Blood-Based Surveillance Biomarkers for Gastroesophageal Cancers
Simple Summary
Abstract
1. Introduction
2. Overview of Blood-Based Biomarker Assays and Classes
| Biomarker (Assay) | Cancer Type | Clinical Context | Sensitivity/Specificity | Key Analytical Limitation | Validation Phase | Source(s) |
|---|---|---|---|---|---|---|
| CEA (Carcinoembryonic antigen, ELISA) Class: Protein (Oncofetal antigen) | Esophageal adenocarcinoma; Gastric adenocarcinoma | Surveillance for recurrence in advanced disease (also diagnostic adjunct) | EC (detection): Se = 27.5% (18.9–35.2%) Sp = 95.4% (94.1–96.8%) Esophageal (recurrence): Se = 54.7% (40.9–67.8%) Sp = 90.0% (73.5–97.9%) GC (detection): Se = 20.1% (18.3–22.1%), Sp = 94.7% (93.6–95.7%) Gastric (recurrence): Se = 73.0% (68.8–77.2%), Sp = 59.0% (56.3–61.7%) | Low sensitivity and specificity in early-stage cancer | Esophagus: Phase 2 Gastric: Phase 3 | [44,45,46,47,48,49] |
| CA19-9 (Carbohydrate Antigen 19-9): ELISA; Class: Protein (glycan antigen) | Gastric adenocarcinoma (subset); | Advanced disease monitoring (especially in pancreatobiliary-type or intestinal-type tumors) | GC (detection): Se = 21.4% (19.3–23.0%) Sp = 96.2% (95.2–97.1%) Gastric (Recurrence): Se = 24.1% (10.3–43.5%), Sp = 93.3% (87.3–97.1%) | Poor sensitivity, low specificity, false-negative results in some patient populations, and a lack of standardized cut-off values further hinders its reliable use | Phase 3 | [48,49] |
| CA72-4 (Carbohydrate Antigen 72-4): ELISA; Class: Protein (glycoprotein antigen) | Gastric adenocarcinoma | Diagnostic adjunct; recurrence monitoring | Detection: Se = 58.0% (40.0–73.0%) Sp = 86.0% (80.0–90.0%) Recurrence: Se = 25.0% (0.63–80.6%) (early GC), 45.5% (30.4–61.1%) (advanced GC) Sp = 88.6% (84.6–92.0%) (early GC), 84.6% (76.9–90.4%) (advanced GC) | Low sensitivity in early stages, poor specificity and low positive predictive value | Phase 2 | [50,51,52] |
| SCC-antigen (Squamous Cell Carcinoma Ag): ELISA; Class: Protein (squamous marker) | Esophageal squamous carcinoma | Response evaluation; recurrence surveillance in ESCC | Detection of EC: Se = 35.1% (32.0–38.3%) Sp = 95.4 (93.8–96.7%); Recurrence: Se = 26.8% (14.2–42.9%), Sp = Not reported; elevated SCC-antigen associated with poor OS | Low sensitivity and lack of diagnostic specificity due to elevation in non-malignant conditions | Phase 2 | [44,53] |
| Pepsinogen I/II + H. pylori serology (“ABC” test) ELISA; Class: Protein enzymes (PGI, PGII) + antibody | Gastric (screening for risk) | Screening risk stratification (detects atrophic gastritis) | GC (detection): Se = 87.9% (71.8–96.6%), Sp = 50.8% (37.9–63.6%) | Low sensitivity, poor performance for early cancer, less effective for cancers of cardia and pylorus, and affected by the use of PPIs and presence of certain H pylori strains | Phase 2 | [54] |
| Methylated Reprimo (RPRM) DNA (MSP assay) Class: ctDNA (methylated tumor DNA) | Gastric adenocarcinoma | Early detection; post-op surveillance | GC (detection): Se = 65.0% (53.5–75.3%), Sp = 75.9% (73.2–78.5%) | Heterogeneity in methylation levels across different stages of cancer, potential for false positives, and difficulty in distinguishing early-stage lesions from normal tissue | Phase 2 | [55] |
| Circulating Tumor DNA—personalized panel (NGS) Class: ctDNA (mutations, INDELs) | Esophagus (EAC, ESCC); Gastric | MRD detection post-surgery; relapse surveillance | Esophagus/Gastric (Natera MRD detection): Se = 85.7% (69.7–95.2%) Sp = 95.5% (88.9–98.8%) | Insufficient sensitivity for very low ctDNA concentrations and high technical variability | Phase 2 | [56] |
| Serum DSG2 (Desmoglein-2) ELISA; Class: Protein (adhesion molecule) | ESCC; EGJ adenocarcinoma | Diagnostic/prognostic marker (ESCC) | ESCC (detection): Se = 58.2% (43.2–70.8%) Sp = 84.7% (73.0–92.8%) EJA (detection): Se = 29.2% (20.6–39.5%) Sp = 90.2% (79.1–96.0%) | Low specificity, varied diagnostic accuracy based on cancer subtype and suboptimal sensitivity for early stage | Phase 1 | [57] |
| Multi-TAA autoantibody panel Muliplex Immunoassay; Class: Autoantibodies | Esophageal adenocarcinoma (Barrett’s) | Risk stratify Barrett’s progression | Detection: Se = 53.5–64.0% Sp = 87.0–93.7% | Inter-assay variability, lack of standardization across different multiplex platforms, and the inherent variability of autoantibody responses | Phase 2 | [58] |
| D-mannose (serum metabolite): LC-MS; Class: Metabolite (sugar) | esophageal adenocarcinoma | Prognostic biomarker (EAC) | Low levels associated with poor prognosis; insufficient literature to report performance metrics | Lack of standardized clinical assays and high pre-analytical variability | Phase 1 | [59] |
| Trial (Study) & ID, Country | Design (Biomarkers) | Sample Size | Study Aim | Endpoints | Status/Key Findings | References |
|---|---|---|---|---|---|---|
| NCT05431621; China | Prospective multicenter case–control; Biomarkers: “GutSeer” (1656-locus methylation + fragmentomics ctDNA panel) | Training/validation: 1057 cancer cases vs. 1415 controls; Testing cohort: 846 patients | To develop GutSeer, a blood-based assay using DNA methylation and fragmentomics for multi-GI cancer detection (colorectal, esophagus, gastric, pancreas, and liver) | Diagnostic performance of GutSeer assay for detecting GI cancers, measured by sensitivity, specificity, and AUC in an independent validation cohort. | Completed; Overall GI cancer cohort: AUC = 92.1% Se = 81.5%, Sp = 94.4%; Gastric: Se = 90.5% Esophagus: Se = 65.2% | [62] |
