SPP1 as a Potential Stage-Specific Marker of Colorectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Clinical Tissue Samples
2.2. RNA Purification and cDNA Preparation
2.3. Targeted RNA Sequencing of Cancer Transcriptome Genes
2.4. Validation of RNA-Seq Data Using qPCR Method
2.5. Data and Statistical Analysis
2.6. Public Data Survival Analysis
3. Results
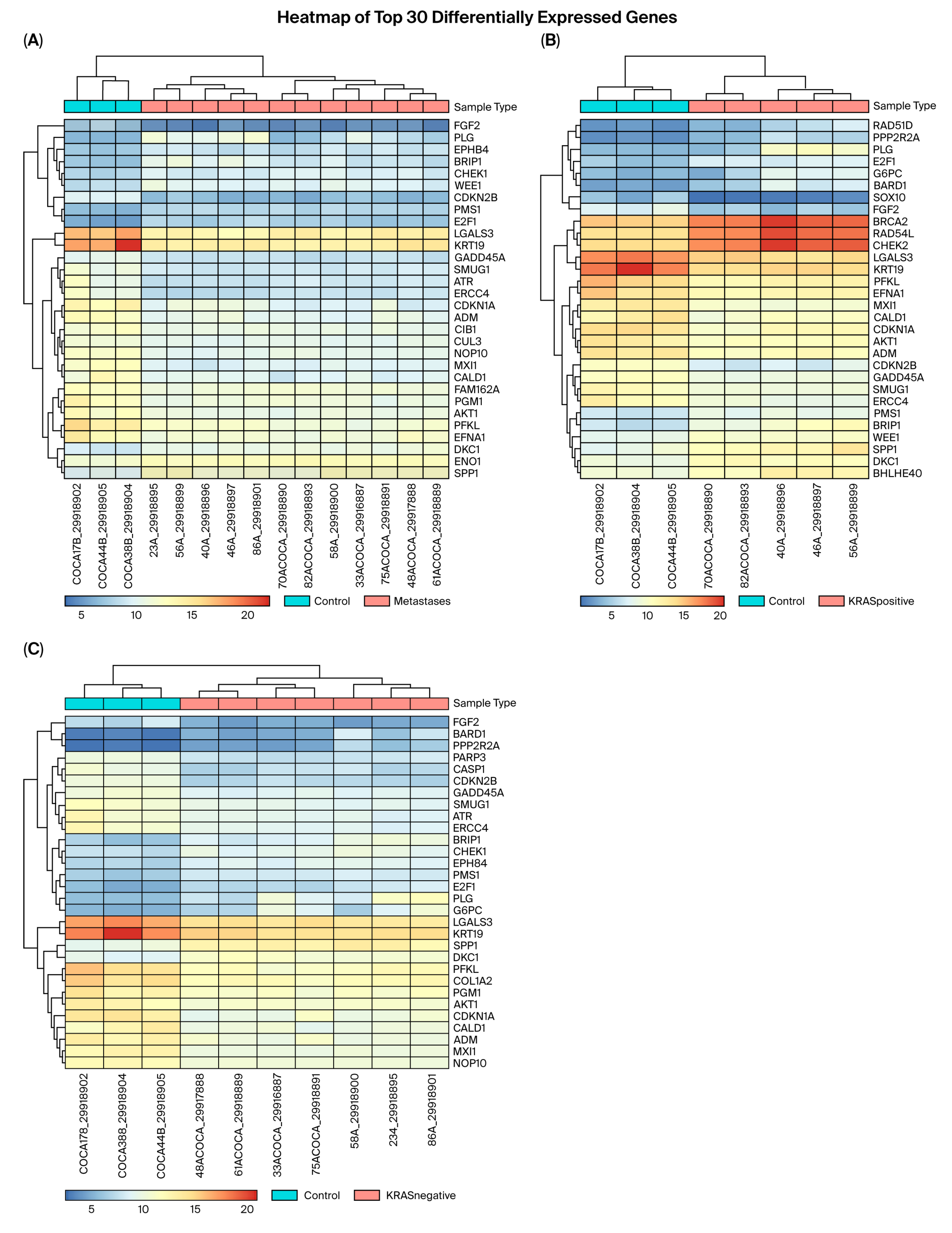
3.1. RNA Sequencing
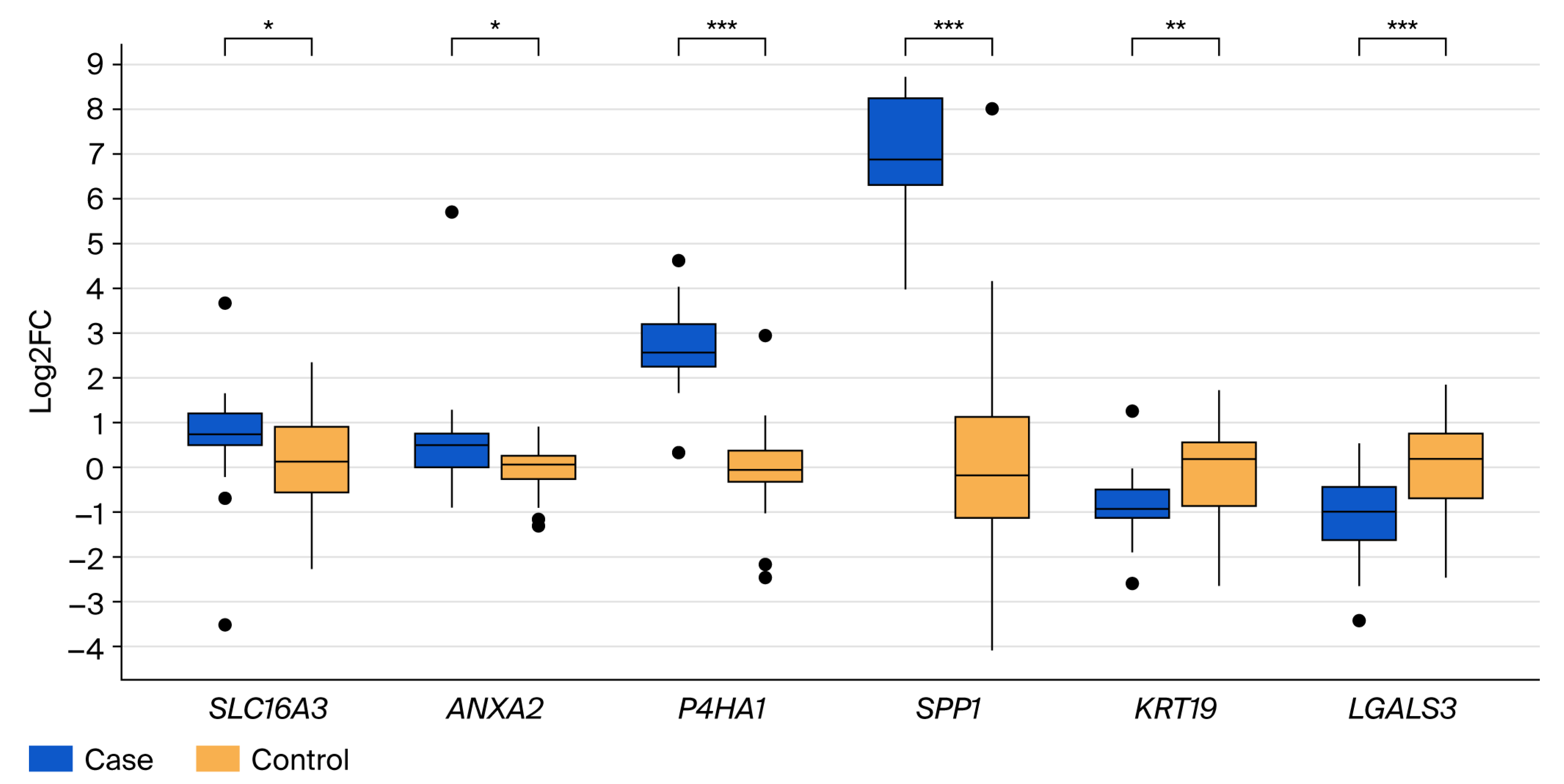
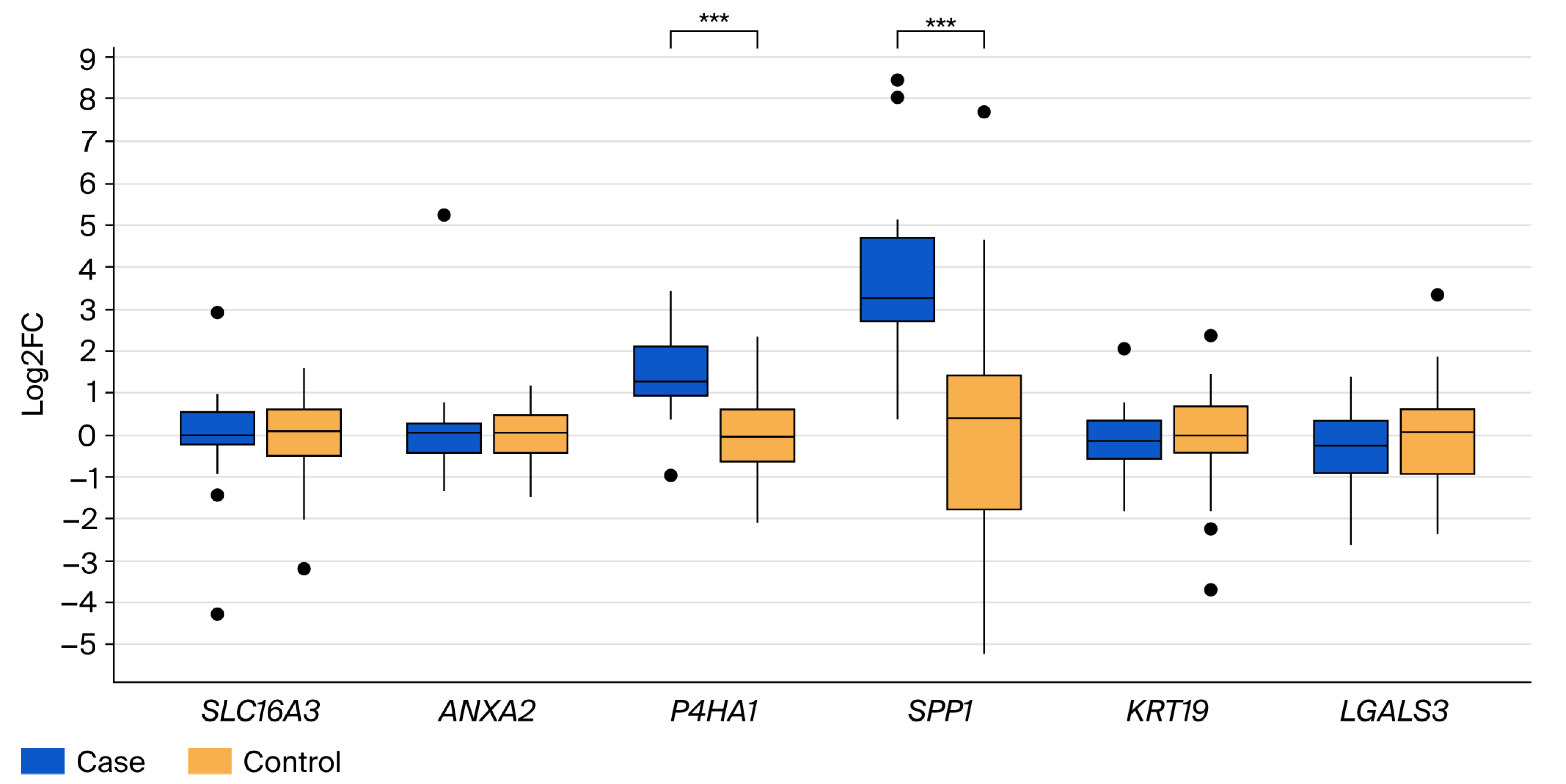
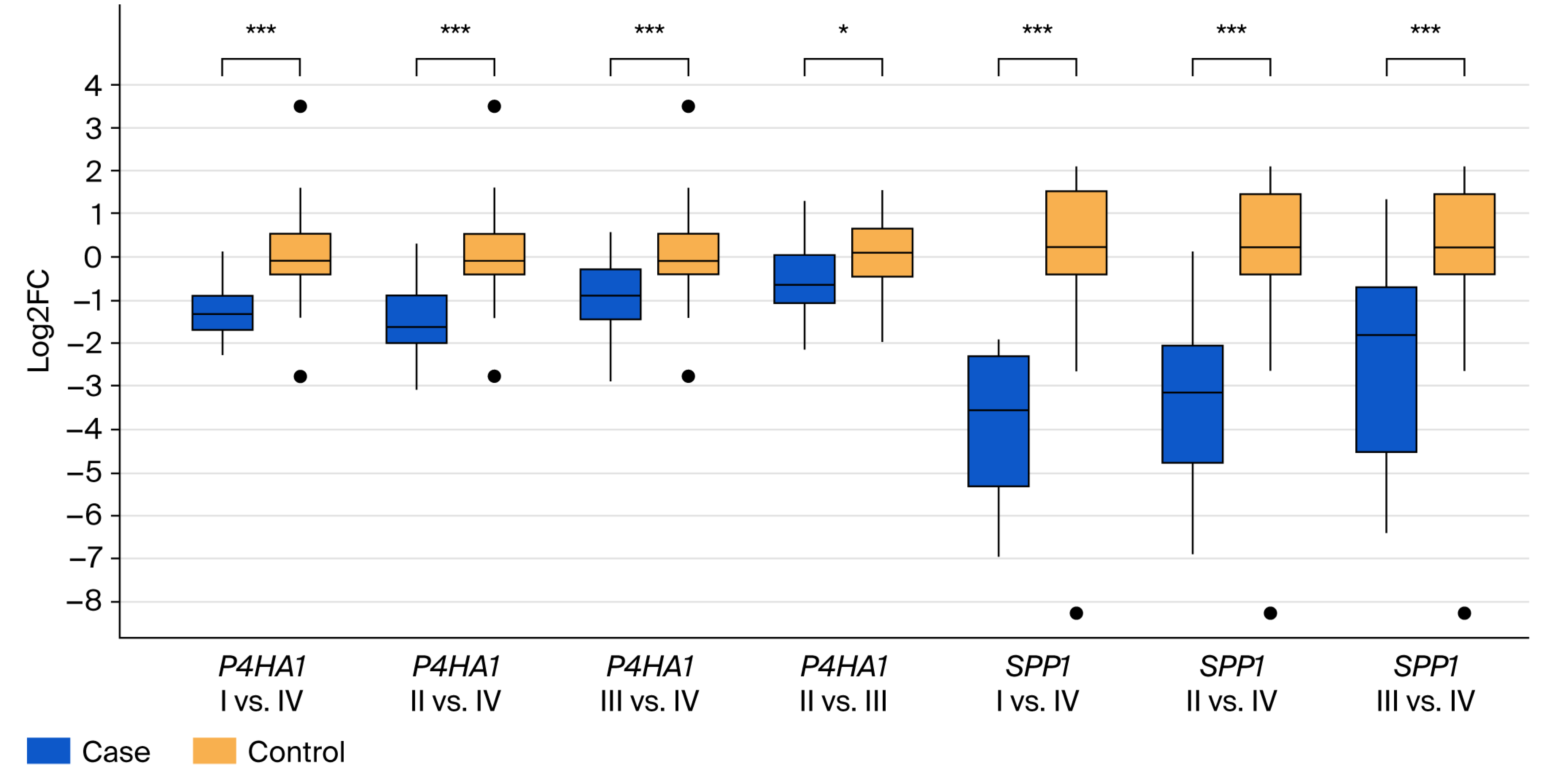
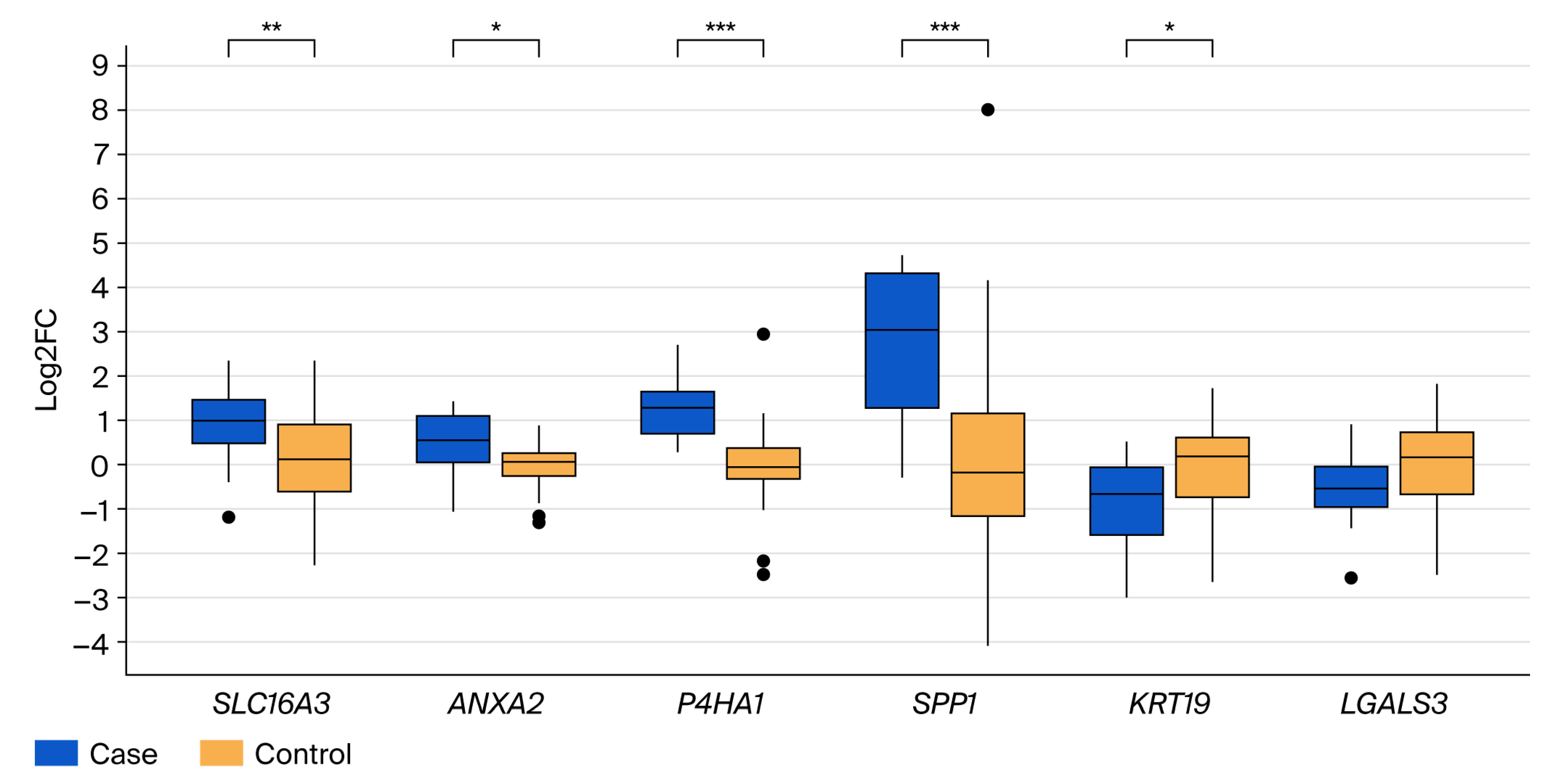
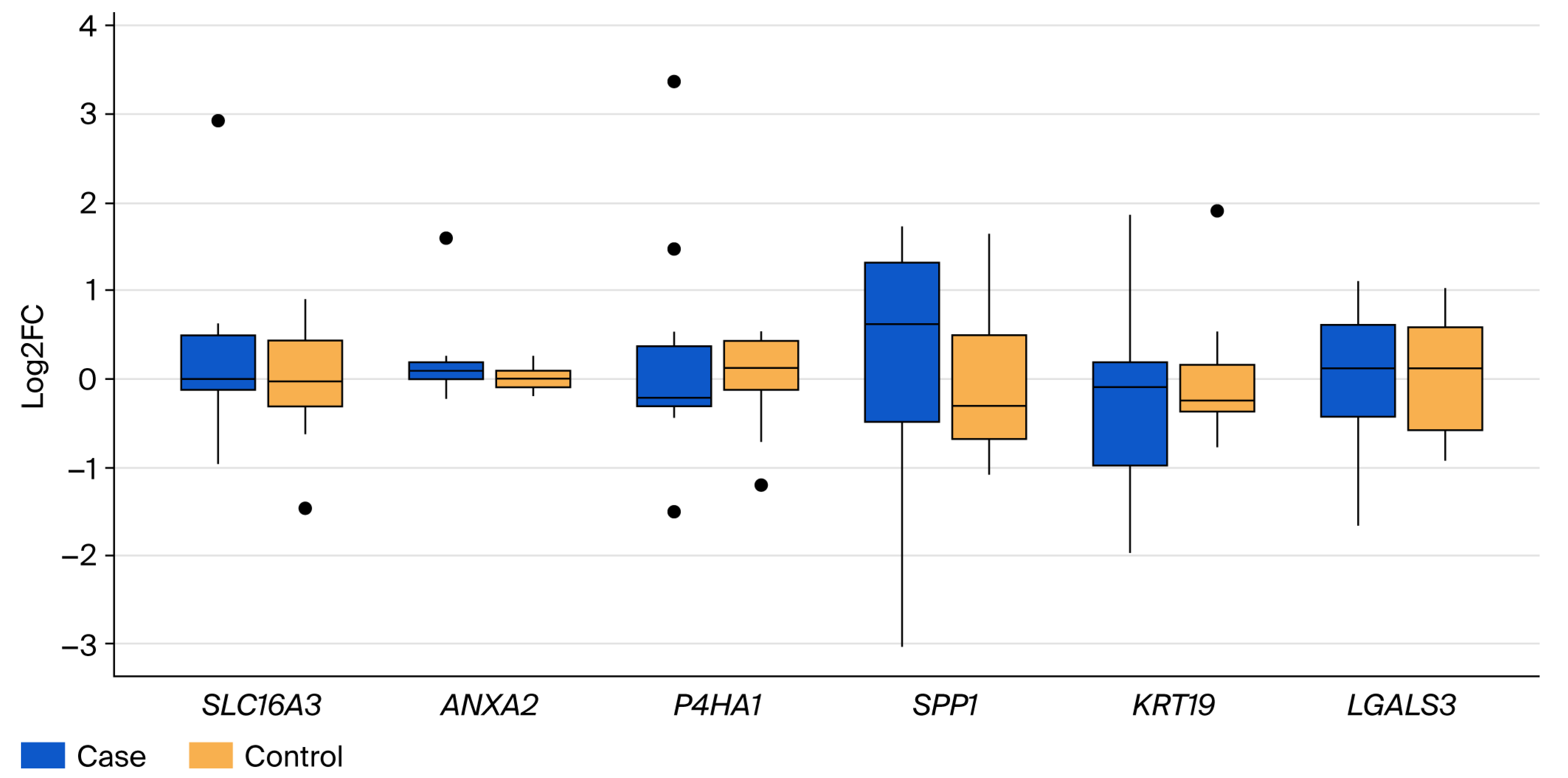
3.2. Validation of Differential Gene Expression
3.3. Survival Association of SPP1 and P4HA1 in Public Colorectal Cancer Datasets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRC | Colorectal cancer |
| DGE | Differential gene expression |
| DEGs | Differentially expressed genes |
| EMT | Epithelial-to-mesenchymal transition |
| PTIL | Peripheral tumor-infiltrating lymphocyte |
| TIL | Tumor-infiltrating lymphocyte |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- O’Brien, M.J.; Yang, S.; Mack, C.; Xu, H.; Huang, C.S.; Mulcahy, E.; Amorosino, M.; Farraye, F.A. Comparison of Microsatellite Instability, CpG Island Methylation Phenotype, BRAF and KRAS Status in Serrated Polyps and Traditional Adenomas Indicates Separate Pathways to Distinct Colorectal Carcinoma End Points. Am. J. Surg. Pathol. 2006, 30, 1491–1501. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, H.; Kuroda, H.; Imai, Y.; Hiraishi, H. Molecular pathogenesis of sporadic colorectal cancers. Chin. J. Cancer 2016, 35, 4. [Google Scholar] [CrossRef]
- Kuo, E.; Wang, K.; Liu, X. A Focused Review on Advances in Risk Stratification of Malignant Polyps. Gastroenterol. Res. 2020, 13, 163–183. [Google Scholar] [CrossRef]
- Qiu, M.; Hu, J.; Yang, D.; Cosgrove, D.P.; Xu, R. Pattern of distant metastases in colorectal cancer: A SEER based study. Oncotarget 2015, 6, 38658–38666. [Google Scholar] [CrossRef]
- Hugen, N.; van de Velde, C.J.H.; de Wilt, J.H.W.; Nagtegaal, I.D. Metastatic pattern in colorectal cancer is strongly influenced by histological subtype. Ann. Oncol. 2014, 25, 651–657. [Google Scholar] [CrossRef]
