Germline Variants in the Immune Response-Related Genes: Possible Modifying Effect on Age-Dependent BRCA1 Penetrance in Breast Cancer Patient
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Slavin, T.P.; Maxwell, K.N.; Lilyquist, J.; Vijai, J.; Neuhausen, S.L.; Hart, S.N.; Ravichandran, V.; Thomas, T.; Maria, A.; Villano, D.; et al. The contribution of pathogenic variants in breast cancer susceptibility genes to familial breast cancer risk. NPJ Breast Cancer 2017, 3, 22. [Google Scholar] [CrossRef]
- Kurian, A.W.; Ward, K.C.; Howlader, N.; Deapen, D.; Hamilton, A.S.; Mariotto, A.; Miller, D.; Penberthy, L.S.; Katz, S.J. Genetic Testing and Results in a Population-Based Cohort of Breast Cancer Patients and Ovarian Cancer Patients. J. Clin. Oncol. 2019, 37, 1305–1315. [Google Scholar] [CrossRef]
- Hu, C.; Hart, S.N.; Gnanaolivu, R.; Huang, H.; Lee, K.Y.; Na, J.; Gao, C.; Lilyquist, J.; Yadav, S.; Boddicker, N.J.; et al. A Population-Based Study of Genes Previously Implicated in Breast Cancer. N. Engl. J. Med. 2021, 384, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Risch, H.A.; McLaughlin, J.R.; Cole, D.E.; Rosen, B.; Bradley, L.; Fan, I.; Tang, J.; Li, S.; Zhang, S.; Shaw, P.A.; et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: A kin-cohort study in Ontario, Canada. J. Natl. Cancer Inst. 2013, 23, 1694–1706. [Google Scholar] [CrossRef]
- Mavaddat, N.; Peock, S.; Frost, D.; Ellis, S.; Platte, R.; Fineberg, E.; Evans, D.G.; Izatt, L.; Eeles, R.A.; Adlard, J.; et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: Results from prospective analysis of EMBRACE. J. Natl. Cancer Inst. 2013, 105, 812–822. [Google Scholar] [CrossRef]
- Kuchenbaecker, K.B.; Hopper, J.L.; Barnes, D.R.; Phillips, K.A.; Mooij, T.M.; Roos-Blom, M.J.; Jervis, S.; van Leeuwen, F.E.; Milne, R.L.; Andrieu, N.; et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA 2017, 317, 2402–2416. [Google Scholar] [CrossRef]
- Laitman, Y.; Michaelson-Cohen, R.; Chen-Shtoyerman, R.; Goldberg, Y.; Reish, O.; Bernstein-Molho, R.; Levy-Lahad, E.; Baruch, N.E.B.; Kedar, I.; Evans, D.G.; et al. Age at diagnosis of cancer in 185delAG BRCA1 mutation carriers of diverse ethnicities: Tentative evidence for modifier factors. Fam. Cancer 2021, 20, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Michailidou, K.; Lindström, S.; Dennis, J.; Beesley, J.; Hui, S.; Kar, S.; Lemaçon, A.; Soucy, P.; Glubb, D.; Rostamianfar, A.; et al. Association analysis identifies 65 new breast cancer risk loci. Nature 2017, 551, 92–94. [Google Scholar] [CrossRef] [PubMed]
- Milne, R.L.; Kuchenbaecker, K.B.; Michailidou, K.; Beesley, J.; Kar, S.; Lindström, S.; Hui, S.; Lemaçon, A.; Soucy, P.; Dennis, J.; et al. Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat. Genet. 2017, 49, 1767–1778. [Google Scholar] [CrossRef]
- Lilyquist, J.; Ruddy, K.J.; Vachon, C.M.; Couch, F.J. Common Genetic Variation and Breast Cancer Risk-Past, Present, and Future. Cancer Epidemiol. Biomark. Prev. 2018, 27, 380–394. [Google Scholar] [CrossRef]
- Hakkaart, C.; Pearson, J.F.; Marquart, L.; Dennis, J.; Wiggins, G.A.R.; Barnes, D.R.; Robinson, B.A.; Mace, P.D.; Aittomäki, K.; Andrulis, I.L.; et al. Copy number variants as modifiers of breast cancer risk for BRCA1/BRCA2 pathogenic variant carriers. Commun. Biol. 2022, 5, 1061. [Google Scholar] [CrossRef]
