Simple Summary
Enhancer of Zeste Homolog 2 (EZH2) is a protein widely recognized for its role in gene silencing and plays a significant role in cancer-related processes, including cell survival, proliferation, invasion, and self-renewal. As the catalytic subunit of Polycomb-Repressive Complex 2 (PRC2), canonically EZH2 mediates the trimethylation of histone H3 at lysine 27 (H3K27), leading to transcriptional repression. Dysregulated EZH2 expression is a hallmark of numerous cancers, both solid and hematologic, and is frequently associated with enhanced metastasis and poor patient prognosis. Notably, EZH2 also exhibits non-canonical functions, including gene activation, which can further promote tumor progression. Due to its significant involvement in oncogenesis and therapy resistance, EZH2 expression is being explored as a diagnostic and/or prognostic biomarker. This review summarizes the function and diverse roles of EZH2 across human cancers, highlighting its potential as both a diagnostic and/or prognostic biomarker. A deeper understanding of EZH2’s intricate regulatory network may enable the development of more effective strategies for managing EZH2-driven malignancies.
Abstract
Enhancer of Zeste Homolog 2 (EZH2) is a key epigenetic regulator known for its role in global gene silencing and is involved in a variety of cellular processes, including cell survival, proliferation, invasion, and self-renewal. As a core component of the Polycomb Repressive Complex 2 (PRC2), EZH2 catalyzes the trimethylation of histone H3 at lysine 27 (H3K27me3), leading to chromatin compaction and transcriptional repression. Dysregulated EZH2 expression is observed in a wide range of solid tumors and hematological malignancies and is frequently associated with increased metastatic potential and poor clinical outcomes. While EZH2 primarily mediates gene silencing through its canonical PRC2-dependent activity, it also exerts oncogenic effects via non-canonical mechanisms. In its non-canonical role, EZH2 acts independently of PRC2, interacting with other signaling molecules as a transcriptional activator or co-activator, thereby promoting the activation of oncogenic pathways. Through both canonical and non-canonical mechanisms, EZH2 significantly contributes to tumor initiation and its subsequent progression. Given its critical role in oncogenesis and cancer progression, EZH2 is under investigation as a potential biomarker for cancer diagnosis and prognosis. This review provides a comprehensive overview of EZH2’s function and oncogenic roles across human cancers. Enhanced insight into EZH2’s complex regulatory network may facilitate the development of more effective strategies to manage EZH2-driven malignancies.
1. Introduction
Enhancer of zeste homolog 2 (EZH2) is an evolutionarily conserved histone methyltransferase that mediates transcriptional silencing by catalyzing the trimethylation of histone H3 at lysine 27 (H3K27me3) [1,2]. EZH2 functions as the catalytic subunit of the polycomb repressive complex 2 (PRC2), a highly conserved polycomb group protein complex composed of core subunits EZH2, Embryonic Ectoderm Development (EED), Suppressor of Zeste 12 homolog (SUZ12), and Retinoblastoma-binding Protein 7 (RBBP7), along with several accessory factors [3,4]. Together, these components regulate chromatin structure and gene expression [5,6,7]. Within PRC2, EZH2 establishes the H3K27me3 mark, while EED binds both EZH2 and histone H3, acting as a scaffold protein [8]. Structural studies have shown that the EZH2 EED-binding domain (EBD) interacts with the WD repeat domain of EED [9]. SUZ12 is essential for nucleosome recognition, catalytic activity, and overall stability of the complex [10], while accessory proteins such as AE Binding Protein 2 (AEBP2), Polycomb-like (PCLs), and Jumonji AT rich Interactive Domain 2 (JARID2) fine-tune PRC2 activity [9,10]. Once established, the H3K27me3 mark recruits the Polycomb Repressive Complex 1 (PRC1), which mono-ubiquitinates histone H2A at lysine 119, promoting chromatin compaction and stable transcriptional silencing [11]. In addition, EZH2 cooperates with other epigenetic regulators, including DNA methyltransferases (DNMTs) and histone deacetylases (HDACs), suggesting cross-talk among different silencing pathways in the control of gene expression [1,2].
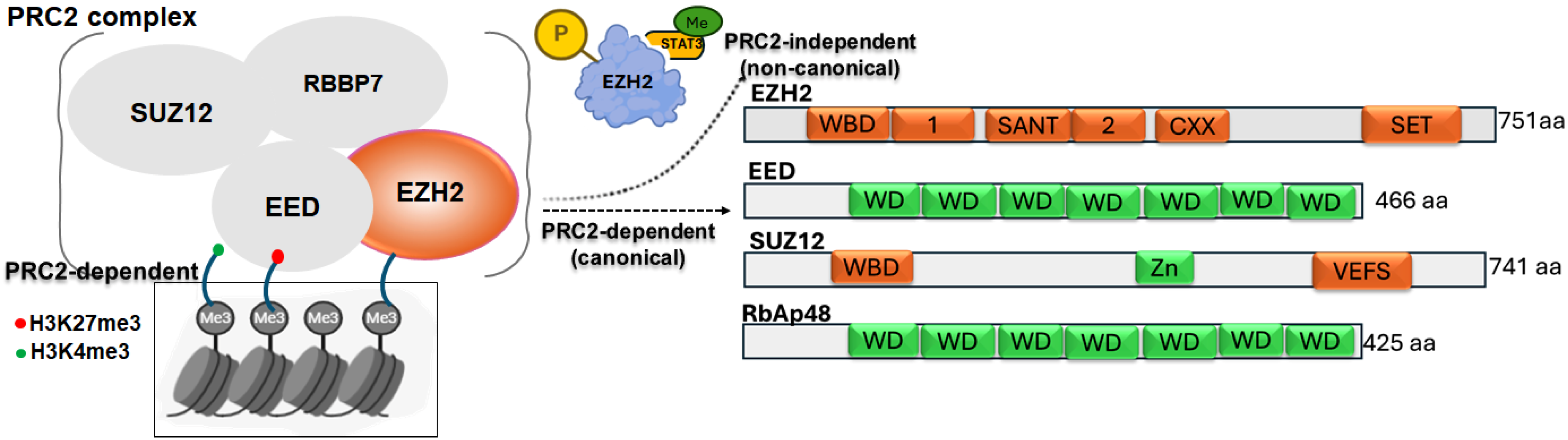
Emerging research has identified a non-canonical role for EZH2 as a transcriptional co-activator, potentially mediated through the methylation of non-histone proteins [3,4]. In this PRC2-independent context, EZH2 directly associates with and modifies non-histone targets, leading to activation of downstream gene expression [12]. Through these mechanisms, EZH2 regulates key cancer-related processes, including cell survival, proliferation, invasion, and senescence [13]. Aberrant EZH2 expression is frequently observed across diverse cancer types highlighting its critical contribution to tumorigenesis [14]. Importantly, the functional impact of EZH2 alterations is highly context-dependent, with gain-of-function changes in EZH2 often drive oncogenic activity, whereas loss-of-function alterations confer tumor suppressive effects in certain malignancies [1,2]. This review examines the multifaceted roles of EZH2’s in human cancers, linking its molecular mechanisms to clinical features, pathological outcomes, and patient prognosis. Figure 1 illustrates both the canonical and non-canonical modes of EZH2 action.
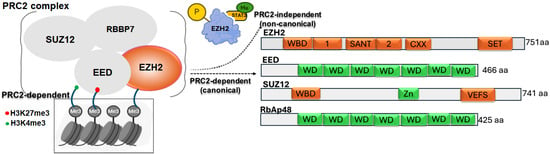
Figure 1.
Schematic representation of the PRC2 complex showing core components (EZH2, EED, SUZ12, RBBP7) and their role in catalyzing H3K27me3-mediated gene silencing. A non-canonical function of EZH2 is also depicted, where its phosphorylation enables STAT3 methylation, promoting tumorigenicity. Domain structures of each PRC2 subunit are shown, highlighting key functional motifs involved in complex assembly and enzymatic activity.
2. EZH2 Structure
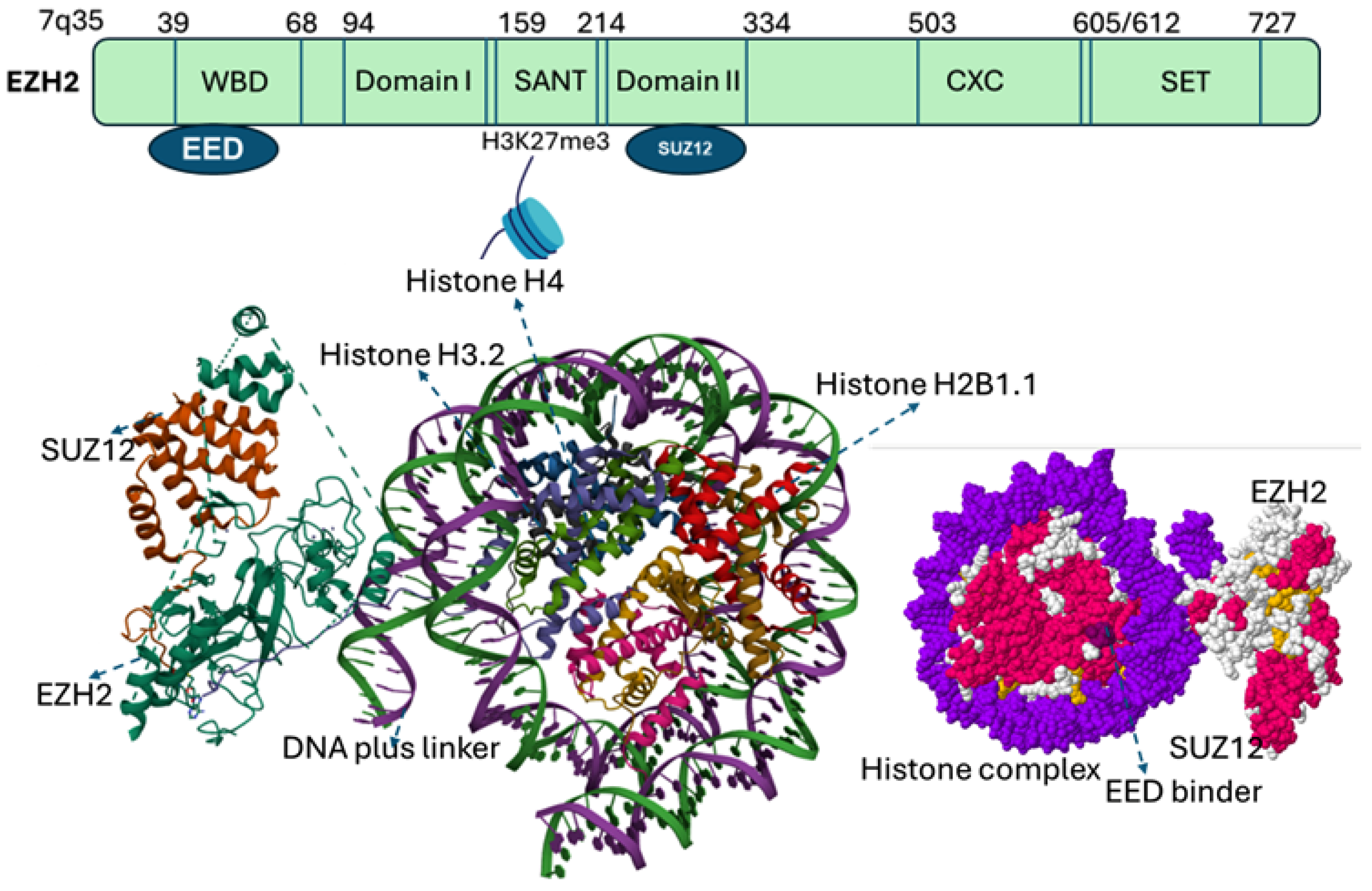
EZH2 gene, located on chromosome 7q35, comprises 20 exons encoding a protein of 746 amino acids. EZH2 contains five major domains, including the EED-interaction domain (EID), Domain I, Domain II, cysteine-rich domain (CXC domain), and C-terminal SET domain (suppressor of variegation 39, enhancer of zeste and trithorax). Structural and biochemical studies of SET domain in histone methyltransferases have elucidated the molecular basis of histone methylation [4,9], revealing a conserved catalytic Asp–His–Ser (NHS) triad essential for recognizing histone peptide tails and binding S-adenosyl-methionine (SAM), the methyl donor [4,15]. Beyond the SET domain, EZH2 harbors additional functional regions. The CXC (cysteine-rich domain) and an ncRBD (non-coding RNA– and a DNA-binding domain) facilitate interactions with regulatory proteins, while N-terminal domain mediate protein–protein interactions critical for PRC2 assembly and function [16] (Figure 2).
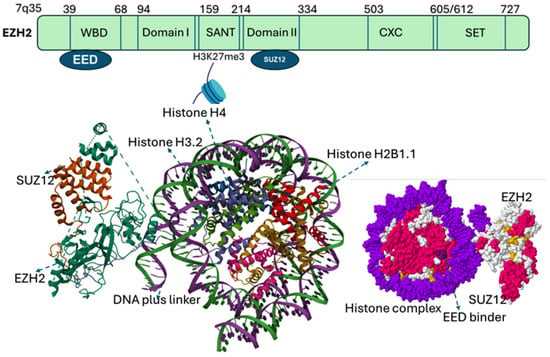
Figure 2.
Categorized domains of EZH2. Five functional domains: a C-terminal SET domain, an adjacent cysteine-rich CXC domain, domain I, domain II, and an EED interaction domain (EID). The bottom panel illustrates the PRC2 complex structure with EZH2 (purple), SUZ12 (orange), and EED (gray surface). EED binds to EZH2 to form the core of the Polycomb Repressive Complex 2 (PRC2). The catalytic SET domain of EZH2 mediates H3K27 methylation (blue). EED serves as a regulatory subunit, and binding sites for EED inhibitors (yellow) as well as the EED–EZH2 interaction inhibitor (boxed) are indicated. EED Binders are small molecule or peptide that binds EED, disrupting its interaction with EZH2, while an EED–EZH2 interaction inhibitor specifically blocks the EZH2–EED binding, destabilizing PRC2 and impairing its histone methyltransferase activity.
3. Molecular Alterations of EZH2 in Cancer
The multifaceted role of EZH2 in cancer has been demonstrated through changes in DNA, posttranslational modifications, and interactions with other epigenetic regulators that collectively modulates its activity. Hyperactivation of EZH2, whether by amplification or mutation, is common in diverse human cancers [17]. A well characterized example is the heterozygous DNA-mediated change at tyrosine 641 (Y641) within the SET domain [18]. Initially thought to be loss-of-function, Y641 alteration (including Y641F, Y641N, Y641S, Y641H, and Y641C) instead confer gain-of-function, shifting substrate preference from unmethylated or monomethylated H3K27 to dimethylated H3K27 (H3K27me2) [17,18,19]. In combination with wild type EZH2, this leads to enhanced accumulation of H3K27me3. Another less frequent amino acid alteration, A677G, also increases catalytic activity on H3K27me2, but unlike Y641 variants, it retains activity toward all three substrates (H3K27, H3K27me1, H3K27me2), reflecting a distinct mechanism [20].
Beyond genomic alterations, posttranslational modifications critically regulate EZH2 activity. Phosphorylation at Ser21 by Akt redirect EZH2 towards non-histone substrates such as androgen receptor (AR), enhancing AR target gene transcription independently of PRC2 [21,22]. Similarly, phosphorylated EZH2 promotes methylation and activation of signal transducer and activator of transcription 3 (STAT3) signaling [12], in part via interaction with SUZ12. Phosphorylation by MELK, a maternal embryonic leucine-zipper kinase, activates NF-κB, driving tumorigenesis and self-renewal [12], while cyclin E/CDK2-mediated phosphorylation at of EZH2 at Thr416 increases EZH2 activity to promote invasion [23]. In contrast, phosphorylation of EZH2 at threonine 311, which is mediated by AMP-activated protein kinase (AMPK), leads to a disruption in the physical interaction between EZH2 and its essential partner SUZ12 [22]. This disruption significantly impairs the histone methyltransferase (HMTase) activity of EZH2. Consequently, this alteration results in the release of the transcriptional silencing that is typically imposed on tumor suppressor genes through the canonical repressive functions of EZH2 [22,23].
