EIF4A3-Mediated circ_0008126 Inhibits the Progression and Metastasis of Gastric Cancer by Modulating the APC/β-Catenin Pathway
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Patient Data
2.2. Cell Culture and Transfection
2.3. Quantitative Reverse Transcription-PCR (qRT-PCR)
2.4. RNA Fluorescence In Situ Hybridization (FISH)
2.5. RNase R Digestion and Actinomycin D Assay
2.6. Cell Counting Kit-8 (CCK-8)
2.7. Colony Formation Assay
2.8. 5-Ethynyl-2′-Deoxyuridine (EdU)
2.9. Wound Healing Assay
2.10. Transwell Assay
2.11. Western Blotting (WB)
2.12. Dual-Luciferase Reporter Assay
2.13. Immunofluorescence (IF)
2.14. RNA Pull-Down
2.15. RNA Immunoprecipitation (RIP)
2.16. Immunohistochemistry (IHC)
2.17. Animal Model Experiments
2.18. Statistical Analysis
3. Results
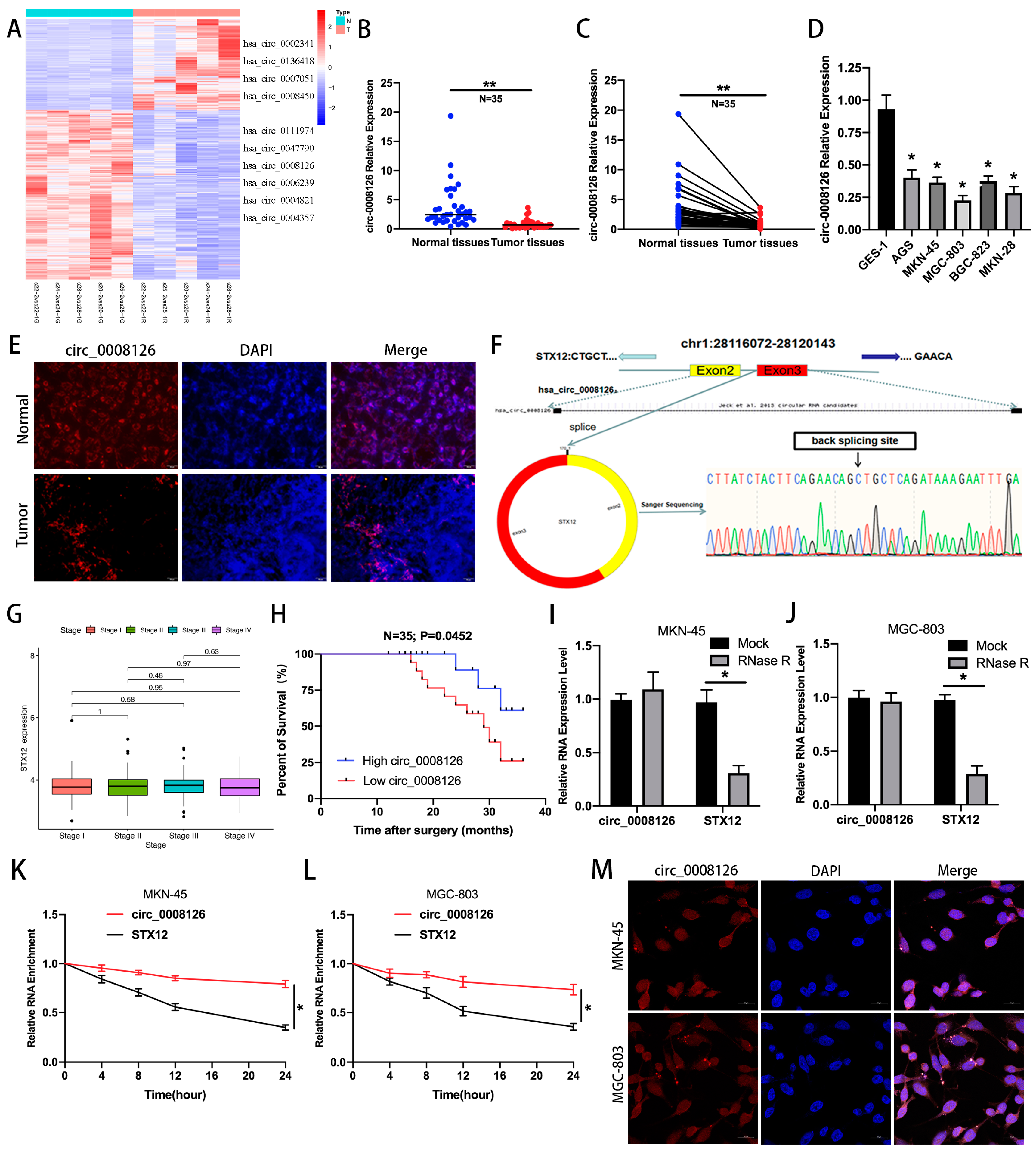
3.1. circ_0008126 Is Verified and Characterized in GC Samples
3.2. EIF4A3-Induced circ_0008126 Suppresses the Proliferation, Metastasis, and EMT Ability of GC Cells
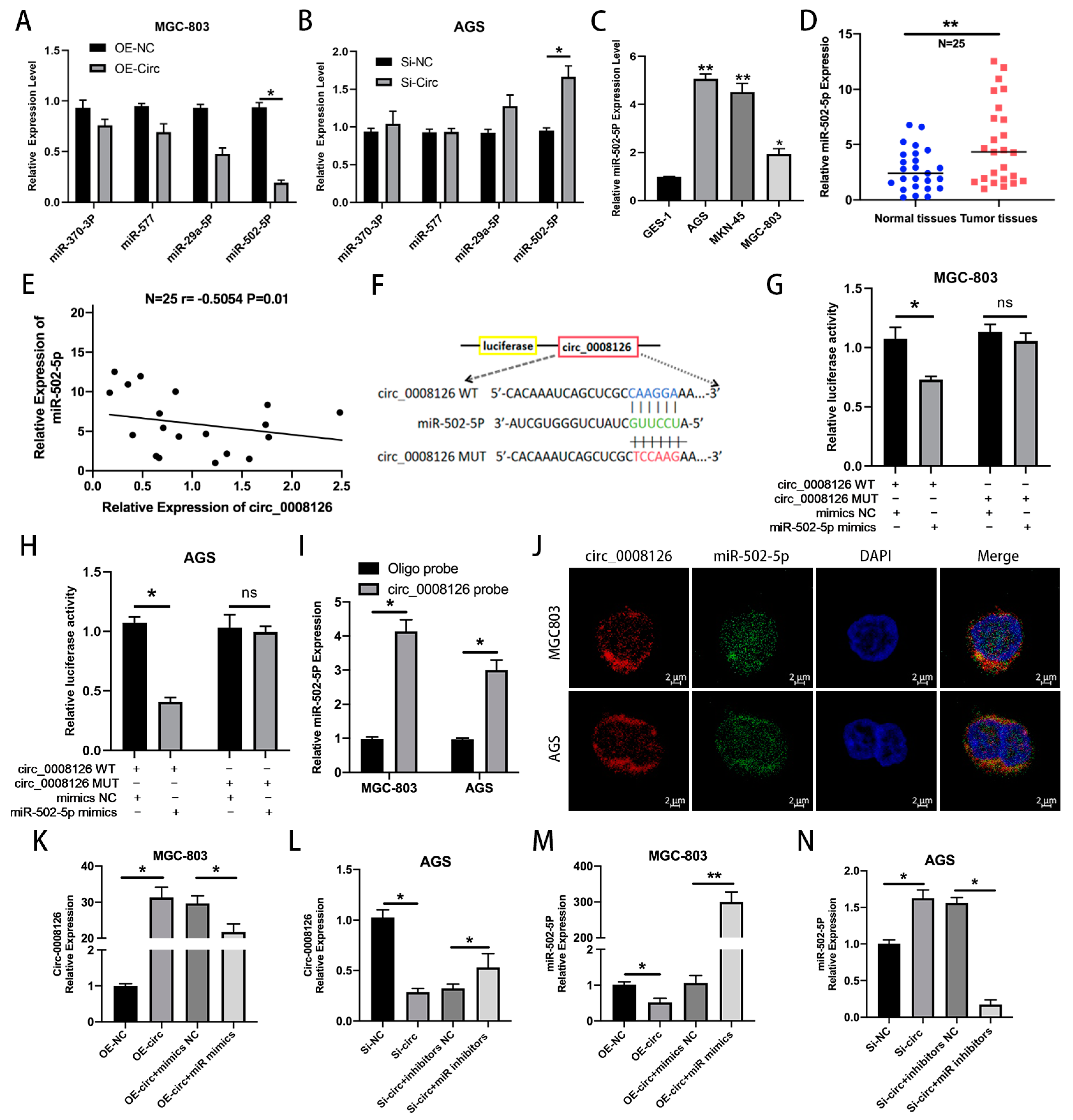
3.3. circ_0008126 Directly Binds to miR-502-5p in GC
3.4. circ_0008126 Reduced GC Progression via Targeting miR-502-5p
3.5. circ_0008126 Inhibits GC Progression Through miR-502-5p/APC/β-Catenin Pathway
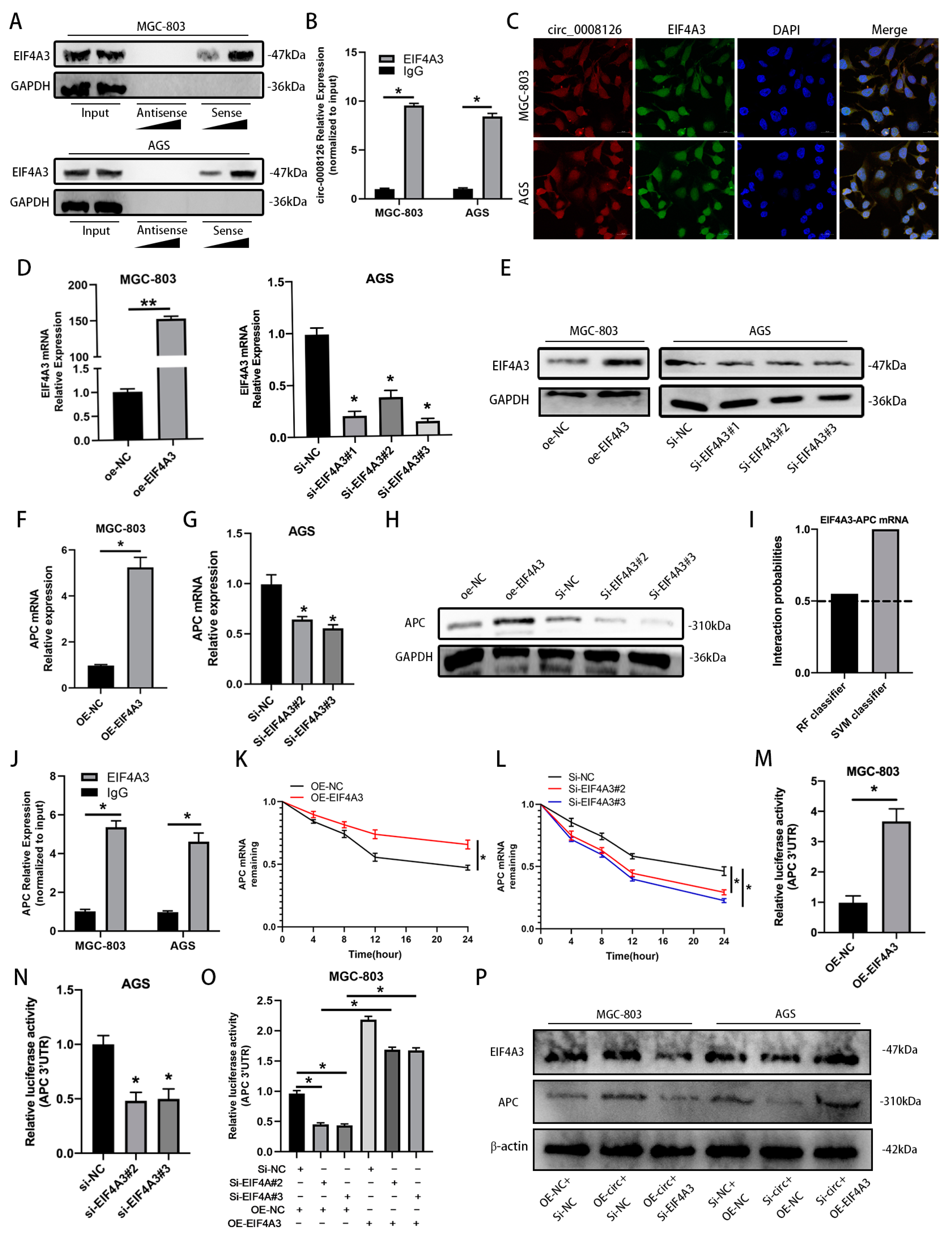
3.6. circ_0008126 Interacts with the Cytoplasmic EIF4A3 and Enhances the Stability of APC in GC
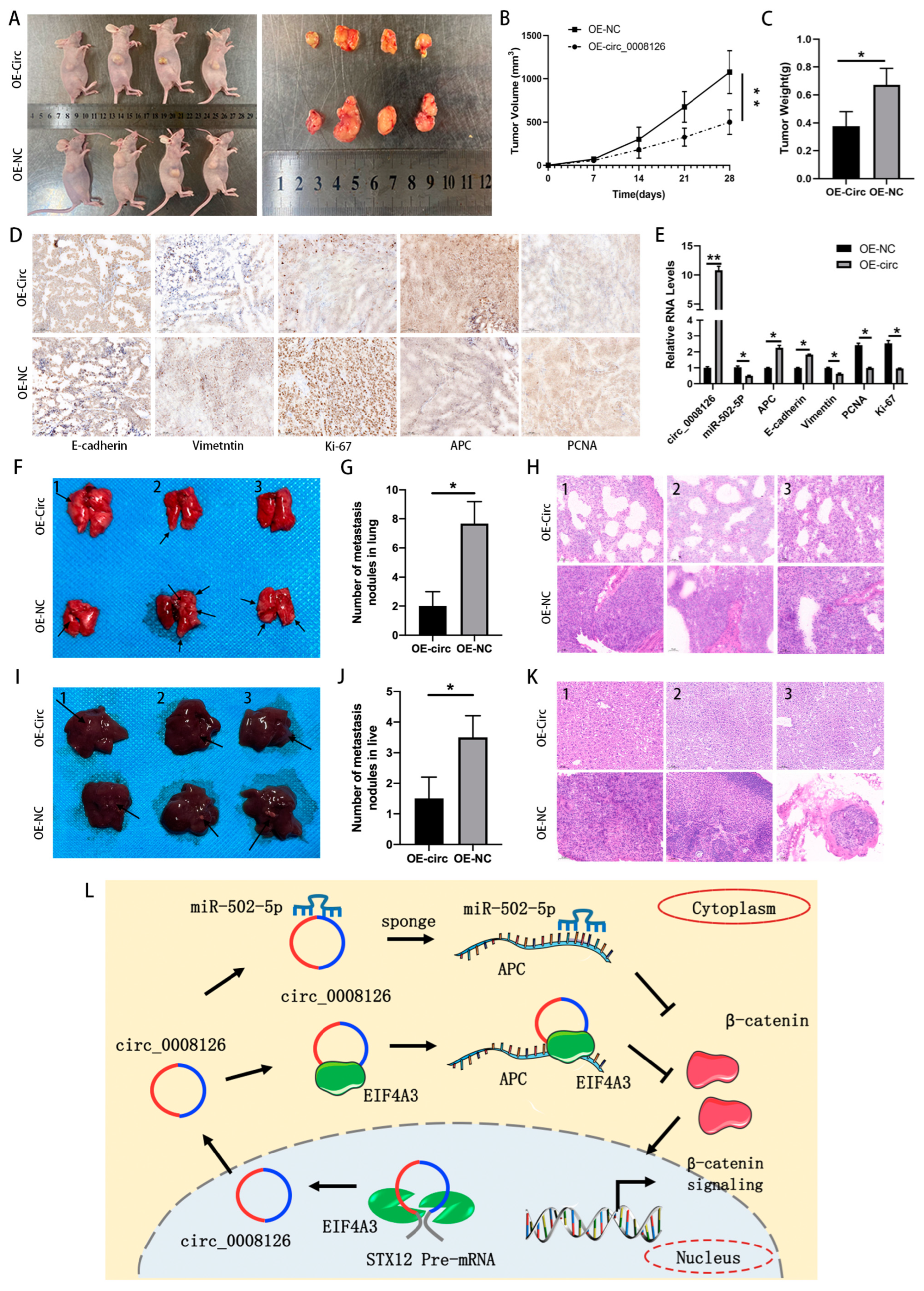
3.7. circ_0008126 Suppresses GC Growth and Metastasis In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef]
