Bacteroides fragilis Promotes Mesenchymal Subtype in Colorectal Cancer
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort and Samples
2.2. Cell Culture and Reagents
2.3. Patient-Derived Tumor Organoid Culture
2.4. Bacterial Culture and Treatment
2.5. Genomic DNA Isolation, Gene Library Amplification and Gel Electrophoresis
2.6. RNA Isolation, Sequencing and Consensus Molecular Subtype Analysis
2.7. 16S rRNA Gene Amplicon Sequencing
2.8. Comparative Analysis of Gut Microbiota in Colorectal Cancer CMSs
2.9. Analysis of TCGA Validation Cohort
2.10. Gene Set and Pathway Analysis
2.11. Statistical Analysis
3. Results
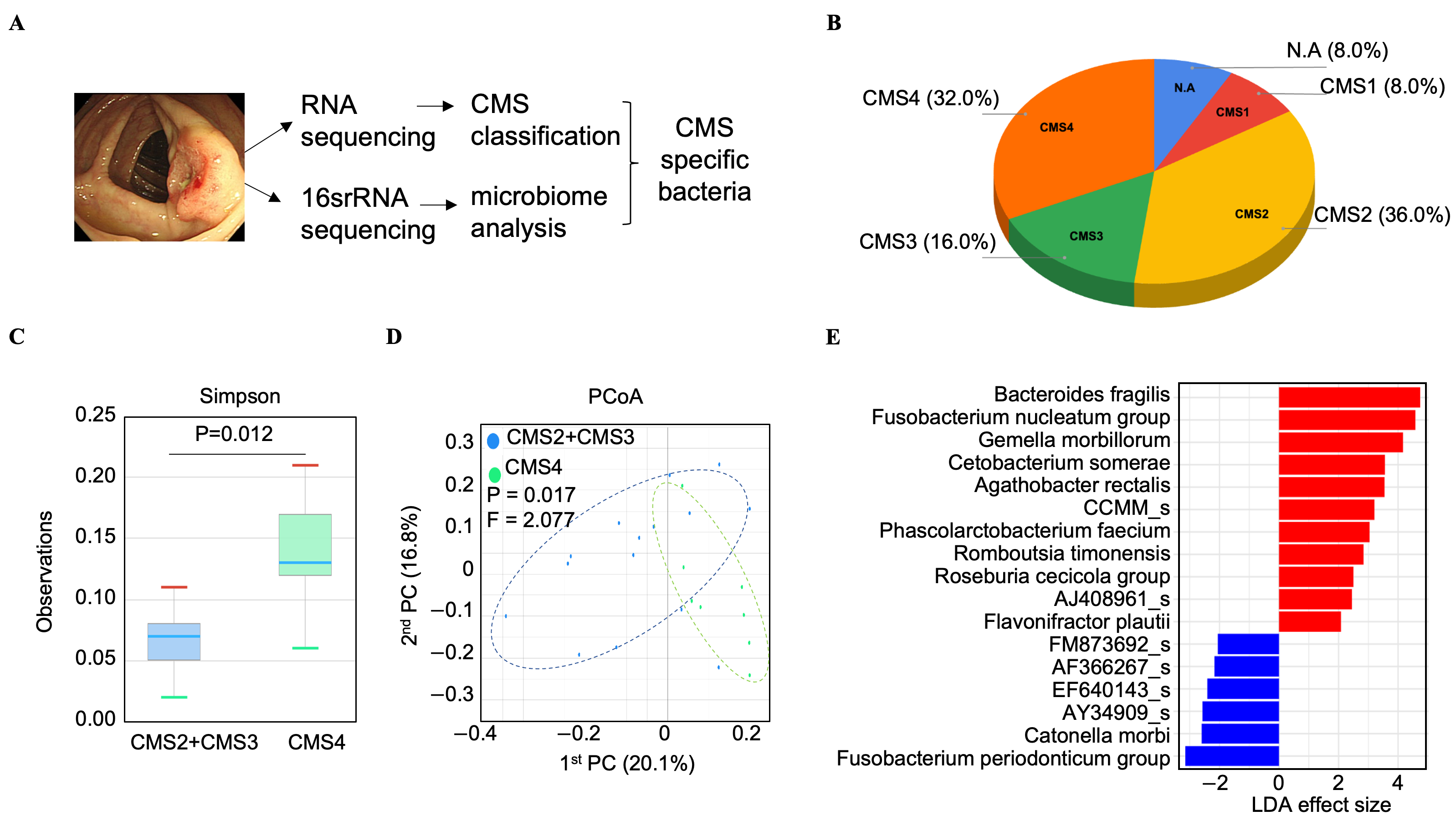
3.1. Bacteroides fragilis Is Significantly Enriched in the Microbiome of CMS4 Tumor Tissues
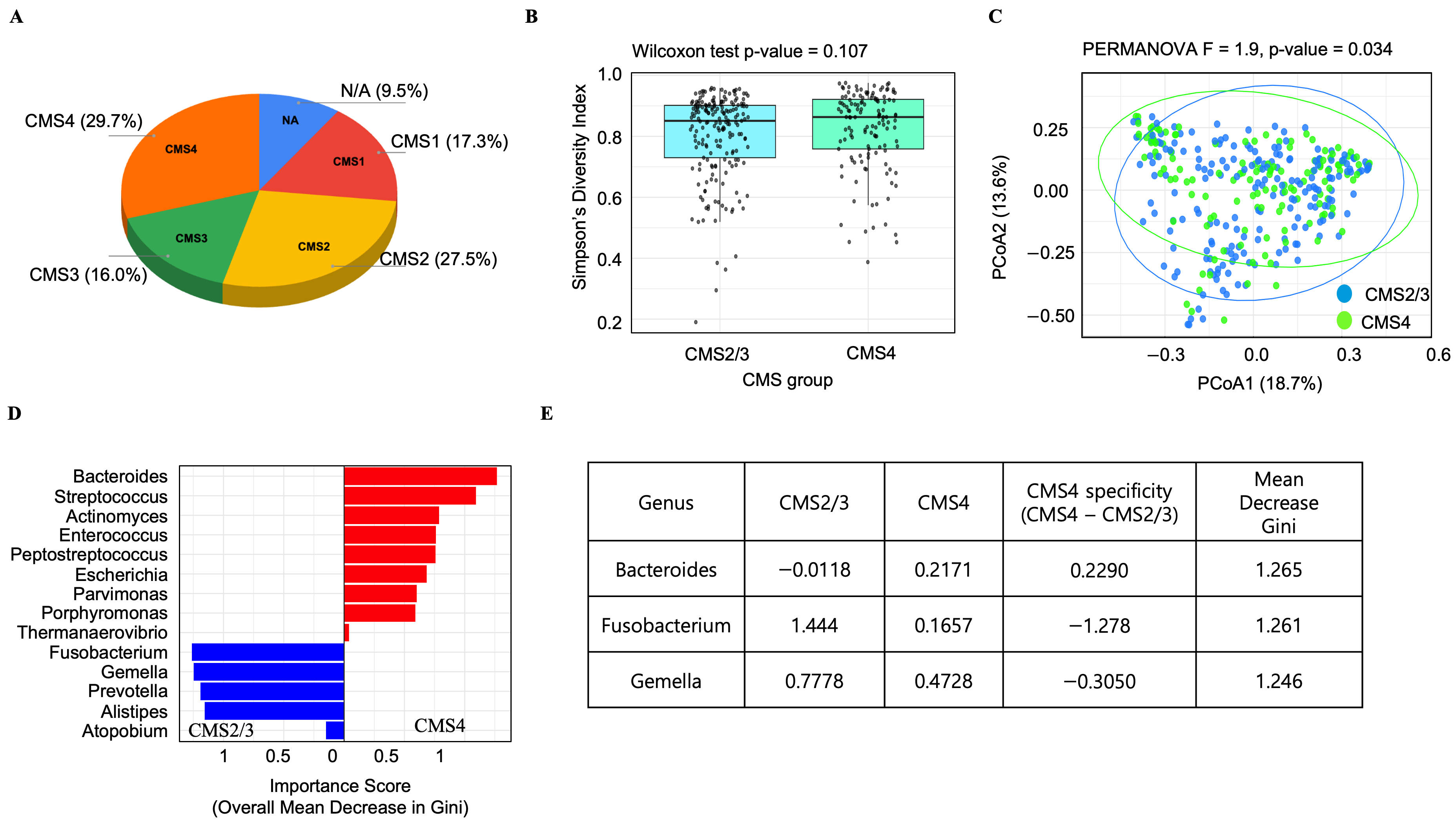
3.2. Analysis of the TCGA-COAD Dataset Reveals a CMS4-Specific Microbial Signature in Colorectal Cancer, Including a Notable Enrichment of Bacteroides
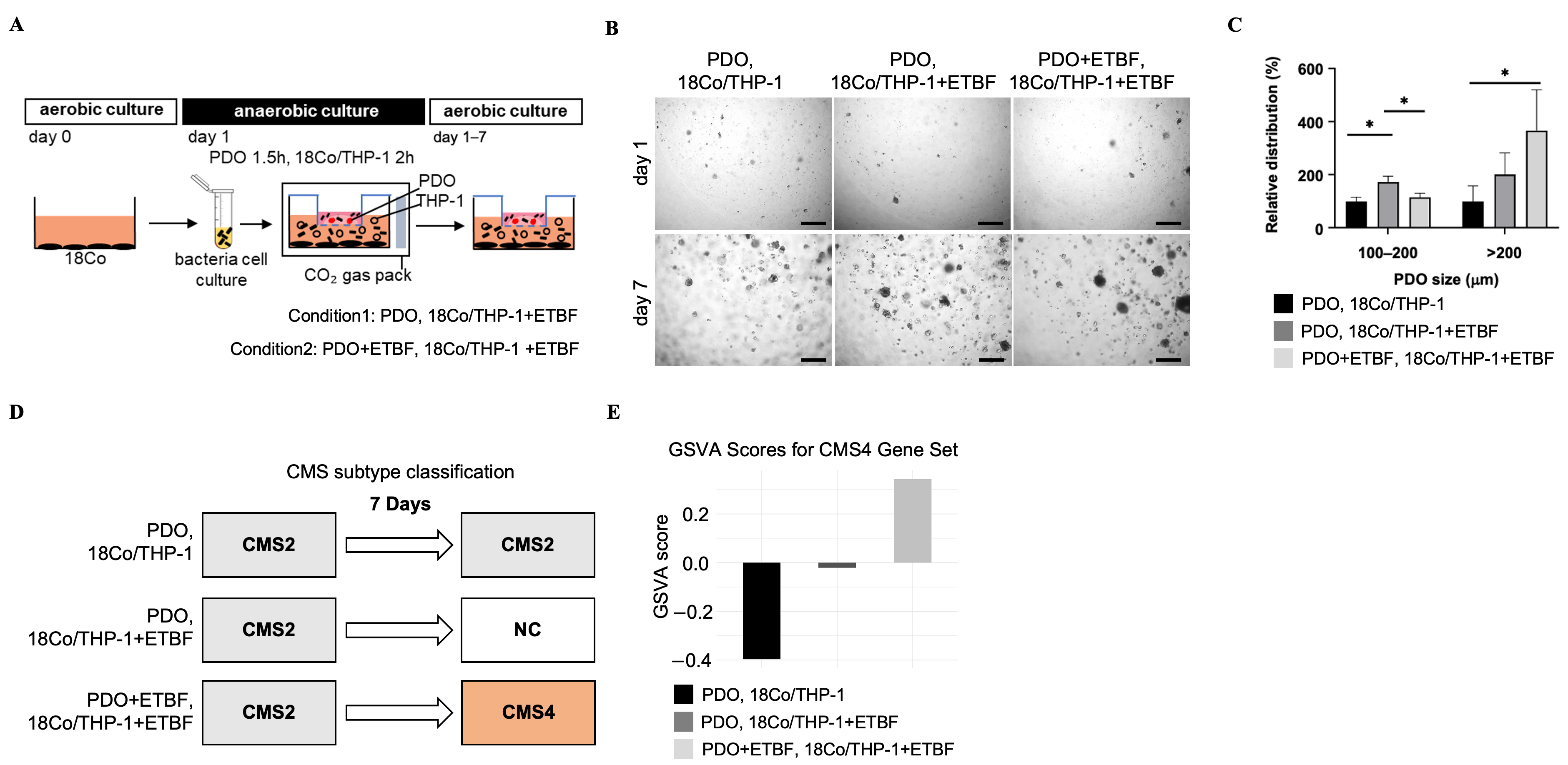
3.3. Bacteroides fragilis Enhances Growth and Changes the Gene Expression Profile into CMS4 in Co-Cultured Patient-Derived Organoids
3.4. ETBF-Treated Co-Cultured PDOs Acquire a Transcriptional Profile Similar to CMS4 Colon Tumors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRC | Colorectal cancer |
| CMS | Consensus molecular subtype |
| TME | Tumor microenvironment |
| ETBF | Enterotoxigenic Bacteroides fragilis |
| ECM | Extracellular matrix |
| PDO | Patient-derived organoid |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| FBS | Fetal bovine serum |
| PBS | Phosphate-buffered saline |