- Riihimäki, M.; Hemminki, A.; Sundquist, J.; Hemminki, K. Patterns of metastasis in colon and rectal cancer. Sci. Rep. 2016, 6, 29765. [Google Scholar] [CrossRef]
- Hapke, R.; Haake, S.M. Hypoxia-induced epithelial to mesenchymal transition in cancer. Cancer Lett. 2020, 487, 10–20. [Google Scholar] [CrossRef]
- Vu, T.; Datta, P.K. Regulation of EMT in Colorectal Cancer: A Culprit in Metastasis. Cancers 2017, 9, 171. [Google Scholar] [CrossRef]
- Tam, S.Y.; Wu, V.W.C.; Law, H.K.W. Hypoxia-Induced Epithelial-Mesenchymal Transition in Cancers: HIF-1α and Beyond. Front. Oncol. 2020, 10, 486. [Google Scholar] [CrossRef]
- Lasabová, Z.; Kalman, M.; Holubeková, V.; Grendár, M.; Kašubová, I.; Jašek, K.; Meršaková, S.; Malicherová, B.; Baranenko, D.; Adamek, M.; et al. Mutation analysis of POLE gene in patients with early-onset colorectal cancer revealed a rare silent variant within the endonuclease domain with potential effect on splicing. Clin. Exp. Med. 2019, 19, 393–400. [Google Scholar] [CrossRef]
- Meršaková, S.; Lasabová, Z.; Strnádel, J.; Kalman, M.; Gabonova, E.; Sabaka, P.; Ciccocioppo, R.; Rodrigo, L.; Kruzliak, P.; Mikolajčík, P. Genomic profile and immune contexture in colorectal cancer-relevance for prognosis and immunotherapy. Clin. Exp. Med. 2021, 21, 195–204. [Google Scholar] [CrossRef]
- Snahnicanova, Z.; Kasubova, I.; Kalman, M.; Grendar, M.; Mikolajcik, P.; Gabonova, E.; Laca, L.; Caprnda, M.; Rodrigo, L.; Ciccocioppo, R.; et al. Genetic and epigenetic analysis of the beta-2-microglobulin gene in microsatellite instable colorectal cancer. Clin. Exp. Med. 2020, 20, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Mikolajcik, P.; Lasabova, Z.; Loderer, D.; Grendar, M.; Kalman, M.; Kasubova, I.; Lucansky, V.; Wiederhold, A.J.; Marcinek, J.; Burjanivova, T.; et al. Detection of therapeutically relevant and concomitant rare somatic variants in colorectal cancer. Neoplasma 2021, 68, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Curion, F.; Handel, A.E.; Attar, M.; Gallone, G.; Bowden, R.; Cader, M.Z.; Clark, M.B. Targeted RNA sequencing enhances gene expression profiling of ultra-low input samples. RNA Biol. 2020, 17, 1741–1753. [Google Scholar] [CrossRef] [PubMed]