- Kuchenbaecker, K.B.; McGuffog, L.; Barrowdale, D.; Lee, A.; Soucy, P.; Dennis, J.; Domchek, S.M.; Robson, M.; Spurdle, A.B.; Ramus, S.J.; et al. Evaluation of Polygenic Risk Scores for Breast and Ovarian Cancer Risk Prediction in BRCA1 and BRCA2 Mutation Carriers. J. Natl. Cancer Inst. 2017, 109, djw302. [Google Scholar] [CrossRef]
- Reddi, H.V.; Wand, H.; Funke, B.; Zimmermann, M.T.; Lebo, M.S.; Qian, E.; Shirts, B.H.; Zou, Y.S.; Zhang, B.M.; Rose, N.C.; et al. Laboratory perspectives in the development of polygenic risk scores for disease: A points to consider statement of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2023, 25, 100804. [Google Scholar] [CrossRef]
- Huntley, C.; Torr, B.; Sud, A.; Rowlands, C.F.; Way, R.; Snape, K.; Hanson, H.; Swanton, C.; Broggio, J.; Lucassen, A.; et al. Utility of polygenic risk scores in UK cancer screening: A modelling analysis. Lancet Oncol. 2023, 24, 658–668. [Google Scholar] [CrossRef]
- Lee, E.C.Y.; Kok, J.S.T.; Teh, B.T.; Lim, K.S. Interplay between the DNA Damage Response and Immunotherapy Response in Cancer. Int. J. Mol. Sci. 2022, 23, 13356. [Google Scholar] [CrossRef]
- Ruangapirom, L.; Sutivijit, N.; Teerapakpinyo, C.; Mutirangura, A.; Doungkamchan, C. Identification of Shared Neoantigens in BRCA1-Related Breast Cancer. Vaccines 2022, 10, 1597. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Qi, L.; Chiang, H.C.; Pan, H.; Zhang, X.; Greenbaum, A.; Stark, E.; Wang, L.J.; Chen, Y.; Haddad, B.R.; et al. BRCA1 deficiency in mature CD8+ T lymphocytes impairs antitumor immunity. J. Immunother. Cancer 2023, 11, e005852. [Google Scholar] [CrossRef]
- Wang, S.M. A global perspective on the ethnic-specific BRCA variation and its implication in clinical application. J. Natl. Cancer Cent. 2022, 3, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Sokolenko, A.; Gorodnova, T.; Enaldieva, D.; Shestakova, A.; Ivantsov, A.; Nyuganen, A.; Berlev, I.; Krivorotko, P.; Belyaev, A.; Imyanitov, E. Comparison of outcomes of neoadjuvant chemotherapy in BRCA1- versus BRCA2-associated breast and ovarian cancers. Explor. Target. Antitumor Ther. 2025, 6, 1002325. [Google Scholar] [CrossRef] [PubMed]
- Antoniou, A.; Pharoah, P.D.; Narod, S.; Risch, H.A.; Eyfjord, J.E.; Hopper, J.L.; Loman, N.; Olsson, H.; Johannsson, O.; Borg, A.; et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: A combined analysis of 22 studies. Am. J. Hum. Genet. 2003, 72, 1117–1130. [Google Scholar] [CrossRef]
- Kuligina, E.S.; Romanko, A.A.; Jankevic, T.; Martianov, A.S.; Ivantsov, A.O.; Sokolova, T.N.; Trofimov, D.; Kashyap, A.; Cybulski, C.; Lubiński, J.; et al. HLA gene polymorphism is a modifier of age-related breast cancer penetrance in carriers of BRCA1 pathogenic alleles. Breast Cancer Res. Treat. 2025, 209, 341–354. [Google Scholar] [CrossRef]
- Bousfiha, A.; Jeddane, L.; Picard, C.; Al-Herz, W.; Ailal, F.; Chatila, T.; Cunningham-Rundles, C.; Etzioni, A.; Franco, J.L.; Holland, S.M.; et al. Human Inborn Errors of Immunity: 2019 Update of the IUIS Phenotypical Classification. J. Clin. Immunol. 2020, 40, 66–81. [Google Scholar] [CrossRef]
- Suspitsin, E.N.; Guseva, M.N.; Kostik, M.M.; Sokolenko, A.P.; Skripchenko, N.V.; Levina, A.S.; Goleva, O.V.; Dubko, M.F.; Tumakova, A.V.; Makhova, M.A.; et al. Next generation sequencing analysis of consecutive Russian patients with clinical suspicion of inborn errors of immunity. Clin. Genet. 2020, 98, 231–239. [Google Scholar] [CrossRef]
