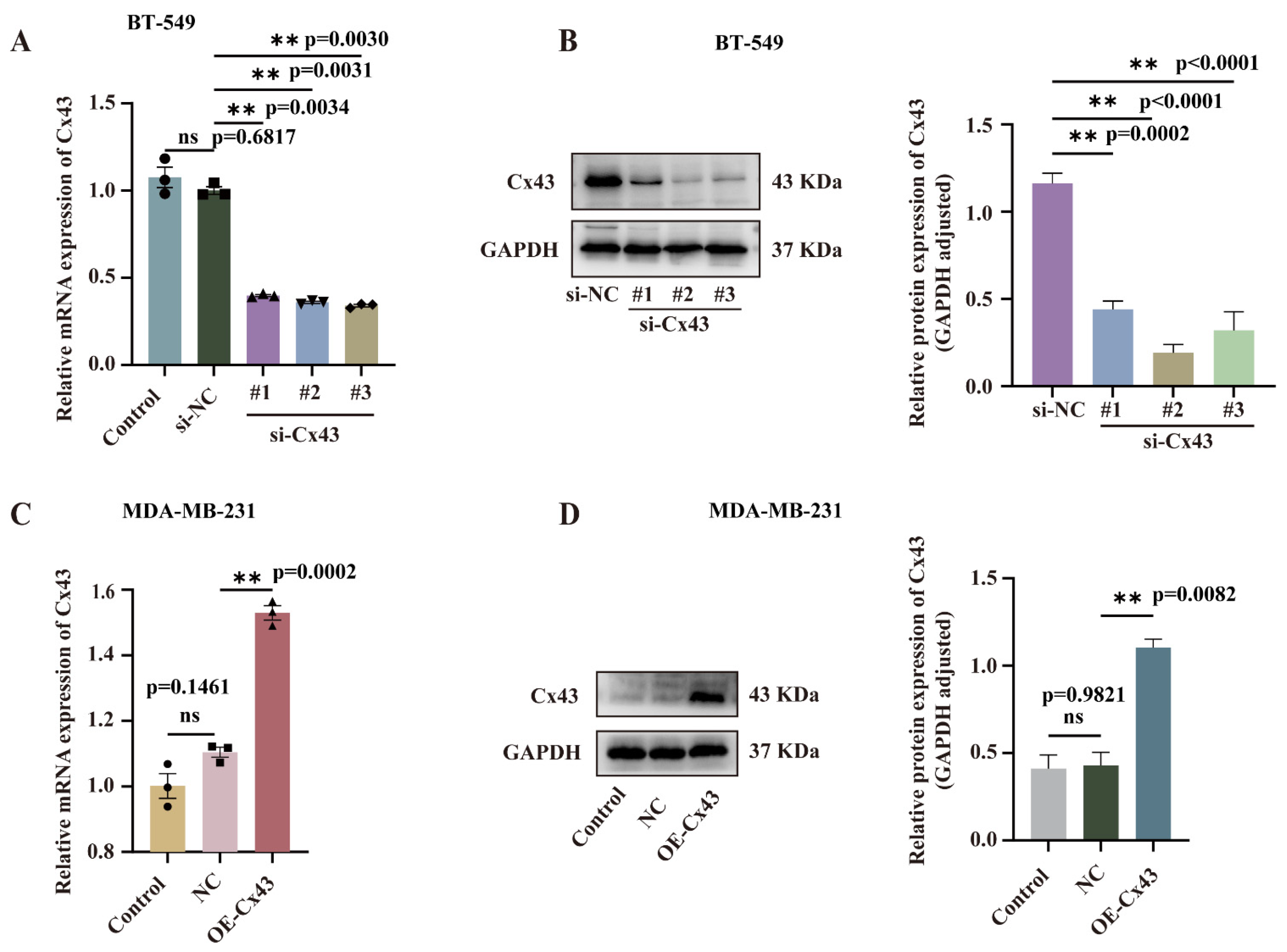
3.2. Validation of the Efficiency of Cx43 Interference and Overexpression
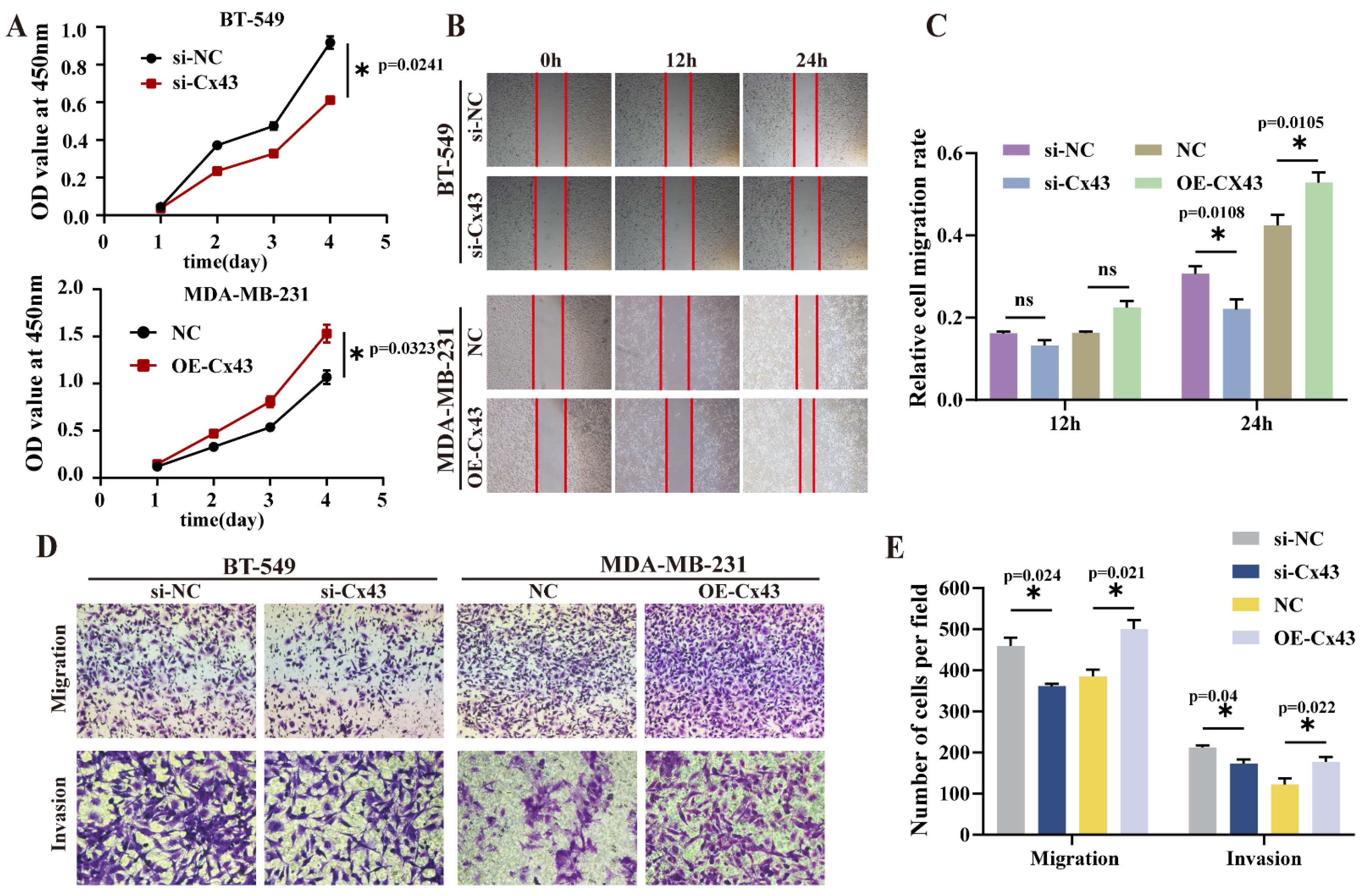
3.3. Inference of Cx43 Expression Significantly Suppressed Cell Proliferation, Migration, and Invasion, Whereas the Overexpression of Cx43 Promoted These Processes
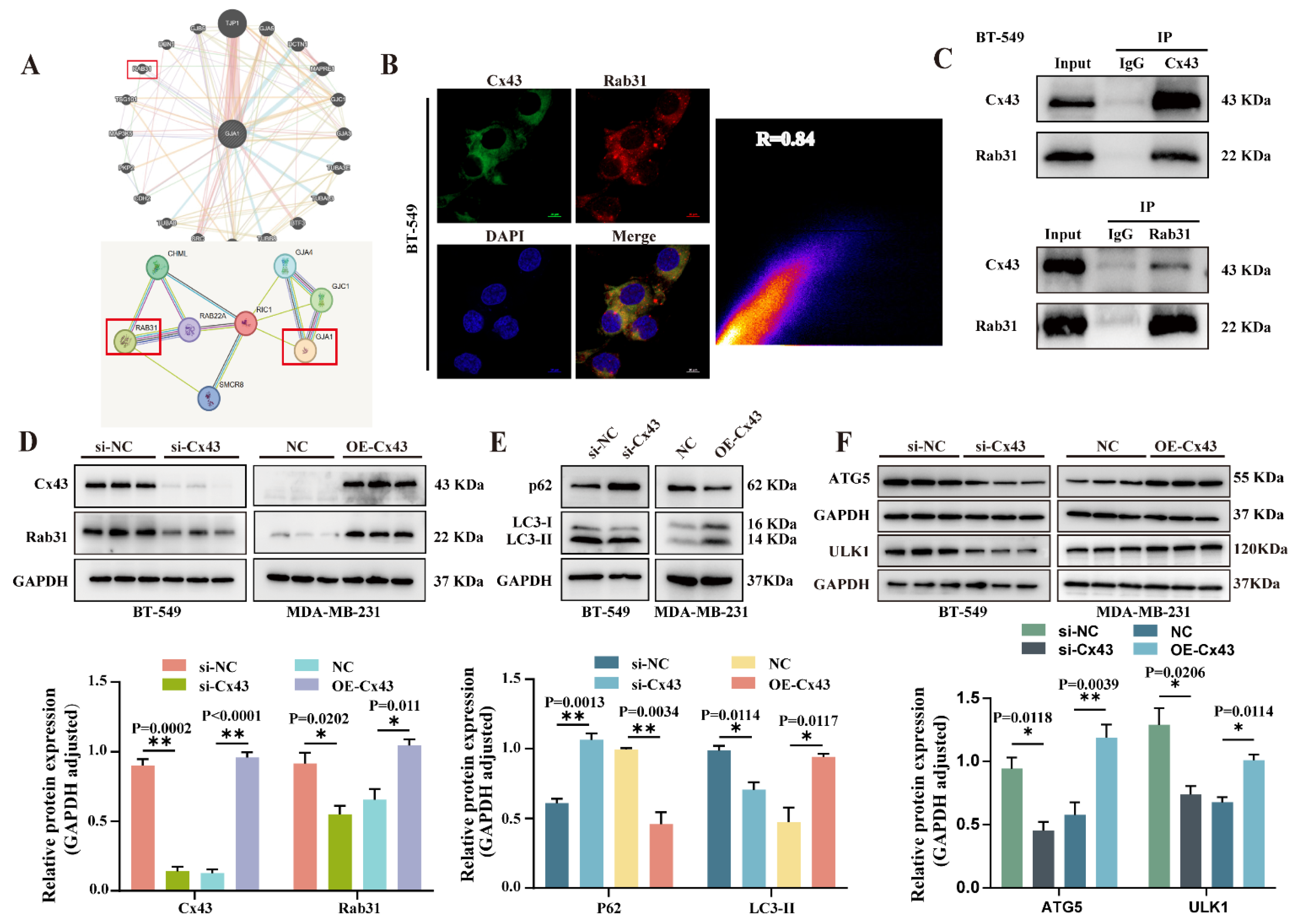
3.4. Cx43 Interacts with Rab31 to Regulate the Autophagy Pathway in Triple-Negative Breast Cancer Cells
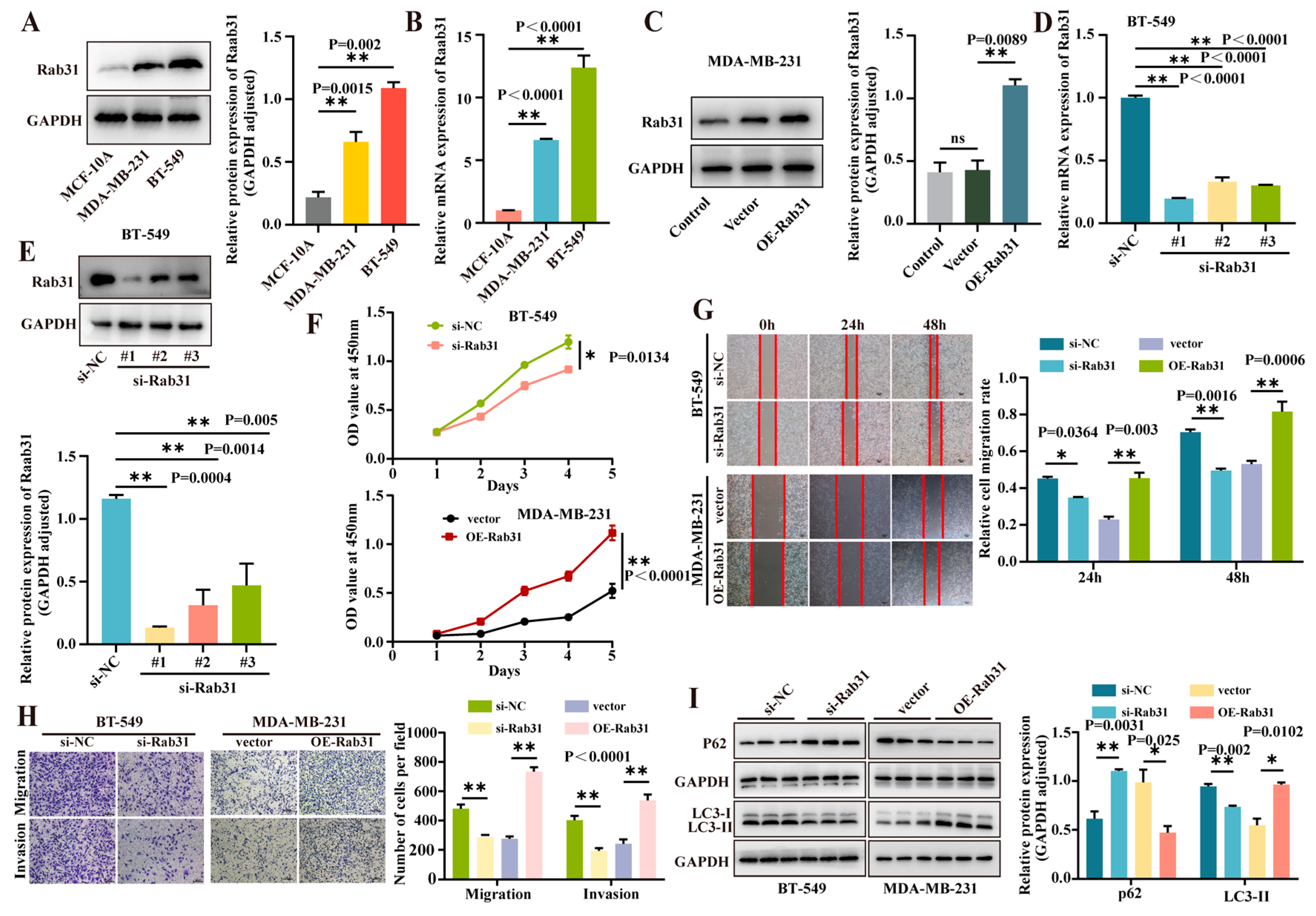
3.5. Rab31 Was Highly Expressed in Triple-Negative Breast Cancer Cells, and Interference of Rab31 Expression Inhibited Cell Proliferation, Migration, and Invasion, While Overexpression of Rab31 Promoted These Processes
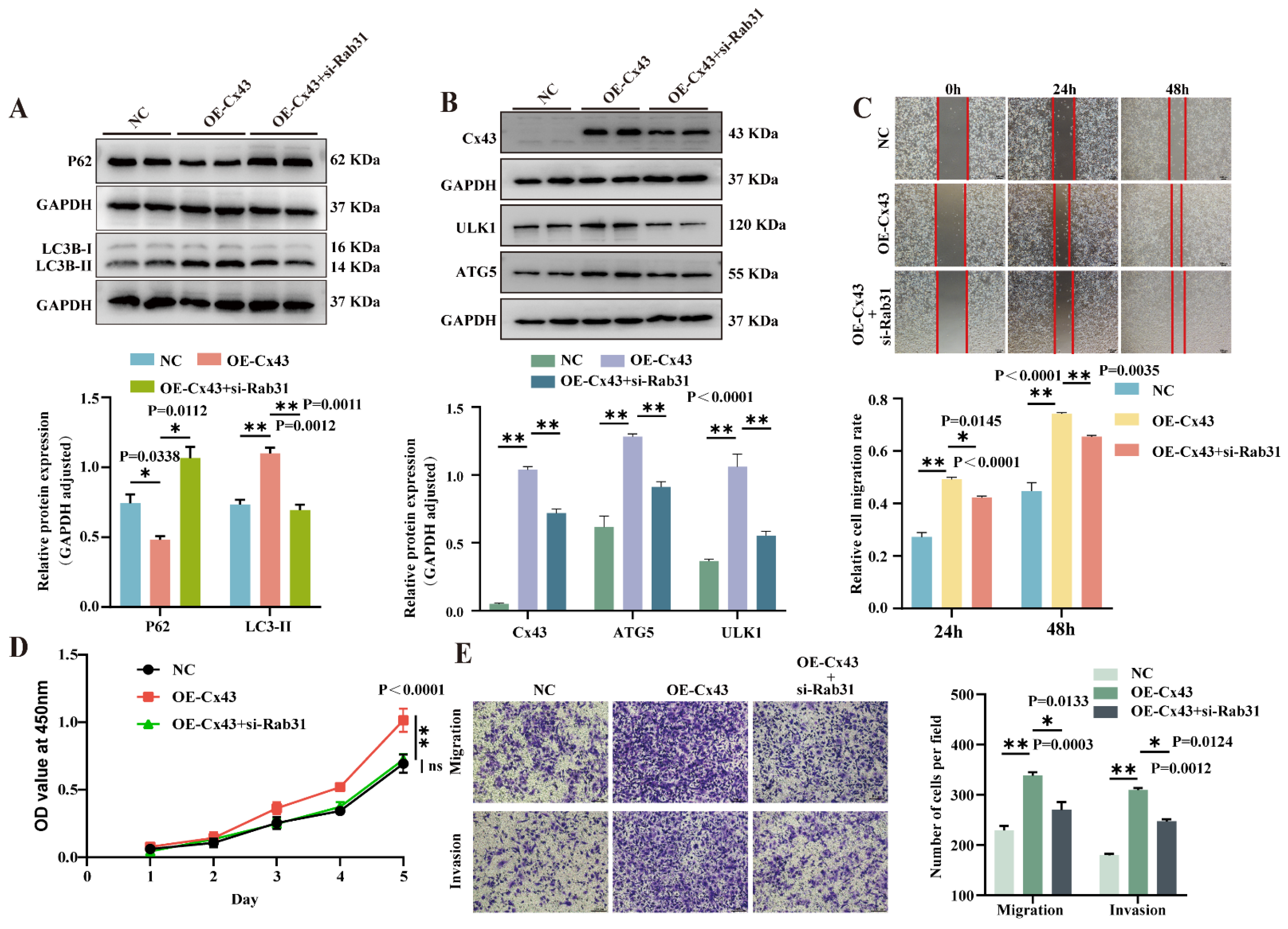
3.6. Knockdown of Rab31 Reversed the Effects of Cx43 Overexpression on Autophagy and Biological Characteristics in Triple-Negative Breast Cancer Cells
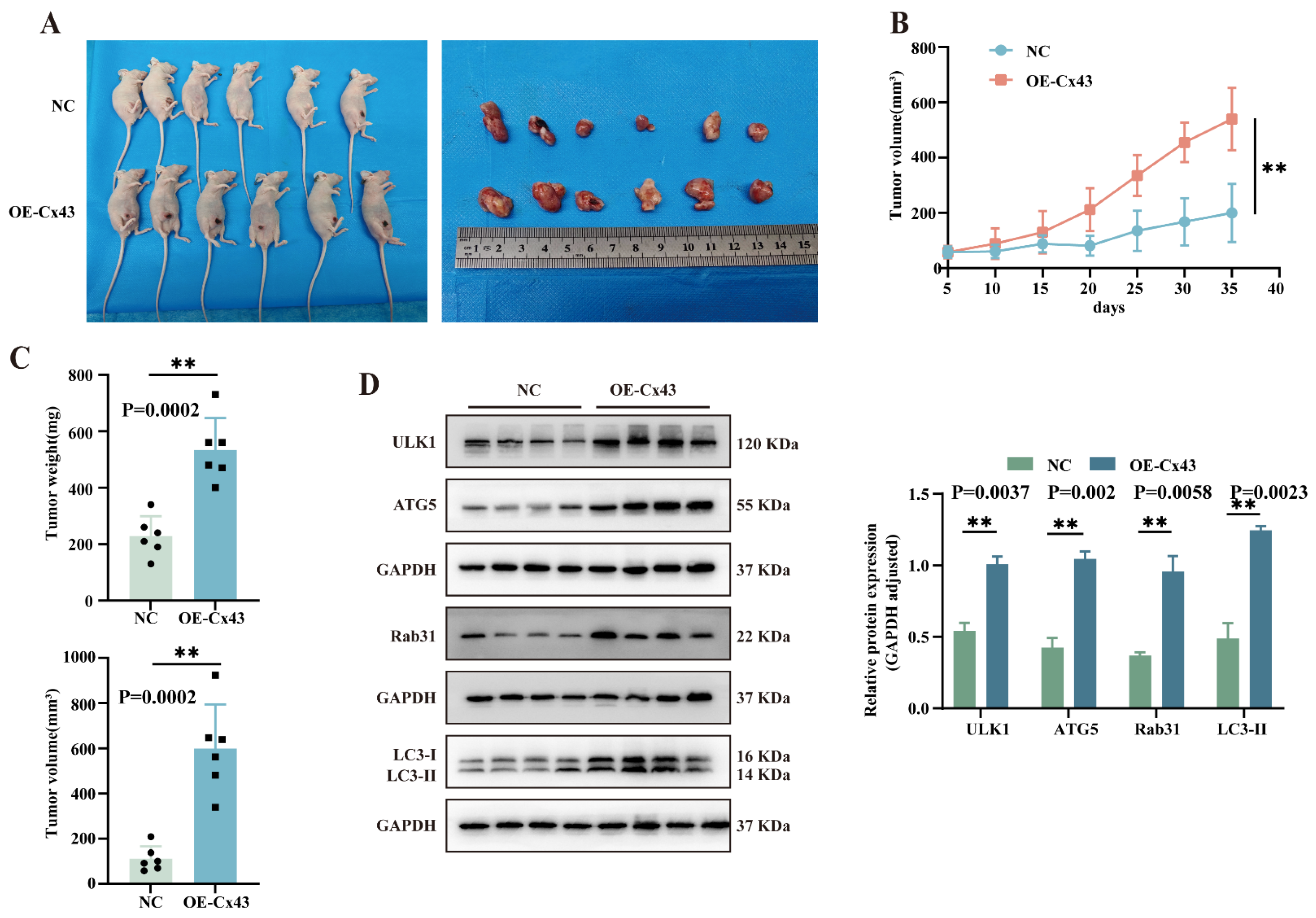
3.7. Overexpression of Cx43 Promotes Triple-Negative Breast Tumor Growth In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviations
| Abbreviation | Full Term |
| TNBC | Triple-negative breast cancer |
| Cx43 | Connexin43 |
| Rab31 | Ras-related protein Rab-31 |
| ULK1 | Unc-51-like autophagy activating kinase 1 |
| ATG5 | Autophagy-related gene 5 |
| LC3 | Microtubule-associated proteins 1A/1B light chain 3 |
| SQSTM1/p62 | Sequestosome 1 |
| CTC | Circulating tumor cell |
| OE-CX43/NC | Over-expression of CX43/ negative control |
| si-CX43/si-NC | siRNA targeting CX43/interference negative control |
| OE-Rab31/vector | Over-expression of RAB31/negative control |
| GJ | Gap junction |
| ER | Estrogen receptor |
| PR | Progesterone receptor |
| HER2 | Human epidermal growth factor |
| WB | Western blot |
| IHC | Immunohistochemistry |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Acs, B.; Hartman, J.; Sönmez, D.; Lindman, H.; Johansson, A.L.V.; Fredriksson, I. Real-world overall survival and characteristics of patients with ER-zero and ER-low HER2-negative breast cancer treated as triple-negative breast cancer: A Swedish population-based cohort study. Lancet Reg. Health Eur. 2024, 40, 100886. [Google Scholar] [CrossRef]