- Kivioja, T.; Vähärautio, A.; Karlsson, K.; Bonke, M.; Enge, M.; Linnarsson, S.; Taipale, J. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 2011, 9, 72–74. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 3, W98–W102. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The Consensus Molecular Subtypes of Colorectal Cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Rachinger, N.; Fischer, S.; Böhme, I.; Linck-Paulus, L.; Kuphal, S.; Kappelmann-Fenzl, M.; Bosserhoff, A.K. Loss of Gene Information: Discrepancies between RNA Sequencing, cDNA Microarray, and qRT-PCR. Int. J. Mol. Sci. 2021, 22, 9349. [Google Scholar] [CrossRef]
- Holubekova, V.; Loderer, D.; Grendar, M.; Mikolajcik, P.; Kolkova, Z.; Turyova, E.; Kudelova, E.; Kalman, M.; Marcinek, J.; Miklusica, J.; et al. Differential gene expression of immunity and inflammation genes in colorectal cancer using targeted RNA sequencing. Front. Oncol. 2023, 13, 1206482. [Google Scholar] [CrossRef]
- Schurch, N.J.; Schofield, P.; Gierlinski, M.; Cole, C.; Sherstnev, A.; Singh, V.; Wrobel, N.; Gharbi, K.; Simpson, G.G.; Owen-Hughes, T.; et al. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 2016, 22, 839–851. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szczesniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, S.; Garcia-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xie, J.; Wu, D.; Chen, L.; Gong, Z.; Wu, R.; Hu, Y.; Zhao, J.; Xu, Y. A pan-cancer analysis revealed the role of the SLC16 family in cancer. Channels 2021, 15, 528–540. [Google Scholar] [CrossRef]
- Wang, J.; Akter, R.; Shahriar, M.F.; Uddin, M.N. Cancer-Associated Stromal Fibroblast-Derived Transcriptomes Predict Poor Clinical Outcomes and Immunosuppression in Colon Cancer. Pathol. Oncol. Res. 2022, 28, 1610350. [Google Scholar] [CrossRef]
- Tao, Q.; Li, X.; Zhu, T.; Ge, X.; Gong, S.; Guo, J.; Ma, R. Lactate Transporter SLC16A3 (MCT4) as an Onco-Immunological Biomarker Associating Tumor Microenvironment and Immune Responses in Lung Cancer. Internat. J. Gen. Med. 2022, 15, 4465–4474. [Google Scholar] [CrossRef]
- Xue, L.; Liu, J.; Xie, J.; Luo, J. Prognostic Value of SLC16A3(MCT4) in Lung Adenocarcinoma and Its Clinical Significance. Int. J. Gen. Med. 2021, 14, 8413–8425. [Google Scholar] [CrossRef]