| NCT03425058; China | Prospective single center; Biomarkers: dMMR/MSI status with dynamic evaluation of CTC and ctDNA | 50 | To verify the value of ctDNA and CTC as biomarkers for tumor response in the neoadjuvant chemotherapy (nCRT) treatment of locally advance gastric adenocarcinoma. | Concordance and accuracy of response evaluation results determined by ctDNA, CTCs compared with imaging and serum tumor biomarkers (CEA, CA19-9, CA72-4) | Completed; ctDNA and CTC alteration during neoadjuvant therapy is consistent with conventional histopathological grading and radiological response assessment. | [63] |
| NCT05227261, Vietnam | Prospective multicenter; Biomarkers: SPOT-MAS multimodal ctDNA panel (methylation, fragmentomics, copy number, end motif) | 9057 | To validate the clinical utility of a multimodal non-invasive ctDNA-based MCED test, SPOT-MAS | PPV, NPV, sensitivity, and specificity of the blood ctDNA test in early detection of cancers (breast, lung, gastric, liver, colorectal) | Completed; PPV = 39.5%, NPV = 99.9%, Se = 70.7%, Sp = 99.7%; performance metrics for detecting various cancer types at 12 month follow up | [13] |
| NCT02159339, Korea | Prospective cohort; Biomarkers: GFRA1, SRF, ZNF382 methylation alterations. P16 and E-cadherin status as well. | 198 | To evaluate the feasibility of predicting GC metastasis using CDH1, GFRA1, P16 and ZNF382 DNA methylation as biomarkers. | HR, PPV and NPV of recurrence/metastasis of gastric cancer based on different methylation status | Completed; GFRA1m and ZNF382m are potential biomarkers for the prediction of pN0M0 GC metastasis | [64] |
| NCT04830618, Korea | Prospective cohort; Biomarkers: MOS methylation | 294 overall; 123 gastric cancer vs. 171 gastric dysplasia | To evaluate if MOS methylation can be used to predict metachronous recurrence after endoscopic resection of gastric neoplasms. | MOS methylation for prediction of metachronous recurrence at least 1 year after diagnosis | Completed; MOS methylation predictive (adjusted HR = 4.76) for metachronous recurrence after endoscopic resection for gastric cancer. Se = 80.0%, sp = 53.2%, | [65] |
| NCT02887612, China | Prospective cohort; Biomarkers: ctDNA (targeted sequencing panel of 425 cancer-related genes) | 100 | Predictive value of ctDNA in Early and intermediate-stage gastric cancer | Positive Predictive Value; The proportions of patients with positive serum ctDNA that have postoperative relapse | Completed; postoperative positive ctDNA, HR = 2.74 for recurrence vs. post-ACT positive ctDNA, HR = 15.0. Post ACT ctDNA, se = 77.8%, sp = 90.6% for recurrence. | [66] |
| NCT02674373, France | Prospective cohort; Biomarkers: ctDNA | 82 | ctDNA to predict response and risk stratification in gastric or GE adenocarcinoma | PFS, OS and tumor response rate | Completed; ctDNA + ve during NAT (HR = 6.2), post NAT (HR = 5.3), and after surgery (HR = 12.9) associated with worst outcomes. Early ctDNA clearance during NAT associated with better outcomes | [67] |
| No NCT number, China | Prospective cohort; Biomarkers: tumor-informed ctDNA (NGS) | 46 | To evaluate MRD detection by ctDNA and its association with clinical outcome in resected gastric cancer | DFS and OS | Completed; ctDNA + ve in post-op period associated with DFS and OS (HR = 14.78 and HR = 7.66, respectively) and preceded radiographic recurrence by a median of 6 months. | [68] |
| NCT04005170, China | Interventional Phase 2 open label; Biomarkers: tumor naïve ctDNA (NGS) | 42 | To evaluate the efficacy and safety of the combination of toripalimab (an anti-PD-1 antibody) combined with definitive CRT in locally advanced ESCC | cCR, OS, PFS, duration of response and QOL | Completed; ctDNA -ve patients had a high a cCR to those with detectable ctDNA during CRT83% vs. 39%) or post CRT (78% vs. 30%). ctDNA + ve, shorter PFS and OS. | [69] |
| NCT04460066, NCT05543057, China | Prospective cohort; Biomarkers: ctDNA | 89 | To develop a MRD profiling approach with enhanced sensitivity and specificity for detecting minimal tumor DNA from cfDNA in ESCC | pCR | Completed; MRD -ve associated with pCR in neoadjuvant, surgical, and adjuvant therapy cohort whereas MRD + ve was associated with non-pCR. All MRD -ve patients stayed progression free while 23/26 MRD + ve developed progression. Similar MRD results for radiotherapy cohort | [70] |
| No NCT number, China | Retrospective cohort; Biomarkers: ctDNA (NGS of 77 genes) | 147 | Clinical utility of longitudinal ctDNA as a prognostic biomarker in ESCC | OS and PFS | Completed; For curative surgical resection, high ctDNA (HR = 7.84) and (HR = 5.71) ctDNA alterations associated with poor OS. NAT group, post NAT ctDNA (HR = 3.16) alterations associated with poor PFS. | [71] |
| No NCT number, United States | Retrospective cohort; Biomarkers: ctDNA (CAPP-Seq) | 45 | To evaluate whether ctDNA analysis can predict recurrence in patients with localized ESCA earlier than standard-of-care imaging | Distant metastases, OS and progression | Completed; Detection of ctDNA was associated with tumor progression, metastasis, and disease-specific survival. | [72] |
| No NCT number, United Kingdom | Prospective cohort; Biomarkers: ctDNA (77 gene panel) | 97 | Prognostic potential of ctDNA dynamics in EAC, taking into account CHIP | Recurrence | Completed; ctDNA in plasma following surgery for EAC is prognostic for relapse | [60] |
| No NCT number, Netherlands, Sweden, and Denmark | Retrospective cohort; Biomarkers: ctDNA | 42 | To detect ctDNA alterations after preoperative chemotherapy and after surgery in patients with resectable gastric cancer | Recurrence | Completed; ctDNA is a predictive biomarker of patient outcome to perioperative cancer therapy and surgical resection in patients with gastric cancer. | [73] |
| NCT03044613, United States | Phase IB, open-label, multicenter trial; Biomarkers: ctDNA | 32 | To assess the safety and feasibility of nivolumab +/− relatlimab prior to chemoradiation with II/III gastro/esophageal Cancer | Safety, feasibility, OS, RFS, MPR and pCR. ctDNA association with RFS and OS. | Completed; undetectable ctDNA post-ICI induction, preoperatively and postoperatively had a significantly longer RFS and OS | [61] |