EZH2 also engages in crosstalk with other epigenetic regulators. Physical interactions with DNA methyltransferases (DNMT1, DNMT3A, and DNMT3B) recruit DNMTs to EZH2 target loci, linking H3K27me3 with CpG hypermethylation in cancer [24,25]. Similarly, EZH2 transiently interacts with histone deacetylases (HDAC1, HDAC2), which may remove acetyl groups from H3K27 or other lysine residues to facilitate PRC2-mediated methylation [26,27,28,29]. Antagonistic histone marks such as H3K27ac, H3K4me3, and H3K36me2 counteract EZH2 function, highlighting the importance of local chromatin context in determining EZH2 activity [30,31,32]. Collectively, these findings underscore the context-dependent roles of EZH2. Furthermore, upregulation or aberrant activation of EZH2 can silence tumor suppressor genes via canonical PRC2-mediated promoter methylation or act as a non-canonical co-activator of oncogenic pathways [32,33].
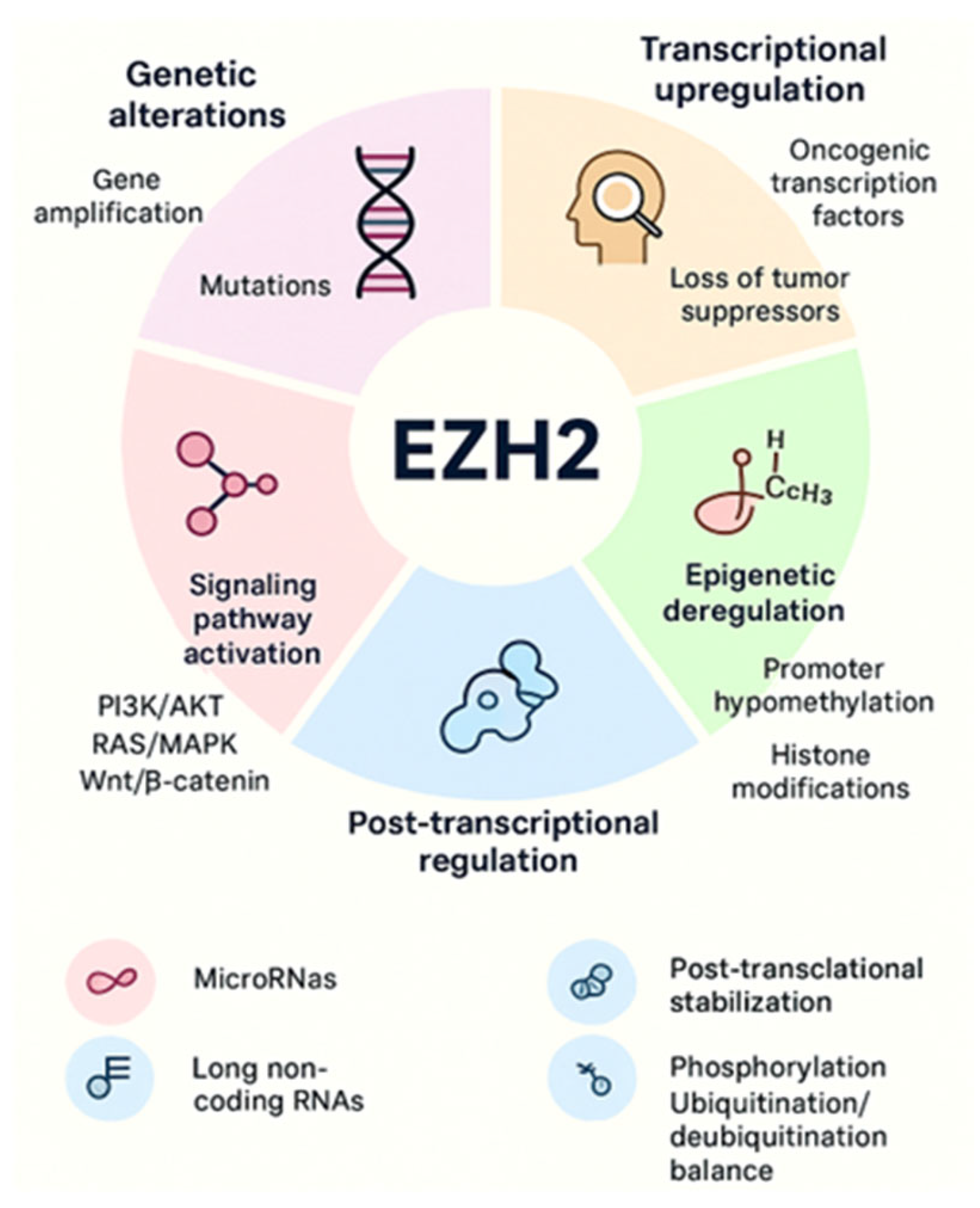
Increased EZH2 activity is consistently associated with tumor initiation, progression, and poor prognosis across both solid and hematologic malignancies. Multiple mechanisms of EZH2 regulation, including transcriptional regulation, mRNA regulation by miRNAs, accessibility to DNA via DNA binding proteins and ncRNAs, and post-translational modifications. EZH2 is overexpressed in cancer due to several factors, including transcriptional activation by oncogenic proteins like MYC and ETS family members, deletion, or downregulation of EZH2-inhibiting miRNAs such as miR-101 and miR-26a, and gene amplification in some solid cancers [34,35]. These mechanisms lead to increased EZH2 protein levels, promoting cancer cell proliferation, invasion, and overall tumor aggressiveness. A schematic illustration is shown in Figure 3. More detailed context-dependent studies on the oncogenic roles of EZH2 in cancer have been previously published by our group [1,6,22].
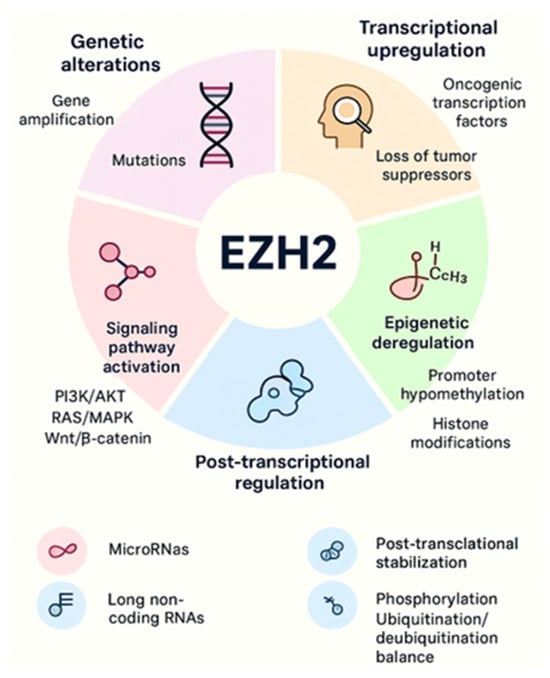
Figure 3.
EZH2 overexpression and aberrant activity arise through multiple mechanisms, including genetic alterations, transcriptional upregulation, epigenetic deregulation, signaling pathway activation, post-transcriptional regulation, and post-translational modifications. Together, these processes enhance EZH2 expression and activity, promoting cancer initiation and progression.
4. EZH2 Protein Interactions in Cancer
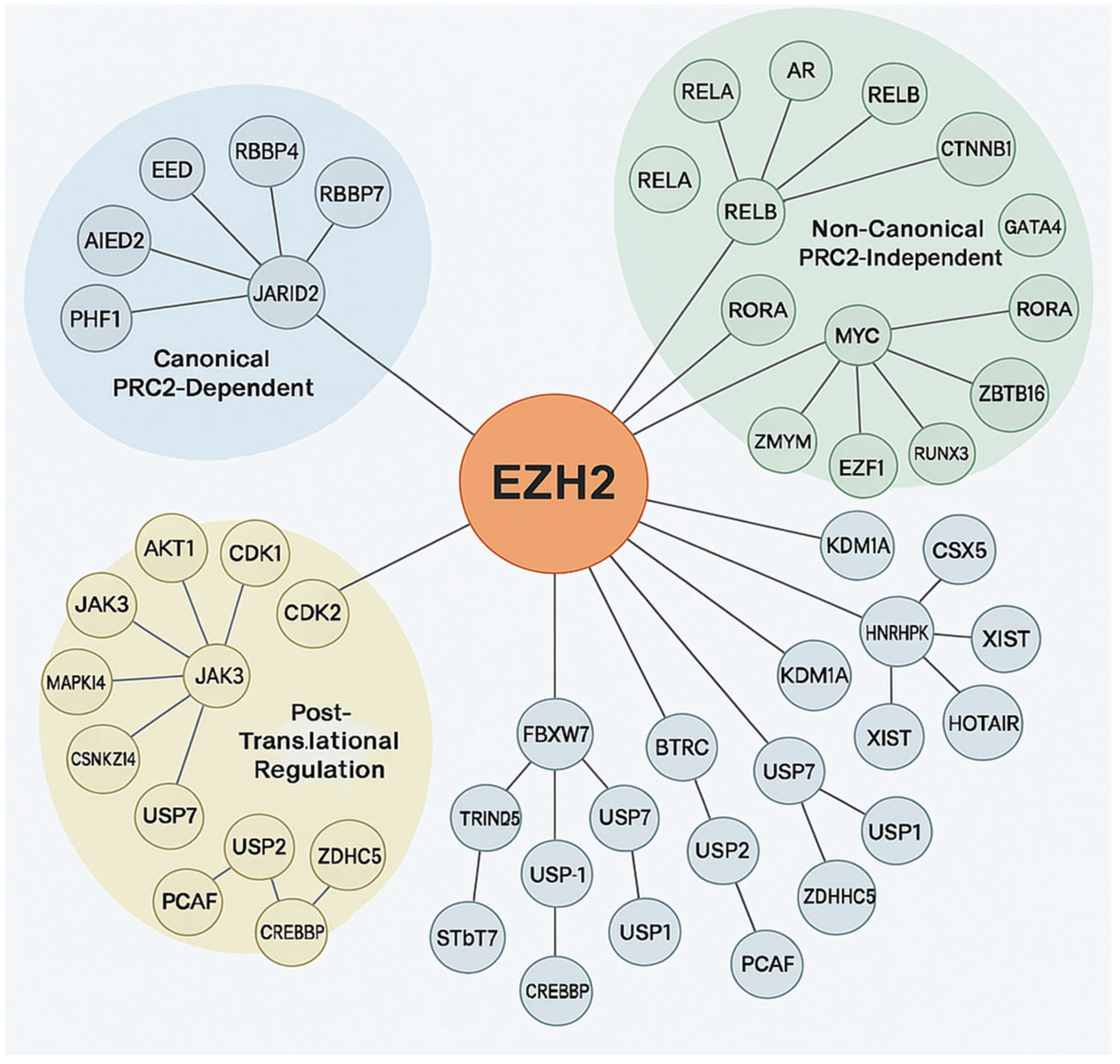
EZH2 is known for its role in transcriptional repression via H3K27me3 and also participates in PRC2-independent, non-epigenetic pathways by interacting with diverse oncogenic and tumor suppressor proteins [34]. Post-translational modifications tightly regulate EZH2’s enzymatic function, stability, and adaptability in cancer [36]. EZH2 protein–protein interaction network highlights its dual role in epigenetic and non-epigenetic oncogenic processes and its complex involvement in tumorigenesis. This involvement includes canonical PRC2-mediated repression by maintaining cancer cell identity and hindering differentiation through interaction with accessory proteins like JARID2, AE Binding Protein 2 (AEBP2), PHD Finger Protein 1 (PHF1) and PHF19, Metal-Response Element Binding Transcription Factor 2 (MTF2), which are critical for modulating chromatin targeting and enhancing H3K27 methylation [37]. Through non-canonical/post-translational mechanisms EZH2 enables dynamic responses to oncogenic stress, immune evasion, and therapy resistance. EZH2 interacts with oncogenic transcription factors such as AR in prostate cancer, MYC in lymphomas and medulloblastomas, STAT3 in glioblastoma and breast cancer, where it can enhance STAT3 activity through methylation, and binds RelA/RelB thereby potentiating NF-κB signaling and CTNNB1 in Wnt-driven tumors [12]. It also represses or destabilizes tumor suppressors viz. Retinoic acid-related Orphan Receptor Alpha (RORA), GATA4, Runt-related transcription factor 3 (RUNX3), Zinc finger and BTB domain containing 16 (ZBTB16/PLZF) through direct methylation or recruitment to repressive complexes. Interactions with Lysine-specific histone demethylase 1A (KDM1A and/or LSD1) and Heterogeneous Nuclear Ribonucleoprotein K (HNRNPK) involve chromatin and RNA processing, while lncRNA scaffolding including X-inactive-specific transcript (XIST), HOX Transcript Antisense RNA (HOTAIR), Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) directs EZH2 to specific genomic loci. This non-canonical activity allows EZH2 to act as a transcriptional co-activator or repressor independently of its methyltransferase activity, promoting oncogenic plasticity [38]. Targeting specific components of this protein–protein interaction (PPI) network offers new avenues for precision cancer therapies beyond enzymatic inhibition. Furthermore, EZH2’s function is modulated by phosphorylation such as AKT1 at Ser21 inhibits H3K27me3, CDK1/2 at Thr345/Thr487 regulates chromatin dynamics, Janus kinase 3/mitogen-activated protein kinase 14 (JAK3/MAPK14) in inflammation and stress and ubiquitination and deubiquitination of E3 ligases FBXW7, BTRC, TRIM28 target for degradation; deubiquitinases USP7, USP21, USP1 stabilization as illustrated in Figure 4 [39]. In conclusion, EZH2 is a key regulator of cancer biology, bridging canonical PRC2-mediated repression with diverse non-canonical functions and post-translational modifications. Its dual capacity to act as both a transcriptional repressor and co-activator, independent of methyltransferase activity, underscores its contribution to oncogenic plasticity and immune evasion. EZH2 regulation involves multiple layers, including phosphorylation, ubiquitination, deubiquitination, and interactions with transcription factors, chromatin modifiers, and lncRNAs, reflecting the complexity of its regulatory network. This multifaceted role establishes EZH2 as a clinically significant diagnostic and/or prognostic biomarker, with therapeutic opportunities extending beyond enzymatic inhibition to targeting its specific interactions and regulatory pathways.
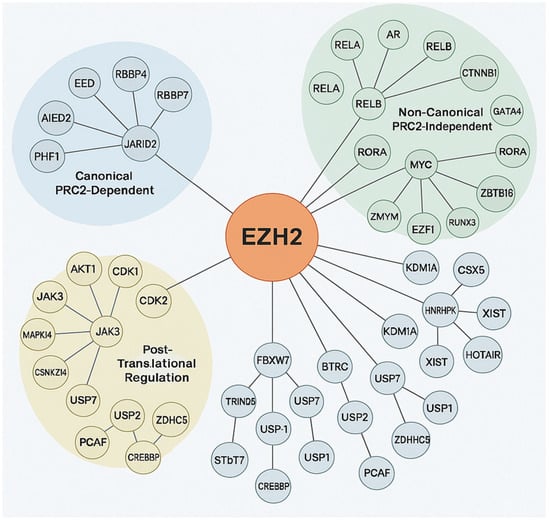
Figure 4.
Protein–protein interactions of EZH2. The central orange node represents EZH2, with connecting lines indicating direct or indirect molecular interactions. The surrounding nodes are grouped and color-coded based on functional categories: Canonical PRC2-Dependent (blue cluster). Non-Canonical PRC2-Independent (green cluster): Post-Translational Regulation (yellow cluster), with expected number of edges: 102, PPI enrichment p-value: <1.0 × 10−16, average node degree: 14.7, avg. local clustering coefficient 0.707.
5. EZH2 Dysregulation in Human Cancers
In normal human tissues, EZH2 expression is generally maintained at low levels, indicative of its specific and tightly controlled roles in cellular homeostasis. For example, studies examining breast epithelium have shown a median percentage of EZH2-positive epithelial cells in normal terminal duct lobular units of approximately 5.88%, with an interquartile range of 1.89–12.46% [40]. This restricted expression suggests that EZH2’s histone methyltransferase activity is only required in a limited population of cells within these normal structures, possibly for maintaining lineage identity or regulating specific developmental programs. Similarly, normal ovarian tissue exhibits negligible EZH2 immunoreactivity, further supporting the notion that EZH2 is not broadly expressed in adulthood or differentiated tissues [41]. This low baseline expression across various normal tissues underscores the importance of maintaining EZH2 activity within a physiological range, preventing aberrant gene silencing or activation that could disrupt normal cellular function and tissue architecture [1,2,3,4]. The tight regulation of EZH2 in normal tissues likely involves intricate mechanisms controlling its transcription, translation, and protein stability, ensuring its potent chromatin-modifying activity is only deployed when and where necessary for proper cellular function and tissue integrity.
In contrast, EZH2 is frequently modified (epigenetically/post-translationally) or genomically altered in a wide array of human cancers [34]. Elevated EZH2 levels have been observed in solid tumors such as prostate, breast, uterine, gastric, and renal cell carcinomas, among others [42]. This aberrant upregulation contributes to oncogenesis by repressing tumor suppressor genes, altering transcriptional programs, and promoting cellular proliferation, invasion, and survival. The dysregulation of EZH2 in cancer often hijacks these normal physiological roles, leading to the acquisition of aggressive cancer hallmarks. In many human cancers, EZH2 upregulation correlates with increased tumor aggressiveness, enhanced metastatic potential, resistance to therapy, and poor clinical outcomes in most of these solid tumors which are described in detail below [43].
5.1. EZH2 and Bladder Cancer
Numerous studies have demonstrated the pivotal role of EZH2 upregulation or dysfunction in the pathogenesis of bladder cancer. In recent work by Li et al. serum samples from bladder cancer patients and normal controls revealed significantly elevated EZH2 levels in the patient cohort [44]. These elevated levels were strongly associated with adverse clinical features, including lymph node metastasis, muscle invasion, increased tumor size, and poor overall prognosis. These findings suggest that serum EZH2 could serve as a promising non-invasive biomarker for assessing disease progression and prognosis in bladder cancer. At the molecular level, EZH2 has been shown to contribute to bladder cancer progression through transcriptional repression of tumor suppressor genes. Specifically, EZH2-mediated silencing of E-cadherin, a key epithelial marker has been implicated in enhancing metastatic potential, particularly in superficial transitional cell carcinoma of the bladder [45]. Further supporting EZH2’s role in tumor aggressiveness, Wang et al. identified an enrichment of cancer stem cell subpopulations with high EZH2 expression during bladder cancer recurrence, highlighting its involvement in tumor relapse and/or treatment resistance [46]. Similarly, Chen et al. demonstrated that pharmacological inhibition of EZH2 significantly reduced tumor growth and invasiveness via suppression of the JAK2/STAT3 signaling pathway, further emphasizing its prognostic relevance [47]. In addition to protein-level regulation, upstream non-coding RNAs also influence EZH2 expression [48]. Min et al. reported that long non-coding RNA SNHG1 [49], for instance, has been found to facilitate bladder cancer progression by upregulating EZH2 expression [50]. These findings reveal a broader regulatory network centered on EZH2 and suggest that both EZH2 and its regulatory partners hold promise as prognostic biomarkers. Table 1 summarizes some of the important studies implicating the role of EZH2 in bladder cancer.