- Rocha, M.R.; Barcellos-de-Souza, P.; Sousa-Squiavinato, A.C.M.; Fernandes, P.V.; de Oliveira, I.M.; Boroni, M.; Morgado-Diaz, J.A. Annexin A2 overexpression associates with colorectal cancer invasiveness and TGF-ß induced epithelial mesenchymal transition via Src/ANXA2/STAT3. Sci. Rep. 2018, 8, 11285. [Google Scholar] [CrossRef]
- Christofidis, K.; Pergaris, A.; Fioretzaki, R.; Charalampakis, N.; Kapetanakis, E.Ι.; Kavantzas, N.; Schizas, D.; Sakellariou, S. Annexin A2 in Tumors of the Gastrointestinal Tract, Liver, and Pancreas. Cancers 2024, 16, 3764. [Google Scholar] [CrossRef]
- Yang, T.; Peng, H.; Wang, J.; Yang, J.; Nice, E.C.; Xie, K.; Huang, C. Prognostic and diagnostic significance of annexin A2 in colorectal cancer. Color. Dis. 2013, 15, e373–e381. [Google Scholar] [CrossRef]
- Mahdi, A.F.; Malacrida, B.; Nolan, J.; McCumiskey, M.E.; Merrigan, A.B.; Lal, A.; Tormey, S.; Lowery, A.J.; McGourty, K.; Kiely, P.A. Expression of Annexin A2 Promotes Cancer Progression in Estrogen Receptor Negative Breast Cancers. Cells 2020, 9, 1582. [Google Scholar] [CrossRef]
- Tan, S.-H.; Young, D.; Chen, Y.; Kuo, H.-C.; Srinivasan, A.; Dobi, A.; Petrovics, G.; Cullen, J.; Mcleod, D.G.; Rosner, I.L.; et al. Prognostic features of Annexin A2 expression in prostate cancer. Pathology 2021, 53, 205–213. [Google Scholar] [CrossRef]
- Li, Y.; Ge, Y.-Z.; Qian, Y.; Chen, K.; Zhao, F.; Qin, Z.; Zhou, L.; Xu, L.; Xu, Z.; Dou, Q.; et al. The Role of P4HA1 in Multiple Cancer Types and its Potential as a Target in Renal Cell Carcinoma. Front. Genet. 2022, 13, 848456. [Google Scholar] [CrossRef]
- Agarwal, S.; Behring, M.; Kim, H.-G.; Bajpai, P.; Chakravarthi, B.V.S.K.; Gupta, N.; Elkholy, A.; Al Diffalha, S.; Varambally, S.; Manne, U. Targeting P4HA1 with a Small Molecule Inhibitor in a Colorectal Cancer PDX Model. Transl. Oncol. 2020, 13, 100754. [Google Scholar] [CrossRef]
- Tanaka, A.; Zhou, Y.; Shia, J.; Ginty, F.; Ogawa, M.; Klimstra, D.S.; Hendrickson, R.C.; Wang, J.Y.; Roehrl, M.H. Prolyl 4-hydroxylase alpha 1 protein expression risk-stratifies early stage colorectal cancer. Oncotarget 2020, 11, 813–824. [Google Scholar] [CrossRef]