- Amendola, L.M.; Muenzen, K.; Biesecker, L.G.; Bowling, K.M.; Cooper, G.M.; Dorschner, M.O.; Driscoll, C.; Foreman, A.K.M.; Golden-Grant, K.; Greally, J.M.; et al. Variant Classification Concordance using the ACMG-AMP Variant Interpretation Guidelines across Nine Genomic Implementation Research Studies. Am. J. Hum. Genet. 2020, 107, 932–941. [Google Scholar] [CrossRef]
- Davidson, A.L.; Kondrashova, O.; Leonard, C.; Wood, S.; Tudini, E.; Hollway, G.E.; Pearson, J.V.; Newell, F.; Spurdle, A.B.; Waddell, N. Analysis of hereditary cancer gene variant classifications from ClinVar indicates a need for regular reassessment of clinical assertions. Hum. Mutat. 2022, 43, 2054–2062. [Google Scholar] [CrossRef] [PubMed]
- Gudmundsson, S.; Singer-Berk, M.; Watts, N.A.; Phu, W.; Goodrich, J.K.; Solomonson, M.; Genome Aggregation Database Consortium; Rehm, H.L.; MacArthur, D.G.; O’Donnell-Luria, A. Variant interpretation using population databases: Lessons from gnomAD. Hum. Mutat. 2022, 43, 1012–1030. [Google Scholar] [CrossRef] [PubMed]
- Kulandaisamy, A.; Binny Priya, S.; Sakthivel, R.; Tarnovskaya, S.; Bizin, I.; Hönigschmid, P.; Frishman, D.; Gromiha, M.M. MutHTP: Mutations in human transmembrane proteins. Bioinformatics 2018, 34, 2325–2326. [Google Scholar] [CrossRef]
- Chang, H.; Schirra, C.; Pattu, V.; Krause, E.; Becherer, U. Lytic granule exocytosis at immune synapses: Lessons from neuronal synapses. Front. Immunol. 2023, 14, 1177670. [Google Scholar] [CrossRef]
- Trapani, J.A.; Smyth, M.J. Functional significance of the perforin/granzyme cell death pathway. Nat. Rev. Immunol. 2002, 2, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.A.; Susanto, O.; Jenkins, M.R.; Lukoyanova, N.; Sutton, V.R.; Law, R.H.; Johnston, A.; Bird, C.H.; Bird, P.I.; Whisstock, J.C.; et al. Perforin forms transient pores on the target cell plasma membrane to facilitate rapid access of granzymes during killer cell attack. Blood 2013, 121, 2659–2668. [Google Scholar] [CrossRef]
- Filipovich, A.; McClain, K.; Grom, A. Histiocytic disorders: Recent insights into pathophysiology and practical guidelines. Biol. Blood Marrow Transplant. 2010, 16, S82–S89. [Google Scholar] [CrossRef]
- Stadermann, A.; Haar, M.; Riecke, A.; Mayer, T.; Neumann, C.; Bauer, A.; Schulz, A.; Nagarathinam, K.; Gebauer, N.; Böhm, S.; et al. Late Onset of Primary Hemophagocytic Lymphohistiocytosis (HLH) with a Novel Constellation of Compound Heterozygosity Involving Two Missense Variants in the PRF1 Gene. Int. J. Mol. Sci. 2024, 25, 2762. [Google Scholar] [CrossRef]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Voskoboinik, I.; Sutton, V.R.; Ciccone, A.; House, C.M.; Chia, J.; Darcy, P.K.; Yagita, H.; Trapani, J.A. Perforin activity and immune homeostasis: The common A91V polymorphism in perforin results in both presynaptic and postsynaptic defects in function. Blood 2007, 110, 1184–1190. [Google Scholar] [CrossRef]
- House, I.G.; Thia, K.; Brennan, A.J.; Tothill, R.; Dobrovic, A.; Yeh, W.Z.; Saffery, R.; Chatterton, Z.; Trapani, J.A.; Voskoboinik, I. Heterozygosity for the common perforin mutation, p.A91V, impairs the cytotoxicity of primary natural killer cells from healthy individuals. Immunol. Cell Biol. 2015, 93, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Sidore, C.; Orrù, V.; Cocco, E.; Steri, M.; Inshaw, J.R.; Pitzalis, M.; Mulas, A.; McGurnaghan, S.; Frau, J.; Porcu, E.; et al. PRF1 mutation alters immune system activation, inflammation, and risk of autoimmunity. Mult. Scler. 2021, 27, 1332–1340. [Google Scholar] [CrossRef] [PubMed]