- Guan, W.L.; He, Y.; Xu, R.H. Gastric cancer treatment: Recent progress and future perspectives. J. Hematol. Oncol. 2023, 16, 57. [Google Scholar] [PubMed]
- Kasamatsu, A.; Nozaki, R.; Kawasaki, K.; Saito, T.; Minemura, C.; Seki, N.; Moss, J.; Uzawa, K. Synthetic Circular RNA for microRNA-1269a Suppresses Tumor Progression in Oral Squamous Cell Carcinoma. Cancers 2024, 16, 1242. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Jiang, J.; Qian, H.; Yan, Y.; Xu, W. Exosomal circRNA: Emerging insights into cancer progression and clinical application potential. J. Hematol. Oncol. 2023, 16, 67. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Waltzer, W.C.; Wilusz, J.E.; Spaliviero, M.; Darras, F.; Romanov, V. Circular STAG2 RNA Modulates Bladder Cancer Progression via miR-145-5p/TAGLN2 and Is Considered as a biomarker and a therapeutic target for Recurrence. Cancers 2024, 16, 978. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Wang, Y.; Zhang, Y.; Zheng, S.F.; Feng, T.; Tian, X.; Abudurexiti, M.; Wang, Z.D.; Zhu, W.K.; Su, J.Q.; et al. The function and mechanisms of action of circular RNAs in Urologic Cancer. Mol. Cancer. 2023, 22, 61. [Google Scholar] [CrossRef]
- Jiang, X.; Peng, M.; Liu, Q.; Peng, Q.; Oyang, L.; Li, S.; Xu, X.; Shen, M.; Wang, J.; Li, H.; et al. Circular RNA hsa_circ_0000467 promotes colorectal cancer progression by promoting eIF4A3-mediated c-Myc translation. Mol. Cancer. 2024, 23, 151. [Google Scholar] [CrossRef]