3. Ongoing Trials
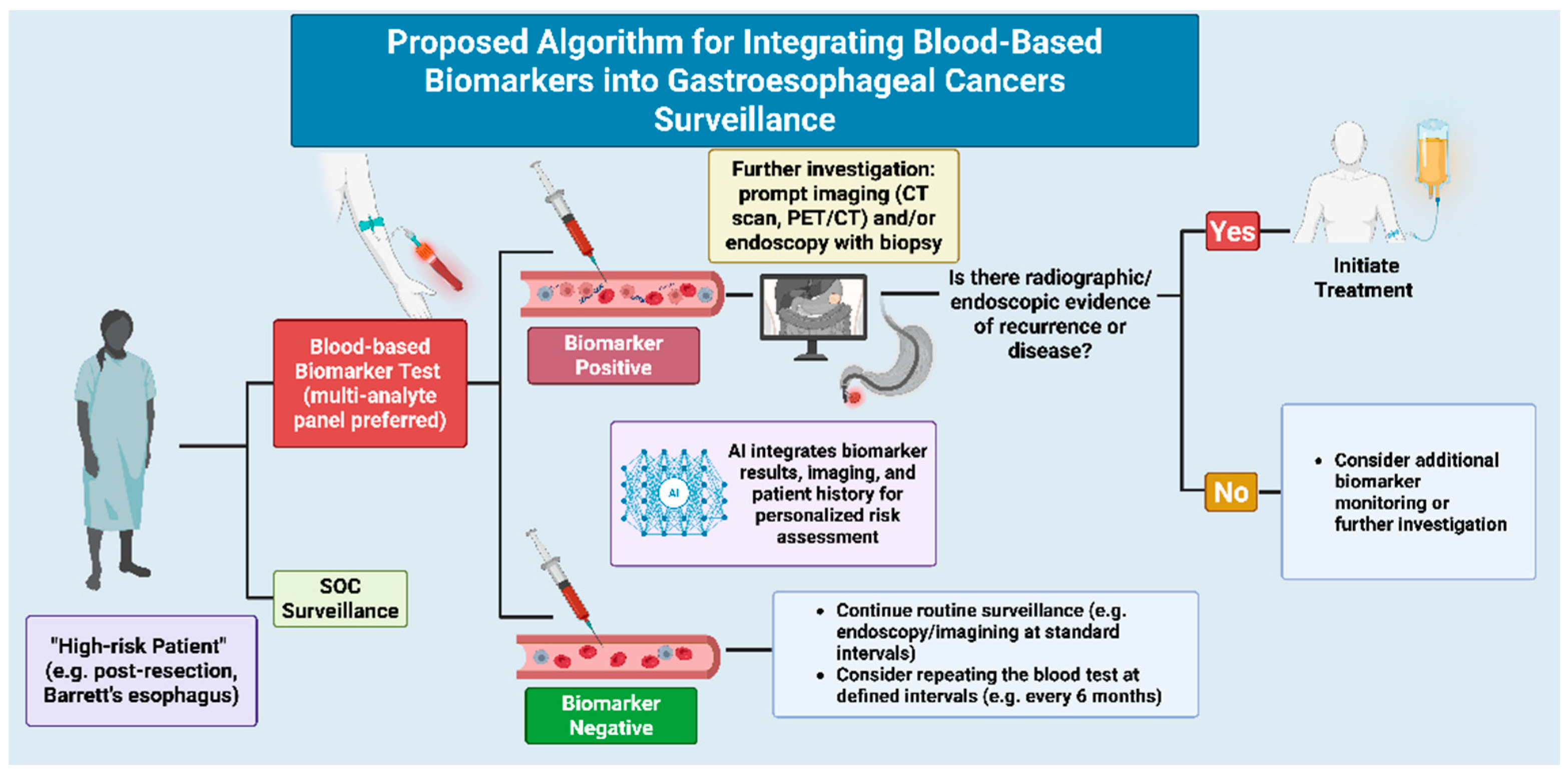
4. Translational Significance and Future Directions
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Qi, L.; Sun, M.; Liu, W.; Zhang, X.; Yu, Y.; Tian, Z.; Ni, Z.; Zheng, R.; Li, Y. Global esophageal cancer epidemiology in 2022 and predictions for 2050: A comprehensive analysis and projections based on GLOBOCAN data. Chin. Med. J. 2024, 137, 3108–3116. [Google Scholar] [CrossRef]
- Alcasid, N.J.; Fink, D.; Banks, K.C.; Susai, C.J.; Barnes, K.; Wile, R.; Sun, A.; Patel, A.; Ashiku, S.; Velotta, J.B. The Impact of Diagnostic Laparoscopy on Upstaging Patients with Siewert II and III Gastroesophageal Junction (GEJ) Cancer. Ann. Surg. Oncol. 2024, 32, 258–264. [Google Scholar] [CrossRef]
- Nassour, I.; Fullington, H.; Hynan, L.S.; Yopp, A.C.; Augustine, M.M.; Polanco, P.M.; Choti, M.A.; Mansour, J.C.; Wang, S.C.; Porembka, M.R. The Yield of Staging Laparoscopy in Gastric Cancer is Affected by Racial and Ethnic Differences in Disease Presentation. Ann. Surg. Oncol. 2017, 24, 1787–1794. [Google Scholar] [CrossRef] [PubMed]
- Schena, C.A.; Laterza, V.; De Sio, D.; Quero, G.; Fiorillo, C.; Gunawardena, G.; Strippoli, A.; Tondolo, V.; De’angelis, N.; Alfieri, S.; et al. The Role of Staging Laparoscopy for Gastric Cancer Patients: Current Evidence and Future Perspectives. Cancers 2023, 15, 3425. [Google Scholar] [CrossRef] [PubMed]
- Bandidwattanawong, C. Multi-disciplinary management of esophageal carcinoma: Current practices and future directions. Crit. Rev. Oncol. 2024, 197, 104315. [Google Scholar] [CrossRef]
- Scheck, M.K.; Hofheinz, R.D.; Lorenzen, S. HER2-Positive Gastric Cancer and Antibody Treatment: State of the Art and Future Developments. Cancers 2024, 16, 1336. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Wang, Y.; Wang, F.; Wang, X. Recurrence pattern and its predictors for advanced gastric cancer after total gastrectomy. Medicine 2020, 99, e23795. [Google Scholar] [CrossRef]
- Wen, X.; Pu, H.; Liu, Q.; Guo, Z.; Luo, D. Circulating Tumor DNA—A Novel Biomarker of Tumor Progression and Its Favorable Detection Techniques. Cancers 2022, 14, 6025. [Google Scholar] [CrossRef]
- Zhu, L.; Xu, R.; Yang, L.; Shi, W.; Zhang, Y.; Liu, J.; Li, X.; Zhou, J.; Bing, P. Minimal residual disease (MRD) detection in solid tumors using circulating tumor DNA: A systematic review. Front. Genet. 2023, 14, 1172108. [Google Scholar] [CrossRef]
- Labiano, I.; Huerta, A.E.; Arrazubi, V.; Hernandez-Garcia, I.; Mata, E.; Gomez, D.; Arasanz, H.; Vera, R.; Alsina, M. State of the Art: ctDNA in Upper Gastrointestinal Malignancies. Cancers 2023, 15, 1379. [Google Scholar] [CrossRef]
- Wang, J.; Huang, J.; Hu, Y.; Guo, Q.; Zhang, S.; Tian, J.; Niu, Y.; Ji, L.; Xu, Y.; Tang, P.; et al. Terminal modifications independent cell-free RNA sequencing enables sensitive early cancer detection and classification. Nat. Commun. 2024, 15, 156. [Google Scholar] [CrossRef]
- Nguyen, L.H.D.; Nguyen, T.H.H.; Van Le, H.; Bui, V.Q.; Nguyen, L.H.; Pham, N.H.; Phan, T.H.; Nguyen, H.T.; Tran, V.S.; Bui, C.V. Prospective validation study: A non-invasive circulating tumor DNA-based assay for simultaneous early detection of multiple cancers in asymptomatic adults. BMC Med. 2025, 23, 90. [Google Scholar] [CrossRef]
- Roy, S.; Kanda, M.; Nomura, S.; Zhu, Z.; Toiyama, Y.; Taketomi, A.; Goldenring, J.; Baba, H.; Kodera, Y.; Goel, A. Diagnostic efficacy of circular RNAs as noninvasive, liquid biopsy biomarkers for early detection of gastric cancer. Mol. Cancer 2022, 21, 42. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Quintero, B. Extracellular MicroRNAs as Intercellular Mediators and Noninvasive Biomarkers of Cancer. Cancers 2020, 12, 3455. [Google Scholar] [CrossRef] [PubMed]
- Roșu, M.C.; Ardelean, A.; Moldovan, S.D.; Faur, F.I.; Nesiu, A.; Totoloci, B.D. The importance of CA 72-4 and CA 19-9 dosing in gastric cancer. J. Med. Life 2023, 16, 186–188. [Google Scholar] [CrossRef]
- Kotzev, A.I.; Draganov, P.V. Carbohydrate Antigen 19-9, Carcinoembryonic Antigen, and Carbohydrate Antigen 72-4 in Gastric Cancer: Is the Old Band Still Playing? Gastrointest. Tumors 2018, 5, 1–13. [Google Scholar] [CrossRef]
- Căinap, C.; Nagy, V.; Gherman, A.; Cetean, S.; Laszlo, I.; Constantin, A.-M.; Căinap, S. Classic Tumor Markers in Gastric Cancer. Current Standards and Limitations. Med. Pharm. Rep. 2015, 88, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Qiu, C.; Wang, B.; Zhang, X.; Wang, X.; Aguilera, R.J.; Zhang, J.-Y. Autoantibody against Tumor-Associated Antigens as Diagnostic Biomarkers in Hispanic Patients with Hepatocellular Carcinoma. Cells 2022, 11, 3227. [Google Scholar] [CrossRef]
- de Jonge, H.; Iamele, L.; Maggi, M.; Pessino, G.; Scotti, C. Anti-Cancer auto-antibodies: Roles, applications and open issues. Cancers 2021, 13, 813. [Google Scholar] [CrossRef]
- Bizzaro, N.; Antico, A.; Villalta, D. Autoimmunity and gastric cancer. Int. J. Mol. Sci. 2018, 19, 377. [Google Scholar] [CrossRef]
- Desmetz, C.; Mange, A.; Maudelonde, T.; Solassol, J. Autoantibody signatures: Progress and perspectives for early cancer detection. J. Cell. Mol. Med. 2011, 15, 2013–2024. [Google Scholar] [CrossRef]
- Wang, Y.; Jiao, Y.; Ding, C.-M.; Sun, W.-Z. The role of autoantibody detection in the diagnosis and staging of lung cancer. Ann. Transl. Med. 2021, 9, 1673. [Google Scholar] [CrossRef] [PubMed]