- Brennan, A.J.; Chia, J.; Trapani, J.A.; Voskoboinik, I. Perforin deficiency and susceptibility to cancer. Cell Death Differ. 2010, 17, 607–615. [Google Scholar] [CrossRef]
- Clementi, R.; Locatelli, F.; Dupré, L.; Garaventa, A.; Emmi, L.; Bregni, M.; Cefalo, G.; Moretta, A.; Danesino, C.; Comis, M.; et al. A proportion of patients with lymphoma may harbor mutations of the perforin gene. Blood 2005, 105, 4424–4428. [Google Scholar] [CrossRef]
- Santoro, A.; Cannella, S.; Trizzino, A.; Lo Nigro, L.; Corsello, G.; Arico, M. A single amino acid change A91V in perforin: A novel, frequent predisposing factor to childhood acute lymphoblastic leukemia? Haematologica 2005, 90, 697–698. [Google Scholar]
- Chaudhry, M.S.; Gilmour, K.C.; House, I.G.; Layton, M.; Panoskaltsis, N.; Sohal, M.; Trapani, J.A.; Voskoboinik, I. Missense mutations in the perforin (PRF1) gene as a cause of hereditary cancer predisposition. Oncoimmunology 2016, 5, e1179415. [Google Scholar] [CrossRef]
- Iranpour, S.; Arif, M.; Szegezdi, E. Disrupting membranes, controlling cell fate: The role of pore-forming proteins in cell death and therapy. Apoptosis 2025, 30, 1961–1988. [Google Scholar] [CrossRef]
- Osińska, I.; Popko, K.; Demkow, U. Perforin: An important player in immune response. Cent. Eur. J. Immunol. 2014, 39, 109–115. [Google Scholar] [CrossRef]
- Comen, E.; Davids, M.; Kirchhoff, T.; Hudis, C.; Offit, K.; Robson, M. Relative contributions of BRCA1 and BRCA2 mutations to “triple-negative” breast cancer in Ashkenazi Women. Breast Cancer Res. Treat. 2011, 129, 185–190. [Google Scholar] [CrossRef]
- Peshkin, B.N.; Alabek, M.L.; Isaacs, C. BRCA1/2 mutations and triple negative breast cancers. Breast Dis. 2010, 32, 25–33. [Google Scholar] [CrossRef]
- Musolino, A.; Bella, M.A.; Bortesi, B.; Michiara, M.; Naldi, N.; Zanelli, P.; Capelletti, M.; Pezzuolo, D.; Camisa, R.; Savi, M.; et al. BRCA mutations, molecular markers, and clinical variables in early-onset breast cancer: A population-based study. Breast 2007, 16, 280–292. [Google Scholar] [CrossRef]
- Ploski, R.; Wozniak, M.; Pawlowski, R.; Monies, D.M.; Branicki, W.; Kupiec, T.; Kloosterman, A.; Dobosz, T.; Bosch, E.; Nowak, M.; et al. Homogeneity and distinctiveness of Polish paternal lineages revealed by Y chromosome microsatellite haplotype analysis. Hum. Genet. 2002, 110, 592–600. [Google Scholar] [CrossRef]
- Anglian Breast Cancer Study Group. Prevalence and penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases. Br. J. Cancer 2000, 83, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Parmigiani, G. Meta-Analysis of BRCA1 and BRCA2 Penetrance. J. Clin. Oncol. 2007, 25, 1329. [Google Scholar] [CrossRef]
- Brohet, R.M.; Velthuizen, M.E.; Hogervorst, F.B.; Meijers-Heijboer, H.E.; Seynaeve, C.; Collée, M.J.; Verhoef, S.; Ausems, M.G.; Hoogerbrugge, N.; van Asperen, C.J.; et al. Breast and ovarian cancer risks in a large series of clinically ascertained families with a high proportion of BRCA1 and BRCA2 Dutch founder mutations. J. Med. Genet. 2014, 51, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Ho, W.K.; Hassan, N.T.; Yoon, S.Y.; Yang, X.; Lim, J.M.C.; Binte Ishak, N.D.; Ho, P.J.; Wijaya, E.A.; Ng, P.P.; Luccarini, C.; et al. Age-specific breast and ovarian cancer risks associated with germline BRCA1 or BRCA2 pathogenic variants—An Asian study of 572 families. Lancet Reg. Health West. Pac. 2024, 44, 101017. [Google Scholar] [CrossRef] [PubMed]
