Artificial Intelligence-Enhanced Liquid Biopsy and Radiomics in Early-Stage Lung Cancer Detection: A Precision Oncology Paradigm
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Review Design
2.2. Information Sources and Search Strategy
2.3. Inclusion and Exclusion Criteria
3. Lung Cancer Overview
3.1. Epidemiology
3.2. Types of Lung Cancer
3.3. Current Screening and Diagnostic Approaches
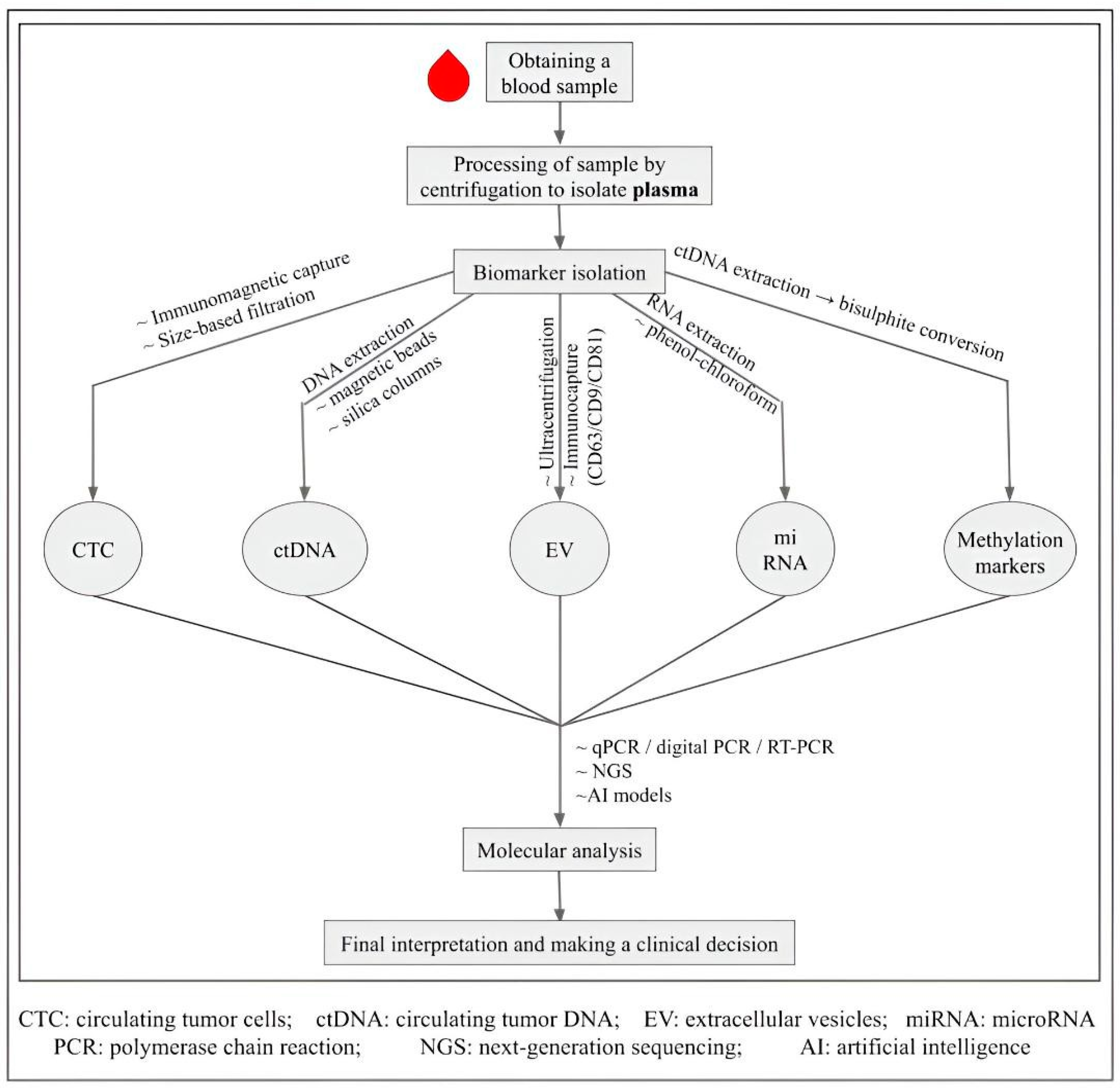
4. Liquid Biopsy in Lung Cancer
4.1. Types of Biomarkers in Liquid Biopsy
- Circulating Tumor Cells (CTCs): These are tumor-derived cells that enter the bloodstream from the primary or metastatic site of the tumor. Although they are present in low concentrations, CTCs offer molecular information (DNA, RNA, protein) and correlate with metastasis. Higher expression of genes such as PIK3CA, AKT2, TWIST, and ALDH1 in CTCs is seen in patients with metastatic lung cancer compared to patients with non-metastatic disease [44].
- Circulating tumor DNA (ctDNA): Cell-free DNA (cfDNA) is defined as non-encapsulated, fragmented DNA found in the blood of an individual. While present in healthy individuals, levels are significantly higher in cancer patients due to increased apoptosis, necrosis, and active secretion from proliferating tumor cells [45]. Plasma is preferred over serum for cfDNA analysis, as serum collection induces leukocyte lysis during clotting, releasing high amounts of genomic DNA that contaminate cfDNA and compromise assay sensitivity [46]. The concentration of circulating tumor DNA (ctDNA) can be extremely low and difficult to detect, but its detection can prove to be beneficial as the molecular alterations present in ctDNA closely resemble the tumor tissue [47,48]. Many techniques are being employed to detect ctDNA with high sensitivity and specificity, including droplet digital PCR, next-generation sequencing, Sanger sequencing, etc. CtDNA is an important biomarker as it can help detect various mutations responsible for the tumor and guide the treatment accordingly [49].
- Extracellular vesicles (EV): Extracellular vesicles are nano-sized, membrane-bound particles released into the extracellular space. The three main subtypes of extracellular vesicles are: microvesicles, exosomes, and apoptotic bodies, which can be distinguished based on multiple factors like their biogenesis, release pathways, size, content, and function [50]. Cancer cells secrete more EVs than normal cells, which promote tumor progression and metastasis through angiogenesis and immune evasion, making it a potential biomarker [51,52]. Exosomal miRNA (exo-miRNA) is another key prognostic biomarker that is being explored and can be easily detected in the bloodstream, as it is protected from degradation by a lipid bilayer [11,53].
- MicroRNAs (miRNAs): Small, non-coding RNAs that regulate gene expression and are found circulating in the body fluids of cancer patients. The microRNAs let-7i-3p and miR-154-5p serve as diagnostic and prognostic biomarkers in lung cancer. They belong to tumor suppressor miRNA families and are downregulated in lung cancer. Low levels of these microRNAs detected via liquid biopsy indicate poor prognosis [54].
- DNA Methylation Markers: Aberrant methylation of CpG islands is common in cancer. Profiling tools such as bisulfite conversion and high-density methylation arrays (e.g., HumanMethylationEPIC BeadChip) are powerful screening and prognostic platforms [55].
- Tumor-educated platelets (TEPs): Platelets reprogrammed by tumors show altered RNA and protein content [56]. Platelet counts, platelet–lymphocyte ratios, and mean platelet volume have long been used in cancer diagnosis; RNA-based assays like ThromboSeq are now being explored as novel biomarkers [57,58]. Table 3 summarizes these biomarkers, their isolation and detection methods, and clinical uses.
4.2. Applications of Liquid Biopsy in Lung Cancer
- Early Detection: Liquid biopsy enables non-invasive screening, particularly valuable where LDCT is unavailable. The ability to detect tumor-specific mutations, DNA methylation markers, and exosomal miRNAs in easily accessible fluids like blood offers a promising approach to identifying lung cancer at an earlier, more treatable stage [72].
- Monitoring Response to Treatment: Liquid biopsy allows for frequent sampling, which helps clinicians monitor dynamic changes in ctDNA levels during systemic therapies such as chemotherapy, targeted therapy, and immunotherapy. These changes can guide treatment modifications, predict resistance patterns, and assist in evaluating therapy effectiveness [73,74,75].
- Prognostic Value: Certain biomarkers offer insight into disease prognosis. Many studies have been done that revealed that patients with eight or more CTCs per 7.5 mL of blood and specific mutations like KRAS G12/G13 are associated with poor prognosis [76,77,78]. Similarly, low levels of tumor-suppressor miRNAs and aberrant DNA methylation patterns also correlate with poor outcomes [79,80].
- Detection of Metastasis: Liquid biopsy can detect early signs of metastasis by identifying biomarkers released during the metastatic cascade. In cases of leptomeningeal metastasis, cerebrospinal fluid analysis via liquid biopsy detects cancer cells earlier than conventional imaging, aiding in timely intervention [81,82].
- Monitoring Minimal Residual Disease (MRD): Following curative treatment, liquid biopsy enables longitudinal surveillance through serial ctDNA analysis. Studies show that ctDNA detection within weeks post-treatment can predict molecular relapse well before clinical or radiologic evidence appears, offering a window for early therapeutic action [83,84,85].
- Tumor Heterogeneity: Liquid biopsy captures spatial and temporal tumor heterogeneity more effectively than a single-site tissue biopsy. It reflects ongoing clonal evolution and acquired resistance mechanisms that emerge under treatment pressure, supporting the personalization of subsequent therapy lines [86,87,88].
- SCLC Mutation Profiling: In SCLC, liquid biopsy has shown unique strengths due to its biology. CTCs are highly abundant and have been used for prognosis, with high baseline counts predicting poor survival [69]. ctDNA analysis can detect hallmark alterations such as TP53 and RB1 loss and track treatment response [91]. Exosomal RNA and protein signatures are also being explored to help distinguish SCLC from NSCLC. While promising, these studies remain limited by small, retrospective cohorts, highlighting the need for larger validation.
