From Slide to Insight: The Emerging Alliance of Digital Pathology and AI in Melanoma Diagnostics
Simple Summary
Abstract
1. Introduction
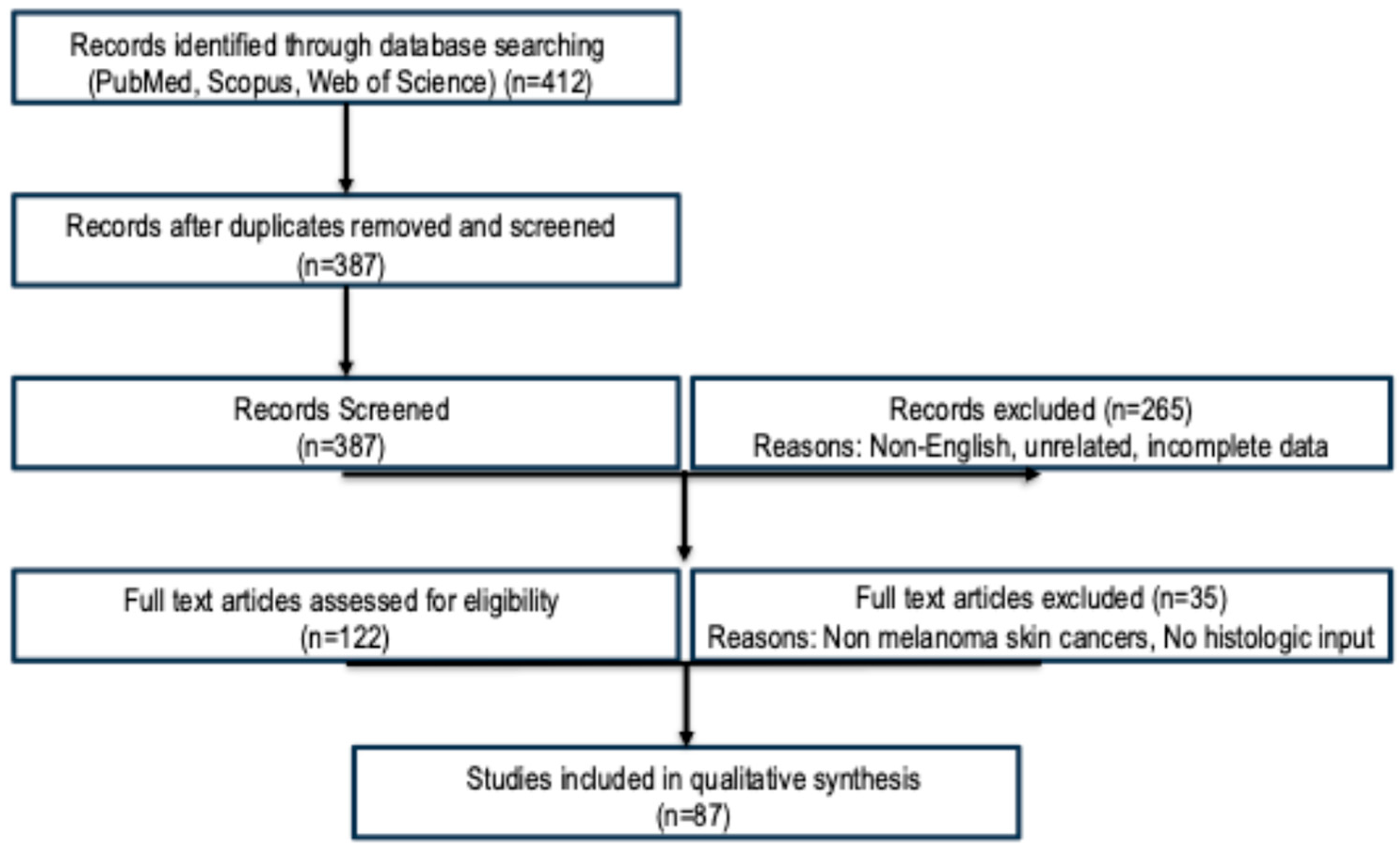
2. Materials and Methods
- PubMed/MEDLINE
- Scopus
- Web of Science
3. Results
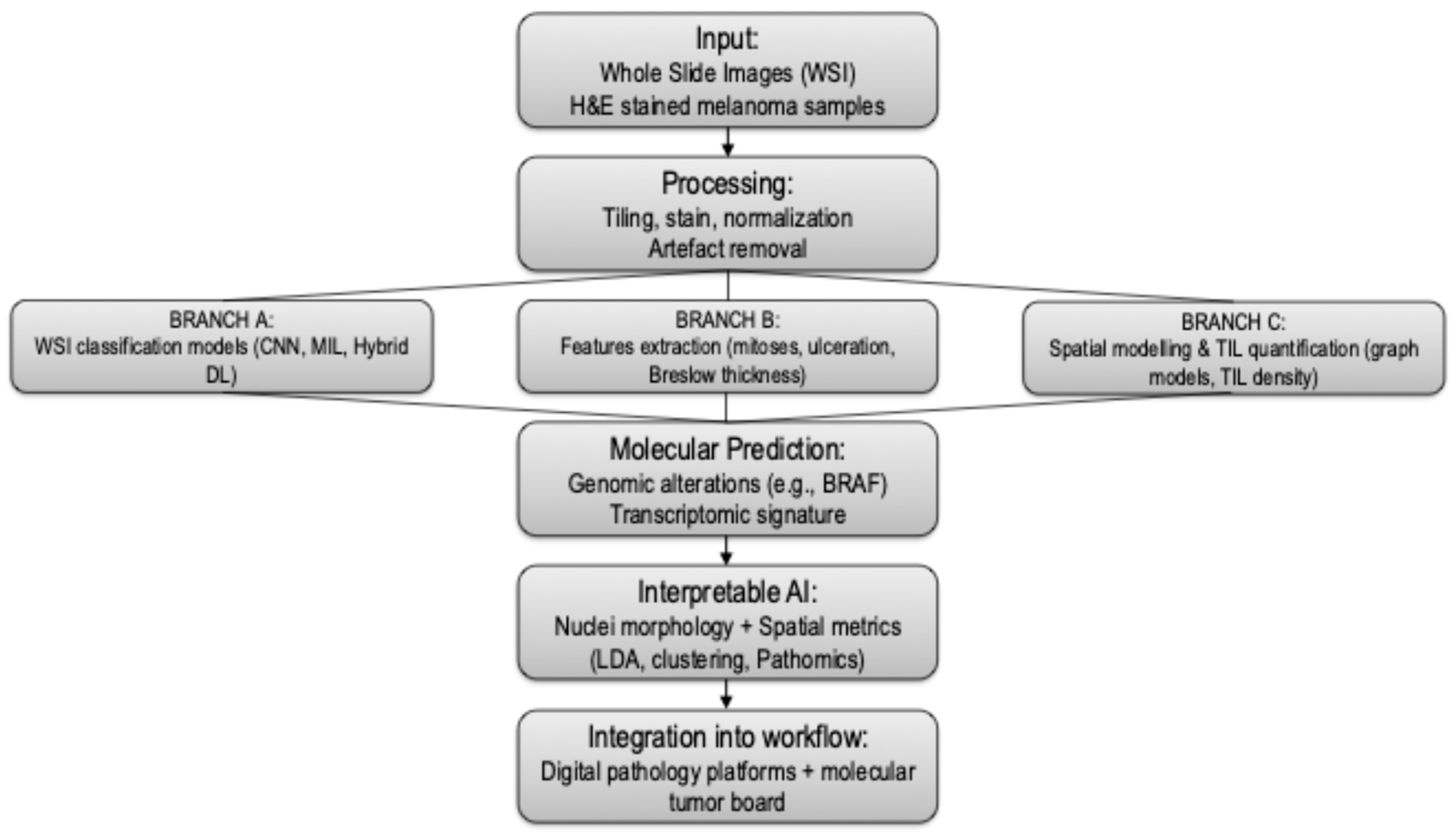
3.1. WSI-Based AI Classification of Melanoma
3.2. Feature Extraction: Mitoses, Ulceration, and Tumor Thickness
3.3. Spatial Modeling and Tumor-Infiltrating Lymphocyte (TIL) Analysis
| Task | Model Types | Accuracy (Range) | AUC (Range) | No. of Studies |
|---|---|---|---|---|
| Melanoma Classification | CNN, MIL | 0.85–0.96 | 0.91–0.98 | 18 |
| Mitosis Detection | U-Net, Mask R-CNN | 0.78–0.88 | 0.83–0.89 | 11 |
| Breslow Thickness Estimation | CNN, Regression | 0.86–0.92 | 0.90–0.95 | 9 |
| Ulceration Detection | Segmentation | 0.78–0.85 | 0.80–0.88 | 7 |
| TIL Analysis | Graph CNN, Spatial Maps | 0.84–0.90 | 0.88–0.93 | 10 |
| Molecular Prediction | CNN, Multimodal Fusion | 0.75–0.82 | 0.80–0.86 | 6 |
3.4. Molecular Prediction from Histopathology
3.5. Interpretable AI and Nuclei-Level Feature Models
3.6. Molecular Tumor Board and AI in Melanoma
3.7. Workflow Integration, Accessibility, and Open-Source Tools
3.8. Workload, Time and Resources
4. Discussion and Conclusions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CM | Cutaneous melanoma |
| WSI | whole slide images |
| ML | machine learning |
| CNN | convolutional neural network |
| AI | artificial intelligence |
| TILs | tumor-infiltrating lymphocytes |
| DP | digital pathology |
| AUC | area under the curve |
| H&E | hematoxylin and eosin |
| LDA | linear discriminant analysis |
| MTB | Molecular Tumor Board |
| NGS | next-generation sequencing |
References
- Shah, M.; Schur, N.; Rosenberg, A.; DeBusk, L.; Burshtein, J.; Zakria, D.; Rigel, D. Trends in Melanoma Incidence and Mortality. Dermatol. Clin. 2025, 43, 373–379. [Google Scholar] [CrossRef]
- De Giorgi, V.; Magnaterra, E.; Zuccaro, B.; Magi, S.; Magliulo, M.; Medri, M.; Mazzoni, L.; Venturi, F.; Silvestri, F.; Tomassini, G.M.; et al. Is Pediatric Melanoma Really That Different from Adult Melanoma? A Multicenter Epidemiological, Clinical and Dermoscopic Study. Cancers 2023, 15, 1835. [Google Scholar] [CrossRef]
- Broseghini, E.; Veronesi, G.; Gardini, A.; Venturi, F.; Scotti, B.; Vespi, L.; Marchese, P.V.; Melotti, B.; Comito, F.; Corti, B.; et al. Defining high-risk patients: Beyond the 8the AJCC melanoma staging system. Arch. Dermatol. Res. 2024, 317, 78. [Google Scholar] [CrossRef]
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef] [PubMed]
- De Giorgi, V.; Scarfì, F.; Gori, A.; Silvestri, F.; Trane, L.; Maida, P.; Venturi, F.; Covarelli, P. Short-term teledermoscopic monitoring of atypical melanocytic lesions in the early diagnosis of melanoma: Utility more apparent than real. J. Eur. Acad. Dermatol. Venereol. JEADV 2020, 34, e398–e399. [Google Scholar] [CrossRef] [PubMed]
- De Giorgi, V.; Silvestri, F.; Cecchi, G.; Venturi, F.; Zuccaro, B.; Perillo, G.; Cosso, F.; Maio, V.; Simi, S.; Antonini, P.; et al. Dermoscopy as a Tool for Identifying Potentially Metastatic Thin Melanoma: A Clinical—Dermoscopic and Histopathological Case—Control Study. Cancers 2024, 16, 1394. [Google Scholar] [CrossRef]
- Stephens, K.R.; Donica, W.R.F.; Philips, P.; McMasters, K.M.; Egger, M.E. Melanoma Deaths by Thickness: Most Melanoma Deaths Are Not Attributable to Thin Melanomas. J. Surg. Res. 2024, 301, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Cazzato, G. Histopathological Diagnosis of Malignant Melanoma at the Dawn of 2023: Knowledge Gained and New Challenges. Dermatopathology 2023, 10, 91–92. [Google Scholar] [CrossRef]