- Tang, D.D.; Ye, Z.J.; Liu, W.W.; Wu, J.; Tan, J.Y.; Zhang, Y.; Xu, Q.; Xiang, Y.B. Survival feature and trend of female breast cancer: A comprehensive review of survival analysis from cancer registration data. Breast 2025, 79, 103862. [Google Scholar] [CrossRef]
- Leybaert, L.; Lampe, P.D.; Dhein, S.; Kwak, B.R.; Ferdinandy, P.; Beyer, E.C.; Laird, D.W.; Naus, C.C.; Green, C.R.; Schulz, R. Connexins in Cardiovascular and Neurovascular Health and Disease: Pharmacological Implications. Pharmacol. Rev. 2017, 69, 396–478. [Google Scholar] [CrossRef]
- Zhang, D.; Kaneda, M.; Nakahama, K.; Arii, S.; Morita, I. Connexin 43 expression promotes malignancy of HuH7 hepatocellular carcinoma cells via the inhibition of cell-cell communication. Cancer Lett. 2007, 252, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Leroy, K.; Silva Costa, C.J.; Pieters, A.; Dos Santos Rodrigues, B.; Van Campenhout, R.; Cooreman, A.; Tabernilla, A.; Cogliati, B.; Vinken, M. Expression and Functionality of Connexin-Based Channels in Human Liver Cancer Cell Lines. Int. J. Mol. Sci. 2021, 22, 12187. [Google Scholar] [CrossRef]
- Sinha, S.; Callow, B.W.; Farfel, A.P.; Roy, S.; Chen, S.; Masotti, M.; Rajendran, S.; Buschhaus, J.M.; Espinoza, C.R.; Luker, K.E.; et al. Breast cancers that disseminate to bone marrow acquire aggressive phenotypes through CX43-related tumor-stroma tunnels. J. Clin. Invest. 2024, 134, e170953. [Google Scholar] [CrossRef]
- Ming, J.; Fan, L.J.; Zhong, L.; Chen, Q.Q.; Jiang, J. Expression changes of microRNA206 and connexin 43 in primary breast cancer lesions and axillary metastatic lymph nodes. Chin. J. Breast Dis. (Electron. Ed.) 2011, 5, 38–41. [Google Scholar]
- Wang, D.Q.; Wang, Y.Y.; Shi, Y.L.; Zeng, B.; Lin, Z.J.; Deng, Q.; Ming, J. Correlation between connexin 43 expression in circulating tumor cells and biological characteristics of breast cancer. Heliyon 2023, 9, e18697. [Google Scholar] [CrossRef] [PubMed]
- Chiou, J.T.; Huang, C.H.; Lee, Y.C.; Wang, L.J.; Shi, Y.J.; Chen, Y.J.; Chang, L.S. Compound C induces autophagy and apoptosis in parental and hydroquinone-selected malignant leukemia cells through the ROS/p38 MAPK/AMPK/TET2/FOXP3 axis. Cell Biol. Toxicol. 2020, 36, 315–331. [Google Scholar] [CrossRef]
- Sun, K.; Deng, W.; Zhang, S.; Cai, N.; Jiao, S.; Song, J.; Wei, L. Paradoxical roles of autophagy in different stages of tumorigenesis: Protector for normal or cancer cells. Cell Biosci. 2013, 3, 35. [Google Scholar] [CrossRef]