- Ju, S.; Chen, C.; Zhang, J.; Xu, L.; Zhang, X.; Li, Z.; Chen, Y.; Zhou, J.; Ji, F.; Wang, L. Detection of circulating tumor cells: Opportunities and challenges. Biomark. Res. 2022, 10, 58. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ge, X.; Ju, M.; Zhang, Y.; Di, X.; Liang, L. Circulating Tumor Cells in Esophageal Squamous Cell Carcinoma—Mini Review. Cancer Manag. Res. 2021, 13, 8355–8365. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhang, N.; Wang, S.; Shi, D.; Zhang, C.; Liu, K.; Xiong, B. Wedge-Shaped microfluidic chip for circulating tumor cells isolation and its clinical significance in gastric cancer. J. Transl. Med. 2018, 16, 139. [Google Scholar] [CrossRef]
- Kanayama, M.; Yoneda, K.; Kuwata, T.; Mori, M.; Manabe, T.; Oyama, R.; Matsumiya, H.; Takenaka, M.; Kuroda, K. Enhanced capture system for mesenchymal-type circulating tumor cells using a polymeric microfluidic device ‘CTC-Chip’incorporating cell-surface vimentin. Oncol. Rep. 2024, 52, 156. [Google Scholar] [CrossRef]
- Doyle, L.; Wang, M. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Q.; Qin, F.; Chen, J. Exosomes: A promising avenue for cancer diagnosis beyond treatment. Front. Cell Dev. Biol. 2024, 12, 1344705. [Google Scholar] [CrossRef]
- Tang, S.; Cheng, J.; Yao, Y.; Lou, C.; Wang, L.; Huang, X.; Zhang, Y. Combination of Four Serum Exosomal MiRNAs as Novel Diagnostic Biomarkers for Early-Stage Gastric Cancer. Front. Genet. 2020, 11, 237. [Google Scholar] [CrossRef]
- Li, L.; Sun, Y. Circulating tumor DNA methylation detection as biomarker and its application in tumor liquid biopsy: Advances and challenges. Medcomm 2024, 5, e766. [Google Scholar] [CrossRef]
- Geissler, F.; Nesic, K.; Kondrashova, O.; Dobrovic, A.; Swisher, E.M.; Scott, C.L.; Wakefield, M.J. The role of aberrant DNA methylation in cancer initiation and clinical impacts. Ther. Adv. Med. Oncol. 2024, 16, 17588359231220511. [Google Scholar] [CrossRef]
- Saliminejad, K.; Soleymani Fard, S.; Khorram Khorshid, H.R.; Yaghmaie, M.; Mahmoodzadeh, H.; Mousavi, S.A.; Ghaffari, S.H. Methylation Analysis of P16, RASSF1A, RPRM, and RUNX3 in Circulating Cell-Free DNA for Detection of Gastric Cancer: A Validation Study. Avicenna J. Med. Biotechnol. 2020, 12, 99–106. [Google Scholar] [PubMed]
- Basharat, S.; Horton, J. Emerging Multi-Cancer Early Detection Technologies. Can. J. Health Technol. 2022, 2. [Google Scholar] [CrossRef]
- Loomans-Kropp, H.A.; Song, Y.; Gala, M.; Parikh, A.R.; Van Seventer, E.E.; Alvarez, R.; Hitchins, M.P.; Shoemaker, R.H.; Umar, A. Methylated Septin9 (mSEPT9): A Promising Blood-Based Biomarker for the Detection and Screening of Early-Onset Colorectal Cancer. Cancer Res. Commun. 2022, 2, 90–98. [Google Scholar] [CrossRef]
- Zhu, L.; Li, X.; Yuan, Y.; Dong, C.; Yang, M. APC Promoter Methylation in Gastrointestinal Cancer. Front. Oncol. 2021, 11, 653222. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, Z.; Pei, B.; Wang, J.; Xue, Y.; Zhao, G. DNA methylation markers in esophageal cancer. Front. Genet. 2024, 15, 1354195. [Google Scholar] [CrossRef]
- Nannini, G.; Meoni, G.; Amedei, A.; Tenori, L. Metabolomics profile in gastrointestinal cancers: Update and future perspectives. World J. Gastroenterol. 2020, 26, 2514–2532. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Li, S.; Xiao, X.; Li, J.; Sun, Y.; Ji, S.; Jin, H.; Hua, Z.; Ma, J.; Wang, X.; et al. Metabolomic profiling of plasma reveals potential biomarkers for screening and early diagnosis of gastric cancer and precancerous stages. MedComm–Oncol. 2023, 2, e32. [Google Scholar] [CrossRef]
- Benfatto, S.; Sill, M.; Jones, D.T.W.; Pfister, S.M.; Sahm, F.; von Deimling, A.; Capper, D.; Hovestadt, V. Explainable artificial intelligence of DNA methylation-based brain tumor diagnostics. Nat. Commun. 2025, 16, 1787. [Google Scholar] [CrossRef]
- Sahoo, K.; Lingasamy, P.; Khatun, M.; Sudhakaran, S.L.; Salumets, A.; Sundararajan, V.; Modhukur, V. Artificial Intelligence in cancer epigenomics: A review on advances in pan-cancer detection and precision medicine. Epigenetics Chromatin 2025, 18, 35. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.; Kim, J.S.; Youk, H.-J.; Kim, H.; Hwang, S.-H.; Oh, H.-B.; Ko, D.-H. Relative immunogenicity of blood group antigens: First report in a Korean population. Transfus. Apher. Sci. 2022, 62, 103585. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Wang, J.; Wang, Y.; Tong, M.; Hu, H.; Huang, C.; Li, D. Diagnostic Value of CA 19-9 and Carcinoembryonic Antigen for Pancreatic Cancer: A Meta-Analysis. Gastroenterol. Res. Pract. 2018, 2018, 8704751. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhu, Z.; Liu, Y.; Jin, X.; Xu, Z.; Yu, Q.; Li, K. Diagnostic Value of Multiple Tumor Markers for Patients with Esophageal Carcinoma. PLoS ONE 2015, 10, e0116951. [Google Scholar] [CrossRef]
- Bagaria, B.; Sood, S.; Sharma, R.; Lalwani, S. Comparative study of CEA and CA19-9 in esophageal, gastric and colon cancers individually and in combination (ROC curve analysis). Cancer Biol. Med. 2013, 10, 148–157. [Google Scholar] [CrossRef]
- Clark, G.W.; Ireland, A.P.; Hagen, J.A.; Collard, J.-M.; Peters, J.H.; DeMeester, T.R. Carcinoembryonic antigen measurements in the management of esophageal cancer: An indicator of subclinical recurrence. Am. J. Surg. 1995, 170, 597–601. [Google Scholar] [CrossRef]
- Zhang, R.; Chen, X.; Chen, G.; Zhao, Z.; Zhang, F.; Chen, Z.; Wei, C.; Lin, J.; Liu, Z.; Liang, Y.; et al. Novel surveillance protocol for gastric cancer based on CEA: A high-volume multi-center study. BMC Cancer 2025, 25, 1396. [Google Scholar] [CrossRef]
- Liang, Y.; Wang, W.; Fang, C.; Raj, S.S.; Hu, W.-M.; Li, Q.-W.; Zhou, Z.-W. Clinical significance and diagnostic value of serum CEA, CA19-9 and CA72-4 in patients with gastric cancer. Oncotarget 2016, 7, 49565–49573. [Google Scholar] [CrossRef]