- Ricci, C.; Dika, E.; Ambrosi, F.; Lambertini, M.; Veronesi, G.; Barbara, C. Cutaneous Melanomas: A Single Center Experience on the Usage of Immunohistochemistry Applied for the Diagnosis. Int. J. Mol. Sci. 2022, 23, 5911. [Google Scholar] [CrossRef]
- De Giorgi, V.; Maida, P.; Salvati, L.; Scarfì, F.; Trane, L.; Gori, A.; Silvestri, F.; Venturi, F.; Covarelli, P. Trauma and foreign bodies may favour the onset of melanoma metastases. Clin. Exp. Dermatol. 2020, 45, 619–621. [Google Scholar] [CrossRef]
- WHO Classification of Tumours Online. Available online: https://tumourclassification.iarc.who.int/welcome/# (accessed on 11 July 2025).
- Zhai, H.; Dika, E.; Goldovsky, M.; Maibach, H.I. Tape-stripping method in man: Comparison of evaporimetric methods. Skin Res. Technol. 2007, 13, 207–210. [Google Scholar] [CrossRef]
- De Giorgi, V.; Scarfì, F.; Silvestri, F.; Maida, P.; Venturi, F.; Trane, L.; Gori, A. Genital piercing: A warning for the risk of vulvar lichen sclerosus. Dermatol. Ther. 2021, 34, e14703. [Google Scholar] [CrossRef]
- Krieger, N.; Hiatt, R.A.; Sagebiel, R.W.; Clark, W.H.; Mihm, M.C. Inter-observer variability among pathologists’ evaluation of malignant melanoma: Effects upon an analytic study. J. Clin. Epidemiol. 1994, 47, 897–902. [Google Scholar] [CrossRef]
- De Giorgi, V.; Venturi, F.; Silvestri, F.; Trane, L.; Savarese, I.; Scarfì, F.; Cencetti, F.; Pecenco, S.; Tramontana, M.; Maio, V.; et al. Atypical Spitz tumours: An epidemiological, clinical and dermoscopic multicentre study with 16 years of follow-up. Clin. Exp. Dermatol. 2022, 47, 1464–1471. [Google Scholar] [CrossRef] [PubMed]
- Berger, D.M.S.; Wassenberg, R.M.; Jóźwiak, K.; van de Wiel, B.A.; Balm, A.J.M.; van den Berg, J.G.; Klop, W.M.C. Inter-observer variation in the histopathology reports of head and neck melanoma; a comparison between the seventh and eighth edition of the AJCC staging system. Eur. J. Surg. Oncol. 2019, 45, 235–241. [Google Scholar] [CrossRef]
- Mosquera-Zamudio, A.; Launet, L.; Tabatabaei, Z.; Parra-Medina, R.; Colomer, A.; Oliver Moll, J.; Monteagudo, C.; Janssen, E.; Naranjo, V. Deep Learning for Skin Melanocytic Tumors in Whole-Slide Images: A Systematic Review. Cancers 2022, 15, 42. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Chen, W.; Li, S.; Quan, H.; Zou, M.; Zheng, S.; Zhao, Y.; Gao, X.; Cui, X. Learning how to detect: A deep reinforcement learning method for whole-slide melanoma histopathology images. Comput. Med. Imaging Graph. 2023, 108, 102275. [Google Scholar] [CrossRef]
- Yin, W.; Zhou, D.; Nie, R. DI-UNet: Dual-branch interactive U-Net for skin cancer image segmentation. J. Cancer Res. Clin. Oncol. 2023, 149, 15511–15524. [Google Scholar] [CrossRef] [PubMed]
- Van Dieren, L.; Amar, J.Z.; Geurs, N.; Quisenaerts, T.; Gillet, C.; Delforge, B.; D’heysselaer, L.D.C.; Thiessen, E.F.; Cetrulo, C.L.; Lellouch, A.G. Unveiling the power of convolutional neural networks in melanoma diagnosis. Eur. J. Dermatol. EJD 2023, 33, 495–505. [Google Scholar] [CrossRef]
- Pérez, E.; Reyes, O.; Ventura, S. Convolutional neural networks for the automatic diagnosis of melanoma: An extensive experimental study. Med. Image Anal. 2021, 67, 101858. [Google Scholar] [CrossRef]
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hassen, A.B.H.; Thomas, L.; Enk, A.; et al. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef]
- Menzies, S.W.; Sinz, C.; Menzies, M.; Lo, S.N.; Yolland, W.; Lingohr, J.; Razmara, M.; Tschandl, P.; Guitera, P.; Scolyer, R.A.; et al. Comparison of humans versus mobile phone-powered artificial intelligence for the diagnosis and management of pigmented skin cancer in secondary care: A multicentre, prospective, diagnostic, clinical trial. Lancet Digit. Health 2023, 5, e679–e691. [Google Scholar] [CrossRef]