- Poole, L.P.; Macleod, K.F. Mitophagy in tumorigenesis and metastasis. Cell Mol. Life Sci. 2021, 78, 3817–3851. [Google Scholar] [CrossRef]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Koh, M.; Lim, H.; Jin, H.; Kim, M.; Hong, Y.; Hwang, Y.K.; Woo, Y.; Kim, E.S.; Kim, S.Y.; Kim, K.M.; et al. ANXA2 (annexin A2) is crucial to ATG7-mediated autophagy, leading to tumor aggressiveness in triple-negative breast cancer cells. Autophagy 2024, 20, 659–674. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Zhu, L.; Luo, Y.; Tang, Y.; Tan, Q.; Zou, Y.; Chen, K.; Deng, X.; Tang, H.; Li, H.; et al. Autophagy Deficiency Induced by SAT1 Potentiates Tumor Progression in Triple-Negative Breast Cancer. Adv. Sci. 2024, 11, e2309903. [Google Scholar] [CrossRef]
- Zhang, W.; Tang, Y.; Yang, P.; Chen, Y.; Xu, Z.; Qi, C.; Huang, H.; Liu, R.; Qin, H.; Ke, H.; et al. TMX2 potentiates cell viability of hepatocellular carcinoma by promoting autophagy and mitophagy. Autophagy 2024, 20, 2146–2163. [Google Scholar] [CrossRef]
- Gielen, P.R.; Aftab, Q.; Ma, N.; Chen, V.C.; Hong, X.; Lozinsky, S.; Naus, C.C.; Sin, W.C. Connexin43 confers Temozolomide resistance in human glioma cells by modulating the mitochondrial apoptosis pathway. Neuropharmacology 2013, 75, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Kutova, O.M.; Pospelov, A.D.; Balalaeva, I.V. The Multifaceted Role of Connexins in Tumor Microenvironment Initiation and Maintenance. Biology 2023, 12, 204. [Google Scholar] [CrossRef]
- Chua, C.E.; Tang, B.L. The role of the small GTPase Rab31 in cancer. J. Cell Mol. Med. 2015, 19, 1–10. [Google Scholar] [CrossRef]
- Yang, C.C.; Meng, G.X.; Dong, Z.R.; Li, T. Role of Rab GTPases in Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2021, 8, 1389–1397. [Google Scholar] [CrossRef]
- Soelch, S.; Beaufort, N.; Loessner, D.; Kotzsch, M.; Reuning, U.; Luther, T.; Kirchner, T.; Magdolen, V. Rab31-dependent regulation of transforming growth factor ß expression in breast cancer cells. Mol. Med. 2021, 27, 158. [Google Scholar] [CrossRef]