| PCR | Polymerase chain reaction |
| MTP | Microbiome taxonomic profiling |
| PCoA | Principal coordinate analysis |
| LDA | Linear discriminant analysis |
| GSVA | Gene set variation analysis |
| GSEA | Gene set enrichment analysis |
| MSigDB | Molecular Signatures Database |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| BFT | B. Fragilis toxin |
| IL | Interleukin |
| MDG | Mean decrease in Gini |
| DW | Distilled water |
| NTBF | Non-toxigenic Bacteroides fragilis |
| PD | Patient-derived |
| P | P value |
| F | PERMANOVA F value |
| PC | Principal coordinate |
| NA | Not applicable |
References
- Thanki, K.; Nicholls, M.E.; Gajjar, A.; Senagore, A.J.; Qiu, S.; Szabo, C.; Hellmich, M.R.; Chao, C. Consensus Molecular Subtypes of Colorectal Cancer and their Clinical Implications. Int. Biol. Biomed. J. 2017, 3, 105–111. [Google Scholar]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reynies, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Lu, Y.; Gu, D.; Zhao, C.; Sun, Y.; Li, W.; He, L.; Wang, X.; Kou, Z.; Su, J.; Guo, F. Genomic landscape and expression profile of consensus molecular subtype four of colorectal cancer. Front. Immunol. 2023, 14, 1160052. [Google Scholar] [CrossRef]
- Ten Hoorn, S.; de Back, T.R.; Sommeijer, D.W.; Vermeulen, L. Clinical Value of Consensus Molecular Subtypes in Colorectal Cancer: A Systematic Review and Meta-Analysis. J. Natl. Cancer Inst. 2022, 114, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.; Tong, L.; Sun, X. Acidic Tumor Microenvironments and Emerging Therapeutic Strategies for Cancer Therapy. Yonsei Med. J. 2025, 66, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Meng, W.J.; Wang, Z.Q. Cancer Stem Cells and the Tumor Microenvironment in Gastric Cancer. Front. Oncol. 2021, 11, 803974. [Google Scholar] [CrossRef]