- Papachristou, P.; Söderholm, M.; Pallon, J.; Taloyan, M.; Polesie, S.; Paoli, J.; Anderson, C.D.; Falk, M. Evaluation of an artificial intelligence-based decision support for the detection of cutaneous melanoma in primary care: A prospective real-life clinical trial. Br. J. Dermatol. 2024, 191, 125–133. [Google Scholar] [CrossRef]
- Acs, B.; Rantalainen, M.; Hartman, J. Artificial intelligence as the next step towards precision pathology. J. Intern. Med. 2020, 288, 62–81. [Google Scholar] [CrossRef]
- Sahni, S.; Wang, B.; Wu, D.; Dhruba, S.R.; Nagy, M.; Patkar, S.; Ferreira, I.; Day, C.-P.; Wang, K.; Ruppin, E. A machine learning model reveals expansive downregulation of ligand-receptor interactions that enhance lymphocyte infiltration in melanoma with developed resistance to immune checkpoint blockade. Nat. Commun. 2024, 15, 8867. [Google Scholar] [CrossRef]
- Chou, M.; Illa-Bochaca, I.; Minxi, B.; Darvishian, F.; Johannet, P.; Moran, U.; Shapiro, R.L.; Berman, R.S.; Osman, I.; Jour, G.; et al. Optimization of an automated tumor-infiltrating lymphocyte algorithm for improved prognostication in primary melanoma. Mod. Pathol. 2021, 34, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Harder, N.; Schönmeyer, R.; Nekolla, K.; Meier, A.; Brieu, N.; Vanegas, C.; Madonna, G.; Capone, M.; Botti, G.; Ascierto, P.A.; et al. Automatic discovery of image-based signatures for ipilimumab response prediction in malignant melanoma. Sci. Rep. 2019, 9, 7449. [Google Scholar] [CrossRef]
- Jartarkar, S.R.; Cockerell, C.J.; Patil, A.; Kassir, M.; Babaei, M.; Weidenthaler-Barth, B.; Grabbe, S.; Goldust, M. Artificial intelligence in Dermatopathology. J. Cosmet. Dermatol. 2023, 22, 1163–1167. [Google Scholar] [CrossRef]
- Gordon, E.R.; Trager, M.H.; Kontos, D.; Weng, C.; Geskin, L.J.; Dugdale, L.S.; Samie, F.H. Ethical considerations for artificial intelligence in dermatology: A scoping review. Br. J. Dermatol. 2024, 190, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Cazzato, G.; Rongioletti, F. Artificial intelligence in dermatopathology: Updates, strengths, and challenges. Clin. Dermatol. 2024, 42, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Virgens, G.S.; Teodoro, J.A.; Iarussi, E.; Rodrigues, T.; Amaral, D.T. Enhancing and advancements in deep learning for melanoma detection: A comprehensive review. Comput. Biol. Med. 2025, 194, 110533. [Google Scholar] [CrossRef]
- Clarke, E.L.; Wade, R.G.; Magee, D.; Newton-Bishop, J.; Treanor, D. Image analysis of cutaneous melanoma histology: A systematic review and meta-analysis. Sci. Rep. 2023, 13, 4774. [Google Scholar] [CrossRef]
- Grant, S.R.; Andrew, T.W.; Alvarez, E.V.; Huss, W.J.; Paragh, G. Diagnostic and Prognostic Deep Learning Applications for Histological Assessment of Cutaneous Melanoma. Cancers 2022, 14, 6231. [Google Scholar] [CrossRef]
- Sauter, D.; Lodde, G.; Nensa, F.; Schadendorf, D.; Livingstone, E.; Kukuk, M. Deep learning in computational dermatopathology of melanoma: A technical systematic literature review. Comput. Biol. Med. 2023, 163, 107083. [Google Scholar] [CrossRef] [PubMed]
- Yee, J.; Rosendahl, C.; Aoude, L.G. The role of artificial intelligence and convolutional neural networks in the management of melanoma: A clinical, pathological, and radiological perspective. Melanoma Res. 2024, 34, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Naddeo, M.; Broseghini, E.; Venturi, F.; Vaccari, S.; Corti, B.; Lambertini, M.; Ricci, C.; Fontana, B.; Durante, G.; Pariali, M.; et al. Association of miR-146a-5p and miR-21-5p with Prognostic Features in Melanomas. Cancers 2024, 16, 1688. [Google Scholar] [CrossRef]