- Shang, Z.; Luo, Z.; Wang, Y.; Liu, Q.; Xin, Y.; Zhang, M.; Li, X.; Zeng, S.; Yu, L.; Zhang, X.; et al. CircHIPK3 contributes to cisplatin resistance in gastric cancer by blocking autophagy-dependent ferroptosis. J. Cell Physiol. 2023, 238, 2407–2424. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.; Wu, H.; Wu, J.; Li, T.; He, J.; Ju, Y.; Liu, Y.; Li, F.; Deng, H.; Gu, R.; et al. N6-methyladenosine modified circPAK2 promotes lymph node metastasis via targeting IGF2BPs/VEGFA signaling in gastric cancer. Oncogene 2024, 43, 2548–2563. [Google Scholar] [CrossRef]
- Asthana, S.; Martin, H.; Rupkey, J.; Patel, S.; Yoon, J.; Keegan, A.; Mao, Y. The Physiological Roles of the Exon Junction Complex in Development and Diseases. Cells 2022, 11, 1192. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Tan, W.; Liu, Z.; Fu, X.; Li, Z.; Lai, S.; Li, Q.; Zhong, X.; Qu, F.; Zhang, H.; et al. EIF4A3-induced circZFAND6 promotes breast cancer proliferation and metastasis through the miR-647/FASN axis. Life Sci. 2023, 324, 121745. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.; Gong, Y.; Weng, J.; Wen, F.; Luo, J.; Hu, C.; Xiao, J.; Shu, J. CircKIF4A combines EIF4A3 to stabilize SDC1 expression to activate c-src/FAK and promotes TNBC progression. Cell Signal. 2023, 108, 110690. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Chen, W.; Dong, X.; Wang, J.; Mei, X.; Deng, J.; Yang, S.; Zhuo, C.; Huang, X.; Shao, L.; et al. CSCD2: An integrated interactional database of cancer-specific circular RNAs. Nucleic Acids Res. 2022, 50, D1179–D1183. [Google Scholar] [CrossRef]