4.3. Limitations of Liquid Biopsy
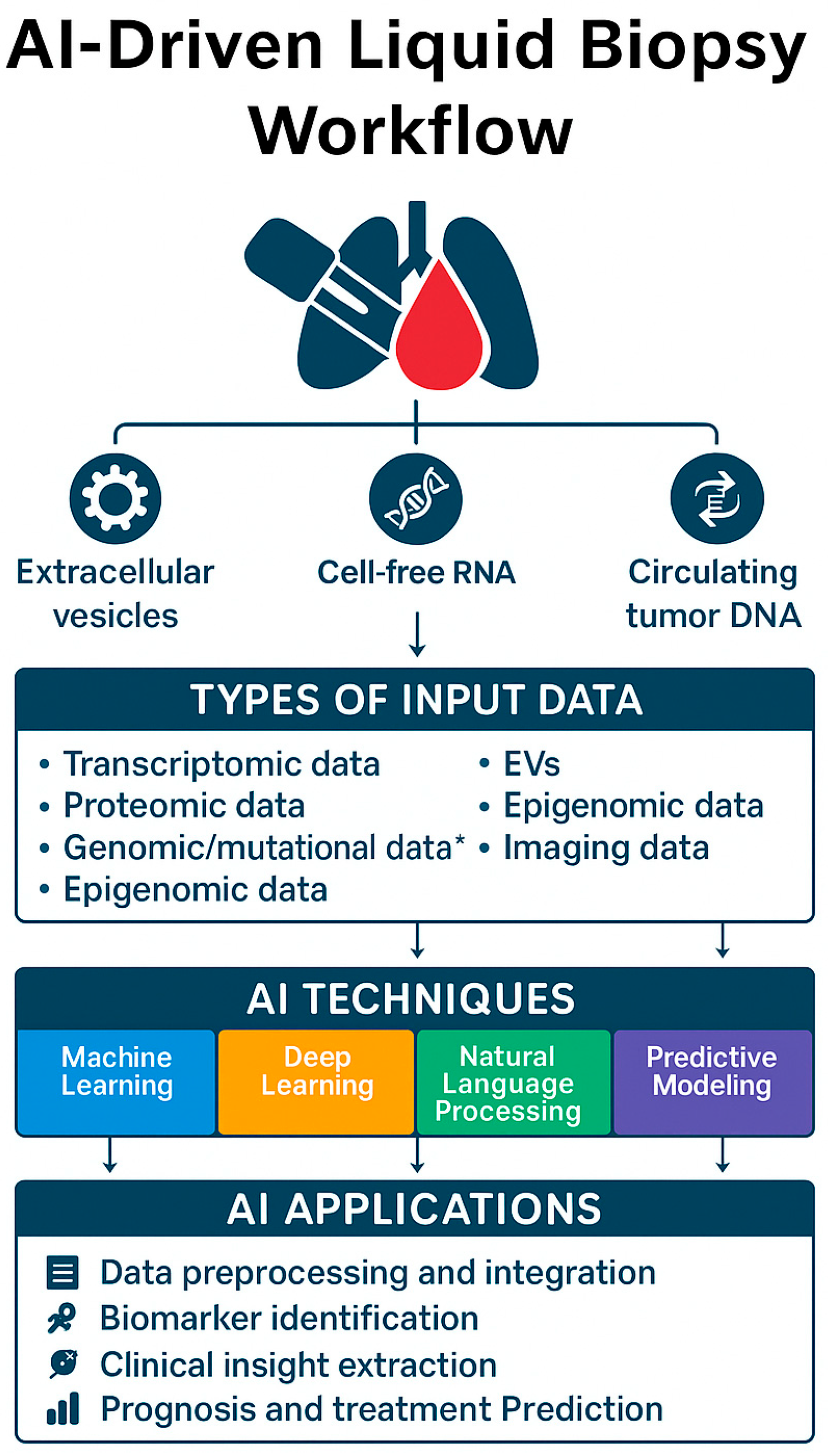
5. Role of Artificial Intelligence in Liquid Biopsy
5.1. AI Techniques in Liquid Biopsy
5.2. AI in Data Integration and Predictive Modeling
| AI Technique | Application in Liquid Biopsy | Key Strengths | Biomarker Detection Role | Predictive Modeling Role | References |
|---|---|---|---|---|---|
| Support Vector Machine (SVM) | Used to classify structured omics datasets and detect disease-linked biomarkers. | Performs well on limited structured data with clear boundaries. | Identifies distinct biomarker profiles from omics-based inputs. | Provides early-stage classification for disease risk. | [96,98] |
| Random Forest (RF) | Selects and ranks important biological features in large-scale biomarker studies. | Handles noise effectively and reduces overfitting through ensemble learning. | Ranks critical features influencing disease classification outcomes. | Delivers robust prediction of outcomes across diverse datasets. | [99,100,101] |
| Convolutional Neural Networks (CNNs) | Learns spatial features from ctDNA arrays and EV signal patterns, supporting diagnosis. | Recognizes patterns in structured omics and image-like biological formats. | Highlights spatial characteristics in liquid biopsy data such as EV signatures. | Supports modeling of treatment effects and diagnostic accuracy. | [102,103] |
| Recurrent Neural Networks (RNNs/LSTM) | Captures time-based changes in biomarker levels for monitoring and prognosis. | Follows sequential biomarker shifts over time, ideal for longitudinal data. | Maps ctDNA fluctuations to biological or clinical changes. | Forecasts recurrence or treatment failure using time-series data. | [105,106] |
| Deep Neural Networks (DNNs) | Combines multi-omics data into integrated models for improved clinical interpretation. | Builds abstract feature relationships across omics platforms for precise modeling. | Links biomarker patterns across different omics layers. | Stratifies patients and models disease progression based on biomarkers. | [107,108,109] |
| Autoencoders | Reduces dimensionality and background noise in high-volume datasets without supervision. | Ideal for discovering hidden structures and reducing complexity in noisy datasets. | Uncovers subtle biomarker trends from complex datasets. | Identifies unusual profiles and assists in survival analysis. | [111] |
| Generative Adversarial Networks (GANs) | Generates synthetic molecular data to improve training where rare biomarker examples exist. | Helps balance datasets through artificial augmentation, especially for rare conditions. | Synthesizes rare variant profiles to enhance model learning. | Improves forecasting when limited real-world data are available. | [112] |
| Natural Language Processing (NLP) | Extracts meaningful terms from clinical notes and links them to structured biomarker data. | Transforms unstructured clinical text into usable insights that enrich model input. | Connects textual mentions of symptoms or markers to structured data. | Enhances personalized prediction by linking notes and omics. | [98,118] |
5.3. NLP and Multi-Modal AI Integration
6. Use of AI in Liquid Biopsy
6.1. For Detection of Specific Biomarkers and Early-Stage Lung Cancer
6.2. For Monitoring Treatment Response and Personalized Treatment
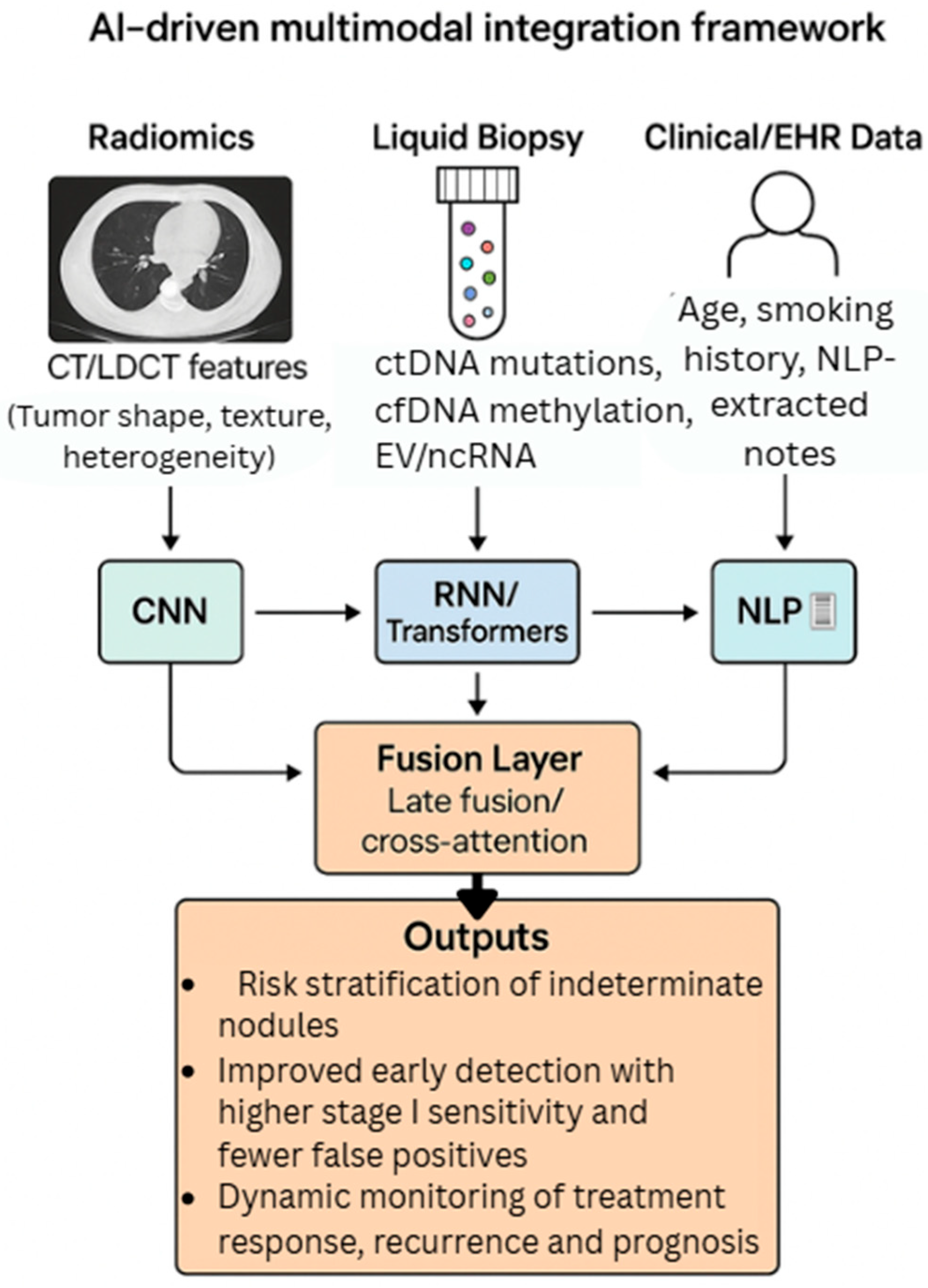
6.3. Integration of AI, Liquid Biopsy, and Radiomics for Multimodal Early Lung Cancer Detection
7. Challenges and Limitations of AI in Liquid Biopsy
7.1. Data Quality and Standardization
7.2. Ethical and Privacy Concerns
7.3. Model Transparency and Interpretability
7.4. Cost and Accessibility
7.5. Population Generalizability and Comparative Effectiveness
8. Future Directions and Research Opportunities
Concluding Remarks
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Cucchiara, F.; Petrini, I.; Romei, C.; Crucitta, S.; Lucchesi, M.; Valleggi, S.; Scavone, C.; Capuano, A.; De Liperi, A.; Chella, A.; et al. Combining liquid biopsy and radiomics for personalized treatment of lung cancer patients. State of the art and new perspectives. Pharmacol. Res. 2021, 169, 105643. [Google Scholar] [CrossRef] [PubMed]
- Howlader, N.; Forjaz, G.; Mooradian, M.J.; Meza, R.; Kong, C.Y.; Cronin, K.A.; Mariotto, A.B.; Lowy, D.R.; Feuer, E.J. The Effect of Advances in Lung-Cancer Treatment on Population Mortality. N. Engl. J. Med. 2020, 383, 640–649. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. (Eds.) SEER Cancer Statistics Review, 1975–2018; National Cancer Institute: Bethesda, MD, USA, 2021. Available online: https://seer.cancer.gov/csr/1975_2018/ (accessed on 9 September 2025).