- Grossarth, S.; Mosley, D.; Madden, C.; Ike, J.; Smith, I.; Huo, Y.; Wheless, L. Recent Advances in Melanoma Diagnosis and Prognosis Using Machine Learning Methods. Curr. Oncol. Rep. 2023, 25, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Querzoli, G.; Veronesi, G.; Corti, B.; Nottegar, A.; Dika, E. Basic Elements of Artificial Intelligence Tools in the Diagnosis of Cutaneous Melanoma. Crit. Rev. Oncog. 2023, 28, 37–41. [Google Scholar] [CrossRef]
- Ertürk Zararsız, G.; Yerlitaş Taştan, S.I.; Çelik Gürbulak, E.; Erakcaoğlu, A.; Yılmaz Işıkhan, S.; Demirbaş, A.; Ertaş, R.; Eroğlu, İ.; Korkmaz, S.; Elmas, Ö.F.; et al. Diagnosis melanoma with artificial intelligence systems: A meta-analysis study and systematic review. J. Eur. Acad. Dermatol. Venereol. JEADV 2025, 39, 1912–1922. [Google Scholar] [CrossRef]
- Karimzadhagh, S.; Ghodous, S.; Robati, R.M.; Abbaspour, E.; Goldust, M.; Zaresharifi, N.; Zaresharifi, S. Performance of Artificial Intelligence in Skin Cancer Detection: An Umbrella Review of Systematic Reviews and Meta-Analyses. Int. J. Dermatol. 2025, 1–17. [Google Scholar] [CrossRef]
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Berking, C.; Klode, J.; Schadendorf, D.; Jansen, P.; Franklin, C.; Holland-Letz, T.; Krahl, D.; et al. Pathologist-level classification of histopathological melanoma images with deep neural networks. Eur. J. Cancer 2019, 115, 79–83. [Google Scholar] [CrossRef]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Berking, C.; Haferkamp, S.; Hauschild, A.; Weichenthal, M.; Klode, J.; Schadendorf, D.; Holland-Letz, T.; et al. Deep neural networks are superior to dermatologists in melanoma image classification. Eur. J. Cancer 2019, 119, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Solass, W.; Schmitt, M.; Klode, J.; Schadendorf, D.; Sondermann, W.; Franklin, C.; Bestvater, F.; et al. Deep learning outperformed 11 pathologists in the classification of histopathological melanoma images. Eur. J. Cancer 2019, 118, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Haggenmüller, S.; Wies, C.; Abels, J.; Winterstein, J.T.; Heinlein, L.; Nogueira Garcia, C.; Utikal, J.S.; Wohlfeil, S.A.; Meier, F.; Hobelsberger, S.; et al. Discordance, accuracy and reproducibility study of pathologists’ diagnosis of melanoma and melanocytic tumors. Nat. Commun. 2025, 16, 789. [Google Scholar] [CrossRef]
- Dika, E.; Curti, N.; Giampieri, E.; Veronesi, G.; Misciali, C.; Ricci, C.; Castellani, G.; Patrizi, A.; Marcelli, E. Advantages of manual and automatic computer-aided compared to traditional histopathological diagnosis of melanoma: A pilot study. Pathol. Res. Pract. 2022, 237, 154014. [Google Scholar] [CrossRef]
- Haggenmüller, S.; Maron, R.C.; Hekler, A.; Utikal, J.S.; Barata, C.; Barnhill, R.L.; Beltraminelli, H.; Berking, C.; Betz-Stablein, B.; Blum, A.; et al. Skin cancer classification via convolutional neural networks: Systematic review of studies involving human experts. Eur. J. Cancer 2021, 156, 202–216. [Google Scholar] [CrossRef]
- Maron, R.C.; Schlager, J.G.; Haggenmüller, S.; von Kalle, C.; Utikal, J.S.; Meier, F.; Gellrich, F.F.; Hobelsberger, S.; Hauschild, A.; French, L.; et al. A benchmark for neural network robustness in skin cancer classification. Eur. J. Cancer 2021, 155, 191–199. [Google Scholar] [CrossRef]
- Maron, R.C.; Haggenmüller, S.; von Kalle, C.; Utikal, J.S.; Meier, F.; Gellrich, F.F.; Hauschild, A.; French, L.E.; Schlaak, M.; Ghoreschi, K.; et al. Robustness of convolutional neural networks in recognition of pigmented skin lesions. Eur. J. Cancer 2021, 145, 81–91. [Google Scholar] [CrossRef]
- Schmitt, M.; Maron, R.C.; Hekler, A.; Stenzinger, A.; Hauschild, A.; Weichenthal, M.; Tiemann, M.; Krahl, D.; Kutzner, H.; Utikal, J.S.; et al. Hidden Variables in Deep Learning Digital Pathology and Their Potential to Cause Batch Effects: Prediction Model Study. J. Med. Internet Res. 2021, 23, e23436. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.I.; Navarrete-Dechent, C.; Daneshjou, R.; Cho, H.S.; Chang, S.E.; Kim, S.H.; Na, J.-I.; Han, S.S. Generation of a Melanoma and Nevus Data Set from Unstandardized Clinical Photographs on the Internet. JAMA Dermatol. 2023, 159, 1223–1231. [Google Scholar] [CrossRef]