- Rebolledo, C.; Silva, J.P.; Saavedra, N.; Maracaja-Coutinho, V. Computational approaches for circRNAs prediction and in silico characterization. Brief. Bioinform. 2023, 24, bbad154. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Li, M.; Xue, C.; Chen, S.; Zheng, L.; Deng, H.; Tang, F.; Li, G.; Xiong, W.; Zeng, Z.; et al. Understanding the roles and regulation patterns of circRNA on its host gene in tumorigenesis and tumor progression. J. Exp. Clin. Cancer Res. 2023, 42, 86. [Google Scholar] [CrossRef] [PubMed]
- Dudekula, D.B.; Panda, A.C.; Grammatikakis, I.; De, S.; Abdelmohsen, K.; Gorospe, M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016, 13, 34–42. [Google Scholar] [CrossRef]
- Liu, M.; Wang, Q.; Shen, J.; Yang, B.B.; Ding, X. Circbank: A comprehensive database for circRNA with standard nomenclature. RNA Biol. 2019, 16, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.C.; Tsay, M.; Lu, R.; Jurisica, I. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018, 46, D360–D370. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Li, B.; Zhu, L.; Lu, C.; Wang, C.; Wang, H.; Jin, H.; Ma, X.; Cheng, Z.; Yu, C.; Wang, S.; et al. circNDUFB2 inhibits non- small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat. Commun. 2021, 12, 295. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Zhao, Q.; Feng, W.; Peng, Y.; Hu, B.; Chen, Q. N6-Methyladenosine Modification of CIRCKRT17 Initiated by METTL3 Promotes Osimertinib Resistance of Lung Adenocarcinoma by EIF4A3 to Enhance YAP1 Stability. Cancers 2022, 14, 5582. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wu, C.; Tang, Y.; Tang, J.; Zhu, D.; Zhang, F.; Xu, Z.; Sun, D.; Tan, Z.; Zhuo, H. CircMYH9 increases KPNA2 mRNA stability to promote hepatocellular carcinoma progression in an EIF4A3-dependent manner. Am. J. Cancer Res. 2022, 12, 4361–4372. [Google Scholar] [PubMed]