- US Preventive Services Task Force; Krist, A.H.; Davidson, K.W.; Mangione, C.M.; Barry, M.J.; Cabana, M.; Caughey, A.B.; Davis, E.M.; Donahue, K.E.; Doubeni, C.A.; et al. Screening for Lung Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2021, 325, 962–970. [Google Scholar] [CrossRef] [PubMed]
- de Koning, H.J.; van der Aalst, C.M.; de Jong, P.A.; Scholten, E.T.; Nackaerts, K.; Heuvelmans, M.A.; Lammers, J.J.; Weenink, C.; Yousaf-Khan, U.; Horeweg, N.; et al. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N. Engl. J. Med. 2020, 382, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Pinsky, P.F.; Gierada, D.S.; Black, W.; Munden, R.; Nath, H.; Aberle, D.; Kazerooni, E. Performance of Lung-RADS in the National Lung Screening Trial: A retrospective assessment. Ann. Intern. Med. 2015, 162, 485–491. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- National Lung Screening Trial Research, Team; Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid Biopsy for Advanced NSCLC: A Consensus Statement from the International Association for the Study of Lung Cancer. J. Thorac. Oncol. 2021, 16, 1647–1662. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabières, C.; Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 2014, 14, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.Ø.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019, 570, 385–389. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vijh, S.; Gaur, D.; Kumar, S. An intelligent lung tumor diagnosis system using whale optimization algorithm and support vector machine. Int. J. Syst. Assur. Eng. Manag. 2019, 11, 374–384. [Google Scholar] [CrossRef]
- Cao, H.; Yan, B.; Dong, L.; Yuan, X. Multipopulation Whale Optimization-Based Feature Selection Algorithm and Its Application in Human Fall Detection Using Inertial Measurement Unit Sensors. Sensors 2024, 24, 7879. [Google Scholar] [CrossRef]
- Costa, J.; Membrino, A.; Zanchetta, C.; Rizzato, S.; Cortiula, F.; Rossetto, C.; Pelizzari, G.; Aprile, G.; Macerelli, M. The Role of ctDNA in the Management of Non-Small-Cell Lung Cancer in the AI and NGS Era. Int. J. Mol. Sci. 2024, 25, 13669. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Siddiqui, F.; Vaqar, S.; Siddiqui, A.H. Lung Cancer. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar] [PubMed]
- Nooreldeen, R.; Bach, H. Current and Future Development in Lung Cancer Diagnosis. Int. J. Mol. Sci. 2021, 22, 8661. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kim, J.; Lee, H.; Huang, B.W. Lung Cancer: Diagnosis, Treatment Principles, and Screening. Am. Fam. Physician 2022, 105, 487–494. [Google Scholar] [PubMed]
- Nicholson, A.G.; Tsao, M.S.; Beasley, M.B.; Borczuk, A.C.; Brambilla, E.; Cooper, W.A.; Dacic, S.; Jain, D.; Kerr, K.M.; Lantuejoul, S.; et al. The 2021 WHO Classification of Lung Tumors: Impact of Advances Since 2015. J. Thorac. Oncol. 2022, 17, 362–387. [Google Scholar] [CrossRef] [PubMed]
- George, J.; Lim, J.S.; Jang, S.J.; Cun, Y.; Ozretić, L.; Kong, G.; Leenders, F.; Lu, X.; Fernández-Cuesta, L.; Bosco, G.; et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015, 524, 47–53. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gazdar, A.F.; Bunn, P.A.; Minna, J.D. Small-cell lung cancer: What we know, what we need to know and the path forward. Nat. Rev. Cancer 2017, 17, 725–737, Erratum in Nat. Rev. Cancer 2017, 17, 765. [Google Scholar] [CrossRef] [PubMed]
- Denny, S.K.; Yang, D.; Chuang, C.H.; Brady, J.J.; Lim, J.S.; Grüner, B.M.; Chiou, S.H.; Schep, A.N.; Baral, J.; Hamard, C.; et al. Nfib Promotes Metastasis through a Widespread Increase in Chromatin Accessibility. Cell 2016, 166, 328–342. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rudin, C.M.; Brambilla, E.; Faivre-Finn, C.; Sage, J. Small-cell lung cancer. Nat. Rev. Dis. Primers 2021, 7, 3. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kalemkerian, G.P.; Akerley, W.; Bogner, P.; Borghaei, H.; Chow, L.Q.; Downey, R.J.; Gandhi, L.; Ganti, A.K.; Govindan, R.; Grecula, J.C.; et al. Small cell lung cancer. J. Natl. Compr. Cancer Netw. 2013, 11, 78–98. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, L.; Soler, J.; Reckamp, K.L.; Sankar, K. Emerging Targets in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2024, 25, 10046. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, D.; Li, Z.; Jiang, T.; Yang, C.; Li, N. Artificial intelligence in lung cancer: Current applications, future perspectives, and challenges. Front. Oncol. 2024, 14, 1486310. [Google Scholar] [CrossRef]
- Czerlanis, C.M.; Singh, N.; Fintelmann, F.J.; Damaraju, V.; Chang, A.E.B.; White, M.; Hanna, N. Broadening the Net: Overcoming Challenges and Embracing Novel Technologies in Lung Cancer Screening. Am. Soc. Clin. Oncol. Educ. Book 2025, 45, e473778. [Google Scholar] [CrossRef] [PubMed]
- Duranti, L.; Tavecchio, L.; Rolli, L.; Solli, P. New Perspectives on Lung Cancer Screening and Artificial Intelligence. Life 2025, 15, 498. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- American Lung Association. State of Lung Cancer 2024. Available online: https://www.lung.org/getmedia/12020193-7fb3-46b8-8d78-0e5d9cd8f93c/SOLC-2024.pdf (accessed on 9 September 2025).