- Gawel, D.R.; Lee, E.J.; Li, X.; Lilja, S.; Matussek, A.; Schäfer, S.; Olsen, R.S.; Stenmarker, M.; Zhang, H.; Benson, M. An algorithm-based meta-analysis of genome- and proteome-wide data identifies a combination of potential plasma biomarkers for colorectal cancer. Sci. Rep. 2019, 9, 15575. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, M.; Xue, Z.; Zhu, X. Comprehensive Analysis of Gene Expression Profiles Identifies a P4HA1-Related Gene Panel as a Prognostic Model in Colorectal Cancer Patients. Cancer Biother. Radiopharm. 2021, 36, 693–704. [Google Scholar] [CrossRef]
- Jiang, N.; Jin, L.; Li, S. Role of SPP1 in the diagnosis of gastrointestinal cancer. Oncol. Lett. 2023, 26, 411. [Google Scholar] [CrossRef]
- Zeng, P.; Zhang, X.; Xiang, T.; Ling, Z.; Lin, C.; Diao, H. Secreted phosphoprotein 1 as a potential prognostic and immunotherapy biomarker in multiple human cancers. Bioengineered 2022, 13, 3221–3239. [Google Scholar] [CrossRef]
- Kazakova, E.; Rakina, M.; Sudarskikh, T.; Iamshchikov, P.; Tarasova, A.; Tashireva, L.; Afanasiev, S.; Dobrodeev, A.; Zhuikova, L.; Cherdyntseva, N.; et al. Angiogenesis regulators S100A4, SPARC and SPP1 correlate with macrophage infiltration and are prognostic biomarkers in colon and rectal cancers. Front. Oncol. 2023, 13, 1058337. [Google Scholar] [CrossRef] [PubMed]
- Loosen, S.H.; Heise, D.; Dejong, C.H.; Roy, S.; Tacke, F.; Trautwein, C.; Roderburg, C.; Luedde, T.; Neumann, U.P.; Binnebösel, M. Circulating Levels of Osteopontin Predict Patients’ Outcome after Resection of Colorectal Liver Metastases. J. Clin. Med. 2018, 7, 390. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Qin, J.; Nie, J.; Gao, R.; Hu, S.; Sun, H.; Wang, S.; Pan, Y. ANGPTL2+cancer-associated fibroblasts and SPP1+macrophages are metastasis accelerators of colorectal cancer. Front. Immunol. 2023, 14, 1185208. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Hu, H.; Jiang, H.; Liu, C.; Zhang, H.; Gong, S.; Wei, D.; Yu, Z. Upregulation of secreted phosphoprotein 1 affects malignant progression, prognosis, and resistance to cetuximab via the KRAS/MEK pathway in head and neck cancer. Mol. Carcinog. 2025, 59, 1147–1158. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, D.; Zhang, X.; Su, P.; Li, X.; Li, Z.; Gai, Y.; Li, J.; Yang, Z.; Ding, Y.; et al. Regulation of Hippo/YAP axis in colon cancer progression by the deubiquitinase JOSD1. Cell Death Disov. 