- Dwornik, R.; Białkowska, K. Insights into genetic modifiers of breast cancer risk in carriers of BRCA1 and BRCA2 pathogenic variants. Hered. Cancer Clin. Pract. 2025, 23, 15. [Google Scholar] [CrossRef]
- Sokolenko, A.P.; Bogdanova, N.; Kluzniak, W.; Preobrazhenskaya, E.V.; Kuligina, E.S.; Iyevleva, A.G.; Aleksakhina, S.N.; Mitiushkina, N.V.; Gorodnova, T.V.; Bessonov, A.A.; et al. Double heterozygotes among breast cancer patients analyzed for BRCA1, CHEK2, ATM, NBN/NBS1, and BLM germ-line mutations. Breast Cancer Res. Treat. 2014, 145, 553–562. [Google Scholar] [CrossRef]
- Vietri, M.T.; D’Elia, G.; Caliendo, G.; Casamassimi, A.; Resse, M.; Passariello, L.; Cioffi, M.; Molinari, A.M. Double mutation of APC and BRCA1 in an Italian family. Cancer Genet. 2020, 244, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Coignard, J.; Lush, M.; Beesley, J.; O’Mara, T.A.; Dennis, J.; Tyrer, J.P.; Barnes, D.R.; McGuffog, L.; Leslie, G.; Bolla, M.K.; et al. A case-only study to identify genetic modifiers of breast cancer risk for BRCA1/BRCA2 mutation carriers. Nat. Commun. 2021, 12, 1078. [Google Scholar] [CrossRef] [PubMed]
- Laish, I.; Friedman, E.; Levi-Reznick, G.; Kedar, I.; Katz, L.; Levi, Z.; Halpern, N.; Parnasa, S.; Abu-Shatya, A.; Half, E.; et al. Double heterozygotes of BRCA1/BRCA2 and mismatch repair gene pathogenic variants: Case series and clinical implications. Breast Cancer Res. Treat. 2021, 188, 685–694. [Google Scholar] [CrossRef]
- Colombo, M.; Mondini, P.; Minenza, E.; Foglia, C.; Mosconi, A.; Molica, C.; Pistola, L.; Ludovini, V.; Radice, P. A novel BRCA1 splicing variant detected in an early onset triple-negative breast cancer patient additionally carrying a pathogenic variant in ATM: A case report. Front. Oncol. 2023, 13, 1102184. [Google Scholar] [CrossRef]
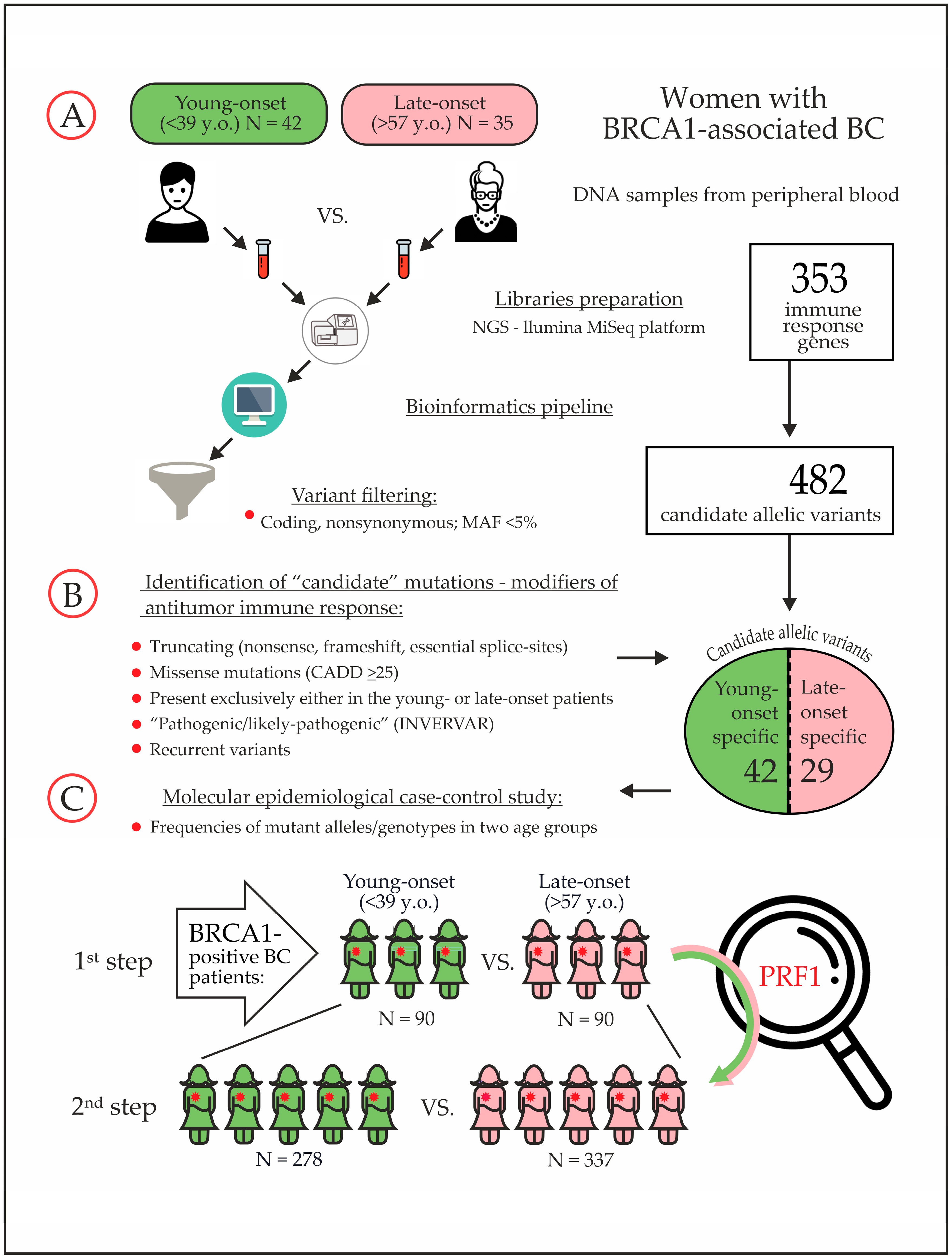
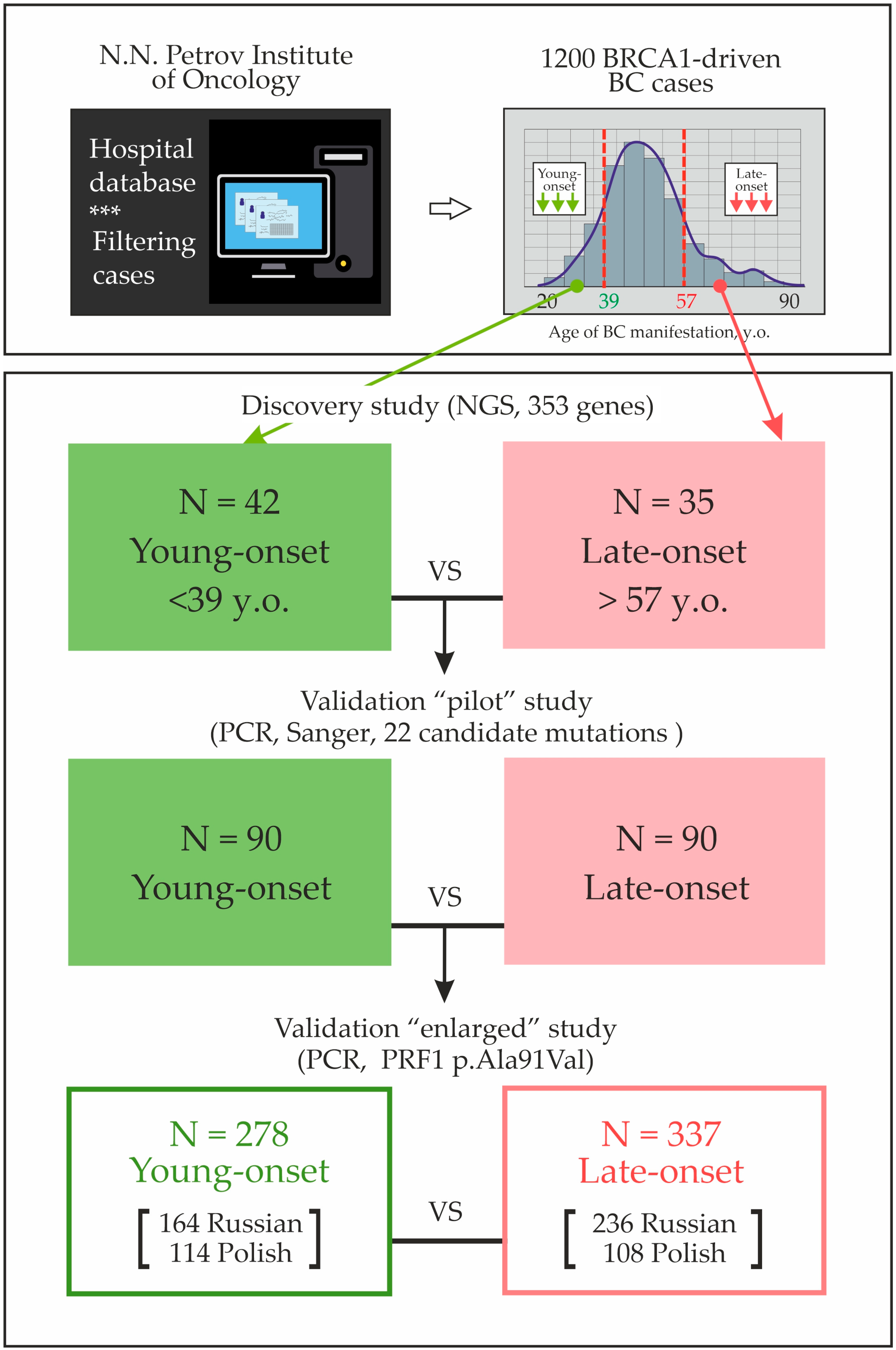
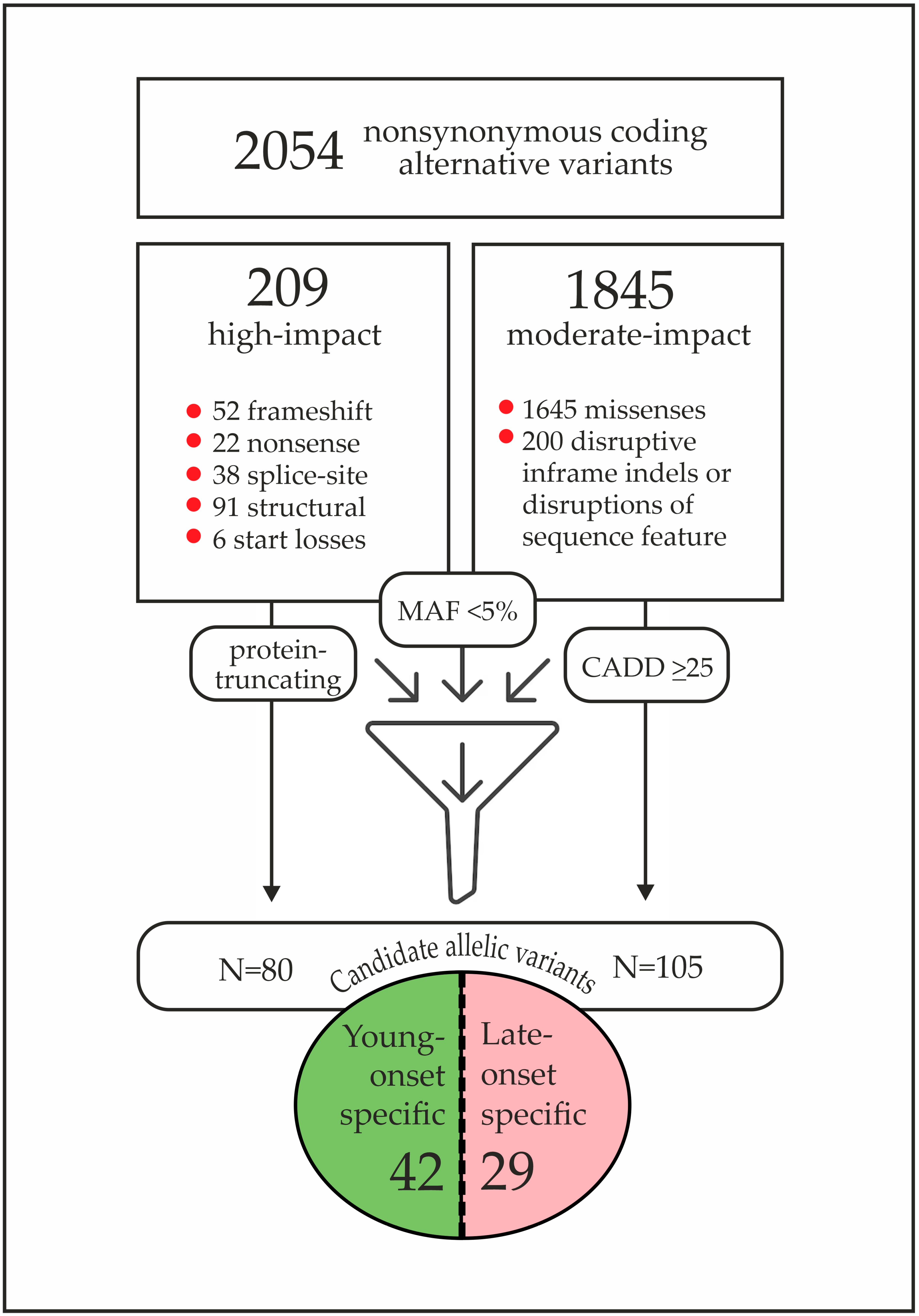
| # | Age Group | Age y.o. | BRCA1 Mutation | Family History | BC Sub Type | Candidate Variant from the “Discovery” Study |
|---|---|---|---|---|---|---|
| 1 | late-onset | 58 | BRCA1 p.Ala457fs | s-BC 49, m-ThC 49 | TNBC | |
| 2 | late-onset | 66 | BRCA1 5382insC | m-OC 50 | nd | |
| 3 | late-onset | 76 | BRCA1 5382insC | d-BC 49 | LumA | |
| 4 | late-onset | 61 | BRCA1 5382insC | m-BC, d-BC | nd | DOCK8 p.Tyr1340Cys |
| 5 | late-onset | 58 | BRCA1 5382insC | m-BC, aunt(f) | TNBC | |
| 6 | late-onset | 83 | BRCA1 5382insC | d-BC(BRCA), s-GaCa | LumA | |
| 7 | late-onset | 64 | BRCA1 5382insC | No | nd | DDX58 rs61752945; DDX41 p.Val408Asp |
| 8 | late-onset | 62 | BRCA1 5382insC | No | nd | |
| 9 | late-onset | 70 | BRCA1 5382insC | f-CRC, m-BC, aunt(f)-BC | LumA | TPP1 p.Arg208Ter |
| 10 | late-onset | 60 | BRCA1 5382insC | No | TNBC | |
| 11 | late-onset | 60 | BRCA1 5382insC | m-OC, s-OC | TNBC | |
| 12 | late-onset | 68 | BRCA1 5382insC | No | LumA | AK2 rs138577419; DNAJC21 p.Arg539Gln |
| 13 | late-onset | 61 | BRCA1 5382insC | aunt(f)-BC | nd | SP110 p.Gly483Arg |
| 14 | late-onset | 64 | BRCA1 5382insC | s-BC 50 | TNBC | |
| 15 | late-onset | 65 | BRCA1 5382insC | f-LC | nd | JAGN1 p.Met1? |
| 16 | late-onset | 61 | BRCA1 5382insC | m-BC, aunt(m)-UtCa | TNBC | |
| 17 | late-onset | 62 | BRCA1 5382insC | No | TNBC | |
| 18 | late-onset | 61 | BRCA1 5382insC | f-EsophCa, aunt (m)-BC 56 | LumA | |
| 19 | late-onset | 61 | BRCA1 4153delA | f-HNSSC | nd | |
