Radiomics-Driven Tumor Prognosis Prediction Across Imaging Modalities: Advances in Sampling, Feature Selection, and Multi-Omics Integration
Simple Summary
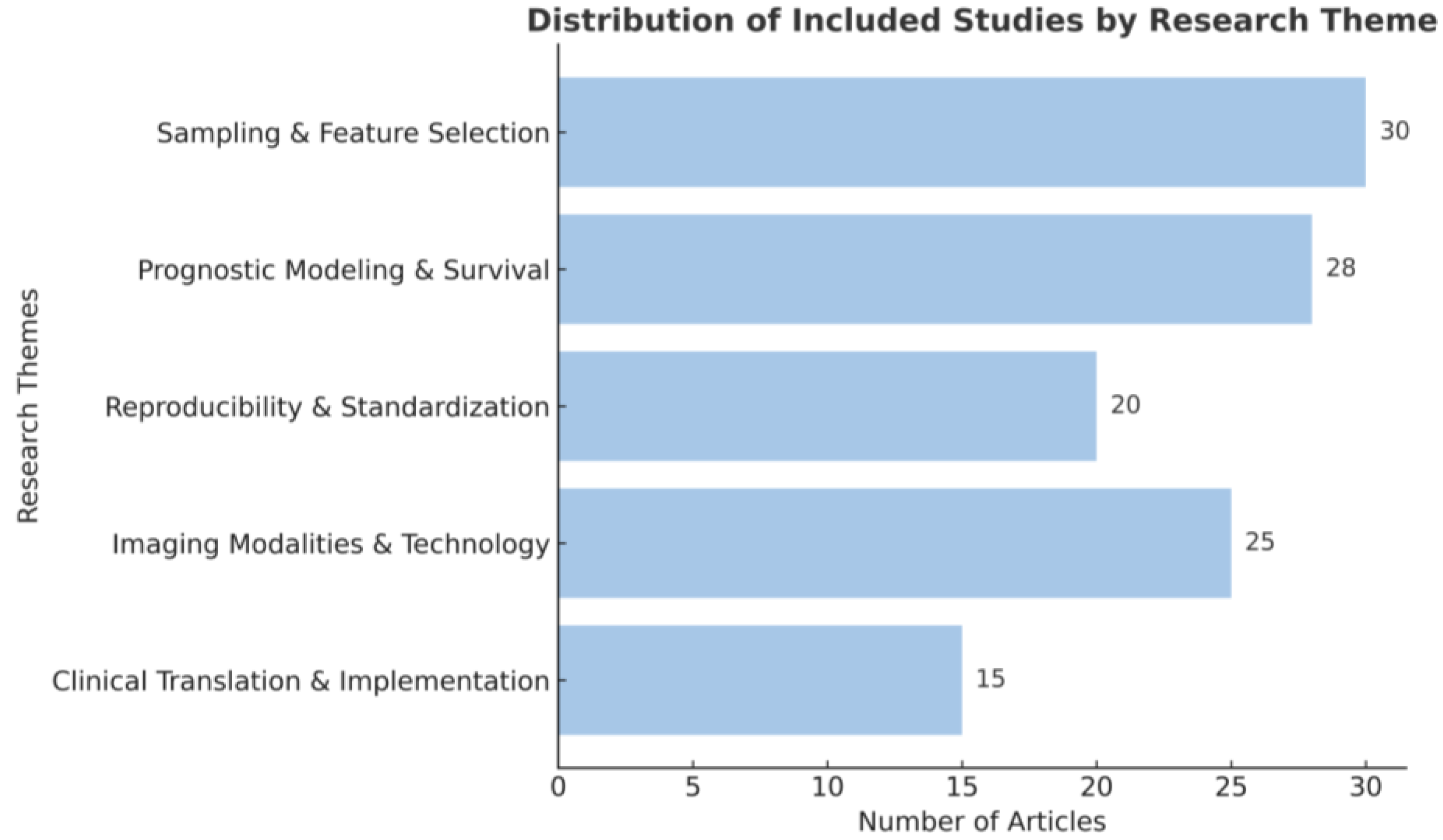
Abstract
1. Introduction
2. Overview of Radiomics in Tumor Prognosis Prediction
3. Barriers to Reproducibility and Feature Stability in Radiomics
4. Radiomics Sampling Methods
5. Recent Advances in Radiomics Feature Selection Methods
6. Application of Radiomics Across Different Imaging Modalities
7. Emerging Trends and Techniques
8. Clinical Translation
9. Challenges and Future Directions
9.1. Reproducibility and Imaging Variability
9.2. Lack of Standardization in Feature Selection Methods
9.3. Limited Prospective Validation in Clinical Settings
9.4. Underexplored Potential of Ensemble and Multi-Omics Integration
9.5. Regulatory Approval and Clinician Acceptance
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CT | Computed tomography |
| MRI | Magnetic resonance imaging |
| PET | Positron emission tomography |
| ROI | Region of interest |
| ANOVA | Analysis of variance |
| LASSO | Least absolute shrinkage and selection operator |
| mRMR | Minimum redundancy maximum relevance |
| NSCLC | Non-small-cell lung cancer |
| HCC | Hepatocellular carcinoma |
| TACE | Transarterial chemoembolization |
| nnU-Net | No-new-net |
| RFE | Recursive Feature Elimination |
| mRMRMSRC | Minimum Redundancy, Maximum Relevance and Maximum Sparse Representation |
| AUC | Area under the curve |
| MO-FS | Multi-objective Feature Selection |
| Coe-Thr-Lasso | Lasso coefficient thresholds |
| 3DCRT | Three-dimensional conformal radiation therapy |
| pCR | Pathological complete response |
| FDG | Fluorodeoxyglucose |
| AI | Artificial intelligence |
| KNN | K-nearest neighbor |
| SVM | Support vector machine |
| IC | Induction chemotherapy |
| IBSI | Image Biomarker Standardization Initiative |
References
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Roelofs, E.; Dekker, A.; Meldolesi, E.; van Stiphout, R.; Valentini, V.; Lambin, P. International data-sharing for radiotherapy research: An open-source based infrastructure for multicentric clinical data mining. Radiother. Oncol. 2014, 110, 370–374. [Google Scholar] [CrossRef]
- Miotto, R.; Li, L.; Kidd, B.A.; Dudley, J.T. Deep Patient: An Unsupervised Representation to Predict the Future of Patients from the Electronic Health Records. Sci. Rep. 2016, 6, 26094. [Google Scholar] [CrossRef]
- Nead, K.T.; Gaskin, G.; Chester, C.; Swisher-McClure, S.; Dudley, J.T.; Leeper, N.J.; Shah, N.H. Androgen Deprivation Therapy and Future Alzheimer’s Disease Risk. J. Clin. Oncol. 2016, 34, 566–571. [Google Scholar] [CrossRef]
- Jiang, M.; Li, C.L.; Luo, X.M.; Chuan, Z.R.; Lv, W.Z.; Li, X.; Cui, X.W.; Dietrich, C.F. Ultrasound-based deep learning radiomics in the assessment of pathological complete response to neoadjuvant chemotherapy in locally advanced breast cancer. Eur. J. Cancer 2021, 147, 95–105. [Google Scholar] [CrossRef]
- Collarino, A.; Feudo, V.; Pasciuto, T.; Florit, A.; Pfaehler, E.; de Summa, M.; Bizzarri, N.; Annunziata, S.; Zannoni, G.F.; de Geus-Oei, L.F.; et al. Is PET Radiomics Useful to Predict Pathologic Tumor Response and Prognosis in Locally Advanced Cervical Cancer? J. Nucl. Med. 2024, 65, 962–970. [Google Scholar] [CrossRef] [PubMed]
- Deng, K.; Chen, T.; Leng, Z.; Yang, F.; Lu, T.; Cao, J.; Pan, W.; Zheng, Y. Radiomics as a tool for prognostic prediction in transarterial chemoembolization for hepatocellular carcinoma: A systematic review and meta-analysis. Radiol. Medica 2024, 129, 1099–1117. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Grove, O.; Gillies, R.J. Quantitative imaging in cancer evolution and ecology. Radiology 2013, 269, 8–15. [Google Scholar] [CrossRef]
- Habibi, M.A.; Rashidi, F.; Habibzadeh, A.; Mehrtabar, E.; Arshadi, M.R.; Mirjani, M.S. Prediction of the treatment response and local failure of patients with brain metastasis treated with stereotactic radiosurgery using machine learning: A systematic review and meta-analysis. Neurosurg. Rev. 2024, 47, 199. [Google Scholar] [CrossRef] [PubMed]
- Vitorino, R. Transforming Clinical Research: The Power of High-Throughput Omics Integration. Proteomes 2024, 12, 25. [Google Scholar] [CrossRef]
- Chaddad, A.; Peng, J.; Xu, J.; Bouridane, A. Survey of Explainable AI Techniques in Healthcare. Sensors 2023, 23, 634. [Google Scholar] [CrossRef]