- Haggenmüller, S.; Schmitt, M.; Krieghoff-Henning, E.; Hekler, A.; Maron, R.C.; Wies, C.; Utikal, J.S.; Meier, F.; Hobelsberger, S.; Gellrich, F.F.; et al. Federated Learning for Decentralized Artificial Intelligence in Melanoma Diagnostics. JAMA Dermatol. 2024, 160, 303–311. [Google Scholar] [CrossRef]
- Onega, T.; Barnhill, R.L.; Piepkorn, M.W.; Longton, G.M.; Elder, D.E.; Weinstock, M.A.; Knezevich, S.R.; Reisch, L.M.; Carney, P.A.; Nelson, H.D.; et al. Accuracy of Digital Pathologic Analysis vs Traditional Microscopy in the Interpretation of Melanocytic Lesions. JAMA Dermatol. 2018, 154, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Curti, N.; Veronesi, G.; Dika, E.; Misciali, C.; Marcelli, E.; Giampieri, E. Breslow thickness: Geometric interpretation, potential pitfalls, and computer automated estimation. Pathol. Res. Pract. 2022, 238, 154117. [Google Scholar] [CrossRef]
- Hu, Y.; Sirinukunwattana, K.; Li, B.; Gaitskell, K.; Domingo, E.; Bonnaffé, W.; Wojciechowska, M.; Wood, R.; Alham, N.K.; Malacrino, S.; et al. Self-interactive learning: Fusion and evolution of multi-scale histomorphology features for molecular traits prediction in computational pathology. Med. Image Anal. 2025, 101, 103437. [Google Scholar] [CrossRef]
- Nofallah, S.; Mehta, S.; Mercan, E.; Knezevich, S.; May, C.J.; Weaver, D.; Witten, D.; Elmore, J.G.; Shapiro, L. Machine learning techniques for mitoses classification. Comput. Med. Imaging Graph. 2021, 87, 101832. [Google Scholar] [CrossRef] [PubMed]
- Papadakis, M.; Paschos, A.; Manios, A.; Lehmann, P.; Manios, G.; Zirngibl, H. Computer-aided clinical image analysis for non-invasive assessment of tumor thickness in cutaneous melanoma. BMC Res. Notes 2021, 14, 232. [Google Scholar] [CrossRef]
- Long, G.V.; Swetter, S.M.; Menzies, A.M.; Gershenwald, J.E.; Scolyer, R.A. Cutaneous melanoma. Lancet Lond. Engl. 2023, 402, 485–502. [Google Scholar] [CrossRef]
- Veronesi, G.; Curti, N.; Gardini, A.; Querzoli, G.; Castellani, G.; Dika, E. Machine learning to detect melanoma exploiting nuclei morphology and Spatial organization. Sci. Rep. 2025, 15, 21594. [Google Scholar] [CrossRef] [PubMed]
- Dika, E.; Riefolo, M.; Porcellini, E.; Broseghini, E.; Ribero, S.; Senetta, R.; Osella-Abate, S.; Scarfì, F.; Lambertini, M.; Veronesi, G.; et al. Defining the Prognostic Role of MicroRNAs in Cutaneous Melanoma. J. Investig. Dermatol. 2020, 140, 2260–2267. [Google Scholar] [CrossRef]
- Neimy, H.; Helmy, J.E.; Snyder, A.; Valdebran, M. Artificial Intelligence in Melanoma Dermatopathology: A Review of Literature. Am. J. Dermatopathol. 2024, 46, 83–94. [Google Scholar] [CrossRef]
- Aung, T.N.; Liu, M.; Su, D.; Shafi, S.; Boyaci, C.; Steen, S.; Tsiknakis, N.; Vidal, J.M.; Maher, N.; Micevic, G.; et al. Pathologist-Read vs AI-Driven Assessment of Tumor-Infiltrating Lymphocytes in Melanoma. JAMA Netw. Open 2025, 8, e2518906. [Google Scholar] [CrossRef]
- Chatziioannou, E.; Roßner, J.; Aung, T.N.; Rimm, D.L.; Niessner, H.; Keim, U.; Serna-Higuita, L.M.; Bonzheim, I.; Kuhn Cuellar, L.; Westphal, D.; et al. Deep learning-based scoring of tumour-infiltrating lymphocytes is prognostic in primary melanoma and predictive to PD-1 checkpoint inhibition in melanoma metastases. eBioMedicine 2023, 93, 104644. [Google Scholar] [CrossRef]
- Moore, M.R.; Friesner, I.D.; Rizk, E.M.; Fullerton, B.T.; Mondal, M.; Trager, M.H.; Mendelson, K.; Chikeka, I.; Kurc, T.; Gupta, R.; et al. Automated digital TIL analysis (ADTA) adds prognostic value to standard assessment of depth and ulceration in primary melanoma. Sci. Rep. 2021, 11, 2809. [Google Scholar] [CrossRef]