Table 1.
EZH2 expression in bladder cancer.
5.2. EZH2 and Breast Cancer
Several studies have established a strong correlation between EZH2 amplification or dysregulation and the initiation, progression, invasion, and metastasis of breast cancer, particularly in its more advanced stages [66]. Breast cancer is broadly classified into three molecular subtypes: hormone receptor-positive (HR+) breast cancer, characterized by the expression of estrogen (ER) and/or progesterone receptors (PgR); HER2-positive breast cancer, defined by amplification or upregulation of the human epidermal growth factor receptor 2 (HER2); and triple-negative breast cancer (TNBC), which lacks ER, PgR, and HER2 expression [67]. High EZH2 expressions in breast cancer have been consistently associated with unfavorable clinicopathological features, including higher histological grades, ER and PgR negativity, HER2 positivity, and elevated p53 expression [68]. Mechanistically, EZH2 contributes to breast tumorigenesis through both its canonical, PRC2-dependent HMT activity, repressing tumor suppressor genes via H3K27 trimethylation, and non-canonical pathways, where EZH2 functions as a transcriptional activator or co-activator in signaling networks [69]. In ER-positive breast cancer cells, EZH2 has been shown to directly interact with ER and β-catenin to activate transcriptional programs driven by estrogen and Wnt signaling pathways [70]. Conversely, research by Lee et al. demonstrated that in ER-positive contexts, EZH2 cooperates with ER to recruit PRC2 to NF-κB gene promoters, leading to EZH2-mediated H3K27me3 and constitutive repression of NF-κB target genes [71]. In ER-negative breast cancer cells, EZH2 engages in non-canonical activation, forming a complex with RelA and RelB to enhance NF-κB signaling, thereby promoting inflammatory and pro-tumorigenic transcriptional programs [72]. These findings reveal a dual role for EZH2, acting as a transcriptional repressor in ER-positive contexts, and as a transcriptional activator in ER-negative environments via non-canonical mechanisms. Feng et al. demonstrated that EZH2 is localized in the cytoplasm and nucleus of breast cancer cells in a site-specific phosphorylation manner [73]. More advanced HER2-positive clinical-stage breast cancers exhibiting metastatic lymph nodes were found to contain elevated levels of EZH2 compared to less aggressive cancers with low EZH2 levels. pEZH2-S21 localization in the nucleus has shown a correlation with invasive and metastatic lymph node HER2-positive breast cancer, potentially establishing it as an indicator of invasive breast cancer.
Enhanced EZH2 activities and EZH2-induced H3K27me3 regulate signaling pathways such as the Forkhead box (FOX) transcription factor family, which can induce tumor cell proliferation, migration, and bone metastasis, contributing to breast cancer progression [74]. EZH2 also targets downstream genes associated with anticancer effects including FOXO3, CDH1, RKIP, and CDKN1C [74,75]. By repressing these tumor suppressor genes, EZH2 can promote the development of malignant breast cancer. To confirm the oncogenic role of EZH2, it has been either inhibited [63] or knock down to reverse EZH2-conferred induction of breast cancer [64]. A study conducted by Li et al. [76], suppresses EZH2 in conjunction with PARP inhibition led to excessive autophagy and synthetic lethality in triple-negative breast cancer cells. Mao et al. were able to combat the proliferation and invasiveness of triple-negative breast cancer cells after CRISPR-Cas9-mediated EZH2 knockdown [77]. Apart from these observations, the literature contains a plethora of evidence substantiating the role of EZH2 in breast cancer which is summarized in Table 2.