- Banworth, M.J.; Liang, Z.; Li, G. A novel membrane targeting domain mediates the endosomal or Golgi localization specificity of small GTPases Rab22 and Rab31. J. Biol. Chem. 2022, 298, 102281. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xu, S.; Li, J.; Wang, N.; Zheng, Y.; Wang, Z. XIAOPI formula inhibits chemoresistance and metastasis of triple-negative breast cancer by suppressing extracellular vesicle/CXCL1-induced TAM/PD-L1 signaling. Phytomedicine 2024, 135, 156039. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Tang, C.; Zhang, Q.W.; Zhuang, Q.; Ye, X.; Xia, J.; Shi, Y.; Ning, M.; Dong, Z.X.; Wan, X.J. Overexpression of RAB31 in gastric cancer is associated with released exosomes and increased tumor cell invasion and metastasis. Cancer Med. 2023, 12, 13497–13510. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Wang, M.; Zhao, C.; Shen, M.; Yu, Y.; He, L.; Zhao, Y.; Chen, H.; Shi, X.; Zhou, M.; et al. TFEB-driven autophagy potentiates TGF-β induced migration in pancreatic cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 340. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Ji, N.; Tang, Z.; Li, J.; Chen, Q. The role of extracellular vesicles from different origin in the microenvironment of head and neck cancers. Mol. Cancer 2019, 18, 83. [Google Scholar] [CrossRef]
- Chen, H.T.; Liu, H.; Mao, M.J.; Tan, Y.; Mo, X.Q.; Meng, X.J.; Cao, M.T.; Zhong, C.Y.; Liu, Y.; Shan, H.; et al. Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy. Mol. Cancer 2019, 18, 101. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.Y.; Chang, C.J.; Cheng, J.S. Survival, treatment regimens and medical costs of women newly diagnosed with metastatic triple-negative breast cancer. Sci. Rep. 2022, 12, 729. [Google Scholar] [CrossRef] [PubMed]
- Strauss, R.E.; Gourdie, R.G. Cx43 and the Actin Cytoskeleton: Novel Roles and Implications for Cell-Cell Junction-Based Barrier Function Regulation. Biomolecules 2020, 10, 1656. [Google Scholar] [CrossRef]
- Ismail, R.; Rashid, R.; Andrabi, K.; Parray, F.Q.; Besina, S.; Shah, M.A.; Ul Hussain, M. Pathological implications of Cx43 down-regulation in human colon cancer. Asian Pac. J. Cancer Prev. 2014, 15, 2987–2991. [Google Scholar] [CrossRef]
- Zhou, J.Z.; Riquelme, M.A.; Gu, S.; Kar, R.; Gao, X.; Sun, L.; Jiang, J.X. Osteocytic connexin hemichannels suppress breast cancer growth and bone metastasis. Oncogene 2016, 35, 5597–5607. [Google Scholar] [CrossRef] [PubMed]