2024, 10, 365. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Huang, H.-Y.; Lin, Z.; Ranieri, M.; Li, S.; Sahu, S.; Liu, Y.; Ban, Y.; Guidry, K.; Hu, H.; et al. Genome-wide CRISPR scrrens that identify multiple synthetic lethal targets that enhance KRASG12C inhibitory efficacy. Cancer Res. 2023, 83, 4095–4111. [Google Scholar] [CrossRef]
- Zhu, G.; Pei, L.; Xia, H.; Tang, Q.; Bi, F. Role of oncogenic KRAS in the prognosis, diagnosis and treatment of colorectal cancer. Mol. Cancer 2021, 20, 143. [Google Scholar] [CrossRef]
- Saha, S.K.; Jeong, Y.; Cho, S.; Cho, S.-G. Systematic expression alteration analysis of master reprogramming factor OCT4 and its three pseudogenes in human cancer and their prognostic outcomes. Sci. Rep. 2018, 8, 14806. [Google Scholar] [CrossRef]
- Mi, L.; Liang, N.; Sun, H. A Comprehensive Analysis of KRT19 Combined with Immune Infiltration to Predict Breast Cancer Prognosis. Genes 2022, 13, 1838. [Google Scholar] [CrossRef]
- Ju, J.-H.; Yang, W.; Lee, K.-M.; Oh, S.; Nam, K.; Shim, S.; Shin, S.Y.; Gye, M.C.; Chu, I.-S.; Shin, I. Regulation of cell proliferation and migration by keratin19-induced nuclear import of early growth response-1 in breast cancer cells. Clin. Cancer Res. 2013, 19, 4335–4346. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, X.; Peng, C.; Qin, Y.; Gao, T.; Jing, J.; Zhao, H. BRAFV600E-induced KRT19 expression in thyroid cancer promotes lymph node metastasis via EMT. Oncol. Lett. 2019, 18, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Yasuchika, K.; Ishii, T.; Katayama, H.; Yoshitoshi, E.Y.; Ogiso, S.; Kita, S.; Yasuda, K.; Fukumitsu, K.; Mizumoto, M.; et al. Keratin 19, a Cancer Stem Cell Marker in Human Hepatocellular Carcinoma. Clin. Cancer Res. 2015, 21, 3081–3091. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.-Q.; Li, X.; Chen, Z.; Dong, S.; Ye, C.; Hou, S.; Fan, D.-A.; Zhang, H.; Zhou, W.-C. KRT19 is regulated by miR-642a-5p and promotes pancreatic cancer progression through the Wnt/β-catenin pathway. iScience 2024, 27, 110782. [Google Scholar] [CrossRef]
- Yuan, X.; Yi, M.; Dong, B.; Chu, Q.; Wu, K. Prognostic significance of KRT19 in Lung Squamous Cancer. J. Cancer 2021, 12, 1240–1248. [Google Scholar] [CrossRef]