- Ugolini, F.; De Logu, F.; Iannone, L.F.; Brutti, F.; Simi, S.; Maio, V.; de Giorgi, V.; Maria di Giacomo, A.; Miracco, C.; Federico, F.; et al. Tumor-Infiltrating Lymphocyte Recognition in Primary Melanoma by Deep Learning Convolutional Neural Network. Am. J. Pathol. 2023, 193, 2099–2110. [Google Scholar] [CrossRef]
- Lapuente-Santana, Ó.; Kant, J.; Eduati, F. Integrating histopathology and transcriptomics for spatial tumor microenvironment profiling in a melanoma case study. npj Precis. Oncol. 2024, 8, 254. [Google Scholar] [CrossRef]
- Van Herck, Y.; Antoranz, A.; Andhari, M.D.; Milli, G.; Bechter, O.; De Smet, F.; Bosisio, F.M. Multiplexed Immunohistochemistry and Digital Pathology as the Foundation for Next-Generation Pathology in Melanoma: Methodological Comparison and Future Clinical Applications. Front. Oncol. 2021, 11, 636681. [Google Scholar] [CrossRef]
- Kurz, A.; Krahl, D.; Kutzner, H.; Barnhill, R.; Perasole, A.; Figueras, M.T.F.; Ferrara, G.; Braun, S.A.; Starz, H.; Llamas-Velasco, M.; et al. A 3-dimensional histology computer model of malignant melanoma and its implications for digital pathology. Eur. J. Cancer 2023, 193, 113294. [Google Scholar] [CrossRef] [PubMed]
- Dika, E.; Veronesi, G.; Altimari, A.; Riefolo, M.; Ravaioli, G.M.; Piraccini, B.M.; Lambertini, M.; Campione, E.; Gruppioni, E.; Fiorentino, M.; et al. BRAF, KIT, and NRAS Mutations of Acral Melanoma in White Patients. Am. J. Clin. Pathol. 2020, 153, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Stenzinger, A.; Alber, M.; Allgäuer, M.; Jurmeister, P.; Bockmayr, M.; Budczies, J.; Lennerz, J.; Eschrich, J.; Kazdal, D.; Schirmacher, P.; et al. Artificial intelligence and pathology: From principles to practice and future applications in histomorphology and molecular profiling. Semin. Cancer Biol. 2022, 84, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Maloberti, T.; De Leo, A.; Coluccelli, S.; Sanza, V.; Gruppioni, E.; Altimari, A.; Comito, F.; Melotti, B.; Marchese, P.V.; Dika, E.; et al. Molecular Characterization of Advanced-Stage Melanomas in Clinical Practice Using a Laboratory-Developed Next-Generation Sequencing Panel. Diagnostics 2024, 14, 800. [Google Scholar] [CrossRef]
- Broseghini, E.; Venturi, F.; Veronesi, G.; Scotti, B.; Migliori, M.; Marini, D.; Ricci, C.; Casadei, R.; Ferracin, M.; Dika, E. Exploring the Common Mutational Landscape in Cutaneous Melanoma and Pancreatic Cancer. Pigment Cell Melanoma Res. 2025, 38, e13210. [Google Scholar] [CrossRef]
- Albahri, M.; Sauter, D.; Nensa, F.; Lodde, G.; Livingstone, E.; Schadendorf, D.; Kukuk, M. A new approach combining a whole-slide foundation model and gradient boosting for predicting BRAF mutation status in dermatopathology. Comput. Struct. Biotechnol. J. 2025, 27, 2503–2514. [Google Scholar] [CrossRef]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef]
- Kim, R.H.; Nomikou, S.; Coudray, N.; Jour, G.; Dawood, Z.; Hong, R.; Esteva, E.; Sakellaropoulos, T.; Donnelly, D.; Moran, U.; et al. Deep Learning and Pathomics Analyses Reveal Cell Nuclei as Important Features for Mutation Prediction of BRAF-Mutated Melanomas. J. Investig. Dermatol. 2022, 142, 1650–1658.e6. [Google Scholar] [CrossRef]
- Schneider, L.; Wies, C.; Krieghoff-Henning, E.I.; Bucher, T.-C.; Utikal, J.S.; Schadendorf, D.; Brinker, T.J. Multimodal integration of image, epigenetic and clinical data to predict BRAF mutation status in melanoma. Eur. J. Cancer 2023, 183, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Failmezger, H.; Muralidhar, S.; Rullan, A.; de Andrea, C.E.; Sahai, E.; Yuan, Y. Topological Tumor Graphs: A Graph-Based Spatial Model to Infer Stromal Recruitment for Immunosuppression in Melanoma Histology. Cancer Res. 2020, 80, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, R.; Koyama, T.; Kouno, N.; Yasuda, T.; Yui, S.; Sudo, K.; Hirata, M.; Sunami, K.; Kubo, T.; Takasawa, K.; et al. Introducing AI to the molecular tumor board: One direction toward the establishment of precision medicine using large-scale cancer clinical and biological information. Exp. Hematol. Oncol. 2022, 11, 82. [Google Scholar] [CrossRef]