Table 2.
EZH2 expression in breast cancer.
5.3. EZH2 and Cervical Cancer
Upregulation of EZH2 in cervical cancer tissues has been consistently associated with advanced disease stage, lymphatic metastasis, deeper tumor infiltration, and reduced overall patient survival [88]. Functionally, EZH2 acts as a primary regulator of cell cycle and an inhibitor of apoptosis, thereby contributing to tumorigenesis and cancer progression. Its upregulation is positively correlated with activation of the Wnt/β-catenin signaling pathway in cervical cancer, which leads to the upregulation of downstream oncogenic effectors such as β-catenin, c-Myc, and Cyclin D1 [89]. These molecules collectively drive uncontrolled cell proliferation and tumor growth. EZH2 also mediates the oncogenic functions of long non-coding RNA SNHG8 in HPV-positive cervical cancers [90]. Through direct interaction, SNHG8 recruits EZH2 to transcriptionally repress RECK (reversion-inducing cysteine-rich protein with kazal motifs), a known tumor suppressor in cervical cancer. This repression promotes cellular proliferation and inhibits apoptosis, enhancing tumor aggressiveness. Furthermore, elevated EZH2 expression in cervical cancer has been linked to hypomethylation of its own promoter region, suggesting an epigenetic feedback mechanism that reinforces its expression [91]. This hypomethylation has been associated with the suppression of senescence-related genes, further contributing to malignant transformation and sustained cancer cell survival. Furthermore, its strong correlation with disease progression and prognosis positions EZH2 as a compelling biomarker in cervical cancer. Table 3 summarizes studies highlighting the oncogenic role of EZH2 in cervical cancer.