- Koh, B.; Jeon, H.; Kim, D.; Kang, D.; Kim, K.R. Effect of fibroblast co-culture on the proliferation, viability and drug response of colon cancer cells. Oncol. Lett. 2019, 17, 2409–2417. [Google Scholar] [CrossRef]
- Peddareddigari, V.G.; Wang, D.; Dubois, R.N. The tumor microenvironment in colorectal carcinogenesis. Cancer Microenviron. 2010, 3, 149–166. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.K.; Cho, Y.S. Pathogenesis and biomarkers of colorectal cancer by epigenetic alteration. Intest. Res. 2024, 22, 131–151. [Google Scholar] [CrossRef]
- Bae, W.Y.; Lee, Y.J.; Jo, S.; Shin, S.L.; Kim, T.R.; Sohn, M.; Seol, H.J. Effects of Lactiplantibacillus plantarum LM1215 on Candida albicans and Gardnerella vaginalis. Yonsei Med. J. 2024, 65, 727–740. [Google Scholar] [CrossRef]
- Cheng, Y.; Ling, Z.; Li, L. The Intestinal Microbiota and Colorectal Cancer. Front. Immunol. 2020, 11, 615056. [Google Scholar] [CrossRef] [PubMed]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Nunez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef] [PubMed]
- Hugon, P.; Dufour, J.C.; Colson, P.; Fournier, P.E.; Sallah, K.; Raoult, D. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect. Dis. 2015, 15, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef]
- Arthur, J.C.; Perez-Chanona, E.; Muhlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef]