- Zhang, Y.P.; Zhang, X.Y.; Cheng, Y.T.; Li, B.; Teng, X.Z.; Zhang, J.; Lam, S.; Zhou, T.; Ma, Z.R.; Sheng, J.B.; et al. Artificial intelligence-driven radiomics study in cancer: The role of feature engineering and modeling. Mil. Med. Res. 2023, 10, 22. [Google Scholar] [CrossRef]
- Dercle, L.; Zhao, B.; Gonen, M.; Moskowitz, C.S.; Connors, D.E.; Yang, H.; Lu, L.; Reidy-Lagunes, D.; Fojo, T.; Karovic, S.; et al. An imaging signature to predict outcome in metastatic colorectal cancer using routine computed tomography scans. Eur. J. Cancer 2022, 161, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Avanzo, M.; Wei, L.; Stancanello, J.; Vallieres, M.; Rao, A.; Morin, O.; Mattonen, S.A.; El Naqa, I. Machine and deep learning methods for radiomics. Med. Phys. 2020, 47, e185–e202. [Google Scholar] [CrossRef]
- Camps, J.; Jimenez-Franco, A.; Garcia-Pablo, R.; Joven, J.; Arenas, M. Artificial intelligence-driven integration of multi-omics and radiomics: A new hope for precision cancer diagnosis and prognosis. Biochim. Biophys. Acta-Mol. Basis Dis. 2025, 1871, 167841. [Google Scholar] [CrossRef]
- Conti, A.; Duggento, A.; Indovina, I.; Guerrisi, M.; Toschi, N. Radiomics in breast cancer classification and prediction. Semin. Cancer Biol. 2021, 72, 238–250. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, Z.; He, L.; Chen, X.; Pan, D.; Ma, Z.; Liang, C.; Tian, J.; Liang, C. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology 2016, 281, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; She, Y.; Yang, Y.; Liu, X.; Chen, S.; Zhong, Y.; Deng, J.; Zhao, M.; Sun, X.; Xie, D.; et al. Radiomics for Survival Risk Stratification of Clinical and Pathologic Stage IA Pure-Solid Non-Small Cell Lung Cancer. Radiology 2022, 302, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Kickingereder, P.; Burth, S.; Wick, A.; Götz, M.; Eidel, O.; Schlemmer, H.P.; Maier-Hein, K.H.; Wick, W.; Bendszus, M.; Radbruch, A.; et al. Radiomic Profiling of Glioblastoma: Identifying an Imaging Predictor of Patient Survival with Improved Performance over Established Clinical and Radiologic Risk Models. Radiology 2016, 280, 880–889. [Google Scholar] [CrossRef]
- Zhang, Y.D.; Wang, J.; Wu, C.J.; Bao, M.L.; Li, H.; Wang, X.N.; Tao, J.; Shi, H.B. An imaging-based approach predicts clinical outcomes in prostate cancer through a novel support vector machine classification. Oncotarget 2016, 7, 78140–78151. [Google Scholar] [CrossRef]
- Negreros-Osuna, A.A.; Parakh, A.; Corcoran, R.B.; Pourvaziri, A.; Kambadakone, A.; Ryan, D.P.; Sahani, D.V. Radiomics Texture Features in Advanced Colorectal Cancer: Correlation with BRAF Mutation and 5-year Overall Survival. Radiol. Imaging Cancer 2020, 2, e190084. [Google Scholar] [CrossRef]
- Kong, C.; Zhao, Z.; Chen, W.; Lv, X.; Shu, G.; Ye, M.; Song, J.; Ying, X.; Weng, Q.; Weng, W.; et al. Prediction of tumor response via a pretreatment MRI radiomics-based nomogram in HCC treated with TACE. Eur. Radiol. 2021, 31, 7500–7511. [Google Scholar] [CrossRef]
- Li, L.; Kan, X.; Zhao, Y.; Liang, B.; Ye, T.; Yang, L.; Zheng, C. Radiomics Signature: A potential biomarker for the prediction of survival in Advanced Hepatocellular Carcinoma. Int. J. Med. Sci. 2021, 18, 2276–2284. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Dong, S.Y.; Bai, X.L.; Song, T.Q.; Zhang, B.H.; Zhou, L.D.; Chen, Y.J.; Zeng, Z.M.; Wang, K.; Zhao, H.T.; et al. Tumor Radiomic Features on Pretreatment MRI to Predict Response to Lenvatinib plus an Anti-PD-1 Antibody in Advanced Hepatocellular Carcinoma: A Multicenter Study. Liver Cancer 2023, 12, 262–276. [Google Scholar] [CrossRef]
- Park, J.E.; Park, S.Y.; Kim, H.J.; Kim, H.S. Reproducibility and generalizability in radiomics modeling: Possible strategies in radiologic and statistical perspectives. Korean J. Radiol. 2019, 20, 1124–1137. [Google Scholar] [CrossRef]
- Zhao, B.; Tan, Y.; Tsai, W.-Y.; Qi, J.; Xie, C.; Lu, L.; Schwartz, L.H. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci. Rep. 2016, 6, 23428. [Google Scholar] [CrossRef]