- Shibata, C.; Nakano, T.; Yasumoto, A.; Mitamura, A.; Sawada, K.; Ogawa, H.; Miura, T.; Ise, I.; Takami, K.; Yamamoto, K.; et al. Comparison of CEA and CA19-9 as a predictive factor for recurrence after curative gastrectomy in gastric cancer. BMC Surg. 2022, 22, 213. [Google Scholar] [CrossRef]
- Wang, H.; Jin, W.; Wan, C.; Zhu, C. Diagnostic value of combined detection of CA72-4, CA19-9, and carcinoembryonic antigen comparing to CA72-4 alone in gastric cancer: A systematic review and meta-analysis. Transl. Cancer Res. 2022, 11, 848–856. [Google Scholar] [CrossRef]
- Hu, P.-J.; Chen, M.-Y.; Wu, M.-S.; Lin, Y.-C.; Shih, P.-H.; Lai, C.-H.; Lin, H.-J. Clinical Evaluation of CA72-4 for Screening Gastric Cancer in a Healthy Population: A Multicenter Retrospective Study. Cancers 2019, 11, 733. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Oh, S.J.; Oh, C.A.; Choi, M.G.; Noh, J.H.; Sohn, T.S.; Bae, J.M.; Kim, S. The relationships between perioperative CEA, CA 19-9, and CA 72-4 and recurrence in gastric cancer patients after curative radical gastrectomy. J. Surg. Oncol. 2011, 104, 585–591. [Google Scholar] [CrossRef]
- Kawaguchi, H.; Ohno, S.; Miyazaki, M.; Hashimoto, K.; Egashira, A.; Saeki, H.; Watanabe, M.; Sugimachi, K. CYFRA 21-1 determination in patients with esophageal squamous cell carcinoma: Clinical utility for detection of recurrences. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2000, 89, 1413–1417. [Google Scholar] [CrossRef]
- In, H.; Adams, A.; Ho, S.; Sarkar, S.; Wang, F.; Oh, A.; Yun, D.J.; Gandhi, A.; Li, S.; Liu, H.; et al. Pepsinogen and Helicobacter pylori: Serum biomarkers for gastric cancer risk in a diverse United States population. Surg. Oncol. Insight 2024, 1, 100086. [Google Scholar] [CrossRef]
- Maturana, M.J.; Padilla, O.; Santoro, P.M.; Alarcón, M.A.; Olivares, W.; Blanco, A.; Armisen, R.; Garrido, M.; Aravena, E.; Barrientos, C.; et al. Methylated Reprimo Cell-Free DNA as a Non-Invasive Biomarker for Gastric Cancer. Int. J. Mol. Sci. 2025, 26, 3333. [Google Scholar] [CrossRef]
- Huffman, B.M.; Aushev, V.N.; Budde, G.L.; Chao, J.; Dayyani, F.; Hanna, D.; Botta, G.P.; Catenacci, D.V.; Maron, S.B.; Krinshpun, S.; et al. Analysis of Circulating Tumor DNA to Predict Risk of Recurrence in Patients With Esophageal and Gastric Cancers. JCO Precis. Oncol. 2022, 6, e2200420. [Google Scholar] [CrossRef]
- Liu, Y.-Q.; Chu, L.-Y.; Yang, T.; Zhang, B.; Zheng, Z.-T.; Xie, J.-J.; Xu, Y.-W.; Fang, W.-K. Serum DSG2 as a potential biomarker for diagnosis of esophageal squamous cell carcinoma and esophagogastric junction adenocarcinoma. Biosci. Rep. 2022, 42, BSR20212612. [Google Scholar] [CrossRef]
- Xu, Y.-W.; Peng, Y.-H.; Xu, L.-Y.; Xie, J.-J.; Li, E.-M. Autoantibodies: Potential clinical applications in early detection of esophageal squamous cell carcinoma and esophagogastric junction adenocarcinoma. World J. Gastroenterol. 2019, 25, 5049–5068. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Liang, D.; Pierzynski, J.A.; Zheng, L.; Ye, Y.; Zhang, J.; Ajani, J.A.; Wu, X. D-Mannose: A Novel Prognostic Biomarker for Patients with Esophageal Adenocarcinoma. Carcinogenesis 2017, 38, 162–167. [Google Scholar] [CrossRef]
- Ococks, E.; Frankell, A.; Soler, N.M.; Grehan, N.; Northrop, A.; Coles, H.; Redmond, A.; Devonshire, G.; Weaver, J.; Hughes, C.; et al. Longitudinal tracking of 97 esophageal adenocarcinomas using liquid biopsy sampling. Ann. Oncol. 2021, 32, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.J.; Landon, B.V.; Zaidi, A.H.; Singh, D.; Canzoniero, J.V.; Balan, A.; Hales, R.K.; Voong, K.R.; Battafarano, R.J.; Jobe, B.A.; et al. Neoadjuvant nivolumab or nivolumab plus LAG-3 inhibitor relatlimab in resectable esophageal/gastroesophageal junction cancer: A phase Ib trial and ctDNA analyses. Nat. Med. 2024, 30, 1023–1034. [Google Scholar] [CrossRef]
- Huang, A.; Guo, D.-Z.; Su, Z.-X.; Zhong, Y.-S.; Liu, L.; Xiong, Z.-G.; He, D.-L.; Yan, B.; Li, Q.-L.; Feng, Z.; et al. GUIDE: A prospective cohort study for blood-based early detection of gastrointestinal cancers using targeted DNA methylation and fragmentomics sequencing. Mol. Cancer 2025, 24, 163. [Google Scholar] [CrossRef]
- Fu, T. Monitoring patient-specific mutation in ctDNA and CTC for tumour response evaluation after neoadjuvant chemotherapy in locally advanced gastric cancer. Ann. Oncol. 2019, 30, v313–v314. [Google Scholar] [CrossRef]
- Liu, Z.; Cheng, X.; Zhang, L.; Zhou, J.; Deng, D.; Ji, J. A panel of DNA methylated markers predicts metastasis of pN0M0 gastric carcinoma: A prospective cohort study. Br. J. Cancer 2019, 121, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Shin, C.M.; Kim, N.; Yoon, H.; Choi, Y.J.; Park, J.H.; Park, Y.S.; Lee, D.H. Aberrant DNA Methylation Maker for Predicting Metachronous Recurrence After Endoscopic Resection of Gastric Neoplasms. Cancer Res. Treat. 2022, 54, 1157–1166. [Google Scholar] [CrossRef]
- Yuan, S.; Nie, R.; Huang, Y.; Chen, Y.; Wang, S.; Sun, X.; Li, Y.; Liu, Z.; Chen, Y.; Yao, Y.; et al. Residual circulating tumor DNA after adjuvant chemotherapy effectively predicts recurrence of stage II-III gastric cancer. Cancer Commun. 2023, 43, 1312–1325. [Google Scholar] [CrossRef]
- Zaanan, A.; Didelot, A.; Broudin, C.; Laliotis, G.; Spickard, E.; Dutta, P.; Saltel-Fulero, A.; Sullo, F.G.; Pizzamiglio, M.; Mariani, A.; et al. Longitudinal circulating tumor DNA analysis during treatment of locally advanced resectable gastric or gastroesophageal junction adenocarcinoma: The PLAGAST prospective biomarker study. Nat. Commun. 2025, 16, 6815. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Gong, Y.; Lam, V.K.; Shi, Y.; Guan, Y.; Zhang, Y.; Ji, L.; Chen, Y.; Zhao, Y.; Qian, F.; et al. Deep sequencing of circulating tumor DNA detects molecular residual disease and predicts recurrence in gastric cancer. Cell Death Dis. 2020, 11, 346. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Liu, S.; Zhu, Y.; Wang, R.; Cheng, X.; Chen, B.; Dragomir, M.P.; Zhang, Y.; Hu, Y.; Liu, M. Predictive role of ctDNA in esophageal squamous cell carcinoma receiving definitive chemoradiotherapy combined with toripalimab. Nat. Commun. 2024, 15, 1919. [Google Scholar] [CrossRef]