- Jefferis, T.M.; Scano, C.; Ashraf, A.; Stevens, D.M.; Sevcik, A.; Bruce, E.; Greathouse, L.; Sayes, C.M. Investigating the activation of the immune response by outer membrane vesicles from Bacteroides fragilis using a human gastrointestinal cell system. J. Immunol. 2025, vkaf257. [Google Scholar] [CrossRef]
- Chan, J.L.; Wu, S.; Geis, A.L.; Chan, G.V.; Gomes, T.A.M.; Beck, S.E.; Wu, X.; Fan, H.; Tam, A.J.; Chung, L.; et al. Non-toxigenic Bacteroides fragilis (NTBF) administration reduces bacteria-driven chronic colitis and tumor development independent of polysaccharide A. Mucosal Immunol. 2019, 12, 164–177. [Google Scholar] [CrossRef]
- Goodwin, A.C.; Destefano Shields, C.E.; Wu, S.; Huso, D.L.; Wu, X.; Murray-Stewart, T.R.; Hacker-Prietz, A.; Rabizadeh, S.; Woster, P.M.; Sears, C.L.; et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 15354–15359. [Google Scholar] [CrossRef]
- Purcell, R.V.; Permain, J.; Keenan, J.I. Enterotoxigenic Bacteroides fragilis activates IL-8 expression through Stat3 in colorectal cancer cells. Gut Pathog. 2022, 14, 16. [Google Scholar] [CrossRef]
- Liu, Q.Q.; Li, C.M.; Fu, L.N.; Wang, H.L.; Tan, J.; Wang, Y.Q.; Sun, D.F.; Gao, Q.Y.; Chen, Y.X.; Fang, J.Y. Enterotoxigenic Bacteroides fragilis induces the stemness in colorectal cancer via upregulating histone demethylase JMJD2B. Gut Microbes 2020, 12, 1788900. [Google Scholar] [CrossRef]
- Silva-Garcia, O.; Valdez-Alarcon, J.J.; Baizabal-Aguirre, V.M. Wnt/beta-Catenin Signaling as a Molecular Target by Pathogenic Bacteria. Front. Immunol. 2019, 10, 2135. [Google Scholar] [CrossRef]