- Ren, Y.; Qian, Y.; Zhang, Q.; Li, X.; Li, M.; Li, W.; Yang, P.; Ren, H.; Li, H.; Weng, Y.; et al. High LGALS3 expression induced by HCP5/hsa-miR-27b-3p correlates with poor prognosis and tumor immune infiltration in hepatocellular carcinoma. Cancer Cell Int. 2024, 24, 142. [Google Scholar] [CrossRef]
- Wang, K.; Fu, S.; Dong, L.; Zhang, D.; Wang, M.; Wu, X.; Shen, E.; Luo, L.; Li, C.; Nice, E.C.; et al. Periplocin suppresses the growth of colorectal cancer cells by triggering LGALS3 (galectin 3)-mediated lysophagy. Autophagy 2023, 19, 3132–3150. [Google Scholar] [CrossRef]
- Tao, L.; Jin, L.; Dechun, L.; Hongqiang, Y.; Changhua, K.; Guijun, L. Galectin-3 Expression in Colorectal Cancer and its Correlation with Clinical Pathological Characteristics and Prognosis. Open Med. 2017, 12, 226–230. [Google Scholar] [CrossRef]
- Kim, S.-J.; Kang, H.-G.; Kim, K.; Kim, H.; Zetterberg, F.; Park, Y.S.; Cho, H.-S.; Hewitt, S.M.; Chung, J.-Y.; Nilsson, U.J.; et al. Crosstalk between WNT and STAT3 is mediated by galectin-3 in tumor progression. Gastric Cancer 2021, 24, 1050–1062. [Google Scholar] [CrossRef]
- Watanabe, T.; Kobunai, T.; Yamamoto, Y.; Matsuda, K.; Ishihara, S.; Nozawa, K.; Iinuma, H.; Ikeuchi, H.; Eshima, K. Differential gene expression signatures between colorectal cancers with and without KRAS mutations: Crosstalk between the KRAS pathway and other signalling pathways. Eur. J. Cancer 2011, 47, 1946–1954. [Google Scholar] [CrossRef] [PubMed]
- Saliani, M.; Jalal, R.; Javadmanesh, A. Differential expression analysis of genes and long non-coding RNAs associated with KRAS mutation in colorectal cancer cells. Sci. Rep. 2022, 12, 7965. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.-T.; Tuersun, A.; Yu, S.-Y.; Zhang, Y.-C.; Feng, W.-Q.; Xu, Z.-Q.; Zhao, J.-K.; Zong, Y.-P.; Lu, A.-G. Leveraging a KRASbased signature to predict the prognosis and drug sensitivity of colon cancer and identifying SPINK4 as a new biomarker. Sci. Rep. 2023, 13, 22230. [Google Scholar] [CrossRef] [PubMed]
| Primary Tumors | Liver Metastases | Non-Tumor Controls | |
|---|---|---|---|
| Patients | 84 | 27 | 31 |
| Female | 39 | 7 | 15 |
| Male | 45 | 20 | 16 |