- Pfaehler, E.; Zhovannik, I.; Wei, L.; Boellaard, R.; Dekker, A.; Monshouwer, R.; El Naqa, I.; Bussink, J.; Gillies, R.; Wee, L.; et al. A systematic review and quality of reporting checklist for repeatability and reproducibility of radiomic features. Phys. Imaging Radiat. Oncol. 2021, 20, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Fiset, S.; Welch, M.L.; Weiss, J.; Pintilie, M.; Conway, J.L.; Milosevic, M.; Fyles, A.; Traverso, A.; Jaffray, D.; Metser, U.; et al. Repeatability and reproducibility of MRI-based radiomic features in cervical cancer. Radiother. Oncol. 2019, 135, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Scalco, E.; Belfatto, A.; Mastropietro, A.; Rancati, T.; Avuzzi, B.; Messina, A.; Valdagni, R.; Rizzo, G. T2w-MRI signal normalization affects radiomics features reproducibility. Med. Phys. 2020, 47, 1680–1691. [Google Scholar] [CrossRef]
- Schwier, M.; van Griethuysen, J.; Vangel, M.G.; Pieper, S.; Peled, S.; Tempany, C.; Aerts, H.J.W.L.; Kikinis, R.; Fennessy, F.M.; Fedorov, A. Repeatability of Multiparametric Prostate MRI Radiomics Features. Sci. Rep. 2019, 9, 9441. [Google Scholar] [CrossRef]
- Laajili, R.; Said, M.; Tagina, M. Application of radiomics features selection and classification algorithms for medical imaging decision: MRI radiomics breast cancer cases study. Inform. Med. Unlocked 2021, 27, 100801. [Google Scholar] [CrossRef]
- Demircioğlu, A. Measuring the bias of incorrect application of feature selection when using cross-validation in radiomics. Insights Into Imaging 2021, 12, 172. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Lu, L.; Dercle, L.; Lichtenstein, P.; Li, Y.; Yin, Q.; Zong, M.; Schwartz, L.; Zhao, B. Interobserver variability in tumor contouring affects the use of radiomics to predict mutational status. J. Med. Imaging 2018, 5, 011005. [Google Scholar] [CrossRef]
- Park, J.; Joo, I.; Jeon, S.K.; Kim, J.M.; Park, S.J.; Yoon, S.H. Automated abdominal organ segmentation algorithms for non-enhanced CT for volumetry and 3D radiomics analysis. Abdom. Radiol. 2025, 50, 1448–1456. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, H.; Lin, F.; Zhang, R.; Ma, H.; Shi, Y.; Yang, P.; Zhang, K.; Zhao, F.; Mao, N.; et al. Intra- and Peritumoral Radiomics of Contrast-Enhanced Mammography Predicts Axillary Lymph Node Metastasis in Patients With Breast Cancer: A Multicenter Study. Acad. Radiol. 2023, 30 (Suppl. S2), S133–S142. [Google Scholar] [CrossRef]
- Khorrami, M.; Prasanna, P.; Gupta, A.; Patil, P.; Velu, P.D.; Thawani, R.; Corredor, G.; Alilou, M.; Bera, K.; Fu, P.; et al. Changes in CT Radiomic Features Associated with Lymphocyte Distribution Predict Overall Survival and Response to Immunotherapy in Non-Small Cell Lung Cancer. Cancer Immunol. Res. 2020, 8, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Zedda, L.; Loddo, A.; Di Ruberto, C. Advancements in radiomics: A comprehensive survey of feature types and their correlation on modalities and regions. Neurocomputing 2025, 656, 131192. [Google Scholar] [CrossRef]
- Wilson, A.; Anwar, M.R. The future of adaptive machine learning algorithms in high-dimensional data processing. Int. Trans. Artif. Intell. 2024, 3, 97–107. [Google Scholar] [CrossRef]
- Cè, M.; Chiriac, M.D.; Cozzi, A.; Macrì, L.; Rabaiotti, F.L.; Irmici, G.; Fazzini, D.; Carrafiello, G.; Cellina, M. Decoding Radiomics: A Step-by-Step Guide to Machine Learning Workflow in Hand-Crafted and Deep Learning Radiomics Studies. Diagnostics 2024, 14, 2473. [Google Scholar] [CrossRef]
- Papanikolaou, N.; Matos, C.; Koh, D.M. How to develop a meaningful radiomic signature for clinical use in oncologic patients. Cancer Imaging 2020, 20, 33. [Google Scholar] [CrossRef]
- Horvat, N.; Papanikolaou, N.; Koh, D.-M. Radiomics Beyond the Hype: A Critical Evaluation Toward Oncologic Clinical Use. Radiol. Artif. Intell. 2024, 6, e230437. [Google Scholar] [CrossRef] [PubMed]
- Gidwani, M.; Chang, K.; Patel, J.B.; Hoebel, K.V.; Ahmed, S.R.; Singh, P.; Fuller, C.D.; Kalpathy-Cramer, J. Inconsistent Partitioning and Unproductive Feature Associations Yield Idealized Radiomic Models. Radiology 2023, 307, e220715. [Google Scholar] [CrossRef]
- Perniciano, A.; Loddo, A.; Di Ruberto, C.; Pes, B. Insights into radiomics: Impact of feature selection and classification. Multimed. Tools Appl. 2024, 84, 31695–31721. [Google Scholar] [CrossRef]
- Ligero, M.; Torres, G.; Sanchez, C.; Diaz-Chito, K.; Perez, R.; Gil, D. Selection of Radiomics Features based on their Reproducibility. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 403–408. [Google Scholar]