- McNulty, W.; Baldwin, D. Management of pulmonary nodules. BJR Open 2019, 1, 20180051. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- PDQ Screening and Prevention Editorial Board. Lung Cancer Screening (PDQ®): Health Professional Version; National Cancer Institute (US): Bethesda, MD, USA, 2025. Available online: https://www.cancer.gov/types/lung/hp/lung-screening-pdq (accessed on 9 September 2025). [PubMed]
- Katsimpoula, S.; Patrinou-Georgoula, M.; Makrilia, N.; Dimakou, K.; Guialis, A.; Orfanidou, D.; Syrigos, K.N. Overexpression of hnRNPA2/B1 in bronchoscopic specimens: A potential early detection marker in lung cancer. Anticancer Res. 2009, 29, 1373–1382. [Google Scholar] [PubMed]
- Molina, R.; Marrades, R.M.; Augé, J.M.; Escudero, J.M.; Viñolas, N.; Reguart, N.; Ramirez, J.; Filella, X.; Molins, L.; Agustí, A. Assessment of a Combined Panel of Six Serum Tumor Markers for Lung Cancer. Am. J. Respir. Crit. Care Med. 2016, 193, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Rivera, M.P.; Mehta, A.C.; Wahidi, M.M. Establishing the diagnosis of lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013, 143 (Suppl. S5), e142S–e165S. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sozzi, G.; Conte, D.; Leon, M.; Ciricione, R.; Roz, L.; Ratcliffe, C.; Roz, E.; Cirenei, N.; Bellomi, M.; Pelosi, G.; et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J. Clin. Oncol. 2003, 21, 3902–3908. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Wang, S.; Zhang, Y.J.; Kappil, M.; Wu, H.C.; Kibriya, M.G.; Wang, Q.; Jasmine, F.; Ahsan, H.; Lee, P.H.; et al. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology 2012, 55, 1799–1808. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, X.J.; Ren, Z.J.; Tang, J.H. MicroRNA-34a: A potential therapeutic target in human cancer. Cell Death Dis. 2014, 5, e1327. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pantel, K.; Alix-Panabières, C. Circulating tumour cells in cancer patients: Challenges and perspectives. Trends Mol. Med. 2010, 16, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Castro-Giner, F.; Gkountela, S.; Donato, C.; Alborelli, I.; Quagliata, L.; Ng, C.K.Y.; Piscuoglio, S.; Aceto, N. Cancer Diagnosis Using a Liquid Biopsy: Challenges and Expectations. Diagnostics 2018, 8, 31. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Connal, S.; Cameron, J.M.; Sala, A.; Brennan, P.M.; Palmer, D.S.; Palmer, J.D.; Perlow, H.; Baker, M.J. Liquid biopsies: The future of cancer early detection. J. Transl. Med. 2023, 21, 118. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hanssen, A.; Wagner, J.; Gorges, T.M.; Taenzer, A.; Uzunoglu, F.G.; Driemel, C.; Stoecklein, N.H.; Knoefel, W.T.; Angenendt, S.; Hauch, S.; et al. Characterization of different CTC subpopulations in non-small cell lung cancer. Sci. Rep. 2016, 6, 28010. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stroun, M.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Beljanski, M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 1989, 46, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Wong, F.C.; Sun, K.; Jiang, P.; Cheng, Y.K.; Chan, K.C.; Leung, T.Y.; Chiu, R.W.; Lo, Y.M. Cell-free DNA in maternal plasma and serum: A comparison of quantity, quality and tissue origin using genomic and epigenomic approaches. Clin. Biochem. 2016, 49, 1379–1386. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.A.; Cristofanilli, M. Detection of Predictive Biomarkers Using Liquid Biopsies. In Predictive Biomarkers in Oncology; Springer: Berlin/Heidelberg, Germany, 2019; pp. 107–117. [Google Scholar]
- Gale, D.; Lawson, A.R.J.; Howarth, K.; Madi, M.; Durham, B.; Smalley, S.; Calaway, J.; Blais, S.; Jones, G.; Clark, J.; et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS ONE 2018, 13, e0194630. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Casagrande, G.M.S.; Silva, M.O.; Reis, R.M.; Leal, L.F. Liquid Biopsy for Lung Cancer: Up-to-Date and Perspectives for Screening Programs. Int. J. Mol. Sci. 2023, 24, 2505. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rizwan, M.N.; Ma, Y.; Nenkov, M.; Jin, L.; Schröder, D.C.; Westermann, M.; Gaßler, N.; Chen, Y. Tumor-derived exosomes: Key players in non-small cell lung cancer metastasis and their implication for targeted therapy. Mol. Carcinog. 2022, 61, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. Exosomes and tumor-mediated immune suppression. J. Clin. Investig. 2016, 126, 1216–1223. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bracht, J.W.P.; Mayo-de-Las-Casas, C.; Berenguer, J.; Karachaliou, N.; Rosell, R. The Present and Future of Liquid Biopsies in Non-Small Cell Lung Cancer: Combining Four Biosources for Diagnosis, Prognosis, Prediction, and Disease Monitoring. Curr. Oncol. Rep. 2018, 20, 70, Erratum in Curr. Oncol. Rep. 2020, 22, 52. [Google Scholar] [CrossRef] [PubMed]
- Takamizawa, J.; Konishi, H.; Yanagisawa, K.; Tomida, S.; Osada, H.; Endoh, H.; Harano, T.; Yatabe, Y.; Nagino, M.; Nimura, Y.; et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004, 64, 3753–3756. [Google Scholar] [CrossRef] [PubMed]
- Souza, V.G.P.; Forder, A.; Brockley, L.J.; Pewarchuk, M.E.; Telkar, N.; de Araújo, R.P.; Trejo, J.; Benard, K.; Seneda, A.L.; Minutentag, I.W.; et al. Liquid Biopsy in Lung Cancer: Biomarkers for the Management of Recurrence and Metastasis. Int. J. Mol. Sci. 2023, 24, 8894. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Roweth, H.G.; Battinelli, E.M. Lessons to learn from tumor-educated platelets. Blood 2021, 137, 3174–3180. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bailey, S.E.; Ukoumunne, O.C.; Shephard, E.; Hamilton, W. How useful is thrombocytosis in predicting an underlying cancer in primary care? A systematic review. Fam. Pract. 2017, 34, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Asher, V.; Lee, J.; Innamaa, A.; Bali, A. Preoperative platelet lymphocyte ratio as an independent prognostic marker in ovarian cancer. Clin. Transl. Oncol. 2011, 13, 499–503. [Google Scholar] [CrossRef] [PubMed]
- van Bussel, M.T.J.; Pluim, D.; Bol, M.; Beijnen, J.H.; Schellens, J.H.M.; Brandsma, D. EpCAM-based assays for epithelial tumor cell detection in cerebrospinal fluid. J. Neurooncol. 2018, 137, 1–10. [Google Scholar] [CrossRef] [PubMed]
- de Wit, S.; van Dalum, G.; Lenferink, A.T.; Tibbe, A.G.; Hiltermann, T.J.; Groen, H.J.; van Rijn, C.J.; Terstappen, L.W. The detection of EpCAM(+) and EpCAM(-) circulating tumor cells. Sci. Rep. 2015, 5, 12270. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Diefenbach, R.J.; Lee, J.H.; Kefford, R.F.; Rizos, H. Evaluation of commercial kits for purification of circulating free DNA. Cancer Genet. 2018, 228–229, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Watanabe, Y.; Itoh, F. Cell-Free DNA. In Biomarkers in Cancer Therapy; Springer: Berlin/Heidelberg, Germany, 2019; pp. 11–24. [Google Scholar]
- Zhao, Z.; Wijerathne, H.; Godwin, A.K.; Soper, S.A. Isolation and analysis methods of extracellular vesicles (EVs). Extracell. Vesicles Circ. Nucl. Acids 2021, 2, 80–103, Erratum in Extracell. Vesicles Circ. Nucl. Acids 2021, 2, 222–223. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Trakunram, K.; Champoochana, N.; Chaniad, P.; Thongsuksai, P.; Raungrut, P. MicroRNA Isolation by Trizol-Based Method and Its Stability in Stored Serum and cDNA Derivatives. Asian Pac. J. Cancer Prev. 2019, 20, 1641–1647. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Miyata, K.; Naito, M.; Miyata, T.; Mokuda, S.; Asahara, H. Bisulfite Sequencing for DNA Methylation Analysis of Primary Muscle Stem Cells. Methods Mol. Biol. 2017, 1668, 3–13. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Best, M.G.; In ‘t Veld, S.G.J.G.; Sol, N.; Wurdinger, T. RNA sequencing and swarm intelligence-enhanced classification algorithm development for blood-based disease diagnostics using spliced blood platelet RNA. Nat. Protoc. 2019, 14, 1206–1234. [Google Scholar] [CrossRef] [PubMed]
- Hong, T.H.; Hwang, S.; Dasgupta, A.; Abbosh, C.; Hung, T.; Bredno, J.; Walker, J.; Shi, X.; Milenkova, T.; Horn, L.; et al. Clinical Utility of Tumor-Naïve Presurgical Circulating Tumor DNA Detection in Early-Stage NSCLC. J. Thorac. Oncol. 2024, 19, 1512–1524. [Google Scholar] [CrossRef] [PubMed]