- Morash, M.; Mitchell, H.; Beltran, H.; Elemento, O.; Pathak, J. The Role of Next-Generation Sequencing in Precision Medicine: A Review of Outcomes in Oncology. J. Pers. Med. 2018, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, R.; Suvarna, K.; Yamada, M.; Kobayashi, K.; Shinkai, N.; Miyake, M.; Takahashi, M.; Jinnai, S.; Shimoyama, R.; Sakai, A.; et al. Application of Artificial Intelligence Technology in Oncology: Towards the Establishment of Precision Medicine. Cancers 2020, 12, 3532. [Google Scholar] [CrossRef]
- Diaz-Ramón, J.L.; Gardeazabal, J.; Izu, R.M.; Garrote, E.; Rasero, J.; Apraiz, A.; Penas, C.; Seijo, S.; Lopez-Saratxaga, C.; De la Peña, P.M.; et al. Melanoma Clinical Decision Support System: An Artificial Intelligence-Based Tool to Diagnose and Predict Disease Outcome in Early-Stage Melanoma Patients. Cancers 2023, 15, 2174. [Google Scholar] [CrossRef]
- Song, Q.; Li, M.; Li, Q.; Lu, X.; Song, K.; Zhang, Z.; Wei, J.; Zhang, L.; Wei, J.; Ye, Y.; et al. DeepAlloDriver: A deep learning-based strategy to predict cancer driver mutations. Nucleic Acids Res. 2023, 51, W129–W133. [Google Scholar] [CrossRef]
- Nardone, V.; Marmorino, F.; Germani, M.M.; Cichowska-Cwalińska, N.; Menditti, V.S.; Gallo, P.; Studiale, V.; Taravella, A.; Landi, M.; Reginelli, A.; et al. The Role of Artificial Intelligence on Tumor Boards: Perspectives from Surgeons, Medical Oncologists and Radiation Oncologists. Curr. Oncol. 2024, 31, 4984–5007. [Google Scholar] [CrossRef]
- Tiwari, A.; Mishra, S.; Kuo, T.-R. Current AI technologies in cancer diagnostics and treatment. Mol. Cancer 2025, 24, 159. [Google Scholar] [CrossRef]
- Annessi, G.; Annessi, E. Considerations on The Biologic Gray Zone of Melanocytic Tumors. Dermatol. Pract. Concept. 2024, 14, e2024154. [Google Scholar] [CrossRef]
- Ferrara, G.; Gualandi, A.; Rizzo, N. Biologic Gray Zone of Melanocytic Tumors in Reality: Defining “Non-Conventional” Melanocytic Tumors. Dermatol. Pract. Concept. 2024, 14, e2024149. [Google Scholar] [CrossRef]
- Dika, E.; Lambertini, M.; Venturi, F.; Veronesi, G.; Mastroeni, S.; Hrvatin Stancic, B.; Bergant-Suhodolcan, A.; Fortes, C. A Comparative Demographic Study of Atypical Spitz Nevi and Malignant Melanoma. Acta Dermatovenerol. Croat. ADC 2023, 31, 165–168. [Google Scholar]
- Kalidindi, S. The Role of Artificial Intelligence in the Diagnosis of Melanoma. Cureus 2024, 16, e69818. [Google Scholar] [CrossRef] [PubMed]
- Brancaccio, G.; Balato, A.; Malvehy, J.; Puig, S.; Argenziano, G.; Kittler, H. Artificial Intelligence in Skin Cancer Diagnosis: A Reality Check. J. Investig. Dermatol. 2024, 144, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Kahraman, F.; Aktas, A.; Bayrakceken, S.; Çakar, T.; Tarcan, H.S.; Bayram, B.; Durak, B.; Ulman, Y.I. Physicians’ ethical concerns about artificial intelligence in medicine: A qualitative study: “The final decision should rest with a human”. Front. Public Health 2024, 12, 1428396. [Google Scholar] [CrossRef]
- Abgrall, G.; Holder, A.L.; Chelly Dagdia, Z.; Zeitouni, K.; Monnet, X. Should AI models be explainable to clinicians? Crit. Care 2024, 28, 301. [Google Scholar] [CrossRef] [PubMed]
- Jairath, N.; Pahalyants, V.; Shah, R.; Weed, J.; Carucci, J.A.; Criscito, M.C. Artificial Intelligence in Dermatology: A Systematic Review of Its Applications in Melanoma and Keratinocyte Carcinoma Diagnosis. Dermatol. Surg. 2024, 50, 791–798. [Google Scholar] [CrossRef]