- Wu, S.; Rhee, K.J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F.; et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Morin, P.J.; Maouyo, D.; Sears, C.L. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology 2003, 124, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Purcell, R.V.; Visnovska, M.; Biggs, P.J.; Schmeier, S.; Frizelle, F.A. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci. Rep. 2017, 7, 11590. [Google Scholar] [CrossRef] [PubMed]
- Aitchison, A.; Frizelle, F.A.; Keenan, J.I. PCR Detection of the Bacteroides fragilis Enterotoxin Gene Relies on Robust Primer Design. J. Clin. Microbiol. 2016, 54, 239–240. [Google Scholar] [CrossRef]
- Odamaki, T.; Sugahara, H.; Yonezawa, S.; Yaeshima, T.; Iwatsuki, K.; Tanabe, S.; Tominaga, T.; Togashi, H.; Benno, Y.; Xiao, J.Z. Effect of the oral intake of yogurt containing Bifidobacterium longum BB536 on the cell numbers of enterotoxigenic Bacteroides fragilis in microbiota. Anaerobe 2012, 18, 14–18. [Google Scholar] [CrossRef]
- Eide, P.W.; Bruun, J.; Lothe, R.A.; Sveen, A. CMScaller: An R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci. Rep. 2017, 7, 16618. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 29 June 2025).
- Chalita, M.; Kim, Y.O.; Park, S.; Oh, H.S.; Cho, J.H.; Moon, J.; Baek, N.; Moon, C.; Lee, K.; Yang, J.; et al. EzBioCloud: A genome-driven database and platform for microbiome identification and discovery. Int. J. Syst. Evol. Microbiol. 2024, 74, 006421. [Google Scholar] [CrossRef] [PubMed]
- jamovi—Open Statistical Software for the Desktop and Cloud. Available online: https://www.jamovi.org/ (accessed on 29 June 2025).