| 20 | late-onset | 68 | BRCA1 5382insC | m-GaCa, aunt(f), uncle(m)-GaCa | TNBC | DDX41 p.Val408Asp |
| 21 | late-onset | 62 | BRCA1 5382insC | m-BC, f-CRC, gm(m)-OC | TNBC | |
| 22 | late-onset | 58 | BRCA1 5382insC | s-BC | LumB | DDX41 p.Val408Asp |
| 23 | late-onset | 64 | BRCA1 4153delA | gm(m)-GaCa 60, uncle(m)-GaCa 60, aunt(f)-UtCa 60 | TNBC | TMC6 p.Pro502Leu |
| 24 | late-onset | 63 | BRCA1 p.L1205fs | m-BC 38 | nd | |
| 25 | late-onset | 61 | BRCA1 4153delA | m-UtCa | LumA | IL12B rs3213119 |
| 26 | late-onset | 59 | BRCA1 5382insC | no | LumA | |
| 27 | late-onset | 61 | BRCA1 p.D435fs | m-BC 53, b-BraCa 29 | LumA | |
| 28 | late-onset | 61 | BRCA1 5382insC | no | nd | DDX58 rs61752945 |
| 29 | late-onset | 59 | BRCA1 4153delA | no | nd | |
| 30 | late-onset | 60 | BRCA1 5382insC | aunt(m)-GaCa | nd | |
| 31 | late-onset | 61 | BRCA1 5382insC | nd | nd | IL12B rs3213119; TMC6 p.Pro502Leu |
| 32 | late-onset | 61 | BRCA1 5382insC | nd | nd | |
| 33 | late-onset | 59 | BRCA1 5382insC | no | nd | |
| 34 | late-onset | 63 | BRCA1 5382insC | no | nd | |
| 35 | late-onset | 62 | BRCA1 5382insC | m-BC | TNBC | ATP6AP1 p.Arg15Ter |
| 36 | young-onset | 27 | BRCA1 5382insC | gm(m)-LC 45 | LumA | NOP10 p.Asp12His; NLRP1 p.Phe629Leu |
| 37 | young-onset | 36 | BRCA1 2080delA | m-BC 62, gm(m)-BC 70 | TNBC | |
| 38 | young-onset | 37 | BRCA1 185delAG | m-BC, s-BC | nd | PEPD p.Arg237Cys |
| 39 | young-onset | 35 | BRCA1 c.3629_3630delAG | nd | TNBC | |
| 40 | young-onset | 38 | BRCA1 5382insC | m-BC 55, gm(m)-OC 48, aunt(f)-OC 50, gf(f)-HCC 80, s-ThC 32 | LumA | |
| 41 | young-onset | 38 | BRCA1 c.5215+1G>T | s-BC 40 | TNBC | |
| 42 | young-onset | 33 | BRCA1 c.3304_3307delAATT | m-BC | TNBC | NOP10 p.Asp12His |
| 43 | young-onset | 36 | BRCA1 4153delA | m-BiBC, OC 30, aunt(m)-CRC, gf(m)-LC 70 | nd | |
| 44 | young-onset | 27 | BRCA1 p.S281fs | m-OC 36, gm(f)-RenC | TNBC | STAT4 p.Thr446Ile |
| 45 | young-onset | 30 | BRCA1 5382insC | no | LumB | NOP10 p.Asp12His |
| 46 | young-onset | 27 | BRCA1 5382insC | m-BC 46, gm(m)-CRC 55, gm(f)-HCC 60 | HER2+++ | |
| 47 | young-onset | 29 | BRCA1 p.R1726fs | no | TNBC | |
| 48 | young-onset | 29 | BRCA1 C61G | m-BC | TNBC | |
| 49 | young-onset | 30 | BRCA1 5382insC | m-BC, gm(m)-BC | TNBC | |
| 50 | young-onset | 30 | BRCA1 5382insC | aunt(m)-CaUt | nd | RORC p.Arg10Ter; NLRC4 p.Arg310Ter |
| 51 | young-onset | 26 | BRCA1 5382insC | aunt(f)-BC, gf(m)-LC | TNBC | |
| 52 | young-onset | 27 | BRCA1 5382insC | gm(f)-small intestine Ca | TNBC | PRF1 p.Ala91Val |
| 53 | young-onset | 34 | BRCA1 5382insC | gm-BC | nd | |
| 54 | young-onset | 28 | BRCA1 5382insC | gm(f)-BC, aunts (m,f)-BC | TNBC | |
| 55 | young-onset | 33 | BRCA1 5382insC | no | LumB | NOD2 p.Gly908Arg |
| 56 | young-onset | 31 | BRCA1 5382insC | m-BC 27, gm(m)-BC | TNBC | NOD2 p.Gly908Arg |
| 57 | young-onset | 37 | BRCA1 5382insC | no | LumA | |
| 58 | young-onset | 35 | BRCA1 5382insC | gm(m)-melanoma 60, gm(f)-BC 50 | TNBC | |
| 59 | young-onset | 36 | BRCA1 4153delA | aunt(f)-OC 45, gm(f)-CRC 68, gf(f)-CaLarynx 62, aunt(cousin,f)-OC 55 | LumA | PRF1 p.Ala91Val (homo) |
| 60 | young-onset | 37 | BRCA1 5382insC | aunt(f)-GaCa 39 | LumA | |
| 61 | young-onset | 36 | BRCA1 5382insC | m-BC 38, aunt(m)-BraCa 42 | nd | |
| 62 | young-onset | 27 | BRCA1 5382insC | no | TNBC | |
| 63 | young-onset | 34 | BRCA1 c.5075-1G>C | m-LC 54, aunt(m)-BC 60 | LumA | |
| 64 | young-onset | 31 | BRCA1 5382insC | gf(m)-LC | TNBC | |