- Kazan, J.M.; El-Saghir, J.; Saliba, J.; Shaito, A.; Jalaleddine, N.; El-Hajjar, L.; Al-Ghadban, S.; Yehia, L.; Zibara, K.; El-Sabban, M. Cx43 Expression Correlates with Breast Cancer Metastasis in MDA-MB-231 Cells In Vitro, In a Mouse Xenograft Model and in Human Breast Cancer Tissues. Cancers 2019, 11, 460. [Google Scholar] [CrossRef]
- Fu, Y.; Sun, X.; He, Q.; Gu, Z.; Jia, X.; Zhuang, Z. Connexin 43 controls metastatic behavior in triple negative breast cancer through TGFβ1-Smad3-intergin αV signaling axis Based on optical image diagnosis. SLAS Technol. 2024, 29, 100190. [Google Scholar] [CrossRef] [PubMed]
- McLachlan, E.; Shao, Q.; Wang, H.L.; Langlois, S.; Laird, D.W. Connexins act as tumor suppressors in three-dimensional mammary cell organoids by regulating differentiation and angiogenesis. Cancer Res. 2006, 66, 9886–9894. [Google Scholar] [CrossRef]
- Hirschi, K.K.; Xu, C.E.; Tsukamoto, T.; Sager, R. Gap junction genes Cx26 and Cx43 individually suppress the cancer phenotype of human mammary carcinoma cells and restore differentiation potential. Cell Growth Differ. 1996, 7, 861–870. [Google Scholar] [PubMed]
- Grek, C.L.; Rhett, J.M.; Bruce, J.S.; Ghatnekar, G.S.; Yeh, E.S. Connexin 43, breast cancer tumor suppressor: Missed connections? Cancer Lett. 2016, 374, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Tishchenko, A.; Azorín, D.D.; Vidal-Brime, L.; Muñoz, M.J.; Arenas, P.J.; Pearce, C.; Girao, H.; Cajal, S.R.Y.; Aasen, T. Cx43 and Associated Cell Signaling Pathways Regulate Tunneling Nanotubes in Breast Cancer Cells. Cancers 2020, 12, 2798. [Google Scholar] [CrossRef]
- Shen, Y.; Li, M.; Liao, L.; Gao, S.; Wang, Y. Plasma exosome-derived connexin43 as a promising biomarker for melanoma patients. BMC Cancer 2023, 23, 242. [Google Scholar] [CrossRef]
- Zibara, K.; Awada, Z.; Dib, L.; El-Saghir, J.; Al-Ghadban, S.; Ibrik, A.; El-Zein, N.; El-Sabban, M. Anti-angiogenesis therapy and gap junction inhibition reduce MDA-MB-231 breast cancer cell invasion and metastasis in vitro and in vivo. Sci. Rep. 2015, 5, 12598. [Google Scholar] [CrossRef]
- Ek-Vitorín, J.F.; Pontifex, T.K.; Burt, J.M. Cx43 Channel Gating and Permeation: Multiple Phosphorylation-Dependent Roles of the Carboxyl Terminus. Int. J. Mol. Sci. 2018, 19, 1659. [Google Scholar] [CrossRef]