- Sun, P.; Wang, D.; Mok, V.C.; Shi, L. Comparison of Feature Selection Methods and Machine Learning Classifiers for Radiomics Analysis in Glioma Grading. IEEE Access 2019, 7, 102010–102020. [Google Scholar] [CrossRef]
- Demircioğlu, A. Benchmarking Feature Selection Methods in Radiomics. Investig. Radiol. 2022, 57, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Li, S.; Qin, G.; Folkert, M.; Jiang, S.; Wang, J. Multi-Objective-Based Radiomic Feature Selection for Lesion Malignancy Classification. IEEE J. Biomed. Health Inform. 2020, 24, 194–204. [Google Scholar] [CrossRef]
- Wang, K.; An, Y.; Zhou, J.; Long, Y.; Chen, X. A novel Multi-Level feature selection method for radiomics. Alex. Eng. J. 2023, 66, 993–999. [Google Scholar] [CrossRef]
- Liu, T.; Wu, G.; Yu, J.; Guo, Y.; Wang, Y.; Shi, Z.; Chen, L. A mRMRMSRC feature selection method for radiomics approach. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju Island, Republic of Korea, 11–15 July 2017; pp. 616–619. [Google Scholar]
- Parmar, C.; Grossmann, P.; Bussink, J.; Lambin, P.; Aerts, H.J.W.L. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci. Rep. 2015, 5, 13087. [Google Scholar] [CrossRef]
- Mishra, P.; Singh, U.; Pandey, C.M.; Mishra, P.; Pandey, G. Application of student’s t-test, analysis of variance, and covariance. Ann. Card. Anaesth. 2019, 22, 407–411. [Google Scholar] [CrossRef]
- McHugh, M.L. Multiple comparison analysis testing in ANOVA. Biochem. Med. 2011, 21, 203–209. [Google Scholar] [CrossRef]
- Barone, M.; Losurdo, G.; Iannone, A.; Leandro, G.; Di Leo, A.; Trerotoli, P. Assessment of body composition: Intrinsic methodological limitations and statistical pitfalls. Nutrition 2022, 102, 111736. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, H.; Wang, J.; Pu, Y.; Pal, N.R. Feature Selection Using a Neural Network With Group Lasso Regularization and Controlled Redundancy. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 1110–1123. [Google Scholar] [CrossRef]
- Li, Z.; Sillanpaa, M.J. Overview of LASSO-related penalized regression methods for quantitative trait mapping and genomic selection. Theor. Appl. Genet. 2012, 125, 419–435. [Google Scholar] [CrossRef]
- Jeon, H.; Oh, S. Hybrid-Recursive Feature Elimination for Efficient Feature Selection. Appl. Sci. 2020, 10, 3211. [Google Scholar] [CrossRef]
- Ding, Y.; Wilkins, D. Improving the performance of SVM-RFE to select genes in microarray data. BMC Bioinform. 2006, 7 (Suppl. S2), S12. [Google Scholar] [CrossRef]
- Mylona, E.; Zaridis, D.I.; Kalantzopoulos, C.; Tachos, N.S.; Regge, D.; Papanikolaou, N.; Tsiknakis, M.; Marias, K.; Pro, C.-I.C.; Fotiadis, D.I. Optimizing radiomics for prostate cancer diagnosis: Feature selection strategies, machine learning classifiers, and MRI sequences. Insights Imaging 2024, 15, 265. [Google Scholar] [CrossRef]
- Ting, W.; Yuanqian, W. Pattern classification with ordered features using mRMR and neural networks. In Proceedings of the 2010 International Conference on Information, Networking and Automation (ICINA), Kunming, China, 18–19 October 2010; pp. V2-128–V2-131. [Google Scholar]
- Fu, Q.; Li, Q.; Li, X.; Wang, H.; Xie, J.; Wang, Q. MOFS-REPLS: A large-scale multi-objective feature selection algorithm based on real-valued encoding and preference leadership strategy. Inf. Sci. 2024, 667, 120483. [Google Scholar] [CrossRef]
- Traverso, A.; Wee, L.; Dekker, A.; Gillies, R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1143–1158. [Google Scholar] [CrossRef]
- Meng, X.-P.; Wang, Y.-C.; Zhou, J.-Y.; Yu, Q.; Lu, C.-Q.; Xia, C.; Tang, T.-Y.; Xu, J.; Sun, K.; Xiao, W.; et al. Comparison of MRI and CT for the Prediction of Microvascular Invasion in Solitary Hepatocellular Carcinoma Based on a Non-Radiomics and Radiomics Method: Which Imaging Modality Is Better? J. Magn. Reson. Imaging 2021, 54, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Zhang, H.; Guo, Y.; Lyu, P.; Yan, J.; Liu, Y.; Liang, P.; Ren, Z.; Gao, J. Comparison of MRI and CT based deep learning radiomics analyses and their combination for diagnosing intrahepatic cholangiocarcinoma. Sci. Rep. 2025, 15, 9629. [Google Scholar] [CrossRef]