| 65 | young-onset | 37 | BRCA1 5382insC | gm(m)-BC 43 | nd | PNP rs104894453 |
| 66 | young-onset | 31 | BRCA1 4153delA | m-BC, s-BC, gm(m)-OC | nd | IL17RC rs148575246 |
| 67 | young-onset | 32 | BRCA1 5382insC | m-OC | TNBC | |
| 68 | young-onset | 34 | BRCA1 5382insC | f-PrC | nd | |
| 69 | young-onset | 34 | BRCA1 5656del20 | no | TNBC | |
| 70 | young-onset | 33 | BRCA1 5382insC | gf(f)-GaCa | LumA | |
| 71 | young-onset | 38 | BRCA1 5382insC | m-BC, s-BC, aunt-BC | TNBC | |
| 72 | young-onset | 32 | BRCA1 5382insC | f-CaLarynx, ggm(m)-BraCa | TNBC | RORC p.Arg10Ter |
| 73 | young-onset | 35 | BRCA1 5382insC | gm(f)-BC | TNBC | |
| 74 | young-onset | 33 | BRCA1 5382insC | gm-CRC, gf(m)-LC | LumA | PRF1 p.Ala91Val |
| 75 | young-onset | 37 | BRCA1 5382insC | gm(f)-PanCa | TNBC | |
| 76 | young-onset | 32 | BRCA1 5382insC | nd | TNBC | |
| 77 | young-onset | 38 | BRCA1 5382insC | m-BC | TNBC |
| Gene | Description | Protein/ rs-id dbSNP | Effect | CADD * | fitCons ** | MAF, % | Discovery Study N Carriers | Validation “Pilot” Study [wt/wt-mut/wt-mut/mut] | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Young- Onset (<39 y.o.) | Late- Onset (>57 y.o.) | Young- Onset (<39 y.o.) | Late- Onset (>57 y.o.) | |||||||
| AK2 | Adenylate kinase 2 | -/ rs138577419 | Structural interaction | 25.8 | 0.73 (del) | 0.221 | 0 | 1 | 90-0-0 | 90-0-0 |
| SP110 | SP110 nuclear body protein | p.Gly483Arg/rs149485401 | Missense | 28.9 | 0.72 (del) | 0.949 | 0 | 1 | 90-0-0 | 90-0-0 |
| JAGN1 | Jagunal homolog 1 | p.Met1?/rs143438463 | Start lost | 25.9 | 0.44 (be) | 0.132 | 0 | 1 | 84-0-0 | 79-0-0 |
| IL12B | Interleukin 12B | -/ rs3213119 | Structural interaction | 25 | 0.53 (del) | 3.002 | 0 | 2 | 88-2-0 | 98-0-0 |
| DOCK8 | Dedicator of cytokinesis 8 | p.Tyr1340Cy/rs116920018 | Missense | 32 | 0.71 (del) | 0.327 | 0 | 1 | 89-1-0 | 98-0-0 |
| DDX58 | DExD/H-box helicase 58 | -/ rs61752945 | Structural interaction | 27.7 | 0.71 (del) | 1.927 | 0 | 2 | 83-1-0 | 86-2-0 |
| TPP1 | Tripeptidyl peptidase 1 | p.Arg208Ter/rs119455955 | Stop gained | 36 | 0.72 (del) | 0.04 | 0 | 1 | 88-2-0 | 90-0-0 |
| TMC6 | Transmembrane channel like 6 | p.Pro502Leu/rs75400929 | Missense | 25.8 | 0.71 (del) | 0.976 | 0 | 2 | 89-0-0 | 90-0-0 |
| ATP6AP1 | ATPase H+ transporting accessory protein 1 | p.Arg15Ter/rs201620814 | Stop gained | 28.8 | n/a | 0.342 | 0 | 1 | 90-0-0 | 90-0-0 |
| DDX41 | DEAD-box helicase 41 | p.Val408Asp/no ID | Missense | 32 | 0.71 (del) | . | 0 | 3 | 87-3-0 | 87-3-0 |
| DNAJC21 | DnaJ heat shock protein family (Hsp40) member C21 | p.Arg539Gln/rs146933471 | Missense | 33 | 0.74 (del) | 0.053 | 0 | 1 | 90-0-0 | 90-0-0 |
| RORC | RAR related orphan receptor C | p.Arg10Ter/rs17582155 | Stop gained | 36 | 0.5 (be) | 0.368 | 2 | 0 | 90-0-0 | 90-0-0 |
| NLRC4 | NLR family CARD domain containing 4 | p.Arg310Ter/rs199475953 | Stop gained | 35 | 0.55 (del) | 0.031 | 1 | 0 | 90-0-0 | 90-0-0 |
| STAT4 | Signal transducer and activator of transcription 4 | p.Thr446Ile/rs141331848 | Missense | 34 | 0.62 (del) | 0.11 | 1 | 0 | 86-0-0 | 88-0-0 |
| IL17RC | Interleukin 17 receptor C | -/ rs148575246 | Splice donor | 29.7 | 0.11 (be) | 1.02 | 1 | 0 | 89-0-0 | 89-1-0 |
| PNP | Purine nucleoside phosphorylase | -/ rs104894453 | Structural interaction | 26.5 | 0.67 (del) | 0.004 | 1 | 0 | 89-0-0 | 86-0-0 |
| NOP10 | NOP10 ribonucleoprotein | p.Asp12His/rs146261631 | Missense | 28 | 0.44 (be) | 1.218 | 3 | 0 | 87-3-0 | 87-3-0 |