- Kristensen, L.S.; Jakobsen, T.; Hager, H.; Kjems, J. The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 2022, 19, 188–206. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z.; Zhuang, J.; Kang, D.; Song, W. CircCOL5A1 is involved in proliferation, invasion, and inhibition of ferroptosis of colorectal cancer cells via miR-1287-5p/SLC7A11. J. Biochem. Mol. Toxicol. 2024, 38, e23772. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Liu, J.; Guo, X.; Cheng, A.; Deng, X.; Guo, L.; Wang, Z. Circular RNA circFGD4 suppresses gastric cancer progression via modulating miR-532-3p/APC/β-catenin signalling pathway. Clin. Sci. 2020, 134, 1821–1839. [Google Scholar] [CrossRef] [PubMed]
- Long, F.; Li, L.; Xie, C.; Ma, M.; Wu, Z.; Lu, Z.; Liu, B.; Yang, M.; Zhang, F.; Ning, Z.; et al. Intergenic CircRNA Circ_0007379 Inhibits Colorectal Cancer Progression by Modulating miR-320a Biogenesis in a KSRP-Dependent Manner. Int. J. Biol. Sci. 2023, 19, 3781–3803. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Tang, J.; Zhang, Q.; Wang, C.; Qi, J.; Wei, Y.; Xu, J.; Yang, K.; Zhou, Z.; Wu, H.; et al. CircHIPK2 Contributes Cell Growth in Intestinal Epithelial of Colitis and Colorectal Cancer through Promoting TAZ Translation. Adv. Sci. 2024, 9, e2401588. [Google Scholar] [CrossRef]