- Yue, P.; Bie, F.; Zhu, J.; Gao, L.-R.; Zhou, Z.; Bai, G.; Wang, X.; Zhao, Z.; Xiao, Z.-F.; Li, Y.; et al. Minimal residual disease profiling predicts pathological complete response in esophageal squamous cell carcinoma. Mol. Cancer 2024, 23, 96. [Google Scholar] [CrossRef]
- Ng, H.Y.; Ko, J.M.Y.; Lam, K.O.; Kwong, D.L.W.; Lo, A.W.I.; Wong, I.Y.H.; Wong, C.L.Y.; Chan, S.Y.; Chan, K.K.; Law, T.T.; et al. Circulating Tumor DNA Dynamics as Prognostic Markers in Locally Advanced and Metastatic Esophageal Squamous Cell Carcinoma. JAMA Surg. 2023, 158, 1141–1150. [Google Scholar] [CrossRef] [PubMed]
- Azad, T.D.; Chaudhuri, A.A.; Fang, P.; Qiao, Y.; Esfahani, M.S.; Chabon, J.J.; Hamilton, E.G.; Yang, Y.D.; Lovejoy, A.; Newman, A.M.; et al. Circulating Tumor DNA Analysis for Detection of Minimal Residual Disease After Chemoradiotherapy for Localized Esophageal Cancer. Gastroenterology 2020, 158, 494–505.e6. [Google Scholar] [CrossRef] [PubMed]
- Leal, A.; van Grieken, N.C.T.; Palsgrove, D.N.; Phallen, J.; Medina, J.E.; Hruban, C.; Broeckaert, M.A.M.; Anagnostou, V.; Adleff, V.; Bruhm, D.C.; et al. White blood cell and cell-free DNA analyses for detection of residual disease in gastric cancer. Nat. Commun. 2020, 11, 525. [Google Scholar] [CrossRef] [PubMed]

| Trial (Study) & ID, Country | Design (Biomarkers) | Sample Size | Study Aim | Endpoints | Status/Key Findings |
|---|---|---|---|---|---|
| NCT05029869; Vietnam | Prospective observational; Biomarkers: ctDNA | 100 | Use of NGS to detect ctDNA in gastric cancer after surgery | Sensitivity/specificity of MRD detection using ctDNA | Ongoing (Active, not recruiting) |
| NCT06232395, China | Prospective multicenter cohort: Biomarkers: ctDNA | 1197 | To develop and validate a new multi-target marker early detection and postoperative monitoring of gastric cancer | Performance of the multi-target panel in diagnosis, detecting postoperative recurrence and metastasis | Ongoing, recruiting |
| NCT04993378, China | Prospective cohort: Biomarkers: extracellular vesicle (EV) protein signature “EV-score” derived from four EV proteins | 40 | Validate whether the EV-score can predict and monitor immunotherapy outcomes in advanced gastric cancer, both at baseline and during treatment | Performance of EV-score at baseline and longitudinally for prediction and monitoring of immunotherapeutic response (accuracy, sensitivity, specificity) | Unknown |
| NCT04053725, China | Prospective cohort; Biomarkers: ctDNA | 200 | Clinical utility of ctDNA in predicting the efficacy of immunotherapy for advanced gastric cancer. | The proportions of patients with positive serum ctDNA that have postoperative recurrence | Unknown |
| NCT06662110, China | Prospective cohort; Biomarkers: PSRscore calculated based on baseline serum immune proteomics | 206 | To validate the predictive value of systemic immune markers in predicting neoadjuvant treatment responses in advanced gastric cancer. | Sensitivity and specificity of PSRscore in predicting tumor regression/objective response/PFS/OS after NAT | Ongoing, recruiting |
| NCT03957564, China | Interventional phase II open label single group trial; Biomarkers: CTC, ctDNA and cfDNA | 40 | Clinical value of dynamic changes in CTC, ctDNA and cfDNA in NAT chemotherapy or and operation of resectable or locally advanced gastric or GEJ cancer | Comparison of biomarker dynamics with CT/RECIST responses and prognosis (e.g., recurrence, survival) | Unknown |
| NCT06893133; China | Prospective observational multicenter; Biomarkers: personalized ctDNA-MRD | 110 | Correlation between ctDNA-MRD status and tumor recurrence and metastasis in gastric cancer patients who have received neoadjuvant therapy followed by curative resection | Sensitivity, Specificity, and Positive predictive value of ctDNA-MRD in predicting postoperative recurrence | Ongoing (Active, not recruiting) |
| NCT01715233, United States | Phase 2, single arm treatment trial; Biomarkers: CHFR methylation | 27 | To estimate and compare the response rates in metastatic GE patients treated with mDCF based on methylation status of CHFR. | objective response rate (PR/SD/PD) stratified by CHFR methylation | Completed; results not reported |
| NCT06979895, China | Prospective cohort; Biomarkers: ctDNA multigene methylation panel | 150 | Correlate methylation dynamics with treatment response for gastric cancer | Association between changes in methylation and objective response | Ongoing; not yet recruiting |
| NCT06335576, China | Prospective single center cohort; Biomarkers: serum proteomics panel | 89 | Establish circulating proteomic subtypes of gastric cancer and explore their clinical applicability | Identification and reproducibility of serum-based proteomic subtypes (e.g., classification accuracy, subtype detection) | Ongoing; not yet recruiting |
| NCT02610218, China | Prospective cohort; Biomarkers: ctDNA (HER2 amplification/mutations) + CTCs | 124 | Evaluate whether changes in cfDNA levels and CTC counts correspond with therapeutic response to HER2-targeted therapy in metastatic HER2-positive gastric cancer | Concordance of ctDNA/CTCs with radiologic treatment response, emergence of HER2 resistance, and detection of progression | Unknown |
| NCT04511559, China | Prospective cohort; Biomarkers: ctDNA methylation | 540 | To describe ctDNA methylation profile in gastric cancer and demonstrate correlation between ctDNA methylation status and diagnosis and prognosis | Analysis of ctDNA methylation status and its correlation to early diagnosis and prognostic evaluation of gastric cancer | Unknown |