- Kang, M.; Lin, N.; Li, C.; Meng, Q.; Zheng, Y.; Yan, X.; Deng, J.; Ou, Y.; Zhang, C.; He, J.; et al. Cx43 phosphorylation on S279/282 and intercellular communication are regulated by IP3/IP3 receptor signaling. Cell Commun. Signal. 2014, 12, 58. [Google Scholar] [CrossRef]
- Stenmark, H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef]
- Yang, T.; Huang, Z.; Wang, Z.; Qian, X.; Yue, L.; Ge, X.; Wei, J.; Shu, Z.; Ding, K. Increased RAB31 Expression in Cancer-Associated Fibroblasts Promotes Colon Cancer Progression Through HGF-MET Signaling. Front. Oncol. 2020, 10, 1747. [Google Scholar] [CrossRef]
- Chen, K.; Xu, J.; Tong, Y.L.; Yan, J.F.; Pan, Y.; Wang, W.J.; Zheng, L.; Zheng, X.X.; Hu, C.; Hu, X.; et al. Rab31 promotes metastasis and cisplatin resistance in stomach adenocarcinoma through Twist1-mediated EMT. Cell Death Dis. 2023, 14, 115. [Google Scholar] [CrossRef]
- Sui, Y.; Zheng, X.; Zhao, D. Rab31 promoted hepatocellular carcinoma (HCC) progression via inhibition of cell apoptosis induced by PI3K/AKT/Bcl-2/BAX pathway. Tumour Biol. 2015, 36, 8661–8670. [Google Scholar] [CrossRef] [PubMed]
- Yoon, D.; Bae, K.; Kim, J.H.; Choi, Y.K.; Yoon, K.A. Oncogenic Effect of the Novel Fusion Gene VAPA-Rab31 in Lung Adenocarcinoma. Int. J. Mol. Sci. 2019, 20, 2309. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, R.; Han, X.; Hou, X.; Tian, Y.; Zhang, W. Rab31 promotes the invasion and metastasis of cervical cancer cells by inhibiting MAPK6 degradation. Int. J. Biol. Sci. 2022, 18, 112–123. [Google Scholar] [CrossRef]
- Sun, Y.; Cao, Y.; Wan, H.; Memetimin, A.; Cao, Y.; Li, L.; Wu, C.; Wang, M.; Chen, S.; Li, Q.; et al. A mitophagy sensor PPTC7 controls BNIP3 and NIX degradation to regulate mitochondrial mass. Mol. Cell 2024, 84, 327–344.e9. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Emr, S.D. Autophagy as a regulated pathway of cellular degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef] [PubMed]
- Mathew, R.; Karantza-Wadsworth, V.; White, E. Role of autophagy in cancer. Nat. Rev. Cancer 2007, 7, 961–967. [Google Scholar] [CrossRef]