- Li, J.; Zhang, H.-l.; Yin, H.-k.; Zhang, H.-k.; Wang, Y.; Xu, S.-n.; Ma, F.; Gao, J.-b.; Li, H.-l.; Qu, J.-r. Comparison of MRI and CT-Based Radiomics and Their Combination for Early Identification of Pathological Response to Neoadjuvant Chemotherapy in Locally Advanced Gastric Cancer. J. Magn. Reson. Imaging 2023, 58, 907–923. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Shi, H.; Lu, M.; Wang, C.; Duan, S.; Xu, Q.; Shi, H. Radiomics Analysis for Predicting Malignant Potential of Intraductal Papillary Mucinous Neoplasms of the Pancreas: Comparison of CT and MRI. Acad. Radiol. 2022, 29, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zheng, J.; Lu, X.; Wang, Y.; Meng, F.; Zhao, J.; Guo, C.; Yu, L.; Zhu, Z.; Zhang, T. Radiomics-based comparison of MRI and CT for differentiating pleomorphic adenomas and Warthin tumors of the parotid gland: A retrospective study. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2021, 131, 591–599. [Google Scholar] [CrossRef]
- Niu, Y.; Yu, X.; Wen, L.; Bi, F.; Jian, L.; Liu, S.; Yang, Y.; Zhang, Y.; Lu, Q. Comparison of preoperative CT- and MRI-based multiparametric radiomics in the prediction of lymph node metastasis in rectal cancer. Front. Oncol. 2023, 13, 1230698. [Google Scholar] [CrossRef]
- Hassaninejad, H.; Abdollahi, H.; Abedi, I.; Amouheidari, A.; Tavakoli, M.B. Radiomics based predictive modeling of rectal toxicity in prostate cancer patients undergoing radiotherapy: CT and MRI comparison. Phys. Eng. Sci. Med. 2023, 46, 1353–1363. [Google Scholar] [CrossRef]
- Liu, H.; Cui, Y.; Chang, C.; Zhou, Z.; Zhang, Y.; Ma, C.; Yin, Y.; Wang, R. Development and validation of a (18)F-FDG PET/CT radiomics nomogram for predicting progression free survival in locally advanced cervical cancer: A retrospective multicenter study. BMC Cancer 2024, 24, 150. [Google Scholar] [CrossRef]
- Hyun, S.H.; Ahn, M.S.; Koh, Y.W.; Lee, S.J. A Machine-Learning Approach Using PET-Based Radiomics to Predict the Histological Subtypes of Lung Cancer. Clin. Nucl. Med. 2019, 44, 956–960. [Google Scholar] [CrossRef]
- Roy, S.; Whitehead, T.D.; Li, S.; Ademuyiwa, F.O.; Wahl, R.L.; Dehdashti, F.; Shoghi, K.I. Co-clinical FDG-PET radiomic signature in predicting response to neoadjuvant chemotherapy in triple-negative breast cancer. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 550–562. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Fang, Q.; Zhang, X.; Hou, P.; Wu, H.; Wang, X. Radiomics analysis of [18F]FDG PET/CT for microvascular invasion and prognosis prediction in very-early- and early-stage hepatocellular carcinoma. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 2599–2614. [Google Scholar] [CrossRef] [PubMed]
- Gómez, O.V.; Herraiz, J.L.; Udías, J.M.; Haug, A.; Papp, L.; Cioni, D.; Neri, E. Analysis of Cross-Combinations of Feature Selection and Machine-Learning Classification Methods Based on [18F]F-FDG PET/CT Radiomic Features for Metabolic Response Prediction of Metastatic Breast Cancer Lesions. Cancers 2022, 14, 2922. [Google Scholar] [CrossRef]
- Ni, B.; Huang, G.; Huang, H.; Wang, T.; Han, X.; Shen, L.; Chen, Y.; Hou, J. Machine Learning Model Based on Optimized Radiomics Feature from 18F-FDG-PET/CT and Clinical Characteristics Predicts Prognosis of Multiple Myeloma: A Preliminary Study. J. Clin. Med. 2023, 12, 2280. [Google Scholar] [CrossRef]
- Hatt, M.; Cheze Le Rest, C.; Antonorsi, N.; Tixier, F.; Tankyevych, O.; Jaouen, V.; Lucia, F.; Bourbonne, V.; Schick, U.; Badic, B.; et al. Radiomics in PET/CT: Current Status and Future AI-Based Evolutions. Semin. Nucl. Med. 2021, 51, 126–133. [Google Scholar] [CrossRef]
- Zhu, F.; Yang, C.; Xia, Y.; Wang, J.; Zou, J.; Zhao, L.; Zhao, Z. CT-based radiomics models may predict the early efficacy of microwave ablation in malignant lung tumors. Cancer Imaging 2023, 23, 60. [Google Scholar] [CrossRef]
- Li, G.; An, C.; Yu, J.; Huang, Q. Radiomics analysis of ultrasonic image predicts sensitive effects of microwave ablation in treatment of patient with benign breast tumors. Biomed. Signal Process. Control. 2022, 76, 103722. [Google Scholar] [CrossRef]
- Ju, C.; Bibaut, A.; van der Laan, M. The Relative Performance of Ensemble Methods with Deep Convolutional Neural Networks for Image Classification. J. Appl. Stat. 2018, 45, 2800–2818. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, S.Y.; Jung, J.Y.; Nam, Y.; Jeon, H.J.; Jung, C.K.; Shin, S.H.; Chung, Y.G. Ensemble learning-based radiomics with multi-sequence magnetic resonance imaging for benign and malignant soft tissue tumor differentiation. PLoS ONE 2023, 18, e0286417. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.H. Greedy Function Approximation: A Gradient Boosting Machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Stodden, V.; Guo, P.; Ma, Z. Toward Reproducible Computational Research: An Empirical Analysis of Data and Code Policy Adoption by Journals. PLoS ONE 2013, 8, e67111. [Google Scholar] [CrossRef]