- Yang, M.; Hu, H.; Wu, S.; Ding, J.; Yin, B.; Huang, B.; Li, F.; Guo, X.; Han, L. EIF4A3-regulated circ_0087429 can reverse EMT and inhibit the progression of cervical cancer via miR-5003-3p-dependent upregulation of OGN expression. J. Exp. Clin. Cancer Res. 2022, 41, 165. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, K.; Bayraktar, R.; Ferracin, M.; Calin, G.A. Non-coding RNAs in disease: From mechanisms to therapeutics. Nat. Rev. Genet. 2024, 25, 211–232. [Google Scholar] [CrossRef]
- Liu, S.; Jiao, B.; Zhao, H.; Liang, X.; Jin, F.; Liu, X.; Hu, J.F. LncRNAs-circRNAs as Rising Epigenetic Binary Superstars in Regulating Lipid Metabolic Reprogramming of Cancers. Adv. Sci. 2024, 11, e2303570. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Liang, Y.; Zhou, L.; Yan, Y.; Liu, N.; Zhang, R.; Huang, Y.; Wang, M.; Tang, Y.; Ali, D.W.; et al. TSPAN1 promotes autophagy flux and mediates cooperation between WNT-CTNNB1 signaling and autophagy via the MIR454-FAM83A-TSPAN1 axis in pancreatic cancer. Autophagy 2021, 17, 3175–3195. [Google Scholar] [CrossRef]
- Lin, X.; Xiaoqin, H.; Jiayu, C.; Li, F.; Yue, L.; Ximing, X. Long non-coding RNA miR143HG predicts good prognosis and inhibits tumor multiplication and metastasis by suppressing mitogen-activated protein kinase and Wnt signaling pathways in hepatocellular carcinoma. Hepatol. Res. 2019, 49, 902–918. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Furth, E.E.; Rustgi, A.K.; Klein, P.S. When You Come to a Fork in the Road, Take It: Wnt Signaling Activates Multiple Pathways through the APC/Axin/GSK-3 Complex. Cells 2023, 12, 2256. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Liu, X.; Xu, Y.; Zhu, S.; Gao, Y. Co-expression of Axin and APC gene fragments inhibits colorectal cancer cell growth via regulation of the Wnt signaling pathway. Mol. Med. Rep. 2017, 16, 3783–3790. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, X.; Wang, J.; Sun, M.; Li, D.; Wang, Z.; Li, J.; Li, Y.; Liu, Y. Exosomal circ-AHCY promotes glioblastoma cell growth via Wnt/β-catenin signaling pathway. Ann. Clin. Transl. Neurol. 2023, 10, 865–878. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Dang, P.; Guo, Y.; Liu, S.; Hu, S.; Sun, H.; Xu, Y.; Wang, W.; Chen, C.; Liu, J.; et al. Targeting CircAURKA prevents colorectal cancer progression via enhancing CTNNB1 protein degradation. Oncogene 2024, 105, 3388–3401. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Chen, W.; Wang, Z.; Dai, X. EIF4A3-Mediated circ_0008126 Inhibits the Progression and Metastasis of Gastric Cancer by Modulating the APC/β-Catenin Pathway. Cancers 2025, 17, 253. https://doi.org/10.3390/cancers17020253
Wang Z, Chen W, Wang Z, Dai X. EIF4A3-Mediated circ_0008126 Inhibits the Progression and Metastasis of Gastric Cancer by Modulating the APC/β-Catenin Pathway. Cancers. 2025; 17(2):253. https://doi.org/10.3390/cancers17020253
Chicago/Turabian StyleWang, Zeen, Wenxing Chen, Ziwei Wang, and Xinglong Dai. 2025. "EIF4A3-Mediated circ_0008126 Inhibits the Progression and Metastasis of Gastric Cancer by Modulating the APC/β-Catenin Pathway" Cancers 17, no. 2: 253. https://doi.org/10.3390/cancers17020253
APA StyleWang, Z., Chen, W., Wang, Z., & Dai, X. (2025). EIF4A3-Mediated circ_0008126 Inhibits the Progression and Metastasis of Gastric Cancer by Modulating the APC/β-Catenin Pathway. Cancers, 17(2), 253. https://doi.org/10.3390/cancers17020253