Table 3.
EZH2 expression in cervical cancer.
5.4. EZH2 and Colorectal Cancer
EZH2 upregulation correlates with poor survival in patients in both early and advanced stage tumors with colorectal cancer (CRC) [95]. In CRC tissues, the long non-coding RNA (lncRNA) LINC01116 is upregulated and promotes tumor cell proliferation by recruiting EZH2, which methylates the Tropomyosin 1 (TPM1) promoter, thereby suppressing its translation [96]. Additionally, EZH2 was found to be inversely associated with miR-31 and in sessile serrated adenomas/polyps in premalignant lesions. EZH2 knockdown in colorectal cancer led to increased miR-31 expression [97]. In CRC, EZH2 has been identified as a potential prognostic marker, with elevated expression associated with reduced overall survival. For instance, its association with KDM2B, a cell cycle regulator, has been demonstrated as downregulation of KDM2B reduces EZH2 expression, suppresses PI3K/AKT pathway components, and delays colorectal cancer cell migration [98]. Conversely, EZH2’s combined expression with other polycomb-group (PcG) proteins BMI1 and SUZ12 and their associated histone modification H3K27me3 were correlated with positive patient survival and greater survival for colorectal cancer [99]. EZH2 may also serve as a predictive marker for chemotherapy response and poor 5-year disease-free survival in patients with rectal cancer [100]. These studies highlight EZH2 as both a biomarker and an oncogene in colorectal cancer, as summarized in Table 4.

Table 4.
EZH2 expression in colorectal cancer.
5.5. EZH2 and Esophageal Cancer
EZH2 has emerged as an independent prognostic factor in esophageal cancer, with elevated expression levels significantly correlating with poor disease outcomes [111]. In esophageal squamous cell carcinoma (ESCC), key independent predictors of poor prognosis include high EZH2 expression, advanced histological grade, and distant lymph node metastasis [112]. Notably, EZH2 is consistently elevated at both mRNA and protein levels in esophageal cancer tissues [113,114]. This upregulation contributes to tumor progression by promoting cellular proliferation and metastasis [115]. Mechanistically, EZH2 drives tumorigenesis through its canonical function as a histone methyltransferase. For instance, LINC00114, a long non-coding RNA, has been shown to promote esophageal cancer development by recruiting EZH2 to DLC1 (Rho GTPase Activating Protein) gene promoter, enhancing H3K27me3 and thereby silencing this tumor suppressor gene [116]. In addition, EZH2 regulates epithelial-to-mesenchymal transition (EMT) in ESCC by modulating the expression of miR-200c and key EMT-related genes, ultimately promoting cancer cell migration and invasiveness [117]. These effects are primarily driven by EZH2’s ability to catalyze H3K27 trimethylation at target gene promoters, altering chromatin structure and gene expression. Forced expression of EZH2 in esophageal cancer cells has been shown to significantly elevate global H3K27me3 levels, emphasizing its role in gene silencing and metastasis [112]. Thus, EZH2 serves as a valuable biomarker for predicting ESCC prognosis and metastatic potential. Evaluation of EZH2 expression may thus aid in stratifying patients and tailoring treatment strategies in ESCC. Key studies implicating EZH2 in esophageal cancer are summarized in Table 5.

Table 5.
EZH2 expression in esophageal cancer.
5.6. EZH2 and Gastric Cancer
EZH2 plays a critical role in promoting tumor cell proliferation and advancing gastric cancer by mediating gene promoter methylation [122]. Inhibition of EZH2 in gastric cancer cells has been shown to induce cellular senescence, primarily through the activation of tumor suppressor genes such as p21 and p16 [123]. Moreover, EZH2 expression is influenced by miRNA dynamics, particularly miR-124 [124]. A decrease in miR-124 levels has been associated with elevated EZH2 expression, while overexpression of miR-124 suppresses EZH2 levels, thereby inhibiting cancer progression in gastric cells [125]. Other miRNAs have also been implicated in the regulation of EZH2. For example, miR-26 interacts with the 3’ untranslated region of EZH2 mRNA and, when suppressed during TET-facilitated gastric carcinogenesis, leads to EZH2 upregulation [126]. Additionally, circular RNAs (circRNAs) modulate EZH2 expression in gastric cancer. circKIF4A has been shown to regulate EZH2 via interaction with miR-144-3p. When miR-144-3p is inhibited, the tumor-suppressive effect of circKIF4A is diminished, resulting in increased EZH2 expression [127]. Similarly, circGSK3B facilitates EZH2 upregulation by blocking its binding to the RORA promoter, thereby reducing EZH2 repression [128]. Clinically, EZH2 upregulation in gastric cancer correlates with aggressive tumor phenotypes, including larger tumor size, lymph node metastasis, and lymphatic invasion [128]. Elevated EZH2 levels have also been associated with advanced clinical stages and poor prognosis. One study reported that 68.6% of gastric cancer patients exhibited an increased EZH2 expression [122]. Collectively, these findings underscore the oncogenic role of EZH2 in gastric cancer and highlight its potential as a prognostic biomarker. Key studies illustrating EZH2’s involvement in gastric cancer is summarized in Table 6.

Table 6.
EZH2 expression in gastric cancer.
5.7. EZH2 and Glioblastoma
Glioblastoma (GBM) is an aggressive brain tumor originating from glial tissue, with a low five-year survival rate of 5.5% and abnormal methylation patterns [140]. EZH2 functions as an oncogene in GBM, contributing to numerous tumor-promoting processes such as cell cycle progression, invasion, glioma stem cell maintenance, resistance to chemotherapy and radiotherapy, angiogenesis, apoptosis inhibition, and tumor proliferation [141]. One key mechanism involves the upregulation of EZH2 leading to increased H3K27 trimethylation, which in turn suppresses the expression of the tumor suppressor PTEN. This suppression activates the PI3K/Akt signaling pathway, promoting enhanced proliferation and migration of GBM cells [142]. Additionally, phosphorylation of EZH2 can lead to increased STAT3 expression through epigenetic methylation, thereby suppressing apoptosis and further advancing GBM progression [12]. EZH2 also cooperates with DNA methyltransferases to regulate miRNA expression, further influencing glioma biology. For example, EZH2 and DNMT1 have been shown to co-mediate the silencing of tumor-suppressive miRNAs, such as miR-200b and miR-429, thereby promoting GBM development [143]. Moreover, higher EZH2 expression facilitates an oncogenic axis by interacting with HP1BP3 and activating WNT7B, a pathway that has been linked to therapeutic resistance [144]. These findings underscore the significant role of EZH2 in the pathogenesis and progression of glioblastoma, supporting its utility as a prognostic biomarker. Additional key studies exploring EZH2’s role in GBM are summarized in Table 7.

Table 7.
EZH2 expression in glioblastoma.
5.8. EZH2 and Head and Neck Cancer
EZH2 upregulation is correlated with aggressive tumor activity and unfavorable patient survival in head and neck squamous cell carcinoma (HNSCC) [150]. Elevated EZH2 expression in HNSCC is associated with enhanced tumor proliferation and metastatic potential. In particular, silencing EZH2 was shown to upregulate E-cadherin expression, a key epithelial marker, thereby reducing cancer cell migration and invasiveness in HNSCC [150]. Moreover, high EZH2 expression has been linked with lymph node metastasis, a critical prognostic indicator often associated with reduced overall survival in HNSCC patients [151]. However, some contradictory findings have emerged. For instance, the same study reporting EZH2 association with lymph node metastasis did not find a statistically significant relationship between EZH2 expression and patient survival outcomes [150]. Additionally, another study revealed that younger HNSCC patients exhibited lower EZH2 expression levels compared to older counterparts, suggesting that age-specific expression patterns may influence disease behavior and prognosis [152]. This observation implies that EZH2 could serve as a prognostic marker. Overall, while many studies suggest EZH2 is involved in the pathogenesis of HNSCC and could serve as a valuable biomarker or treatment target, conflicting evidence underscores the need for further research. Understanding the context-dependent roles of EZH2 in head and neck cancers will be essential for developing precision therapies. Key studies elucidating EZH2’s role in HNSCC are summarized in Table 8.

Table 8.
EZH2 expression in head and neck cancer.
5.9. EZH2 and Kidney Cancer
Types of kidney cancer include renal cell carcinoma (RCC), transitional cell cancer (TCC), clear cell renal carcinoma (ccRCC), and Wilms tumor. Among these, RCC is the most prevalent, and numerous studies have demonstrated a strong association between EZH2 upregulation and poor clinical outcome [154]. Elevated EZH2 levels have been shown to enhance proliferation and invasion of the RCC cell line ACHN through activation of the Wnt/β-catenin signaling pathway [155]. Additionally, high EZH2 levels represses E-cadherin, a key tumor suppressor gene, and correlates with advanced disease stages and reduced survival in RCC patients [156]. In ccRCC specifically, higher EZH2 level is linked to increased expression of vascular endothelial growth factor, augmented tumor cell proliferation, and reduced apoptosis, aligning with more aggressive clinicopathological features and shorter patient survival [157]. Beyond its pro-proliferative effects, EZH2 can epigenetically silence various tumor suppressor genes and signaling pathways. For example, EZH2-mediated methylation of the Runt-related transcription factor 3 (RUNX3) promoter leads to transcriptional silencing of RUNX3, thereby promoting cancer cell proliferation [158]. Furthermore, high EZH2 expression has been associated with the presence and activation of tumor-infiltrating immune cells, suggesting a broader role in modulating the tumor microenvironment. Moreover, EZH2 depletion results in the re-expression of the cell cycle inhibitor p27/Kip1 and reduced proliferation of RCC cells [159]. EZH2 knockdown has been shown to decrease global levels of histone H3 trimethylation in ACHN cells, reinforcing its role as a key epigenetic regulator in RCC progression. Collectively, these findings underscore EZH2 as a novel prognostic marker in kidney cancer. Additional relevant studies are summarized in Table 9.