- Black, J.R.M.; Bartha, G.; Abbott, C.W.; Boyle, S.M.; Karasaki, T.; Li, B.; Chen, R.; Harris, J.; Veeriah, S.; Colopi, M.; et al. Ultrasensitive ctDNA detection for preoperative disease stratification in early-stage lung adenocarcinoma. Nat. Med. 2025, 31, 70–76. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hodgkinson, C.L.; Morrow, C.J.; Li, Y.; Metcalf, R.L.; Rothwell, D.G.; Trapani, F.; Polanski, R.; Burt, D.J.; Simpson, K.L.; Morris, K.; et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat. Med. 2014, 20, 897–903. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Wang, P.; Gu, L.; Liu, J. Use of four genes in exosomes as biomarkers for the identification of lung adenocarcinoma and lung squamous cell carcinoma. Oncol. Lett. 2021, 21, 249. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, W.; Wang, X.; Li, C.; Chen, T.; Yang, Q. Exosomal non-coding RNAs: Emerging roles in bilateral communication between cancer cells and macrophages. Mol. Ther. 2022, 30, 1036–1053. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, K.Z.; Lou, F.; Yang, F.; Zhang, J.B.; Ye, H.; Chen, W.; Guan, T.; Zhao, M.Y.; Su, X.X.; Shi, R.; et al. Circulating Tumor DNA Detection in Early-Stage Non-Small Cell Lung Cancer Patients by Targeted Sequencing. Sci. Rep. 2016, 6, 31985. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zou, W.; Yaung, S.J.; Fuhlbrück, F.; Ballinger, M.; Peters, E.; Palma, J.F.; Shames, D.S.; Gandara, D.; Jiang, Y.; Patil, N.S. ctDNA Predicts Overall Survival in Patients with NSCLC Treated With PD-L1 Blockade or With Chemotherapy. JCO Precis. Oncol. 2021, 5, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Buder, A.; Hochmair, M.J.; Setinek, U.; Pirker, R.; Filipits, M. EGFR mutation tracking predicts survival in advanced EGFR-mutated non-small cell lung cancer patients treated with osimertinib. Transl. Lung Cancer Res. 2020, 9, 239–245. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cheng, M.L.; Lau, C.J.; Milan, M.S.D.; Supplee, J.G.; Riess, J.W.; Bradbury, P.A.; Jänne, P.A.; Oxnard, G.R.; Paweletz, C.P. Plasma ctDNA Response Is an Early Marker of Treatment Effect in Advanced NSCLC. JCO Precis. Oncol. 2021, 5, PO.20.00419. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, R.Y.; Liang, Z.Y. Circulating tumor DNA in lung cancer: Real-time monitoring of disease evolution and treatment response. Chin. Med. J. 2020, 133, 2476–2485. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, Z.; Xiao, Y.; Zhao, J.; Chen, M.; Xu, Y.; Zhong, W.; Xing, J.; Wang, M. Relationship between circulating tumour cell count and prognosis following chemotherapy in patients with advanced non-small-cell lung cancer. Respirology 2016, 21, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Michaelidou, K.; Koutoulaki, C.; Mavridis, K.; Vorrias, E.; Papadaki, M.A.; Koutsopoulos, A.V.; Mavroudis, D.; Agelaki, S. Detection of KRAS G12/G13 Mutations in Cell Free-DNA by Droplet Digital PCR, Offers Prognostic Information for Patients with Advanced Non-Small Cell Lung Cancer. Cells 2020, 9, 2514. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wen, S.W.C.; Andersen, R.F.; Hansen, T.F.; Nyhus, C.H.; Hager, H.; Hilberg, O.; Jakobsen, A. The prognostic impact of circulating homeobox A9 methylated DNA in advanced non-small cell lung cancer. Transl. Lung Cancer Res. 2021, 10, 855–865. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Khandelwal, A.; Seam, R.K.; Gupta, M.; Rana, M.K.; Prakash, H.; Vasquez, K.M.; Jain, A. Circulating microRNA-590-5p functions as a liquid biopsy marker in non-small cell lung cancer. Cancer Sci. 2020, 111, 826–839. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Eslami-S, Z.; Cortés-Hernández, L.E.; Alix-Panabières, C. The Metastatic Cascade as the Basis for Liquid Biopsy Development. Front. Oncol. 2020, 10, 1055. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nakasu, Y.; Deguchi, S.; Nakasu, S.; Yamazaki, M.; Notsu, A.; Mitsuya, K.; Hayashi, N. Diagnostic accuracy of cerebrospinal fluid liquid biopsy and MRI for leptomeningeal metastases in solid cancers: A systematic review and meta-analysis. Neurooncol. Adv. 2023, 5, vdad002. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gale, D.; Heider, K.; Ruiz-Valdepenas, A.; Hackinger, S.; Perry, M.; Marsico, G.; Rundell, V.; Wulff, J.; Sharma, G.; Knock, H.; et al. Residual ctDNA after treatment predicts early relapse in patients with early-stage non-small cell lung cancer. Ann. Oncol. 2022, 33, 500–510. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Verzè, M.; Pluchino, M.; Leonetti, A.; Corianò, M.; Bonatti, F.; Armillotta, M.P.; Perrone, F.; Casali, M.; Minari, R.; Tiseo, M. Role of ctDNA for the detection of minimal residual disease in resected non-small cell lung cancer: A systematic review. Transl. Lung Cancer Res. 2022, 11, 2588–2600. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stergiopoulou, D.; Markou, A.; Strati, A.; Zavridou, M.; Tzanikou, E.; Mastoraki, S.; Kallergi, G.; Georgoulias, V.; Lianidou, E. Comprehensive liquid biopsy analysis as a tool for the early detection of minimal residual disease in breast cancer. Sci. Rep. 2023, 13, 1258. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Black, J.R.M.; McGranahan, N. Genetic and non-genetic clonal diversity in cancer evolution. Nat. Rev. Cancer. 2021, 21, 379–392. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef] [PubMed]
- Bonanno, L.; Dal Maso, A.; Pavan, A.; Zulato, E.; Calvetti, L.; Pasello, G.; Guarneri, V.; Conte, P.; Indraccolo, S. Liquid biopsy and non-small cell lung cancer: Are we looking at the tip of the iceberg? Br. J. Cancer. 2022, 127, 383–393. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Heitzer, E.; van den Broek, D.; Denis, M.G.; Hofman, P.; Hubank, M.; Mouliere, F.; Paz-Ares, L.; Schuuring, E.; Sültmann, H.; Vainer, G.; et al. Recommendations for a practical implementation of circulating tumor DNA mutation testing in metastatic non-small-cell lung cancer. ESMO Open 2022, 7, 100399. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Devarakonda, S.; Sankararaman, S.; Herzog, B.H.; Gold, K.A.; Waqar, S.N.; Ward, J.P.; Raymond, V.M.; Lanman, R.B.; Chaudhuri, A.A.; Owonikoko, T.K.; et al. Circulating Tumor DNA Profiling in Small-Cell Lung Cancer Identifies Potentially Targetable Alterations. Clin. Cancer Res. 2019, 25, 6119–6126. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hu, Y.; Ulrich, B.C.; Supplee, J.; Kuang, Y.; Lizotte, P.H.; Feeney, N.B.; Guibert, N.M.; Awad, M.M.; Wong, K.K.; Jänne, P.A.; et al. False-Positive Plasma Genotyping Due to Clonal Hematopoiesis. Clin. Cancer Res. 2018, 24, 4437–4443. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Ngiam, K.Y.; Teo, H.H. Role of Artificial Intelligence Applications in Real-Life Clinical Practice: Systematic Review. J. Med. Internet Res. 2021, 23, e25759. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bhattamisra, S.K.; Banerjee, P.; Gupta, P.; Mayuren, J.; Patra, S.; Candasamy, M. Artificial intelligence in pharmaceutical and healthcare research. Big Data Cogn. Comput. 2023, 7, 10. [Google Scholar] [CrossRef]
- Mueller, S.T.; Hoffman, R.R.; Clancey, W.; Emrey, A.; Klein, G. Explanation in human-AI systems: A literature meta-review, synopsis of key ideas and publications, and bibliography for explainable AI. arXiv 2019, arXiv:1902.01876. [Google Scholar]
- Greener, J.G.; Kandathil, S.M.; Moffat, L.; Jones, D.T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 2022, 23, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Dou, B.; Zhu, Z.; Merkurjev, E.; Ke, L.; Chen, L.; Jiang, J.; Zhu, Y.; Liu, J.; Zhang, B.; Wei, G.W. Machine Learning Methods for Small Data Challenges in Molecular Science. Chem. Rev. 2023, 123, 8736–8780. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Camacho, D.M.; Collins, K.M.; Powers, R.K.; Costello, J.C.; Collins, J.J. Next-Generation Machine Learning for Biological Networks. Cell 2018, 173, 1581–1592. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061.e18. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Derry, A.; Krzywinski, M.; Altman, N. Convolutional neural networks. Nat. Methods 2023, 20, 1269–1270. [Google Scholar] [CrossRef] [PubMed]
- Qiao, M.; Suo, S.; Cheng, F.; Hua, J.; Xue, D.; Guo, Y.; Xu, J.; Wang, Y. Three-dimensional breast tumor segmentation on DCE-MRI with a multilabel attention-guided joint-phase-learning network. Comput. Med. Imaging Graph. 2021, 90, 101909. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; He, Z.; Jia, Y. AFPNet: A 3D fully convolutional neural network with atrous-convolution feature pyramid for brain tumor segmentation via MRI images. Neurocomputing 2020, 402, 235–244. [Google Scholar] [CrossRef]
- Das, S.; Tariq, A.; Santos, T.; Kantareddy, S.S.; Banerjee, I. Recurrent Neural Networks (RNNs): Architectures, Training Tricks, and Introduction to Influential Research. In Machine Learning for Brain Disorders [Internet]; Colliot, O., Ed.; Humana: New York, NY, USA, 2023; Chapter 4. [Google Scholar] [PubMed]
- Zheng, J.; Lin, D.; Gao, Z.; Wang, S.; He, M.; Fan, J. Deep learning assisted efficient AdaBoost algorithm for breast cancer detection and early diagnosis. IEEE Access 2020, 8, 96946–96954. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Min, S.; Lee, B.; Yoon, S. Deep learning in bioinformatics. Brief Bioinform. 2017, 18, 851–869. [Google Scholar] [CrossRef] [PubMed]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Helmbold, D.P.; Long, P.M. Surprising properties of dropout in deep networks. J. Mach. Learn. Res. 2018, 18, 1–28. [Google Scholar] [CrossRef]