| NCT05513144, China | Prospective cohort; Biomarkers: ctDNA | 30 | To evaluate the use of next generation sequencing (NGS) to detect circulating tumor DNA in advanced HER2 negative gastric cancer patients | Prognostic molecular markers; The sensitivity and specificity of ctDNA detection | Unknown |
| NCT05208372, China | Prospective case–control; Biomarkers: | 200 | Value of CTCs and ctDNA in the diagnosis of metastasis in ascites/peritoneal flushing fluid and blood | Quantity of CTCs; Expression of ctDNA | Unknown |
| NCT04576858, Denmark | Prospective cohort; Biomarkers: ctDNA | 1950 | To evaluate the treatment effect as well as predictive and prognostic factors with special emphasis on the clinical utility of ctDNA in plasma in patients with GE cancer | Time to recurrence | Unknown |
| NCT05348161, China | Interventional non-randomized parallel: Biomarkers: HER2/PD-L1-positive CTCs; ctDNA genomic events | 100 | To evaluate how HER2-targeted therapy and immunotherapy affect molecular profiles in HER2-positive gastric cancer patients via multi-omics liquid biopsy markers | Proportions of HER2- and PD-L1-positive CTCs Incidence rates of various ctDNA genomic alterations (e.g., copy number changes, insertions/deletions) | Unknown |
| NCT07076979, China | Prospective case–control; Biomarkers: Metabolic markers | 250 | To develop and validate metabolic biomarkers for early diagnosis, prognosis, and prediction of recurrence and metastasis in gastric cancer | OS, DFS, HR, PPV and NPV | Ongoing, recruiting |
| NCT04000425, China | Prospective cohort; Biomarkers: AVENIO ctDNA surveillance kit | 55 | Evaluate ctDNA as an indicator of MRD and as a marker of adjuvant chemotherapy response after radical gastrectomy. | Disease recurrence risk; DFS; ctDNA changing to adjuvant chemotherapy response; Time of first negative ctDNA detection from positive ctDNA detection | Unknown |
| NCT05366881, United States | Prospective multicenter case–control; Biomarkers: Genome-wide cfDNA methylome enrichment | 7000 | Train and validate methylation-based ctDNA test for early cancer detection and MRD (including esophagus and gastric) | Sensitivity & specificity of assay vs. controls | Ongoing, recruiting |
| NCT07035587, Korea | Prospective-retrospective cohort; Biomarkers: Serial cfDNA (ctDNA), RNA, protein profiles | 1200 | Early diagnosis & post-treatment MRD monitoring for multiple cancers (including esophagus and gastric) | Sensitivity & specificity for cancer detection; VAF correlation with recurrence | Ongoing, recruiting |
| NCT05059444, United States, Germany, France, Italy and Spain | Prospective multicenter cohort; Biomarkers: Guardant reveal assay using methylated ctDNA | 2020 | Using a novel ctDNA approach to detect recurrence in early-stage solid tumors (including esophagus and gastric cancer) | Distant recurrence free interval. Lead time, sensitivity, and specificity and of ctDNA in detecting recurrence | Ongoing, recruiting |
| NCT06227728, Vietnam | Prospective multicenter cohort; Biomarkers: ctDNA using targeted sequencing and multiplex PCR approaches. | 50 | Assess if changes in ctDNA can predict early response to ICIs in patients with advanced-stage cancer (including gastric cancer) | Association between ctDNA dynamics and clinical response; comparison with RECIST; prognostic value of ctDNA clearance and with PFS/OS | Ongoing, recruiting |
| NCT04168931, Brazil | Interventional Phase II open label; Biomarkers: HER-2 positive CTCs | 85 | To investigate whether HER2-expressing CTCs may be suitable for prediction of response in patients with relapsed or metastatic gastric cancer who are histologically HER2-negative and treated with trastuzumab combination chemotherapy | Radiological response rate, frequency of HER 2 expression among CTCs of patients with recurrence or metastasis with negative expression in tumor tissue | Terminated, recruitment failure |
| NCT03023436, China | Interventional Phase III open label trial; Biomarkers: ctDNA and CTCs | 220 | To assess ctDNA and CTC alterations as potential biomarkers for debulking surgery combined with HIPEC and systemic chemotherapy in patients with gastric cancer and peritoneal dissemination (as a secondary outcome measure) | Median survival time, OS, PFS, morbidity and mortality, QOL, CTCs changes, ctDNA changes, and molecular biomarker (including 14 genes) alteration | Unknown status |
| NCT04510285, United States | Interventional Phase II open label; Biomarkers: ctDNA | 48 | To evaluate differences in 6-month ctDNA clearance rate in HER2+ esophagogastric cancer with persistent ctDNA following curative surgery when treated with “second adjuvant” trastuzumab with or without pembrolizumab | Rate of ctDNA clearance at 6 months | Terminated, recruitment failure |
| NCT04665687, Korea | Prospective cohort; Biomarkers: ctDNA | 1730 | To differentiate early gastric cancer and precancerous adenoma and predict recurrence by finding biomarkers through molecular profiling | Biomarker-based differentiation between adenoma and early GC; prognostic biomarkers for recurrence | Unknown status |
| NCT05594381, China | Interventional Phase II open label; Biomarkers: ctDNA | 90 | To dynamically detect gene mutations, protein expressions and tumor images in G/GEJ tumor tissues and blood samples before, under and after PD-1 monoclonal antibody (sintilimab) combined with SOX neoadjuvant therapy by using ctDNA targeted sequencing combined with multi-omics technology | pCR, ORR, DCR, MPR, TRG, R0 resection rate, OS, tumor downstaging, DFS, treatment-emergent adverse events and 30-day postoperative mortality | Not yet recruiting |