Table 9.
EZH2 expression in kidney cancer.
5.10. EZH2 and Liver Cancer
EZH2 is highly expressed in hepatocellular carcinoma (HCC) and hepatoblastoma tumor tissues and plays a critical role in promoting tumor progression through the regulation of various oncogenic and epigenetic mechanisms [166]. In HCC, one study demonstrated that EZH2 suppresses miR-381 by catalyzing H3K27me3 deposition at its promoter region, thereby enhancing SETDB1 expression and activating the AKT signaling pathway to drive tumorigenesis [167]. Furthermore, EZH2 has been shown to epigenetically silence PD-L1 by increasing H3K27me3 levels at the CD274 and IRF1 promoter regions, undermining immune checkpoint regulation and contributing to immune evasion in HCC [168]. EZH2 is also characterized by a high tumor transformation in liver cancers, and its genomic status has been associated with reduced progression-free and overall survival [169]. Conversely, suppression of EZH2 expression in liver cells leads to the upregulation of tumor suppressor proteins such as p16 and p27, contributing to inhibited tumor growth [170]. Moreover, O-linked N-acetylglucosamine transferase (OGT) expression, which is normally repressed by p53, indirectly promotes miR-15a activity, destabilizing EZH2 and attenuating HCC progression [171]. Similarly, forced expression of miR-101 in HCC cells suppresses EZH2 levels, leading to reduced oncogenic potential [172]. Beyond HCC, EZH2 has also been implicated in cholangiocarcinoma. EZH2 silencing in cholangiocarcinoma cells reduced DNA methylation at the RUNX3 promoter, thereby restoring its tumor-suppressive activity and contributing to decreased liver tumor cell proliferation [173]. These findings emphasize the critical role of EZH2 in liver cancer development and progression, highlighting its promise as a prognostic biomarker. Key supporting studies are summarized in Table 10.

Table 10.
EZH2 expression in liver cancer.
5.11. EZH2 and Lung Cancer
EZH2 exhibits oncogenic activity in lung cancer primarily by inhibiting gene transcription via promoter methylation. This epigenetic silencing contributes to tumor cell proliferation and cancer progression. A key pathway involves the immune checkpoint protein programmed death-ligand 1 (PD-L1), whose expression has been shown to correlate positively with EZH2 levels in lung adenocarcinomas [183]. Elevated expression of thyroid transcription factor-1 (TTF-1) a diagnostic marker for metastatic lung tumors-combined with low EZH2 expression, was associated with significantly improved recurrence-free survival in patients [184]. Higher EZH2 expression has also been linked to lung cancers characterized by increased KRAS and BRAF activity, particularly in lung squamous cell carcinoma [185]. Functional studies have demonstrated that silencing EZH2 in parental H2087 lung cancer cells lead to reduced expression of VEGF-A, decreased phosphorylation of AKT at Ser473, and suppression of cell proliferation, migration, and metastasis [186]. In contrast, higher levels of EZH2 in A549 cells promoted these oncogenic traits, suggesting that EZH2 facilitates lung cancer progression via the VEGF-A/AKT signaling pathway [186]. EZH2 is also highly specific to malignant phenotypes. For instance, high EZH2 expression is more frequently observed in malignant mesothelioma, a rare cancer of the pleural lining than in benign proliferative conditions [187]. In non-small cell lung cancer (NSCLC), aberrant EZH2 expression has been associated with poor disease-free survival outcomes [188]. Moreover, its expression is elevated in bronchial preneoplastic lesions, with levels increasing as lesions progress toward malignancy [185]. These findings strongly support EZH2 as a viable prognostic biomarker and therapeutic target in various forms of lung cancer. Further supporting studies are detailed in Table 11.

Table 11.
EZH2 expression in lung cancer.
5.12. EZH2 and Nasopharyngeal/Oral Cancer
EZH2 upregulation has been shown to promote the proliferation and migration of endothelial and nasopharyngeal carcinoma (NPC) cells through multiple mechanisms. One pathway involves EZH2-mediated inhibition of miR-1, resulting in increased expression of endothelin-1 (ET-1), a molecule known to promote tumor cell migration and angiogenesis [198]. Additionally, high EZH2 expression has been correlated with p63, a protein involved in epithelial regeneration and associated with significantly lower five-year disease-free survival in patients with NPC [199]. EZH2 has also been implicated in impairing the DNA repair response in NPC. Elevated levels of EZH2 expression were found to suppress the XPA gene, a key component of the nucleotide excision repair pathway [200]. In advanced-stage NPC, this inverse relationship between EZH2 and XPA was evident, and EZH2 inhibition led to increased XPA expression, thereby enhancing DNA repair and accelerating the removal of UV-induced 6-4PP and CPD-DNA adducts [200]. Moreover, EZH2 has been shown to counteract tumor suppressive mechanisms. For instance, miR-506 promotes apoptosis and inhibits proliferation and migration of NPC cells while concurrently downregulating EZH2 [200,201]. In addition, long non-coding RNA H19 has been shown to regulate EZH2 expression by suppressing miR-630, thereby activating the miR-630/EZH2 axis, which enhances NPC cell migration and oncogenic activity [202]. Modulating EZH2, either directly or via regulatory RNAs such as miR-506 or H19, offers a promising strategy for prognostication [203]. These findings highlight the multifaceted role of EZH2 in nasopharyngeal carcinoma as a prognostic marker. Additional supporting studies are summarized in Table 12.

Table 12.
EZH2 expression in nasopharyngeal carcinoma/oral cancer.
5.13. EZH2 and Ovarian Cancer
EZH2 is closely associated with increased malignancy and progression in ovarian cancer, primarily due to its ability to downregulate tumor suppressor genes and repress cell cycle inhibitors, thereby preventing cellular senescence [41]. Specifically, EZH2 has been shown to inhibit the expression of p53, a crucial tumor suppressor gene that normally functions to slow tumor formation, in ovarian cancer tissues [210]. Higher EZH2 expression correlates with therapeutic resistance by promoting DNA replication and cell proliferation [211]. Conversely, EZH2 knockdown results in decreased levels of TGF-β1, a cytokine involved in pathological suppression of normal cellular functions, and an increase in E-cadherin expression, a key component of adherens junctions with tumor-suppressive properties [212]. By inhibiting EZH2, E-cadherin-mediated cellular adhesion and normal cell function are preserved, thereby reducing the proliferation of abnormal ovarian cells. EZH2’s role also extends to the regulation of ferroptosis, a form of programmed cell death recently implicated in ovarian cancer. Upregulation of EZH2 prevents ferroptosis induction, whereas blocking EZH2 expression increases ferroptotic cell death [213]. Furthermore, elevated EZH2 expression is consistently associated with advanced clinical stages of ovarian cancer and is implicated in the progression of diverse subtypes, including ovary granulosa cell tumors [41], small cell carcinoma of the ovary hypercalcemic type (SCCOHT), and high-grade ovarian serous carcinoma (TIL-HGOSC) [214]. Collectively, these findings underscore the significant potential of EZH2 as a prognostic biomarker in ovarian cancer. Additional relevant studies are summarized in Table 13.

Table 13.
EZH2 expression in ovarian cancer.
5.14. EZH2 and Pancreatic Cancer
EZH2 signaling and methylation significantly contribute to the accelerated progression of pancreatic cancer cells. It plays a critical role in regulating cancer cell proliferation, migration, invasion, apoptosis, and cell cycle progression by modulating key signaling pathways such as Wnt, RAS, NF-κB, and NOTCH [222]. Additionally, EZH2 expression induces silencing of E-cadherin via hypermethylation of its promoter, a hallmark associated with metastasis and the development of pancreatic ductal adenocarcinoma (PDAC) [223]. EZH2 also interacts with tumor-suppressive miRNAs, including miR-218 and miR-26a, which are essential for inhibiting tumor proliferation and metastasis. By collaborating with polycomb repressive complexes PRC1 and PRC2, EZH2 promotes methylation of these miRNA promoter regions, silencing their expression in pancreatic cancer [224,225]. Similarly, EZH2 represses tumor suppressor genes like p16INK4, which normally functions to limit tumor proliferation and regeneration, thereby facilitating invasive and metastatic tumor growth [1,2]. Moreover, EZH2 activity is linked to suppression of chemokine signaling and cytotoxic lymphocyte function, correlating with reduced survival in PDAC patients [226]. Given these effects, EZH2 serves as an independent prognostic factor, with higher expression levels predicting poorer clinical outcomes. Combining EZH2 inhibition with senescence-inducing therapies may enhance immune-mediated tumor control in PDAC [227]. Overall, these findings highlight EZH2 as a valuable biomarker for pancreatic cancer prognosis. Additional studies are summarized in Table 14.

Table 14.
EZH2 expression in pancreatic cancer.
5.15. EZH2 and Prostate Cancer
EZH2 upregulation is observed throughout most stages of prostate cancer and is strongly associated with aggressive and metastatic disease. Its upregulation promotes oncogenic behaviors largely through the epigenetic silencing of tumor suppressor genes. The androgen receptor (AR), a hormone-activated transcriptional activator critical for prostate-specific cytodifferentiation, plays dual roles: it stimulates prostatic differentiation by promoting transcription of prostate-specific genes while concurrently repressing non-prostatic differentiation through cooperation with EZH2 to inhibit developmental regulators [236,237]. Prostate cancer cell invasion, angiogenesis, and stem cell-like characteristics are linked to EZH2-mediated suppression of interferon-gamma signaling via the PRC2 complex [238]. Beyond its canonical repressive functions, EZH2 also acts as a transcriptional activator or coactivator by binding other transcription factors to promote oncogene expression. For instance, deregulated phosphorylation of EZH2 can switch its function from a PRC2-dependent transcriptional repressor to a coactivator that cooperates with AR, contributing to castration-resistant prostate cancer (CRPC) [237]. Moreover, EZH2 contributes to CRPC through non-canonical mechanisms, such as directly occupying the AR promoter or methylating AR itself, enhancing AR-mediated transcription without the need for other PRC2 subunits [239]. Conversely, EZH2 can suppress AR expression in a PRC2-dependent manner [237]. EZH2 also methylates FOXA1, which recruits deubiquitinases that prevent FOXA1 degradation, elevating its protein levels [240]. Since EZH2 and FOXA1 co-regulate cell cycle progression and prostate cancer growth, their elevated expression correlates with poor prognosis [240]. Additionally, EZH2 affects DNA methylation by directly interacting with DNA methyltransferases, promoting hypermethylation of target genes like GSTP1 and RARB2—epigenetic changes frequently observed in advanced prostate cancer stages [241].
Increased EZH2 expression also facilitates the emergence of more lethal neuroendocrine prostate cancer subtypes, independent of AR signaling, characterized by poorly differentiated small-cell neuroendocrine carcinoma phenotypes [242,243]. Loss of AR and its binding to androgen-response elements following PRC2 complex displacement increases lncRNA-p21 transactivation, which promotes EZH2 release from chromatin [244]. Free EZH2 then switches roles from histone methyltransferase to non-histone methyltransferase, methylating STAT3 to promote neuroendocrine differentiation. Concurrently, EZH2 acts as a co-repressor with N-Myc to drive neuroendocrine differentiation in CRPC cells [245]. Multiple studies have investigated EZH2’s involvement in prostate cancer; a selection of key findings is summarized in Table 15.

Table 15.
EZH2 expression in prostate cancer.
5.16. EZH2 Expression and Sarcoma
Aberrant EZH2 expression has been associated with poor prognosis, distant metastasis, and tumor necrosis in synovial sarcoma [262]. In pediatric soft tissue sarcoma patients, high EZH2 expression correlated with lymph node involvement and distant metastasis at diagnosis, and those with elevated EZH2 levels showed reduced survival probabilities [263]. Similarly, EZH2 expression was found to be elevated in osteosarcoma tissues and cells. Notably, downregulation of lncRNA-ANCR led to decreased EZH2 levels and increased apoptosis of cancer cells, suggesting a potential regulatory relationship that could inhibit tumor proliferation [264]. Moreover, EZH2 inhibition sensitizes retinoic acid-driven senescence in synovial sarcoma [265]. Overall, these studies highlight the potential of EZH2 as a prognostic biomarker in sarcomas. Additional studies are summarized in Table 16.

Table 16.
EZH2 expression in sarcoma.
5.17. EZH2 and Skin Cancer
EZH2 has been implicated in the progression and prognosis of various skin cancers. A study found that higher levels of EZH2 correlated with a BCL2-negative phenotype, which is often observed in advanced disease stages and is associated with shorter event-free survival [269]. In Merkel cell carcinoma (MCC), a type of skin cancer, lower EZH2 expression in primary tumors was linked to improved prognosis and survival compared to moderate or strong EZH2 expression [270]. Additionally, EZH2 dysregulation through somatic activating mutations, copy number amplifications, or transcriptional upregulation has been associated with epigenetic silencing of tumor suppressor genes and melanoma immune responses, negatively affecting patient survival. Knockdown of T antigen in MCC cells reduced EZH2 expression, inducing selective cytotoxicity in virus-positive MCC [271]. In uveal melanoma, forced knockdown of the long non-coding RNA PVT1 suppressed tumor growth and increased apoptosis by regulating EZH2 expression [272]. Furthermore, the combined inhibition of EZH2 and BRAF in melanoma cells-especially those harboring the BRAF V600E mutation and EZH2 demonstrated enhanced therapeutic efficacy, highlighting the potential of this approach in melanoma treatment [273]. Overall, these findings underscore the critical role of EZH2 in skin cancer progression. Additional studies are summarized in Table 17.