- Wang, T.; Gong, J.; Duan, H.H.; Wang, L.J.; Ye, X.D.; Nie, S.D. Correlation between CT based radiomics features and gene expression data in non-small cell lung cancer. J. X-Ray Sci. Technol. 2019, 27, 773–803. [Google Scholar] [CrossRef]
- Luan, J.; Zhang, D.; Liu, B.; Yang, A.; Lv, K.; Hu, P.; Yu, H.; Shmuel, A.; Zhang, C.; Ma, G. Exploring the prognostic value and biological pathways of transcriptomics and radiomics patterns in glioblastoma multiforme. Heliyon 2024, 10, e33760. [Google Scholar] [CrossRef]
- Chen, W.; Qiao, X.; Yin, S.; Zhang, X.; Xu, X. Integrating Radiomics with Genomics for Non-Small Cell Lung Cancer Survival Analysis. J. Oncol. 2022, 2022, 5131170. [Google Scholar] [CrossRef] [PubMed]
- Bernatowicz, K.; Grussu, F.; Ligero, M.; Garcia, A.; Delgado, E.; Perez-Lopez, R. Robust imaging habitat computation using voxel-wise radiomics features. Sci. Rep. 2021, 11, 20133. [Google Scholar] [CrossRef]
- Shafiq-Ul-Hassan, M.; Zhang, G.G.; Latifi, K.; Ullah, G.; Hunt, D.C.; Balagurunathan, Y.; Abdalah, M.A.; Schabath, M.B.; Goldgof, D.G.; Mackin, D.; et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med. Phys. 2017, 44, 1050–1062. [Google Scholar] [CrossRef]
- Lim, H.; Kim, D.H.; Jung, H.Y.; Gong, E.J.; Na, H.K.; Ahn, J.Y.; Kim, M.Y.; Lee, J.H.; Choi, K.S.; Choi, K.D.; et al. Clinical significance of early detection of esophageal cancer in patients with head and neck cancer. Gut Liver 2015, 9, 159–165. [Google Scholar] [CrossRef]
- Mostafa, G.; Mahmoud, H.; Abd El-Hafeez, T.; ElAraby, M.E. Feature reduction for hepatocellular carcinoma prediction using machine learning algorithms. J. Big Data 2024, 11, 88. [Google Scholar] [CrossRef]
- Bibault, J.E.; Giraud, P.; Housset, M.; Durdux, C.; Taieb, J.; Berger, A.; Coriat, R.; Chaussade, S.; Dousset, B.; Nordlinger, B.; et al. Deep Learning and Radiomics predict complete response after neo-adjuvant chemoradiation for locally advanced rectal cancer. Sci. Rep. 2018, 8, 12611. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, Z.; Luo, T.; Yan, M.; Dekker, A.; De Ruysscher, D.; Traverso, A.; Wee, L.; Zhao, L. Computed tomography and radiation dose images-based deep-learning model for predicting radiation pneumonitis in lung cancer patients after radiation therapy. Radiother. Oncol. 2023, 182, 109581. [Google Scholar] [CrossRef] [PubMed]
- Torrents-Barrena, J.; Monill, N.; Piella, G.; Gratacos, E.; Eixarch, E.; Ceresa, M.; Gonzalez Ballester, M.A. Assessment of Radiomics and Deep Learning for the Segmentation of Fetal and Maternal Anatomy in Magnetic Resonance Imaging and Ultrasound. Acad. Radiol. 2021, 28, 173–188. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Qian, L.C.; Cao, Y.Y.; Daniels, M.J.; Song, L.N.; Tian, Y.; Wang, Z.Q. Computed tomography-based radiomics diagnostic approach for differential diagnosis between early- and late-stage pancreatic ductal adenocarcinoma. World J. Gastrointest. Oncol. 2024, 16, 1256–1267. [Google Scholar] [CrossRef]
- Li, M.; Chen, T.; Zhao, W.; Wei, C.; Li, X.; Duan, S.; Ji, L.; Lu, Z.; Shen, J. Radiomics prediction model for the improved diagnosis of clinically significant prostate cancer on biparametric MRI. Quant. Imaging Med. Surg. 2020, 10, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Wang, W. Radiomic Analysis of CT Predicts Tumor Response in Human Lung Cancer with Radiotherapy. J. Digit. Imaging 2020, 33, 1401–1403. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Ge, X.; Ji, H.; Wang, L.; Duan, S.; Chen, H.; Wang, M.; Hu, H.; Jiang, F.; Ding, Z. Prediction of Response to Induction Chemotherapy Plus Concurrent Chemoradiotherapy for Nasopharyngeal Carcinoma Based on MRI Radiomics and Delta Radiomics: A Two-Center Retrospective Study. Front. Oncol. 2022, 12, 824509. [Google Scholar] [CrossRef] [PubMed]
- Isaksson, L.J.; Pepa, M.; Zaffaroni, M.; Marvaso, G.; Alterio, D.; Volpe, S.; Corrao, G.; Augugliaro, M.; Starzyńska, A.; Leonardi, M.C.; et al. Machine Learning-Based Models for Prediction of Toxicity Outcomes in Radiotherapy. Front. Oncol. 2020, 10, 790. [Google Scholar] [CrossRef]