- Folkerts, H.; Hilgendorf, S.; Vellenga, E.; Bremer, E.; Wiersma, V.R. The multifaceted role of autophagy in cancer and the microenvironment. Med. Res. Rev. 2019, 39, 517–560. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Wang, J.; Wang, X.; Cai, S.; Guo, Y.; Ye, L.; Li, D.; Hu, A.; Jin, S.; Yuan, B.; et al. Therapeutic targeting of the USP2-E2F4 axis inhibits autophagic machinery essential for zinc homeostasis in cancer progression. Autophagy 2022, 18, 2615–2635. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, S.; Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19, 12. [Google Scholar] [CrossRef] [PubMed]
- Prashar, A.; Bussi, C.; Fearns, A.; Capurro, M.I.; Gao, X.; Sesaki, H.; Gutierrez, M.G.; Jones, N.L. Lysosomes drive the piecemeal removal of mitochondrial inner membrane. Nature 2024, 632, 1110–1117. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, H.; Song, X.; Yang, Q. Metastatic heterogeneity of breast cancer: Molecular mechanism and potential therapeutic targets. Semin. Cancer Biol. 2020, 60, 14–27. [Google Scholar] [CrossRef]
- Hu, X.; Pan, G.; Luo, J.; Gao, X.; Mu, Y.; Wang, Z.; Hu, X.; Li, C.; Abbas, M.N.; Zhang, K.; et al. Kuwanon H Inhibits Melanoma Growth through Cytotoxic Endoplasmic Reticulum Stress and Impaired Autophagy Flux. J. Agric. Food Chem. 2023, 71, 13768–13782. [Google Scholar] [CrossRef]
- Luo, J.; Gong, L.; Yang, Y.; Zhang, Y.; Liu, Q.; Bai, L.; Fang, X.; Zhang, B.; Huang, J.; Liu, M.; et al. Enhanced mitophagy driven by ADAR1-GLI1 editing supports the self-renewal of cancer stem cells in HCC. Hepatology 2024, 79, 61–78. [Google Scholar] [CrossRef] [PubMed]
- Catalano, M.; D’Alessandro, G.; Lepore, F.; Corazzari, M.; Caldarola, S.; Valacca, C.; Faienza, F.; Esposito, V.; Limatola, C.; Cecconi, F.; et al. Autophagy induction impairs migration and invasion by reversing EMT in glioblastoma cells. Mol. Oncol. 2015, 9, 1612–1625. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef]
- Zachari, M.; Ganley, I.G. The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 2017, 61, 585–596. [Google Scholar] [CrossRef]