Table 17.
EZH2 expression in skin cancers.
5.18. EZH2 and Thyroid Cancer
EZH2 upregulation is associated with malignant potential in thyroid cancer, promoting it through transcriptional repression of tumor suppressors and maintenance of cells in a stem-cell-like state [286]. EZH2 has been shown to repress the expression of classic tumor suppressor genes such as CDKN2A and p53 directly and reduces the levels of RAD51, leading to the activation of Raf1/ERK and beta-catenin signaling, leading to thyroid cancer progression [286]. EZH2 can also directly control the differentiation of anaplastic thyroid carcinoma cells by silencing the thyroid-specific transcription factor paired-box gene 8 [287]. Furthermore, EZH2 is important in medullary thyroid cancer by affecting ERK and AKT signaling pathways, as well as controlling genes of the Wnt/beta-catenin [288]. Increased EZH2 expression in papillary thyroid cancer upregulates cellular proliferation and migration by affecting the E2-ERɑ signaling pathway [289]. Beyond this, EZH2 can interact with other pathways to drive gene repression. One example of this is EZH2’s interaction with the HOTAIR (HOXA transcript antisense RNA) pathway, which together encourages an immunosuppressive microenvironment [290]. Because of these traits, EZH2 may be a useful prognostic biomarker for aggressive thyroid cancer. Studies show that certain miRNAs could directly target EZH2 and suppress its expression in thyroid cancer, such as miR-124/506 through decreased H3K27me3 and increased H3K27Ac [291]. Further studies show the inhibition of EZH2 in papillary thyroid cancer downregulates cellular proliferation and migration [289]. EZH2 inhibitors can also favorably modify the immune microenvironment. Additional studies are summarized in Table 18.

Table 18.
EZH2 expression in thyroid cancer.
5.19. EZH2 Expression in Hematological Malignancies
Hematological malignancies include a broad group of blood cancers such as leukemia, lymphoma, and myeloproliferative neoplasms (MPNs). MPNs are rare disorders characterized by the uncontrolled production of abnormal red blood cells, white blood cells, and platelets in the bone marrow. Studies have shown that EZH2 genomic alterations are frequently detected in patients with MPNs and are associated with poor clinical outcomes and early events in leukemogenesis [293,294]. Additionally, EZH2 upregulation is correlated with progression to blast phase MPN, and EZH2 aberration may play a critical role in leukemic transformation in these disorders. These findings underline the importance of EZH2 as a prognostic marker in hematological malignancies. Additional studies are summarized in Table 19.

Table 19.
EZH2 expression in myeloproliferative neoplasm.
Genomic alterations in EZH2 result in reduced mRNA expression levels in patients with acute myeloid leukemia (AML), myelodysplastic syndromes (MDS), and myelodysplastic/myeloproliferative neoplasms (MPN) [293]. Studies show that EZH2 expression can induce H3K27me3 trimethylation and confer chronic lymphocytic leukemia (CLL) cells a survival advantage [294]. This occurs through the upregulation of the PI3K/AKT pathway by way of IGF1R and MYC [297]. Therefore, higher EZH2 expression contributes to an increased growth potential of leukemic cells. Furthermore, MDS is characterized by clonal hematopoiesis and impaired differentiation and can develop into AML [294]. One study exploring the mechanism of histone methyltransferase EZH2/EHMT2 during the transformation of MDS into AML showed that NHD13 mice with higher levels of EZH2 transformed into AML. This is because EZH2 catalyzes H3K27me3/H3K9me2 to inhibit the transcription of DLX5, thus promoting the transformation from MDS to AML [298]. Beyond other functions, EZH2 is elevated in most T-cell neoplasms, suggesting that EZH2 could function as an oncogenic protein in T-cell tumorigenesis in adult T-cell leukemia [299]. EZH2 inactivation results in significantly reduced leukemia-initiating cells and enhanced differentiation through the silencing of PRC2 target genes [300]. Furthermore, low EZH2 levels resulted in a decrease in HOX genes and ultimately HOXB7 and HOXA9 knockdown in resistance cells, as shown in Table 20.

Table 20.
EZH2 expression in leukemia.
EZH2 plays an oncogenic role in lymphoma due to its ability to promote transcriptional repression of target genes [303]. EZH2 upregulation was associated with poor survival outcome, high Ki-67 proliferation rate and p53 mutant patterns caused by tumors [304]. EZH2 presence combined with p53 tumor aberrations causes a poor outcome for MCL patients [305]. Increased EZH2 expression was also correlated with poor overall survival in peripheral T-cell lymphoma (PTCL) patients [305]. EZH2 expression is also higher in aggressive B-cell lymphomas, indicating that it may act as an oncogenic protein in these tumors. EZH2 regulations may differ across various signaling pathways in aggressive B-cell lymphomas, highlighting its potential as a prognostic marker [306]. Alterations in the EZH2 gene may also contribute to its increased expression, since one study found a sizable number of follicular lymphoma patients with an altered EZH2 gene [307]. These findings highlight the importance of investigating specific genomic alterations of EZH2, which may serve as prognostic biomarkers. The results as summarized in Table 21.