- Chen, S.B.; Novoa, R.A. Artificial intelligence for dermatopathology: Current trends and the road ahead. Semin. Diagn. Pathol. 2022, 39, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Primiero, C.A.; Kulkarni, V.; Soyer, H.P.; Betz-Stablein, B. Artificial Intelligence for the Classification of Pigmented Skin Lesions in Populations with Skin of Color: A Systematic Review. Dermatology 2023, 239, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Hosler, G.A.; Murphy, K.M. Ancillary testing for melanoma: Current trends and practical considerations. Hum. Pathol. 2023, 140, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Herrero Colomina, J.; Johnston, E.; Duffus, K.; Zaïr, Z.M.; Thistlethwaite, F.; Krebs, M.; Carter, L.; Graham, D.; Cook, N. Real-world experience of Molecular Tumour Boards for clinical decision-making for cancer patients. npj Precis. Oncol. 2025, 9, 87. [Google Scholar] [CrossRef]
- Tran, M.; Schmidle, P.; Guo, R.R.; Wagner, S.J.; Koch, V.; Lupperger, V.; Novotny, B.; Murphree, D.H.; Hardway, H.D.; D’Amato, M.; et al. Generating dermatopathology reports from gigapixel whole slide images with HistoGPT. Nat. Commun. 2025, 16, 4886. [Google Scholar] [CrossRef]
- Schwen, L.O.; Kiehl, T.-R.; Carvalho, R.; Zerbe, N.; Homeyer, A. Digitization of Pathology Labs: A Review of Lessons Learned. Lab. Investig. 2023, 103, 100244. [Google Scholar] [CrossRef]
- Walsh, E.; Orsi, N.M. The current troubled state of the global pathology workforce: A concise review. Diagn. Pathol. 2024, 19, 163. [Google Scholar] [CrossRef]
- Risko, E.F.; Gilbert, S.J. Cognitive Offloading. Trends Cogn. Sci. 2016, 20, 676–688. [Google Scholar] [CrossRef]
- Nakagawa, K.; Moukheiber, L.; Celi, L.A.; Patel, M.; Mahmood, F.; Gondim, D.; Hogarth, M.; Levenson, R. AI in Pathology: What could possibly go wrong? Semin. Diagn. Pathol. 2023, 40, 100–108. [Google Scholar] [CrossRef]
- Stiff, K.M.; Franklin, M.J.; Zhou, Y.; Madabhushi, A.; Knackstedt, T.J. Artificial intelligence and melanoma: A comprehensive review of clinical, dermoscopic, and histologic applications. Pigment Cell Melanoma Res. 2022, 35, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Wells, A.; Patel, S.; Lee, J.B.; Motaparthi, K. Artificial intelligence in dermatopathology: Diagnosis, education, and research. J. Cutan. Pathol. 2021, 48, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venturi, F.; Veronesi, G.; Gualandi, A.; Magnaterra, E.; Scotti, B.; Sotiri, I.; Baraldi, C.; Alessandrini, A.M.; Veneziano, L.; Vaccari, S.; et al. From Slide to Insight: The Emerging Alliance of Digital Pathology and AI in Melanoma Diagnostics. Cancers 2025, 17, 3696. https://doi.org/10.3390/cancers17223696
Venturi F, Veronesi G, Gualandi A, Magnaterra E, Scotti B, Sotiri I, Baraldi C, Alessandrini AM, Veneziano L, Vaccari S, et al. From Slide to Insight: The Emerging Alliance of Digital Pathology and AI in Melanoma Diagnostics. Cancers. 2025; 17(22):3696. https://doi.org/10.3390/cancers17223696
Chicago/Turabian StyleVenturi, Federico, Giulia Veronesi, Alberto Gualandi, Elisabetta Magnaterra, Biagio Scotti, Ina Sotiri, Carlotta Baraldi, Aurora Maria Alessandrini, Leonardo Veneziano, Sabina Vaccari, and et al. 2025. "From Slide to Insight: The Emerging Alliance of Digital Pathology and AI in Melanoma Diagnostics" Cancers 17, no. 22: 3696. https://doi.org/10.3390/cancers17223696
APA StyleVenturi, F., Veronesi, G., Gualandi, A., Magnaterra, E., Scotti, B., Sotiri, I., Baraldi, C., Alessandrini, A. M., Veneziano, L., Vaccari, S., Cama, E. M., Tassone, D., Corti, B., & Dika, E. (2025). From Slide to Insight: The Emerging Alliance of Digital Pathology and AI in Melanoma Diagnostics. Cancers, 17(22), 3696. https://doi.org/10.3390/cancers17223696