| NOD2 | Nucleotide binding oligomerization domain containing 2 | p.Gly908Arg/rs2066845 | Missense | 29.8 | 0.56 (del) | 1.427 | 2 | 0 | 89-1-0 | 82-5-0 |
| NLRP1 | NLR family pyrin domain containing 1 | p.Phe629Leu/rs149035689 | Missense | 25.9 | 0.71 (del) | 1.225 | 1 | 0 | 90-0-0 | 90-0-0 |
| PEPD | Peptidase D | p.Arg237Cys/rs766107449 | Missense | 33 | 0.71 (del) | 0.002 | 1 | 0 | 89-0-0 | 90-0-0 |
| PRF1 | Perforin 1 | p.Ala91Val/rs35947132 | Missense | 25 | 0.55 (del) | 4.662 | 3 | 0 | 67-7-0 *** | 78-0-0 |
| AIRE | Autoimmune regulator | p.Arg257Ter/rs121434254 | Stop gained | 39 | n/a | 0.061 | 1 | 0 | 88-2-0 | 89-1-0 |
| Groups | Age at Onset | PRF1 p.Ala91Val Carriers (%) | Total (Patients) | p- Value * | OR (95% CI) per Carrier | PRF1 p.Ala91Val mut/mut (%) | Total (Patients) | p-Value * | OR (95% CI) per Homozygote | PRF1 p.Ala91Val alleles (%) | Total (Alleles) | p- Value * | OR (95% CI) per Allele |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BC SPb | <39 y.o. young- onset | 14 (8.5%) | 164 | 0.042 | 2.7 [1.08–6.51] p = 0.032 | 2 *** (1.2%) | 164 | 0.047 | 7.1 [0.35–152.58] p = 0.201 | 16 (4.9%) | 328 | 0.01 | 3.0 [1.25–7.03] p = 0.013 |
| >57 y.o. late- onset | 8 (3.4%) | 236 | 0 (0%) | 236 | 8 (1.7%) | 427 | |||||||
| BC PUM | <39 y.o. young- onset | 10 (8.8%) | 114 | 0.800 | 1.3 [0.47–3.52] p = 0.618 | 1 *** (0.9%) | 114 | 1.000 | 2.7 [0.11–66.60] p = 0.540 | 11 (4.8%) | 228 | 0.630 | 1.4 [0.54–3.71] p = 0.484 |
| >57 y.o. late- onset | 7 (7.6%) | 101 | 0 (0%) | 101 | 7 (3.5%) | 202 | |||||||
| BC PUM + SPb | <39 y.o. young- onset | 24 (8.6%) | 278 | 0.045 | 1.9 ** [1.00–3.76] p = 0.068 | 3 *** (1.1%) | 278 | 0.273 | 8.6 [0.44–146.55] p = 0.156 | 27 (4.9%) | 556 | 0.017 | 2.14 ** [1.13–4.07] p = 0.024 |
| >57 y.o. late- onset | 15 (4.4%) | 337 | 0 (0%) | 337 | 15 (2.2%) | 674 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuligina, E.S.; Martianov, A.S.; Yanus, G.A.; Gorgul, Y.A.; Suspitsin, E.N.; Romanko, A.A.; Tumakova, A.V.; Togo, A.V.; Kashyap, A.; Cybulski, C.; et al. Germline Variants in the Immune Response-Related Genes: Possible Modifying Effect on Age-Dependent BRCA1 Penetrance in Breast Cancer Patient. Cancers 2025, 17, 3756. https://doi.org/10.3390/cancers17233756
Kuligina ES, Martianov AS, Yanus GA, Gorgul YA, Suspitsin EN, Romanko AA, Tumakova AV, Togo AV, Kashyap A, Cybulski C, et al. Germline Variants in the Immune Response-Related Genes: Possible Modifying Effect on Age-Dependent BRCA1 Penetrance in Breast Cancer Patient. Cancers. 2025; 17(23):3756. https://doi.org/10.3390/cancers17233756
Chicago/Turabian StyleKuligina, Ekaterina S., Aleksandr S. Martianov, Grigory A. Yanus, Yuliy A. Gorgul, Evgeny N. Suspitsin, Alexandr A. Romanko, Anastasia V. Tumakova, Alexandr V. Togo, Aniruddh Kashyap, Cezary Cybulski, and et al. 2025. "Germline Variants in the Immune Response-Related Genes: Possible Modifying Effect on Age-Dependent BRCA1 Penetrance in Breast Cancer Patient" Cancers 17, no. 23: 3756. https://doi.org/10.3390/cancers17233756
APA StyleKuligina, E. S., Martianov, A. S., Yanus, G. A., Gorgul, Y. A., Suspitsin, E. N., Romanko, A. A., Tumakova, A. V., Togo, A. V., Kashyap, A., Cybulski, C., Lubiński, J., & Imyanitov, E. N. (2025). Germline Variants in the Immune Response-Related Genes: Possible Modifying Effect on Age-Dependent BRCA1 Penetrance in Breast Cancer Patient. Cancers, 17(23), 3756. https://doi.org/10.3390/cancers17233756