| Age | 69.5 ± 9.9 | 65.0 ± 7.9 | 72.8 ± 6.3 |
| Clinical stage I/II/III/IV | 15/25/26/7 | 0/0/0/27 | |
| TIL-positive/-negative/n.a | 13/27/44 | 12/1/14 | |
| PTIL-positive/-negative/n.a | 27/17/40 | 5/1/21 | |
| KRAS-positive/-negative/n.a | 24/28/32 | 11/14/2 | |
| PIK3CA-positive/-negative/n.a | 8/44/32 | 3/22/2 | |
| SMAD4-positive/-negative/n.a | 2/50/32 | 2/23/2 | |
| APC-positive/-negative/n.a | 21/31/32 | 10/15/2 | |
| TP53-positive/-negative/n.a | 24/28/32 | 13/12/2 | |
| Mutations in driver genes 0/1/2/3 or more/n.a | 7/18/17/10/32 | 3/7/13/2/2 | |
| MSI/MSS/n.a | 9/36/39 | 2/16/9 |
| Tumors | Clinical stage III | Mutational status of APC |
| Primary tumors | Clinical stage IV | Mutational status of TP53 |
| Liver metastases | MSI status | TIL-positive tumors |
| Control samples | Mutational status of KRAS | PTIL-positive tumors |
| Clinical stage I | Mutational status of PIK3CA | Tumors with 0, 1, 2, 3, and more mutations in driver genes |
| Clinical stage II | Mutational status of SMAD4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turyova, E.; Mikolajcik, P.; Kalman, M.; Loderer, D.; Slezak, M.; Skerenova, M.; Johnston, E.; Burjanivova, T.; Miklusica, J.; Strnadel, J.; et al. SPP1 as a Potential Stage-Specific Marker of Colorectal Cancer. Cancers 2025, 17, 3200. https://doi.org/10.3390/cancers17193200
Turyova E, Mikolajcik P, Kalman M, Loderer D, Slezak M, Skerenova M, Johnston E, Burjanivova T, Miklusica J, Strnadel J, et al. SPP1 as a Potential Stage-Specific Marker of Colorectal Cancer. Cancers. 2025; 17(19):3200. https://doi.org/10.3390/cancers17193200
Chicago/Turabian StyleTuryova, Eva, Peter Mikolajcik, Michal Kalman, Dusan Loderer, Miroslav Slezak, Maria Skerenova, Emile Johnston, Tatiana Burjanivova, Juraj Miklusica, Jan Strnadel, and et al. 2025. "SPP1 as a Potential Stage-Specific Marker of Colorectal Cancer" Cancers 17, no. 19: 3200. https://doi.org/10.3390/cancers17193200
APA StyleTuryova, E., Mikolajcik, P., Kalman, M., Loderer, D., Slezak, M., Skerenova, M., Johnston, E., Burjanivova, T., Miklusica, J., Strnadel, J., & Lasabova, Z. (2025). SPP1 as a Potential Stage-Specific Marker of Colorectal Cancer. Cancers, 17(19), 3200. https://doi.org/10.3390/cancers17193200