- Bourbonne, V.; Da-Ano, R.; Jaouen, V.; Lucia, F.; Dissaux, G.; Bert, J.; Pradier, O.; Visvikis, D.; Hatt, M.; Schick, U. Radiomics analysis of 3D dose distributions to predict toxicity of radiotherapy for lung cancer. Radiother. Oncol. 2021, 155, 144–150. [Google Scholar] [CrossRef]
- Ibrahim, A.; Primakov, S.; Beuque, M.; Woodruff, H.C.; Halilaj, I.; Wu, G.; Refaee, T.; Granzier, R.; Widaatalla, Y.; Hustinx, R.; et al. Radiomics for precision medicine: Current challenges, future prospects, and the proposal of a new framework. Methods 2021, 188, 20–29. [Google Scholar] [CrossRef]
- Zhang, Y. Enhancing rectal cancer liver metastasis prediction: Magnetic resonance imaging-based radiomics, bias mitigation, and regulatory considerations. World J. Gastrointest. Oncol. 2025, 17, 102151. [Google Scholar] [CrossRef]
- Stefano, A.; Bini, F.; Lauciello, N.; Pasini, G.; Marinozzi, F.; Russo, G. Implementation of Automatic Segmentation Framework as Preprocessing Step for Radiomics Analysis of Lung Anatomical Districts. BioMedInformatics 2024, 4, 2309–2320. [Google Scholar] [CrossRef]
- Cobo, M.; Menéndez Fernández-Miranda, P.; Bastarrika, G.; Lloret Iglesias, L. Enhancing radiomics and Deep Learning systems through the standardization of medical imaging workflows. Sci. Data 2023, 10, 732. [Google Scholar] [CrossRef]
- Mali, S.A.; Ibrahim, A.; Woodruff, H.C.; Andrearczyk, V.; Müller, H.; Primakov, S.; Salahuddin, Z.; Chatterjee, A.; Lambin, P. Making Radiomics More Reproducible across Scanner and Imaging Protocol Variations: A Review of Harmonization Methods. J. Pers. Med. 2021, 11, 842. [Google Scholar] [CrossRef]
- Gupta, S.; Nayak, K.; Pendem, S. Impact of slice thickness on reproducibility of CT radiomic features of lung tumors. F1000Res 2023, 12, 1319. [Google Scholar] [CrossRef] [PubMed]
- Whybra, P.; Zwanenburg, A.; Andrearczyk, V.; Schaer, R.; Apte, A.P.; Ayotte, A.; Baheti, B.; Bakas, S.; Bettinelli, A.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Convolutional Filters for Reproducible Radiomics and Enhanced Clinical Insights. Radiology 2024, 310, e231319. [Google Scholar] [CrossRef]
- Hua, J.; Xiong, Z.; Lowey, J.; Suh, E.; Dougherty, E.R. Optimal number of features as a function of sample size for various classification rules. Bioinformatics 2005, 21, 1509–1515. [Google Scholar] [CrossRef]
- Libling, W.A.; Korn, R.; Weiss, G.J. Review of the use of radiomics to assess the risk of recurrence in early-stage non-small cell lung cancer. Transl. Lung Cancer Res. 2023, 12, 1575–1589. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.; Amstutz, F.; Vuong, D.; Bogowicz, M.; Hullner, M.; Foerster, R.; Basler, L.; Schroder, C.; Eboulet, E.I.; Pless, M.; et al. Preselection of robust radiomic features does not improve outcome modelling in non-small cell lung cancer based on clinical routine FDG-PET imaging. EJNMMI Res. 2021, 11, 79. [Google Scholar] [CrossRef]
- Park, J.E.; Kim, H.S.; Kim, D.; Park, S.Y.; Kim, J.Y.; Cho, S.J.; Kim, J.H. A systematic review reporting quality of radiomics research in neuro-oncology: Toward clinical utility and quality improvement using high-dimensional imaging features. BMC Cancer 2020, 20, 29. [Google Scholar] [CrossRef] [PubMed]
- Ballard, J.L.; Wang, Z.; Li, W.; Shen, L.; Long, Q. Deep learning-based approaches for multi-omics data integration and analysis. BioData Min. 2024, 17, 38. [Google Scholar] [CrossRef]
- Chaudhary, K.; Poirion, O.B.; Lu, L.; Garmire, L.X. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin. Cancer Res. 2018, 24, 1248–1259. [Google Scholar] [CrossRef]
- Zhang, L.; Lv, C.; Jin, Y.; Cheng, G.; Fu, Y.; Yuan, D.; Tao, Y.; Guo, Y.; Ni, X.; Shi, T. Deep Learning-Based Multi-Omics Data Integration Reveals Two Prognostic Subtypes in High-Risk Neuroblastoma. Front. Genet. 2018, 9, 477. [Google Scholar] [CrossRef]
- Song, H.; Ruan, C.; Xu, Y.; Xu, T.; Fan, R.; Jiang, T.; Cao, M.; Song, J. Survival stratification for colorectal cancer via multi-omics integration using an autoencoder-based model. Exp. Biol. Med. 2022, 247, 898–909. [Google Scholar] [CrossRef]
- Munquad, S.; Das, A.B. DeepAutoGlioma: A deep learning autoencoder-based multi-omics data integration and classification tools for glioma subtyping. BioData Min. 2023, 16, 32. [Google Scholar] [CrossRef]