- Ayub, S.; Kannan, R.J.; S, S.; Alsini, R.; Hasanin, T.; Sasidhar, C. LSTM-Based RNN Framework to Remove Motion Artifacts in Dynamic Multicontrast MR Images with Registration Model. Wirel. Commun. Mob. Comput. 2022, 2022, 5906877. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, J.; Liao, B.; Wu, F.X.; Bi, X.A. Deep Canonical Correlation Fusion Algorithm Based on Denoising Autoencoder for ASD Diagnosis and Pathogenic Brain Region Identification. Interdiscip. Sci. Comput. Life Sci. 2024, 16, 455–468. [Google Scholar] [CrossRef]
- Mutepfe, F.; Kalejahi, B.K.; Meshgini, S.; Danishvar, S. Generative Adversarial Network Image Synthesis Method for Skin Lesion Generation and Classification. J. Med. Signals Sens. 2021, 11, 237–252. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Berlin/Heidelberg, Germany, 2018; Volume 11045, pp. 3–11. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nayarisseri, A.; Khandelwal, R.; Tanwar, P.; Madhavi, M.; Sharma, D.; Thakur, G.; Speck-Planche, A.; Singh, S.K. Artificial Intelligence, Big Data and Machine Learning Approaches in Precision Medicine & Drug Discovery. Curr. Drug Targets 2021, 22, 631–655. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems 25 (NIPS 2012); Pereira, F., Burges, C.J.C., Bottou, L., Weinberger, K.Q., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2012; pp. 1097–1105. [Google Scholar]
- Haug, C.J.; Drazen, J.M. Artificial Intelligence and Machine Learning in Clinical Medicine, 2023. N. Engl. J. Med. 2023, 388, 1201–1208. [Google Scholar] [CrossRef] [PubMed]
- Alowais, S.A.; Alghamdi, S.S.; Alsuhebany, N.; Alqahtani, T.; Alshaya, A.I.; Almohareb, S.N.; Aldairem, A.; Alrashed, M.; Bin Saleh, K.; Badreldin, H.A.; et al. Revolutionizing healthcare: The role of artificial intelligence in clinical practice. BMC Med. Educ. 2023, 23, 689. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lisberg, A.; Cummings, A.; Goldman, J.W.; Bornazyan, K.; Reese, N.; Wang, T.; Coluzzi, P.; Ledezma, B.; Mendenhall, M.; Hunt, J.; et al. A Phase II Study of Pembrolizumab in EGFR-Mutant, PD-L1+, Tyrosine Kinase Inhibitor Naïve Patients with Advanced NSCLC. J. Thorac. Oncol. 2018, 13, 1138–1145. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mazieres, J.; Drilon, A.; Lusque, A.; Mhanna, L.; Cortot, A.B.; Mezquita, L.; Thai, A.A.; Mascaux, C.; Couraud, S.; Veillon, R.; et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: Results from the IMMUNOTARGET registry. Ann. Oncol. 2019, 30, 1321–1328. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mok, T.S.K.; Wu, Y.L.; Kudaba, I.; Kowalski, D.M.; Cho, B.C.; Turna, H.Z.; Castro, G., Jr.; Srimuninnimit, V.; Laktionov, K.K.; Bondarenko, I.; et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): A randomised, open-label, controlled, phase 3 trial. Lancet 2019, 393, 1819–1830. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, C.; Rolfo, C.D.; Oxnard, G.R.; Gray, J.E.; Sholl, L.M.; Gandara, D.R. Strategies for the successful implementation of plasma-based NSCLC genotyping in clinical practice. Nat. Rev. Clin. Oncol. 2021, 18, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.L.; Zhou, C.; Hu, C.P.; Feng, J.; Lu, S.; Huang, Y.; Li, W.; Hou, M.; Shi, J.H.; Lee, K.Y.; et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): An open-label, randomised phase 3 trial. Lancet Oncol. 2014, 15, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, S.; Yang, J.C.; Ramalingam, S.S.; Yu, K.; Patel, S.; Weston, S.; Hodge, R.; Cantarini, M.; Jänne, P.A.; Mitsudomi, T.; et al. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non-Small Cell Lung Cancer. J. Thorac. Oncol. 2017, 12, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.E.; Okamoto, I.; Sriuranpong, V.; Vansteenkiste, J.; Imamura, F.; Lee, J.S.; Pang, Y.K.; Cobo, M.; Kasahara, K.; Cheng, Y.; et al. Tissue and Plasma EGFR Mutation Analysis in the FLAURA Trial: Osimertinib versus Comparator EGFR Tyrosine Kinase Inhibitor as First-Line Treatment in Patients with EGFR-Mutated Advanced Non-Small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 6644–6652. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chiu, H.Y.; Chao, H.S.; Chen, Y.M. Application of Artificial Intelligence in Lung Cancer. Cancers 2022, 14, 1370. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liang, N.; Li, B.; Jia, Z.; Wang, C.; Wu, P.; Zheng, T.; Wang, Y.; Qiu, F.; Wu, Y.; Su, J.; et al. Ultrasensitive detection of circulating tumour DNA via deep methylation sequencing aided by machine learning. Nat. Biomed. Eng. 2021, 5, 586–599, Erratum in Nat. Biomed. Eng. 2021, 5, 1402. [Google Scholar] [CrossRef] [PubMed]
- Bie, Z.; Ping, Y.; Li, X.; Lan, X.; Wang, L. Accurate early detection and EGFR mutation status prediction of lung cancer using plasma cfDNA coverage patterns: A proof-of-concept study. Biomolecules 2024, 14, 716. [Google Scholar] [CrossRef]
- Asleh, K.; Dery, V.; Taylor, C.; Davey, M.; Djeungoue-Petga, M.A.; Ouellette, R.J. Extracellular vesicle-based liquid biopsy biomarkers and their application in precision immuno-oncology. Biomark. Res. 2023, 11, 99. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, Y.; Li, C.; Yang, Y.; Zhang, T.; Wang, J.; Tang, W.; Li, N.; Bao, H.; Wang, X.; Bi, N. Predicting Disease Progression in Inoperable Localized NSCLC Patients Using ctDNA Machine Learning Model. Cancer Med. 2024, 13, e70316. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yolchuyeva, S.; Giacomazzi, E.; Tonneau, M.; Lamaze, F.; Orain, M.; Coulombe, F.; Malo, J.; Belkaid, W.; Routy, B.; Joubert, P.; et al. Radiomics approaches to predict PD-L1 and PFS in advanced non-small cell lung patients treated with immunotherapy: A multi-institutional study. Sci. Rep. 2023, 13, 11065. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Karimzadeh, M.; Momen-Roknabadi, A.; Cavazos, T.B.; Fang, Y.; Chen, N.C.; Multhaup, M.; Yen, J.; Ku, J.; Wang, J.; Zhao, X.; et al. Deep generative AI models analyzing circulating orphan non-coding RNAs enable detection of early-stage lung cancer. Nat. Commun. 2024, 15, 10090. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mathios, D.; Johansen, J.S.; Cristiano, S.; Medina, J.E.; Phallen, J.; Larsen, K.R.; Bruhm, D.C.; Niknafs, N.; Ferreira, L.; Adleff, V.; et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat. Commun. 2021, 12, 5060. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bahado-Singh, R.; Vlachos, K.T.; Aydas, B.; Gordevicius, J.; Radhakrishna, U.; Vishweswaraiah, S. Precision Oncology: Artificial Intelligence and DNA Methylation Analysis of Circulating Cell-Free DNA for Lung Cancer Detection. Front. Oncol. 2022, 12, 790645. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, Z.; Xie, K.; Zhu, G.; Ma, C.; Cheng, C.; Li, Y.; Xiao, X.; Li, C.; Tang, J.; Wang, H.; et al. Early detection and stratification of lung cancer aided by a cost-effective assay targeting circulating tumor DNA (ctDNA) methylation. Respir. Res. 2023, 24, 163. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Purohit, L.; Kiamos, A.; Ali, S.; Alvarez-Pinzon, A.M.; Raez, L. Incidental Pulmonary Nodule (IPN) Programs Working Together with Lung Cancer Screening and Artificial Intelligence to Increase Lung Cancer Detection. Cancers 2025, 17, 1143. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ji, G.; Luo, W.; Zhu, Y.; Chen, B.; Wang, M.; Jiang, L.; Yang, M.; Song, W.; Yao, P.; Zheng, T.; et al. Early-stage lung cancer detection via thin-section low-dose CT reconstruction combined with AI in non-high risk populations: A large-scale real-world retrospective cohort study. Precis. Clin. Med. 2025, 8, pbaf007. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, B.T.; Chen, Z.; Ye, N.; Mambetsariev, I.; Fricke, J.; Daniel, E.; Wang, G.; Wong, C.W.; Rockne, R.C.; Colen, R.R.; et al. Differentiating Peripherally-Located Small Cell Lung Cancer From Non-small Cell Lung Cancer Using a CT Radiomic Approach. Front. Oncol. 2020, 10, 593. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, S.; Liu, S.; Zhang, C.; Yu, H.; Liu, X.; Hu, Y.; Xu, W.; Tang, X.; Fu, Q. Exploratory Study of a CT Radiomics Model for the Classification of Small Cell Lung Cancer and Non-small-Cell Lung Cancer. Front. Oncol. 2020, 10, 1268. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Giroux Leprieur, E.; Hélias-Rodzewicz, Z.; Takam Kamga, P.; Costantini, A.; Julie, C.; Corjon, A.; Dumenil, C.; Dumoulin, J.; Giraud, V.; Labrune, S.; et al. Sequential ctDNA whole-exome sequencing in advanced lung adenocarcinoma with initial durable tumor response on immune checkpoint inhibitor and late progression. J. Immunother. Cancer 2020, 8, e000527. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jee, J.; Lebow, E.S.; Yeh, R.; Das, J.P.; Namakydoust, A.; Paik, P.K.; Chaft, J.E.; Jayakumaran, G.; Rose Brannon, A.; Benayed, R.; et al. Overall survival with circulating tumor DNA-guided therapy in advanced non-small-cell lung cancer. Nat. Med. 2022, 28, 2353–2363. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chaudhuri, A.A.; Chabon, J.J.; Lovejoy, A.F.; Newman, A.M.; Stehr, H.; Azad, T.D.; Khodadoust, M.S.; Esfahani, M.S.; Liu, C.L.; Zhou, L.; et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017, 7, 1394–1403. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hellmann, M.D.; Nabet, B.Y.; Rizvi, H.; Chaudhuri, A.A.; Wells, D.K.; Dunphy, M.P.S.; Chabon, J.J.; Liu, C.L.; Hui, A.B.; Arbour, K.C.; et al. Circulating Tumor DNA Analysis to Assess Risk of Progression after Long-term Response to PD-(L)1 Blockade in NSCLC. Clin. Cancer Res. 2020, 26, 2849–2858. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chabon, J.J.; Hamilton, E.G.; Kurtz, D.M.; Esfahani, M.S.; Moding, E.J.; Stehr, H.; Schroers-Martin, J.; Nabet, B.Y.; Chen, B.; Chaudhuri, A.A.; et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 2020, 580, 245–251. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sujit, S.J.; Aminu, M.; Karpinets, T.V.; Chen, P.; Saad, M.B.; Salehjahromi, M.; Boom, J.D.; Qayati, M.; George, J.M.; Allen, H.; et al. Enhancing NSCLC recurrence prediction with PET/CT habitat imaging, ctDNA, and integrative radiogenomics-blood insights. Nat. Commun. 2024, 15, 3152. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Widman, A.J.; Shah, M.; Frydendahl, A.; Halmos, D.; Khamnei, C.C.; Øgaard, N.; Rajagopalan, S.; Arora, A.; Deshpande, A.; Hooper, W.F.; et al. Ultrasensitive plasma-based monitoring of tumor burden using machine-learning-guided signal enrichment. Nat. Med. 2024, 30, 1655–1666. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Helzer, K.T.; Sharifi, M.N.; Sperger, J.M.; Shi, Y.; Annala, M.; Bootsma, M.L.; Reese, S.R.; Taylor, A.; Kaufmann, K.R.; Krause, H.K.; et al. Fragmentomic analysis of circulating tumor DNA-targeted cancer panels. Ann. Oncol. 2023, 34, 813–825. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Assaf, Z.J.F.; Zou, W.; Fine, A.D.; Socinski, M.A.; Young, A.; Lipson, D.; Freidin, J.F.; Kennedy, M.; Polisecki, E.; Nishio, M.; et al. A longitudinal circulating tumor DNA-based model associated with survival in metastatic non-small-cell lung cancer. Nat. Med. 2023, 29, 859–868. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Abbosh, C.; Frankell, A.M.; Harrison, T.; Kisistok, J.; Garnett, A.; Johnson, L.; Veeriah, S.; Moreau, M.; Chesh, A.; Chaunzwa, T.L.; et al. Tracking early lung cancer metastatic dissemination in TRACERx using ctDNA. Nature 2023, 616, 553–562. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, Y.; Sun, B.; Yu, Y.; Lu, J.; Lou, Y.; Qian, F.; Chen, T.; Zhang, L.; Yang, J.; Zhong, H.; et al. Multimodal fusion of liquid biopsy and CT enhances differential diagnosis of early-stage lung adenocarcinoma. NPJ Precis. Oncol. 2024, 8, 50. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lafata, K.J.; Corradetti, M.N.; Gao, J.; Jacobs, C.D.; Weng, J.; Chang, Y.; Wang, C.; Hatch, A.; Xanthopoulos, E.; Jones, G.; et al. Radiogenomic Analysis of Locally Advanced Lung Cancer Based on CT Imaging and Intratreatment Changes in Cell-Free DNA. Radiol. Imaging Cancer 2021, 3, e200157. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yousefi, B.; LaRiviere, M.J.; Cohen, E.A.; Buckingham, T.H.; Yee, S.S.; Black, T.A.; Chien, A.L.; Noël, P.; Hwang, W.T.; Katz, S.I.; et al. Combining radiomic phenotypes of non-small cell lung cancer with liquid biopsy data may improve prediction of response to EGFR inhibitors. Sci. Rep. 2021, 11, 9984. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gevaert, O.; Xu, J.; Hoang, C.D.; Leung, A.N.; Xu, Y.; Quon, A.; Rubin, D.L.; Napel, S.; Plevritis, S.K. Non-small cell lung cancer: Identifying prognostic imaging biomarkers by leveraging public gene expression microarray data–methods and preliminary results. Radiology 2012, 264, 387–396. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Čeri, A.; Somborac-Bačura, A.; Fabijanec, M.; Hulina-Tomašković, A.; Matusina, M.; Detel, D.; Verbanac, D.; Barišić, K. Establishment of liquid biopsy procedure for the analysis of circulating cell free DNA, exosomes, RNA and proteins in colorectal cancer and adenoma patients. Sci. Rep. 2024, 14, 26925, Erratum in Sci. Rep. 2024, 14, 28633. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients with Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef] [PubMed]
- Jamal-Hanjani, M.; Wilson, G.A.; McGranahan, N.; Birkbak, N.J.; Watkins, T.B.K.; Veeriah, S.; Shafi, S.; Johnson, D.H.; Mitter, R.; Rosenthal, R.; et al. Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 2017, 376, 2109–2121. [Google Scholar] [CrossRef] [PubMed]
- Orlhac, F.; Boughdad, S.; Philippe, C.; Stalla-Bourdillon, H.; Nioche, C.; Champion, L.; Soussan, M.; Frouin, F.; Frouin, V.; Buvat, I. A Postreconstruction Harmonization Method for Multicenter Radiomic Studies in PET. J. Nucl. Med. 2018, 59, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Hasan, H.E.; Jaber, D.; Khabour, O.F.; Alzoubi, K.H. Ethical considerations and concerns in the implementation of AI in pharmacy practice: A cross-sectional study. BMC Med. Ethics 2024, 25, 55. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lundberg, S.M.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. Adv. Neural Inf. Process. Syst. NeurIPS 2017, 30, 4765–4774. [Google Scholar]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD’16), San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 1135–1144. [Google Scholar] [CrossRef]
- Ghassemi, M.; Oakden-Rayner, L.; Beam, A.L. The false hope of current approaches to explainable artificial intelligence in health care. Lancet Digit. Health 2021, 3, e745–e750. [Google Scholar] [CrossRef]
- Ronquillo, C.E.; Peltonen, L.M.; Pruinelli, L.; Chu, C.H.; Bakken, S.; Beduschi, A.; Cato, K.; Hardiker, N.; Junger, A.; Michalowski, M.; et al. Artificial intelligence in nursing: Priorities and opportunities from an international invitational think-tank of the Nursing and Artificial Intelligence Leadership Collaborative. J. Adv. Nurs. 2021, 77, 3707–3717. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Abbas, Q.; Jeong, W.; Lee, S.W. Explainable AI in Clinical Decision Support Systems: A Meta-Analysis of Methods, Applications, and Usability Challenges. Healthcare 2025, 13, 2154. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ucci, A.; Rucci, N.; Ponzetti, M. Liquid biopsies in primary and secondary bone cancers. Cancer Drug Resist. 2022, 5, 541–559. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Godsey, J.H.; Silvestro, A.; Barrett, J.C.; Bramlett, K.; Chudova, D.; Deras, I.; Dickey, J.; Hicks, J.; Johann, D.J.; Leary, R.; et al. Generic Protocols for the Analytical Validation of Next-Generation Sequencing-Based ctDNA Assays: A Joint Consensus Recommendation of the BloodPAC’s Analytical Variables Working Group. Clin. Chem. 2020, 66, 1156–1166. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Krebs, M.G.; Malapelle, U.; André, F.; Paz-Ares, L.; Schuler, M.; Thomas, D.M.; Vainer, G.; Yoshino, T.; Rolfo, C. Practical Considerations for the Use of Circulating Tumor DNA in the Treatment of Patients with Cancer: A Narrative Review. JAMA Oncol. 2022, 8, 1830–1839. [Google Scholar] [CrossRef] [PubMed]
- Chartrand, G.; Cheng, P.M.; Vorontsov, E.; Drozdzal, M.; Turcotte, S.; Pal, C.J.; Kadoury, S.; Tang, A. Deep Learning: A Primer for Radiologists. Radiographics 2017, 37, 2113–2131. [Google Scholar] [CrossRef] [PubMed]
- Loken, E.; Gelman, A. Measurement error and the replication crisis. Science 2017, 355, 584–585. [Google Scholar] [CrossRef] [PubMed]
- Eledkawy, A.; Hamza, T.; El-Metwally, S. Precision cancer classification using liquid biopsy and advanced machine learning techniques. Sci. Rep. 2024, 14, 5841. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pavithran, S.; Chakrabortty, S. Lung cancer risk prediction using augmented machine learning pipelines with explainable AI. Front. Artif. Intell. 2025, 8, 1602775. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Sanap, G.; Shenoy, S.; Kalyane, D.; Kalia, K.; Tekade, R.K. Artificial intelligence in drug discovery and development. Drug Discov. Today 2021, 26, 80–93. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Said, R.; Guibert, N.; Oxnard, G.R.; Tsimberidou, A.M. Circulating tumor DNA analysis in the era of precision oncology. Oncotarget 2020, 11, 188–211. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Quanyang, W.; Yao, H.; Sicong, W.; Linlin, Q.; Zewei, Z.; Donghui, H.; Hongjia, L.; Shijun, Z. Artificial intelligence in lung cancer screening: Detection, classification, prediction, and prognosis. Cancer Med. 2024, 13, e7140. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhong, F.; Liu, Z.; Wang, B.; An, W.; Zhang, H.; Liao, M.; Tian, S. A Predictive Model to Differentiate Between Second Primary Lung Cancers and Pulmonary Metastasis. Acad. Radiol. 2022, 29 (Suppl. S2), S137–S144. [Google Scholar] [CrossRef] [PubMed]