- Kim, S.; Choung, S.; Sun, R.X.; Ung, N.; Hashemi, N.; Fong, E.J.; Lau, R.; Spiller, E.; Gasho, J.; Foo, J.; et al. Comparison of Cell and Organoid-Level Analysis of Patient-Derived 3D Organoids to Evaluate Tumor Cell Growth Dynamics and Drug Response. SLAS Discov. 2020, 25, 744–754. [Google Scholar] [CrossRef]
- Zeng, X.; Liu, M. Editorial: Role of fibroblast in cancer. Front. Mol. Biosci. 2025, 12, 1574538. [Google Scholar] [CrossRef]
- Mouillet-Richard, S.; Cazelles, A.; Sroussi, M.; Gallois, C.; Taieb, J.; Laurent-Puig, P. Clinical Challenges of Consensus Molecular Subtype CMS4 Colon Cancer in the Era of Precision Medicine. Clin. Cancer Res. 2024, 30, 2351–2358. [Google Scholar] [CrossRef] [PubMed]
- Peters, N.A.; Constantinides, A.; Ubink, I.; van Kuik, J.; Bloemendal, H.J.; van Dodewaard, J.M.; Brink, M.A.; Schwartz, T.P.; Lolkema, M.; Lacle, M.M.; et al. Consensus molecular subtype 4 (CMS4)-targeted therapy in primary colon cancer: A proof-of-concept study. Front. Oncol. 2022, 12, 969855. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.G.; Hwang, S.; Gwon, S.Y.; Park, C.; Jo, M.; Hong, J.E.; Rhee, K.J. Bacteroides fragilis Toxin Induces Intestinal Epithelial Cell Secretion of Interleukin-8 by the E-Cadherin/beta-Catenin/NF-kappaB Dependent Pathway. Biomedicines 2022, 10, 827. [Google Scholar] [CrossRef]
- Li, A.; Varney, M.L.; Singh, R.K. Expression of interleukin 8 and its receptors in human colon carcinoma cells with different metastatic potentials. Clin. Cancer Res. 2001, 7, 3298–3304. [Google Scholar]
- Wu, X.; Yang, C.; Sun, F.; Zhang, Y.; Wang, Y.; Li, X.; Zheng, F. Enterotoxigenic Bacteroides fragilis (ETBF) Enhances Colorectal Cancer Cell Proliferation and Metastasis Through HDAC3/miR-139-3p Pathway. Biochem. Genet. 2024, 62, 3904–3919. [Google Scholar] [CrossRef]

| Characteristics | N or Mean | % or Range | |
|---|---|---|---|
| Age | 64 years | 26–92 years | |
| Gender | Males | 16 | 64 |
| Females | 9 | 36 | |
| Biopsy Method | Colonoscopy | 17 | 68 |
| Surgery | 8 | 32 | |
| Site | Right | 9 | 36 |
| Left | 7 | 28 | |
| Rectum | 9 | 36 | |
| T stage | T1 | 2 | 8 |
| T2 | 5 | 20 | |
| T3 | 14 | 56 | |
| T4 | 4 | 16 | |
| N stage | N0 | 15 | 60 |
| N1 | 4 | 16 | |
| N2 | 5 | 20 | |
| unspecified | 1 | 4 | |
| M stage | M0 | 20 | 80 |
| M1 | 2 | 8 | |
| unspecified | 3 | 12 | |
| AJCC Stage | 0 | 1 | 4 |
| 1 | 6 | 24 | |
| 2 | 7 | 28 | |
| 3 | 8 | 32 | |
| 4 | 3 | 12 | |
| Histology | Well Differentiated | 8 | 32 |
| Moderately Differentiated | 15 | 60 | |
| Poorly Differentiated | 2 | 8 | |
| MSI status | MSI-H | 2 | 8 |
| MSI-L | 0 | 0 | |
| MSS | 19 | 76 | |
| N/A | 4 | 16 | |
| KRAS mutation status | positive | 8 | 32 |
| negative | 13 | 52 | |
| N/A | 4 | 16 | |
| NRAS mutation status | positive | 0 | 0 |
| negative | 21 | 84 | |
| N/A | 4 | 16 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, S.Y.; Park, J.; Park, S.J.; Park, J.J.; Cheon, J.H.; Kim, D.K.; Kim, T.I. Bacteroides fragilis Promotes Mesenchymal Subtype in Colorectal Cancer. Cancers 2025, 17, 3822. https://doi.org/10.3390/cancers17233822
Chang SY, Park J, Park SJ, Park JJ, Cheon JH, Kim DK, Kim TI. Bacteroides fragilis Promotes Mesenchymal Subtype in Colorectal Cancer. Cancers. 2025; 17(23):3822. https://doi.org/10.3390/cancers17233822
Chicago/Turabian StyleChang, Shin Young, Jihye Park, Soo Jung Park, Jae Jun Park, Jae Hee Cheon, Dong Keon Kim, and Tae Il Kim. 2025. "Bacteroides fragilis Promotes Mesenchymal Subtype in Colorectal Cancer" Cancers 17, no. 23: 3822. https://doi.org/10.3390/cancers17233822
APA StyleChang, S. Y., Park, J., Park, S. J., Park, J. J., Cheon, J. H., Kim, D. K., & Kim, T. I. (2025). Bacteroides fragilis Promotes Mesenchymal Subtype in Colorectal Cancer. Cancers, 17(23), 3822. https://doi.org/10.3390/cancers17233822