- Bera, K.; Braman, N.; Gupta, A.; Velcheti, V.; Madabhushi, A. Predicting cancer outcomes with radiomics and artificial intelligence in radiology. Nat. Rev. Clin. Oncol. 2022, 19, 132–146. [Google Scholar] [CrossRef]
- Funingana, I.-G.; Piyatissa, P.; Reinius, M.; McCague, C.; Basu, B.; Sala, E. Radiomic and Volumetric Measurements as Clinical Trial Endpoints—A Comprehensive Review. Cancers 2022, 14, 5076. [Google Scholar] [CrossRef]
- Lohmann, P.; Franceschi, E.; Vollmuth, P.; Dhermain, F.; Weller, M.; Preusser, M.; Smits, M.; Galldiks, N. Radiomics in neuro-oncological clinical trials. Lancet Digit. Health 2022, 4, e841–e849. [Google Scholar] [CrossRef] [PubMed]
- Pesapane, F. Regulatory and Ethical Issues in the New Era of Radiomics and Radiogenomics. Radiology 2020, 1, 48–53. [Google Scholar]

| Sampling Method | Key Features | Benefits | Limitations |
|---|---|---|---|
| Manual ROI Segmentation [34] | Widely used; prone to inter-observer variability | Simple and interpretable | Observer-dependent; inconsistent |
| Automatic/Deep Learning-Based Segmentation [35] | Uses models like U-Net/nnU-Net; reduces variability; consistent results | High accuracy; reproducibility | Requires computational resources and model training |
| Multi-Regional Extended ROI [36] | Includes tumor and peritumoral regions; captures microenvironment | Improved prediction accuracy; reflects tumor microenvironment | Complex ROI definition; more data required |
| Adaptive ROI (Delta-Radiomics) [37] | Dynamically adjusts ROI based on tumor evolution; supports delta-radiomics | Captures temporal changes; enhances prediction of treatment response | Needs longitudinal data; complex implementation |
| Method | Benefits | Limitations |
|---|---|---|
| ANOVA | Simple, interpretable; good for identifying statistically significant features [52] | Ignores feature interdependencies [53] |
| Pearson Correlation | Easy to compute; identifies linear relationships [54] | Only captures linear relationships; may miss nonlinear patterns [54] |
| Least Absolute Shrinkage and Selection Operator (LASSO) | Performs both variable selection and regularization; enhances generalization [55] | Exclude weakly informative features [56] |
| Recursive Feature Elimination (RFE) | Systematic backward feature elimination; effective in small feature sets [57] | Computationally expensive for large datasets [58] |
| Minimum Redundancy Maximum Relevance (mRMR) | Balances relevance and redundancy; commonly used in radiomics [59] | Time-consuming; insufficient for coping with high-dimensional pattern classification [60] |
| Multi-objective Feature Selection (MO-FS) | Considers multiple objectives (sensitivity, specificity); promising classification | May require large datasets; balancing objectives can be complex [61] |
| Lasso coefficient thresholds (Coe-Thr-Lasso) | Reduces dimensionality and redundancy; enhances subset quality | Threshold selection sensitive; model-specific tuning needed [49] |
| Minimum Redundancy, Maximum Relevance and Maximum Sparse Representation Coefficient (mRMRMSRC) | Improves sparse representation and relevance; high AUC performance | High model complexity; computational burden [50] |
| Wilcoxon + Random Forest | Stable and high-performing; good for prognostic modeling | Classifier-dependent; may not generalize across all datasets [51] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, M.; Law, H.K.W.; Tam, S.Y. Radiomics-Driven Tumor Prognosis Prediction Across Imaging Modalities: Advances in Sampling, Feature Selection, and Multi-Omics Integration. Cancers 2025, 17, 3121. https://doi.org/10.3390/cancers17193121
Huang M, Law HKW, Tam SY. Radiomics-Driven Tumor Prognosis Prediction Across Imaging Modalities: Advances in Sampling, Feature Selection, and Multi-Omics Integration. Cancers. 2025; 17(19):3121. https://doi.org/10.3390/cancers17193121
Chicago/Turabian StyleHuang, Mohan, Helen K. W. Law, and Shing Yau Tam. 2025. "Radiomics-Driven Tumor Prognosis Prediction Across Imaging Modalities: Advances in Sampling, Feature Selection, and Multi-Omics Integration" Cancers 17, no. 19: 3121. https://doi.org/10.3390/cancers17193121
APA StyleHuang, M., Law, H. K. W., & Tam, S. Y. (2025). Radiomics-Driven Tumor Prognosis Prediction Across Imaging Modalities: Advances in Sampling, Feature Selection, and Multi-Omics Integration. Cancers, 17(19), 3121. https://doi.org/10.3390/cancers17193121