| Subtype | Dominant Transcription Factor | Neuroendocrine Features |
|---|---|---|
| SCLC-A | ASCL1 | High [25] |
| SCLC-N | NEUROD1 | Intermediate [25] |
| SCLC-P | POU2F3 | Non-neuroendocrine [25] |
| SCLC-Y | YAP1 | Associated with Chemoresistance [25] |
| Mutation/Target | Pathway | Clinical Implication |
|---|---|---|
| KRAS (G12C/D/V) | RAS–RAF–MEK–ERK | Risk factor associated with tobacco exposure [27] |
| BRAF V600E | MAPK pathway | Predictive biomarker; targetable with BRAF/MEK inhibitors [27] |
| HER3 | PI3K signaling via dimerization | Predictive biomarker; linked to resistance to EGFR inhibitors [17] |
| Nectin-4, ITGB6 | Adhesion, immune suppression | Prognostic biomarker; promotes migration and immune evasion [17] |
| FRα | JAK/STAT3 and ERK | Predictive biomarker; enhances proliferation [17] |
| PRMT5 | Epigenetic silencing | Therapeutic target; especially active in MTAP-deleted tumors [28] |
| S. No. | Biomarkers | Isolation Methods | Detection Methods | Uses |
|---|---|---|---|---|
| 1. | Circulating tumor cells (CTCs) | CellSearch system (EpCAM-based) [59,60] | Immunocytochemistry, FISH [60] | metastasis |
| 2. | Circulating tumor DNA (ctDNA) | DNA extraction (Qiagen Circulating Nucleic Acid Kit) [61] | Droplet digital, PCR, NGS [47,62] | detecting mutations, prognostic |
| 3. | Extracellular vesicles (EV) | Precipitation (miRCURY), Size-exclusion chromatography (qEV, Exo-spin) [63] | Quantitative PCR, NGS [63] | metastasis |
| 4. | MicroRNAs (miRNAs) | Phenol-chloroform extraction (TRIzol method) [64] | Quantitative reverse transcription PCR | prognostic |
| 5. | DNA methylation markers | Bisulphite conversion, The Infinium HumanMethylation450 BeadChip array, The HumanMethylationEPIC BeadChip [55,65]. | Methylation-specific PCR | screening, diagnostic |
| 6. | Tumor-educated platelets (TEPs) | Slow and fast centrifugation | ThromboSeq [66] | Emerging biomarker |
| Year & Author | Biomarker/Target | Method/AI Approach | Validation Cohort Characteristics | Clinical Application | Results & Limitations |
|---|---|---|---|---|---|
| 2024, Bie et al. [131] | cfDNA methylation | cfDNA + ML (XGBoost, SVM) | 196 pts (96 LC, 100 HC); external 142 samples; train/valid split | Early NSCLC detection | High accuracy incl. early stage; benign specificity underexplored; multi-center validation needed |
| 2020, Hoshino et al. [101] | EV proteins | Plasma EV + ML signature | 497 EVP samples; 426 human; 152 control, 274 cancer across fluids | Pan-cancer incl. lung | 13-protein EV panel; high sensitivity/specificity; lung-specific metrics lacking |
| 2023, Asleh et al. [132] | PD-L1 in NSCLC | CT radiomics + Logistic Regression | Retrospective imaging dataset; no ext. validation | PD-L1 prediction | Accurate vs. radiologists; retrospective only |
| 2023, Wu et al. [133] | ncRNAs in NSCLC | Orion AI (deep learning) | 59 NSCLC pts, 97 plasma samples, 3 timepoints, China | Early detection & subtyping | Sens 94%, Spec 87%; prospective validation needed |
| 2023, Yolchuyeva et al. [134] | EGFR, KRAS, ALK, BRAF | cfDNA + AI classifier | 385 NSCLC pts; stratified by gender, age, smoking, ECOG, PD-L1, PFS | Mutation detection | High concordance with tissue; rare mutations less reliable |
| 2024, Karimzadeh et al. [135] | cfDNA fragments + mutations | Fragmentomics + UMI-NGS + ML ensemble | 1050 (419 NSCLC, 631 control); 80% training | Early detection & subtyping | High specificity; reduced false positives; requires deep seq/infra |
| 2024, Purohit et al. [139] | Indeterminate nodules | LDCT CNN reanalysis | Retrospective LDCT dataset; no external validation | Early lung CA diagnosis | Sens 92%, Spec 87%; ext. validation pending |
| 2021, Mathios et al. [136] | cfDNA methylation | Bisulfite seq + ML | 365 at-risk indiv.; external validation 385 controls + 46 LC pts | Early NSCLC detection | High sensitivity; distinguishes benign vs. cancer |
| 2022, Bahado-Singh et al. [137] | cfDNA methylation | Multi-ML ensemble | 10 cases vs. 20 controls, all Caucasian; 10-fold cross-validation | Risk prediction | Improved early detection; retrospective; small |
| 2023, Wang et al. [138] | cfDNA methylation (LunaCAM) | ML classifier + LDCT | Discovery 429, training 513, validation 172 | Risk stratification | Improved specificity; real-world validation needed |
| 2025, Ji et al. [140] | Early-stage lung CA | AI-augmented LDCT-TRAI | 259,121 screened; 87,260 positive; 728 LC dx (634 non-smokers) | Detection & outcomes | Enhanced Stage I detection, survival benefit; multi-site perf. unclear |
| 2021, Liang et al. [130] | ctDNA in lung cancer | NGS profiling | Training/validation details not reported | Tumor burden & relapse | ctDNA decline = response; early rise = progression; subtype coverage lacking |
| 2020, Hellmann et al. [146] | ctDNA in NSCLC | NGS + ML risk model | 31 advanced NSCLC pts; no external validation | ICI response prediction | ctDNA clearance = durable benefit; retrospective only |
| 2020, Giroux et al. [143] | cfDNA methylation | Targeted profiling + ML | 79 NSCLC pts; no separate validation set | Immunotherapy monitoring | Profiles distinguished responders early; small sample |
| 2020, Jee et al. [144] | cfDNA methylation | cfMeDIP-seq + ML | 1127 NSCLC pts; no external validation subset | MRD & relapse detection | Sens ~85%, Spec ~95%; lung-specific validation absent |
| 2019, Chaudhuri et al. [145] | ctDNA (lung & colorectal) | Deep targeted seq + ML | 40 lung CA pts + 54 HC; internal validation only | Response & relapse | 93% concordance w/imaging; relapse detected ~5 mo earlier |
| 2020, Chabon et al. [147] | ctDNA (resected NSCLC) | Tumor-informed NGS | Validation details not reported | MRD & relapse | Relapse detected ~5 mo pre-imaging; requires tumor tissue |
| 2024, Sujit et al. [148] | cfDNA fragmentation | Fragmentomics + ML | 394 pts: discovery 199, validation 195; external cohort | Early NSCLC detection | Acc 93% (AUC = 0.93); incl. small tumors; specificity varies |
| 2021, Widman et al. [149] | ctDNA in NSCLC | Serial NGS ctDNA | Validation details not reported | ICI monitoring | ctDNA decline = better survival; early rise = relapse |
| 2021, Helzer et al. [150] | ctDNA + CNVs | Whole-genome cfDNA + ML | UW (n = 320), GRAIL (n = 198); train/valid split | Broad monitoring | Captured burden & resistance pre-imaging; costly/complex |
| 2023, Assaf et al. [151] | cfDNA + proteins | Multi-analyte ML blood test | 1954 samples; 466 NSCLC pts; ext. validation in 73 OAK trial pts | Early lung CA detection | Sens 89%, Spec 99%; better than single markers; validation pending |
| 2017, Abbosh et al. [152] | ctDNA mutations | Phylogenetic tracking | Validation details not reported | Post-op relapse detection | Relapse ID up to 11 mo earlier; resource-intensive |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cherukuri, S.P.; Kaur, A.; Goyal, B.; Kukunoor, H.R.; Sahito, A.F.; Sachdeva, P.; Yerrapragada, G.; Elangovan, P.; Shariff, M.N.; Natarajan, T.; et al. Artificial Intelligence-Enhanced Liquid Biopsy and Radiomics in Early-Stage Lung Cancer Detection: A Precision Oncology Paradigm. Cancers 2025, 17, 3165. https://doi.org/10.3390/cancers17193165
Cherukuri SP, Kaur A, Goyal B, Kukunoor HR, Sahito AF, Sachdeva P, Yerrapragada G, Elangovan P, Shariff MN, Natarajan T, et al. Artificial Intelligence-Enhanced Liquid Biopsy and Radiomics in Early-Stage Lung Cancer Detection: A Precision Oncology Paradigm. Cancers. 2025; 17(19):3165. https://doi.org/10.3390/cancers17193165
Chicago/Turabian StyleCherukuri, Swathi Priya, Anmolpreet Kaur, Bipasha Goyal, Hanisha Reddy Kukunoor, Areesh Fatima Sahito, Pratyush Sachdeva, Gayathri Yerrapragada, Poonguzhali Elangovan, Mohammed Naveed Shariff, Thangeswaran Natarajan, and et al. 2025. "Artificial Intelligence-Enhanced Liquid Biopsy and Radiomics in Early-Stage Lung Cancer Detection: A Precision Oncology Paradigm" Cancers 17, no. 19: 3165. https://doi.org/10.3390/cancers17193165
APA StyleCherukuri, S. P., Kaur, A., Goyal, B., Kukunoor, H. R., Sahito, A. F., Sachdeva, P., Yerrapragada, G., Elangovan, P., Shariff, M. N., Natarajan, T., Janarthanan, J., Richard, S., Pallikaranai Venkatesaprasath, S., Karuppiah, S. S., Iyer, V. N., Helgeson, S. A., & Arunachalam, S. P. (2025). Artificial Intelligence-Enhanced Liquid Biopsy and Radiomics in Early-Stage Lung Cancer Detection: A Precision Oncology Paradigm. Cancers, 17(19), 3165. https://doi.org/10.3390/cancers17193165