- Bejarano, E.; Yuste, A.; Patel, B.; Stout, R.F., Jr.; Spray, D.C.; Cuervo, A.M. Connexins modulate autophagosome biogenesis. Nat. Cell Biol. 2014, 16, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Tittarelli, A.; Janji, B.; Van Moer, K.; Noman, M.Z.; Chouaib, S. The Selective Degradation of Synaptic Connexin 43 Protein by Hypoxia-induced Autophagy Impairs Natural Killer Cell-mediated Tumor Cell Killing. J. Biol. Chem. 2015, 290, 23670–23679. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.F.; Varghese, R.T.; Lamouille, S.; Guo, S.; Pridham, K.J.; Kanabur, P.; Osimani, A.M.; Sharma, S.; Jourdan, J.; Rodgers, C.M.; et al. Connexin 43 Inhibition Sensitizes Chemoresistant Glioblastoma Cells to Temozolomide. Cancer Res. 2016, 76, 139–149. [Google Scholar] [CrossRef] [PubMed]
| F (5′→3′) | R (5′→3′) | ||
|---|---|---|---|
| Cx43 | AGTTCAATCACTTGGCGTGACTTC | GTTTGCCTAAGGCGCTCCAG | |
| Rab31 | GACCACAACATCAGCCCTACTAT | GCCAATGAATGAAACCGTTCCT | |
| GAPDH | GCACCGTCAAGGCTGAGAAC | TGGTGAAGACGCCAGTGGA | |
| si-NC | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT | |
| si-Cx43 | #1 | UGAAGCAGAUUGAGAUAAATT | UUUAUCUCAAUCUGCUUCATT |
| #2 | CAAGCAAGCAAGUGAGCAATT | UUGCUCACUUGCUUGCUUGTT | |
| #3 | CUGAUGACCUGGAGAUCUATT | UAGAUCUCCAGGUCAUCAGTT | |
| si-Rab31 | #1 | UGAAGGAUGCUAAGGAAUATT | UAUUCCUUAGCAUCCUUCATT |
| #2 | GGACACUGGGGUUGGGAAATT | UUUCCCAACCCCAGUGUCCTT | |
| #3 | GGUUGAGACAAGUGCAAAATT | UUUUGCACUUGUCUCAACCTT | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Wu, D.; Yang, T.; Lin, Z.-J.; Jiang, P.-Y.; Wang, T.-R.; Lu, Z.-J.; Wang, L.; Ming, J. The Cx43-Mediated Autophagy Mechanism Influences Triple-Negative Breast Cancer Through the Regulation of Rab31. Cancers 2025, 17, 3923. https://doi.org/10.3390/cancers17243923
Yang J, Wu D, Yang T, Lin Z-J, Jiang P-Y, Wang T-R, Lu Z-J, Wang L, Ming J. The Cx43-Mediated Autophagy Mechanism Influences Triple-Negative Breast Cancer Through the Regulation of Rab31. Cancers. 2025; 17(24):3923. https://doi.org/10.3390/cancers17243923
Chicago/Turabian StyleYang, Jiao, Die Wu, Ting Yang, Zi-Jing Lin, Pei-Yao Jiang, Ting-Rui Wang, Zheng-Jia Lu, Lu Wang, and Jia Ming. 2025. "The Cx43-Mediated Autophagy Mechanism Influences Triple-Negative Breast Cancer Through the Regulation of Rab31" Cancers 17, no. 24: 3923. https://doi.org/10.3390/cancers17243923
APA StyleYang, J., Wu, D., Yang, T., Lin, Z.-J., Jiang, P.-Y., Wang, T.-R., Lu, Z.-J., Wang, L., & Ming, J. (2025). The Cx43-Mediated Autophagy Mechanism Influences Triple-Negative Breast Cancer Through the Regulation of Rab31. Cancers, 17(24), 3923. https://doi.org/10.3390/cancers17243923