| NCT04929015, United States | Interventional open label; Biomarkers: tumor-informed personalized ctDNA assay (Signatera) | 30 | Utility of ctDNA as a sensitive biomarker in patients with Peritoneal Carcinomatosis treated with chemotherapy, CRS and/or HIPEC | Clearance rate of ctDNA with cytoreductive surgery (CRS), comparing with clinical staging of CRS and activity of chemotherapy in this disease | Ongoing, recruiting |
| NCT05482516, United States | Interventional Phase III open label; Biomarkers: tumor-informed personalized ctDNA-MRD assay (Signatera) | 20 | Guide atezolizumab and bevacizumab therapy by MRD status in GI cancer | Rate of Signatera ctDNA positivity, rate of enrollment, rate of ctDNA complete response, rate if ctDNA partial disease and rate of ctDNA progressive disease | Ongoing, recruiting |
| NCT05661110, China | Prospective cohort; Biomarkers: AmoyDx® Master Panel (559 genes for DNA mutation and 1813 genes for RNA expression) | 46 | To analyze the correlation between genomic alterations, gene expression characteristics and the efficacy of HIPEC combined with PD1/PDL1 inhibitor conversion therapy in patients with peritoneal metastasis of gastric cancer | Relationship between the status, numerical changes in ctDNA during HIPEC combined with PD1/PDL1 inhibitor conversion therapy and postoperative R0 resection rate. Correlation between genomic changes of ctDNA and ORR, OS, RFA and event-free survival | Not yet recruiting |
| NCT04943406, Italy | Prospective cohort; Biomarkers: ctDNA | 150 | Prognostic role of ctDNA in patients with locally advanced gastric cancer | Impact of ctDNA (in peritoneal lavage and peripheral blood) positivity on OS and DFS | Ongoing, recruiting |
| NCT06253650, Italy | Interventional phase II open label; Biomarkers: ctDNA | 46 | To investigate the activity, efficacy and safety of trastuzumab-deruxtecan (T-DXd) plus capecitabine/5-fluorouracil as a postoperative treatment in localized/locally advanced gastric or GE junction cancer (GC/GEJC)/esophageal adenocarcinoma patients with HER2 overexpression/amplification and positive postoperative ctDNA after preoperative 5-fluorouracil plus leucovorin, oxaliplatin, and docetaxel (FLOT) regimen followed by radical surgery | ctDNA clearance, DFS, OS, metastases-free survival, and QOL | Ongoing, recruiting |
| NCT05494060, China | Interventional Phase II open label; Biomarkers: ctDNA | 80 | To assess safety and anti-tumor activity Penpulimab in combination with Anlotinib and standard chemotherapy as adjuvant treatment for ctDNA-positive G/GEJ cancer | DFS at different time points, OS and toxicity | Ongoing, recruiting |
| NCT05965479, United Kingdom | Interventional Phase II open label; Biomarkers: ctDNA (Signatera assay) | 25 | To assess the efficacy of trastuzumab deruxtecan in reducing micrometastatic disease burden in HER2 positive GEA patients who are ctDNA positive after chemotherapy and surgery | ctDNA clearance, DFS, OS, and QOL | Ongoing, recruiting |
| NCT05067842, United States | Prospective case–control; Biomarkers: ctDNA (Signatera assay) | 30 | To determine the feasibility of assessing tumor response utilizing ctDNA in patients of locally advanced esophageal and GE junction (LA-EA/GEJ) cancer | Tumor response measured by ctDNA, R0 surgical resection, OS and RFS | Withdrawn |
| NCT06498752, China | Interventional Phase II open label; Biomarkers: ctDNA | 102 | To validate whether ctDNA MRD status after radical radiotherapy can stratify prognosis and guide consolidation therapy with PD-1 inhibitors in patients with ESCC | Median PFS, OS, cancer-specific survival, toxicity, swallowing function, dynamic ctDNA changes and correlation with recurrence | Ongoing, recruiting |
| NCT05759325, China | Prospective cohort; Biomarkers: ctDNA MRD | 100 | To observe and evaluate the correlation between ctDNA-MRD and the therapeutic effect and prognosis of stage II-IVA operable ESCC | PFS rate of ESCC patients with different MRD status during perioperative period | Not yet recruiting |
| NCT06103890, China | Prospective cohort; Biomarkers: ctDNA | 100 | To explore the clinical value of MRD as a biomarker for assessing treatment efficacy, predicting recurrence risk, and evaluating prognosis in ESCC | pCR, R0 resection rate, ctDNA clearance, MPR, RFS, and OS | Ongoing, recruiting |
| NCT05426850, China | Prospective cohort; Biomarkers: ctDNA | 100 | To analyze the relationship between the dynamic changes in ctDNA and tumor relapse of ESCC treated by concurrent chemoradiotherapy | Changes in ctDNA status and recurrence, OS, and RFS. | Unknown status |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dadgar, N.; Anees, M.; Sherry, C.; Park, H.Y.; Grayhack, E.E.; Goel, A.; Khan, A.F.; Omstead, A.; Bartlett, D.L.; Wagner, P.L.; et al. Blood-Based Surveillance Biomarkers for Gastroesophageal Cancers. Cancers 2025, 17, 3552. https://doi.org/10.3390/cancers17213552
Dadgar N, Anees M, Sherry C, Park HY, Grayhack EE, Goel A, Khan AF, Omstead A, Bartlett DL, Wagner PL, et al. Blood-Based Surveillance Biomarkers for Gastroesophageal Cancers. Cancers. 2025; 17(21):3552. https://doi.org/10.3390/cancers17213552
Chicago/Turabian StyleDadgar, Neda, Muhammad Anees, Christopher Sherry, Hyun Young Park, Erin E. Grayhack, Arul Goel, Alisha F. Khan, Ashten Omstead, David L. Bartlett, Patrick L. Wagner, and et al. 2025. "Blood-Based Surveillance Biomarkers for Gastroesophageal Cancers" Cancers 17, no. 21: 3552. https://doi.org/10.3390/cancers17213552
APA StyleDadgar, N., Anees, M., Sherry, C., Park, H. Y., Grayhack, E. E., Goel, A., Khan, A. F., Omstead, A., Bartlett, D. L., Wagner, P. L., & Zaidi, A. H. (2025). Blood-Based Surveillance Biomarkers for Gastroesophageal Cancers. Cancers, 17(21), 3552. https://doi.org/10.3390/cancers17213552