Table 21.
EZH2 expression in lymphoma.
6. Conclusions and Future Perspectives
The role of EZH2 in human malignancies is well established, with its expression frequently linked to clinical outcomes across diverse cancer types. EZH2 upregulation is commonly observed in prostate, breast, lung, and hematologic cancers, where it correlates with poor prognosis and aggressive disease. Conversely, reduced EZH2 expression has been reported in certain myeloid malignancies, underscoring its context-dependent functions. Dysregulated EZH2 influences both oncogenic and tumor-suppressive pathways, reinforcing its importance as a central regulator in cancer biology.
EZH2 regulation is a multifaceted process that is highly dependent on cellular context and cancer type. Traditionally, EZH2 functions as an oncogene through its role as a transcriptional repressor, silencing tumor suppressor genes via H3K27me3 and thereby driving tumorigenesis. However, studies in hormone-regulated cancers reveal that EZH2 is more versatile, capable of acting as a transcriptional activator independent of PRC2 by targeting non-histone substrates. Its regulation is highly context-dependent and influenced by mechanisms such as transcriptional activation by oncogenic factors (e.g., MYC, ETS family), the loss of inhibitory microRNAs (e.g., miR-101, miR-26a), gene amplification, and interactions with DNA-binding proteins and ncRNAs. Moreover, post-translational modifications, interactions with cofactors, and crosstalk with other epigenetic regulators further diversify their activity (Figure 3). Given this heterogeneity, it is essential to identify the precise EZH2 target genes, whether activated or repressed, in each cancer type. Nonetheless, further studies are needed to define its precise mechanisms and downstream targets, which will be crucial for establishing EZH2 as a reliable, potentially cancer-specific diagnostic or prognostic biomarker.
Gene mapping studies have been crucial in identifying EZH2’s involvement in various cancers by characterizing genomic alterations like overexpression, mutations, and fusions. Such gene-specific mapping would help elucidate the molecular basis of EZH2’s oncogenic or tumor-suppressive roles while also supporting the development of more precise biomarkers tailored to the distinct landscapes of different cancers. However, the interpretation of its prognostic biomarkers remains limited by methodological variability, differences in patient populations, and the absence of standardized longitudinal studies. More consistent approaches and larger cohorts are needed to validate EZH2 as a robust prognostic biomarker. Given the complex regulation of EZH2—shaped by cellular context, cancer type, and specific target genes—defining which genes are activated or repressed by EZH2 in individual malignancies will be essential for tailoring management strategies. At the same time, an important but often overlooked challenge lies in malignancies where loss-of-function EZH2 might contribute to disease progression. Addressing these contrasting roles will be critical for the development of effective EZH2-based biomarkers and precision management strategies in oncology.
Author Contributions
Conceptualization, S.V., P.K. and S.G. (Sanjay Gupta); software, S.V.; formal analysis, S.V.; resources, S.G. (Sanjay Gupta); data curation, S.V., N.G. and S.G. (Suhani Goyal); writing—original draft preparation, S.V., P.K. and S.G. (Sanjay Gupta); writing—review and editing, S.V. and S.G. (Sanjay Gupta); visualization, S.G. (Sanjay Gupta); supervision, S.G. (Sanjay Gupta); funding acquisition, S.G. (Sanjay Gupta). All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Department of Defense Grants W81XWH-18-1-0618, W81XWH-19-1-0720, and Endowment funds to S.G.
Data Availability Statement
No new data were created or analyzed in this study.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
AEBP2: AE binding protein 2; AML, acute myeloid leukemia; AMPK, AMP-activated protein kinase; AR, androgen receptor; BLACAT1, BLACAT1 overlapping LEMD1 locus; BTRC, beta-transducin repeat containing E3 ubiquitin protein ligase; ccRCC, clear cell renal carcinoma; CDKN1C, cyclin-dependent kinase inhibitor 1C; circRNAs, circular RNAs; CLL, chronic lymphocytic leukemia; CRPC, castration-resistant prostate cancer; CXC domain, cysteine-rich domain; DFS, disease free survival; DNMTs, DNA methyltransferases; EBD, EED-binding domain of EZH2; EED, embryonic ectoderm development; EID, EED-interaction domain; ELISA, enzyme-linked immunosorbent assay; ER, estrogen receptor; ESCC, esophageal squamous cell carcinoma; EZH2, enhancer of zeste homolog 2; FBXW7, F-box/WD repeat-containing protein 7; FOX, Forkhead box; GBM, glioblastoma; GSTP1, glutathione S-transferase pi 1; H3K27me3, trimethylation of histone H3 at lysine 27; HCC, hepatocellular carcinoma; HDACs, histone deacetylases; HER2, human epidermal growth factor receptor 2; HMT, histone methyltransferase; HOTAIR, HOX transcript antisense RNA; IGF1R, insulin-like growth factor 1 receptor; IHC, immunohistochemistry; JARID2, jumonji, AT-rich interactive domain 2; KDM1A/LSD1, lysine-specific demethylase 1A; lncRNA, long non-coding RNA; MALAT1, metastasis associated lung adenocarcinoma transcript 1; MCC, Merkel cell carcinoma; MDS, myelodysplastic syndrome; MELK, maternal embryonic leucine-zipper kinase; MPN, myelodysplastic/myeloproliferative neoplasms; MPN, myeloproliferative neoplasm; MTF2, metal response element binding transcription factor 2; NGS, next gen sequencing; NPC, nasopharyngeal carcinoma; NSCLC, non-small cell lung cancer; OS, overall survival; PDAC, pancreatic ductal adenocarcinoma; PD-L1, programmed death-ligand 1; PgR, progesterone receptor; PHF1, PHD finger protein 1; PRC2, polycomb-repressive complex 2; qRT-PCR, quantitative reverse transcriptase PCR; RARα, retinoic acid receptor α; RBBP7, RB binding protein 7; RCC, renal cell carcinoma; RECK, reversion-inducing cysteine-rich protein with kazal motifs; RNA, ribonucleic acid; RORA, RAR-related orphan receptor alpha; RUNX3, RUNX family transcription factor 3; SAM, S-adenosyl-methionine; SCCOHT, small cell carcinoma of the ovary hypercalcemic type; SET domain, enhancer of zeste and trithorax domain; STAT3, signal transducer and activator of transcription 3; TCC, transitional cell cancer; TGF, transforming growth factor; TNBC, triple-negative breast cancer; TRIM28, tripartite motif containing 28; TTF-1, thyroid transcription factor-1; USP, ubiquitin-specific-processing protease; VEGF, vascular endothelial growth factor; WB, Western blotting; XIST, X inactive specific transcript; ZBTB16/PLZF, zinc finger and BTB domain containing 16/promyelocytic leukemia zinc finger.
References
- Deb, G.; Singh, A.K.; Gupta, S. EZH2: Not EZHY (easy) to deal. Mol. Cancer Res. 2014, 12, 639–653. [Google Scholar] [CrossRef]
- Duan, R.; Du, W.; Guo, W. EZH2: A novel target for cancer treatment. J. Hematol. Oncol. 2020, 13, 104. [Google Scholar] [CrossRef]
- Nichol, J.N.; Dupéré-Richer, D.; Ezponda, T.; Licht, J.D.; Miller, W.H., Jr. H3K27 Methylation: A Focal Point of Epigenetic Deregulation in Cancer. Adv. Cancer Res. 2016, 131, 59–95. [Google Scholar] [CrossRef]
- Chammas, P.; Mocavini, I.; Di Croce, L. Engaging chromatin: PRC2 structure meets function. Br. J. Cancer 2020, 122, 315–328. [Google Scholar] [CrossRef]
- Gan, L.; Yang, Y.; Li, Q.; Feng, Y.; Liu, T.; Guo, W. Epigenetic regulation of cancer progression by EZH2: From biological insights to therapeutic potential. Biomark. Res. 2018, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Shankar, E.; Gupta, S. EZH2-mediated development of therapeutic resistance in cancer. Cancer Lett. 2024, 586, 216706. [Google Scholar] [CrossRef] [PubMed]
- Tamburri, S.; Rustichelli, S.; Amato, S.; Pasini, D. Navigating the complexity of Polycomb repression: Enzymatic cores and regulatory modules. Mol. Cell 2024, 84, 3381–3405. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Liu, Y.; Hsu, Y.-J.; Fujiwara, Y.; Kim, J.; Mao, X.; Yuan, G.-C.; Orkin, S.H. EZH1 Mediates Methylation on Histone H3 Lysine 27 and Complements EZH2 in Maintaining Stem Cell Identity and Executing Pluripotency. Mol. Cell 2008, 32, 491–502. [Google Scholar] [CrossRef]
- Han, Z.; Xing, X.; Hu, M.; Zhang, Y.; Liu, P.; Chai, J. Structural Basis of EZH2 Recognition by EED. Structure 2007, 15, 1306–1315. [Google Scholar] [CrossRef][Green Version]
- Laugesen, A.; Højfeldt, J.W.; Helin, K. Molecular Mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol. Cell 2019, 74, 8–18. [Google Scholar] [CrossRef]
- Cao, R.; Zhang, Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 2004, 14, 155–164. [Google Scholar] [CrossRef]
- Kim, E.; Kim, M.; Woo, D.H.; Shin, Y.; Shin, J.; Chang, N.; Oh, Y.T.; Kim, H.; Rheey, J.; Nakano, I.; et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 2013, 23, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Li, Y.; Meng, Q.; Li, Q.; Li, F.; Lu, B.; Shen, J.; Fazli, L.; Zhao, D.; Li, C.; et al. A PRC2-independent function for EZH2 in regulating rRNA 2′-O methylation and IRES-dependent translation. Nat. Cell Biol. 2021, 23, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.; Tan, D.; Zhu, M.; Qu, Y.; Ma, X.; Song, B.-L.; Qi, W. EZH2 W113C is a gain-of-function mutation in B-cell lymphoma enabling both PRC2 methyltransferase activation and tazemetostat resistance. J. Biol. Chem. 2023, 299, 103073. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Shubbar, M.; Yang, X.; Zhang, Q.; Chen, S.; Wu, Q.; Chen, Z.; Rizo, J.; Liu, X. A partially disordered region connects gene repression and activation functions of EZH2. Proc. Natl. Acad. Sci. USA 2020, 117, 16992–17002. [Google Scholar] [CrossRef]
- Wu, H.; Zeng, H.; Dong, A.; Li, F.; He, H.; Senisterra, G.; Seitova, A.; Duan, S.; Brown, P.J.; Vedadi, M.; et al. Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations. PLoS ONE 2013, 8, e83737. [Google Scholar] [CrossRef]
- Tan, J.-z.; Yan, Y.; Wang, X.-x.; Jiang, Y.; Xu, H.E. EZH2: Biology, disease, and structure-based drug discovery. Acta Pharmacol. Sin. 2014, 35, 161–174. [Google Scholar] [CrossRef]
- McCabe, M.T.; Graves, A.P.; Ganji, G.; Diaz, E.; Halsey, W.S.; Jiang, Y.; Smitheman, K.N.; Ott, H.M.; Pappalardi, M.B.; Allen, K.E.; et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc. Natl. Acad. Sci. USA 2012, 109, 2989–2994. [Google Scholar] [CrossRef]
- Majer, C.R.; Jin, L.; Scott, M.P.; Knutson, S.K.; Kuntz, K.W.; Keilhack, H.; Smith, J.J.; Moyer, M.P.; Richon, V.M.; Copeland, R.A.; et al. A687V EZH2 is a gain-of-function mutation found in lymphoma patients. FEBS Lett. 2012, 586, 3448–3451. [Google Scholar] [CrossRef]
- Sneeringer, C.J.; Scott, M.P.; Kuntz, K.W.; Knutson, S.K.; Pollock, R.M.; Richon, V.M.; Copeland, R.A. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl. Acad. Sci. USA 2010, 107, 20980–20985. [Google Scholar] [CrossRef]
- Cha, T.L.; Zhou, B.P.; Xia, W.; Wu, Y.; Yang, C.C.; Chen, C.T.; Ping, B.; Otte, A.P.; Hung, M.C. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 2005, 310, 306–310. [Google Scholar] [CrossRef]
- Kaur, P.; Verma, S.; Kushwaha, P.P.; Gupta, S. EZH2 and NF-κB: A context-dependent crosstalk and transcriptional regulation in cancer. Cancer Lett. 2023, 560, 216143. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.C.; LaBaff, A.; Wei, Y.; Nie, L.; Xia, W.; Huo, L.; Yamaguchi, H.; Hsu, Y.H.; Hsu, J.L.; Liu, D.; et al. Phosphorylation of EZH2 at T416 by CDK2 contributes to the malignancy of triple negative breast cancers. Am. J. Transl. Res. 2015, 7, 1009–1020. [Google Scholar] [PubMed]
- Hernández-Muñoz, I.; Taghavi, P.; Kuijl, C.; Neefjes, J.; van Lohuizen, M. Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1. Mol. Cell Biol. 2005, 25, 11047–11058. [Google Scholar] [CrossRef] [PubMed]
- Viré, E.; Brenner, C.; Deplus, R.; Blanchon, L.; Fraga, M.; Didelot, C.; Morey, L.; Van Eynde, A.; Bernard, D.; Vanderwinden, J.-M.; et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006, 439, 871–874. [Google Scholar] [CrossRef]
- Ohm, J.E.; McGarvey, K.M.; Yu, X.; Cheng, L.; Schuebel, K.E.; Cope, L.; Mohammad, H.P.; Chen, W.; Daniel, V.C.; Yu, W.; et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 2007, 39, 237–242. [Google Scholar] [CrossRef]
- Schlesinger, Y.; Straussman, R.; Keshet, I.; Farkash, S.; Hecht, M.; Zimmerman, J.; Eden, E.; Yakhini, Z.; Ben-Shushan, E.; Reubinoff, B.E.; et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007, 39, 232–236. [Google Scholar] [CrossRef]
- Kuzmichev, A.; Nishioka, K.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002, 16, 2893–2905. [Google Scholar] [CrossRef]
- van der Vlag, J.; Otte, A.P. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat. Genet. 1999, 23, 474–478. [Google Scholar] [CrossRef]
- Lee, E.R.; Murdoch, F.E.; Fritsch, M.K. High histone acetylation and decreased polycomb repressive complex 2 member levelsregulate gene specific transcriptional changes during early embryonic stem cell differentiation induced by retinoic acid. Stem Cells 2007, 25, 2191–2199. [Google Scholar] [CrossRef]
- Tie, F.; Banerjee, R.; Stratton, C.A.; Prasad-Sinha, J.; Stepanik, V.; Zlobin, A.; Diaz, M.O.; Scacheri, P.C.; Harte, P.J. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 2009, 136, 3131–3141. [Google Scholar] [CrossRef]
- Kim, J.; Lee, H.; Yi, S.-J.; Kim, K. Gene regulation by histone-modifying enzymes under hypoxic conditions: A focus on histone methylation and acetylation. Exp. Mol. Med. 2022, 54, 878–889. [Google Scholar] [CrossRef]
- Cookis, T.; Lydecker, A.; Sauer, P.; Kasinath, V.; Nogales, E. Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications. Nat. Struct. Mol. Biol. 2025, 32, 393–404. [Google Scholar] [CrossRef]
- Kim, K.H.; Roberts, C.W. Targeting EZH2 in cancer. Nat. Med. 2016, 22, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Porazzi, P.; Nason, S.; Yang, Z.; Carturan, A.; Ghilardi, G.; Guruprasad, P.; Patel, R.P.; Tan, M.; Padmanabhan, A.A.; Lemoine, J.; et al. EZH1/EZH2 inhibition enhances adoptive T cell immunotherapy against multiple cancer models. Cancer Cell 2025, 43, 537–551.e7. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, M.; Wang, D.; Hou, P.; Chen, X.; Chu, S.; Chai, D.; Zheng, J.; Bai, J. Post-translational modifications of EZH2 in cancer. Cell Biosci. 2020, 10, 143. [Google Scholar] [CrossRef] [PubMed]
- Kempkes, R.W.M.; Prinjha, R.K.; de Winther, M.P.J.; Neele, A.E. Novel insights into the dynamic function of PRC2 in innate immunity. Trends Immunol. 2024, 45, 1015–1030. [Google Scholar] [CrossRef]
- Zimmerman, S.M.; Lin, P.N.; Souroullas, G.P. Non-canonical functions of EZH2 in cancer. Front. Oncol. 2023, 13, 1233953. [Google Scholar] [CrossRef]
- Guo, Y.; Cheng, R.; Wang, Y.; Gonzalez, M.E.; Zhang, H.; Liu, Y.; Kleer, C.G.; Xue, L. Regulation of EZH2 protein stability: New mechanisms, roles in tumorigenesis, and roads to the clinic. EBioMedicine 2024, 100, 104972. [Google Scholar] [CrossRef]
- Beca, F.; Kensler, K.; Glass, B.; Schnitt, S.J.; Tamimi, R.M.; Beck, A.H. EZH2 protein expression in normal breast epithelium and risk of breast cancer: Results from the Nurses’ Health Studies. Breast Cancer Res. 2017, 19, 21. [Google Scholar] [CrossRef]
- Jones, B.A.; Varambally, S.; Arend, R.C. Histone Methyltransferase EZH2: A Therapeutic Target for Ovarian Cancer. Mol. Cancer Ther. 2018, 17, 591–602. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Q. The roles of EZH2 in cancer and its inhibitors. Med. Oncol. 2023, 40, 167. [Google Scholar] [CrossRef] [PubMed]
- Goleij, P.; Heidari, M.M.; Tabari, M.A.K.; Hadipour, M.; Rezaee, A.; Javan, A.; Sanaye, P.M.; Larsen, D.S.; Daglia, M.; Khan, H. Polycomb repressive complex 2 (PRC2) pathway’s role in cancer cell plasticity and drug resistance. Funct. Integr. Genom. 2025, 25, 53. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, P.; Ye, J.; Xie, G.; Yang, J.; Liu, W. Serum EZH2 is a novel biomarker for bladder cancer diagnosis and prognosis. Front. Oncol. 2024, 14, 1303918. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Yu, J.; Dhanasekaran, S.M.; Kim, J.H.; Mani, R.S.; Tomlins, S.A.; Mehra, R.; Laxman, B.; Cao, X.; Yu, J.; et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 2008, 27, 7274–7284. [Google Scholar] [CrossRef]
- Wang, H.; Mei, Y.; Luo, C.; Huang, Q.; Wang, Z.; Lu, G.-M.; Qin, L.; Sun, Z.; Huang, C.-W.; Yang, Z.-W.; et al. Single-Cell Analyses Reveal Mechanisms of Cancer Stem Cell Maintenance and Epithelial–Mesenchymal Transition in Recurrent Bladder Cancer. Clin. Cancer Res. 2022, 27, 6265–6278. [Google Scholar] [CrossRef]
- Chen, Z.; Du, Y.; Liu, X.; Chen, H.; Weng, X.; Guo, J.; Wang, M.; Wang, X.; Wang, L. EZH2 inhibition suppresses bladder cancer cell growth and metastasis via the JAK2/STAT3 signaling pathway. Oncol. Lett. 2019, 18, 907–915. [Google Scholar] [CrossRef]
- Mirzaei, S.; Gholami, M.H.; Hushmandi, K.; Hashemi, F.; Zabolian, A.; Canadas, I.; Zarrabi, A.; Nabavi, N.; Aref, A.R.; Crea, F.; et al. The long and short non-coding RNAs modulating EZH2 signaling in cancer. J. Hematol. Oncol. 2022, 15, 18. [Google Scholar] [CrossRef]
- Min, J.; Ma, J.; Wang, Q.; Yu, D. Long non-coding RNA SNHG1 promotes bladder cancer progression by upregulating EZH2 and repressing KLF2 transcription. Clinics 2022, 77, 100081. [Google Scholar] [CrossRef]
- Xiang, W.; Lyu, L.; Huang, T.; Zheng, F.; Yuan, J.; Zhang, C.; Jiang, G. The long non-coding RNA SNHG1 promotes bladder cancer progression by interacting with miR-143-3p and EZH2. J. Cell. Mol. Med. 2020, 24, 11858–11873. [Google Scholar] [CrossRef]
- Weikert, S.; Christoph, F.; Köllermann, J.; Müller, M.; Schrader, M.; Miller, K.; Krause, H. Expression levels of the EZH2 polycomb transcriptional repressor correlate with aggressiveness and invasive potential of bladder carcinomas. Int. J. Mol. Med. 2005, 16, 349–353. [Google Scholar] [CrossRef]
- Mohamedali, R.; Mitra, S.; Mandal, S.; Nayak, P.; Adhya, A.K.; Purkait, S. Expression of EZH2 and H3K27me3 predicts tumor biology of urothelial carcinoma. Indian J. Pathol. Microbiol. 2023, 66, 488–494. [Google Scholar]
- Sameh, R.; Mostafa, N.; Ramadan, M.; AbdelRaouf, S.; Abdelwahab, K. Prognostic significance of EZH2 and ARID1A expression in urothelial carcinoma: An immunohistochemical study. J. Histotechnol. 2022, 45, 21–28. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, X.; Wang, Q.; Kong, Z. EZH2 targeting to improve the sensitivity of acquired radio-resistance bladder cancer cells. Transl. Oncol. 2022, 16, 101316. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, N.; Zhang, J.; Ji, H.; Liu, Y.; Yang, J.; Chen, Z. Increased expression of EZH2 indicates aggressive potential of urothelial carcinoma of the bladder in a Chinese population. Sci. Rep. 2018, 8, 17792. [Google Scholar] [CrossRef]
- Bi, H.; Zhang, Z.; Guo, L.; Fu, C. Effect of wound fluid on chemotherapy sensitivity of T24 bladder cancer cells with different enhancer of zeste homolog 2 status. Oncotarget 2017, 8, 63258–63264. [Google Scholar] [CrossRef] [PubMed]
- Warrick, J.I.; Raman, J.D.; Kaag, M.; Bruggeman, T.; Cates, J.; Clark, P.; DeGraff, D.J. Enhancer of zeste homolog 2 (EZH2) expression in bladder cancer. Urol. Oncol. Semin. Orig. Investig. 2016, 34, e251–e258. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.S.; Liao, C.H.; Tsai, C.W.; Hu, P.S.; Wu, H.C.; Hsu, S.W.; Hsiao, C.L.; Hsu, C.H.; Hung, Y.W.; Bau, D.T. Association of Enhancer of Zeste 2 (EZH2) Genotypes with Bladder Cancer Risk in Taiwan. Anticancer. Res. 2016, 36, 4509–4514. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.L.; Su, K.J.; Hsieh, M.J.; Wang, S.S.; Wang, P.H.; Weng, W.C.; Yang, S.F. Impact of EZH2 polymorphisms on urothelial cell carcinoma susceptibility and clinicopathologic features. PLoS ONE 2014, 9, e93635. [Google Scholar] [CrossRef]
- Hayashi, A.; Morikawa, T.; Kawai, T.; Kume, H.; Ishikawa, S.; Homma, Y.; Fukayama, M. Clinicopathological and prognostic significance of EZH2 expression in upper urinary tract carcinoma. Virchows Arch. 2014, 464, 463–471. [Google Scholar] [CrossRef]
- Wang, H.; Albadine, R.; Magheli, A.; Guzzo, T.J.; Ball, M.W.; Hinz, S.; Schoenberg, M.P.; Netto, G.J.; Gonzalgo, M.L. Increased EZH2 protein expression is associated with invasive urothelial carcinoma of the bladder. Urol. Oncol. Semin. Orig. Investig. 2012, 30, 428–433. [Google Scholar] [CrossRef]
- Hinz, S.; Kempkensteffen, C.; Christoph, F.; Krause, H.; Schrader, M.; Schostak, M.; Miller, K.; Weikert, S. Expression parameters of the polycomb group proteins BMI1, SUZ12, RING1 and CBX7 in urothelial carcinoma of the bladder and their prognostic relevance. Tumour Biol. 2008, 29, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Hinz, S.; Kempkensteffen, C.; Christoph, F.; Hoffmann, M.; Krause, H.; Schrader, M.; Schostak, M.; Miller, K.; Weikert, S. Expression of the polycomb group protein EZH2 and its relation to outcome in patients with urothelial carcinoma of the bladder. J. Cancer Res. Clin. Oncol. 2008, 134, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Raman, J.D.; Mongan, N.P.; Tickoo, S.K.; Boorjian, S.A.; Scherr, D.S.; Gudas, L.J. Increased expression of the polycomb group gene, EZH2, in transitional cell carcinoma of the bladder. Clin. Cancer Res. 2005, 11 Pt 1, 8570–8576. [Google Scholar] [CrossRef] [PubMed]
- Arisan, S.; Buyuktuncer, E.D.; Palavan-Unsal, N.; Caşkurlu, T.; Cakir, O.O.; Ergenekon, E. Increased expression of EZH2, a polycomb group protein, in bladder carcinoma. Urol. Int. 2005, 75, 252–257. [Google Scholar] [CrossRef]
- Adibfar, S.; Elveny, M.; Kashikova, H.S.; Mikhailova, M.V.; Farhangnia, P.; Vakili-Samiani, S.; Tarokhian, H.; Jadidi-Niaragh, F. The molecular mechanisms and therapeutic potential of EZH2 in breast cancer. Life Sci. 2021, 286, 120047. [Google Scholar] [CrossRef]
- Yersal, O.; Barutca, S. Biological subtypes of breast cancer: Prognostic and therapeutic implications. World J. Clin. Oncol. 2014, 5, 412–424. [Google Scholar] [CrossRef]
- Jang, S.H.; Lee, J.E.; Oh, M.H.; Lee, J.H.; Cho, H.D.; Kim, K.J.; Kim, S.Y.; Han, S.W.; Kim, H.J.; Bae, S.B.; et al. High EZH2 Protein Expression Is Associated with Poor Overall Survival in Patients with Luminal A Breast Cancer. J. Breast Cancer 2016, 19, 53–60. [Google Scholar] [CrossRef]
- Chen, Y.; Zhu, H.; Luo, Y.; Tong, S.; Liu, Y. EZH2: The roles in targeted therapy and mechanisms of resistance in breast cancer. Biomed. Pharmacother. 2024, 175, 116624. [Google Scholar] [CrossRef]
- Shi, B.; Liang, J.; Yang, X.; Wang, Y.; Zhao, Y.; Wu, H.; Sun, L.; Zhang, Y.; Chen, Y.; Li, R.; et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol. Cell. Biol. 2007, 27, 5105–5119. [Google Scholar] [CrossRef]
- Lee, S.T.; Li, Z.; Wu, Z.; Aau, M.; Guan, P.; Karuturi, R.K.M.; Liou, Y.C.; Yu, Q. Context-Specific Regulation of NF-κB Target Gene Expression by EZH2 in Breast Cancers. Mol. Cell 2011, 43, 798–810. [Google Scholar] [CrossRef]
- Anwar, T.; Gonzalez, M.E.; Kleer, C.G. Noncanonical Functions of the Polycomb Group Protein EZH2 in Breast Cancer. Am. J. Pathol. 2021, 191, 774–783. [Google Scholar] [CrossRef]
- Yu, F.; Li, L.; Zhang, M.; Sun, S. Phosphorylation of EZH2 differs HER2-positive breast cancer invasiveness in a site-specific manner. BMC Cancer 2023, 23, 948. [Google Scholar] [CrossRef]
- Zhou, L.; Yu, C.-W. Epigenetic modulations in triple-negative breast cancer: Therapeutic implications for tumor microenvironment. Pharmacol. Res. 2024, 204, 107205. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Lee, S.T.; Qiao, Y.; Li, Z.; Lee, P.L.; Lee, Y.J.; Jiang, X.; Tan, J.; Aau, M.; Lim, C.Z.H.; et al. Polycomb protein EZH2 regulates cancer cell fate decision in response to DNA damage. Cell Death Differ. 2011, 18, 1771–1779. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, C.; Li, S.; Yin, F.; Luo, H.; Zhang, Y.; Luo, Z.; Chen, Y.; Wan, S.; Kong, L.; et al. Dual target PARP1/EZH2 inhibitors inducing excessive autophagy and producing synthetic lethality for triple-negative breast cancer therapy. Eur. J. Med. Chem. 2024, 265, 116054. [Google Scholar] [CrossRef]
- Mao, Q.; Wu, P.; Li, H.; Fu, X.; Gao, X.; Yang, L. CRISPR/Cas9-mediated EZH2 knockout suppresses the proliferation and migration of triple-negative breast cancer cells. Oncol. Lett. 2023, 26, 343. [Google Scholar] [CrossRef]
- Gan, Y.; Lo, Y.; Makower, D.; Kleer, C.; Lu, J.; Fineberg, S. EZH2 Protein Expression in Estrogen Receptor Positive Invasive Breast Cancer Treated with Neoadjuvant Endocrine Therapy: An Exploratory Study of Association with Tumor Response. Appl. Immunohistochem. Mol. Morphol. 2022, 30, 614–622. [Google Scholar] [CrossRef]
- Wang, Y.F.; Yu, L.; Hu, Z.L.; Fang, Y.F.; Shen, Y.Y.; Song, M.F.; Chen, Y. Regulation of CCL2 by EZH2 affects tumor-associated macrophages polarization and infiltration in breast cancer. Cell Death Dis. 2022, 13, 748. [Google Scholar] [CrossRef]
- Liu, L.C.; Chien, Y.C.; Wu, G.W.; Hua, C.H.; Tsai, I.C.; Hung, C.C.; Wu, T.K.; Pan, Y.R.; Yang, S.F.; Yu, Y.L. Analysis of EZH2 Genetic Variants on Triple-Negative Breast Cancer Susceptibility and Pathology. Int. J. Med. Sci. 2022, 19, 1023–1028. [Google Scholar] [CrossRef]
- McMullen, E.R.; Skala, S.L.; Gonzalez, M.E.; Djomehri, S.; Chandrashekar, D.S.; Varambally, S.; Kleer, C.G. Subcellular localization of EZH2 phosphorylated at T367 stratifies metaplastic breast carcinoma subtypes. Breast Cancer 2021, 28, 496–505. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhao, S.; Shi, Z.; Cao, L.; Liu, J.; Pan, T.; Zhou, D.; Zhang, J. Chemotherapy-elicited exosomal miR-378a-3p and miR-378d promote breast cancer stemness and chemoresistance via the activation of EZH2/STAT3 signaling. J. Exp. Clin. Cancer Res. 2021, 40, 120. [Google Scholar] [CrossRef]
- Zhou, X.; Jiao, D.; Dou, M.; Zhang, W.; Lv, L.; Chen, J.; Li, L.; Wang, L.; Han, X. Curcumin inhibits the growth of triple-negative breast cancer cells by silencing EZH2 and restoring DLC1 expression. J. Cell. Mol. Med. 2020, 24, 10648–10662. [Google Scholar] [CrossRef]
- Dou, D.; Ge, X.; Wang, X.; Xu, X.; Zhang, Z.; Seng, J.; Cao, Z.; Gu, Y.; Han, M. EZH2 Contributes To Cisplatin Resistance In Breast Cancer by Epigenetically Suppressing miR-381 Expression. OncoTargets Ther. 2019, 12, 9627–9637. [Google Scholar] [CrossRef]
- Anwar, T.; Arellano-Garcia, C.; Ropa, J.; Chen, Y.-C.; Kim, H.S.; Yoon, E.; Grigsby, S.; Basrur, V.; Nesvizhskii, A.I.; Muntean, A.; et al. p38-mediated phosphorylation at T367 induces EZH2 cytoplasmic localization to promote breast cancer metastasis. Nat. Commun. 2018, 9, 2801. [Google Scholar] [CrossRef]
- Boostani, F.; Dolatkhah, R.; Fakhrjou, A.; Farassati, F.; Sanaat, Z. Association of clinicopathologic characteristics and outcomes with EZH2 expression in patients with breast cancer in East Azerbaijan, Iran. OncoTargets Ther. 2018, 11, 449–457. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, Z.; Cenciarini, M.E.; Proietti, C.J.; Amasino, M.; Hong, T.; Yang, M.; Liao, Y.; Chiang, H.C.; Kaklamani, V.G.; et al. Tamoxifen Resistance in Breast Cancer Is Regulated by the EZH2-ERα-GREB1 Transcriptional Axis. Cancer Res. 2018, 78, 671–684. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, T.; Bao, X.; He, M.; Li, L.; Yang, X. Increased EZH2 expression is associated with proliferation and progression of cervical cancer and indicates a poor prognosis. Int. J. Gynecol. Pathol. 2014, 33, 218–224. [Google Scholar] [CrossRef]
- Zhang, L.; Tian, S.; Chang, J.; Quan, S.; Yang, T.; Zhao, M.; Wang, L.; Yang, X. Activation of the CCL22/CCR4 causing EMT process remodeling under EZH2-mediated epigenetic regulation in cervical carcinoma. J. Cancer 2024, 15, 6299–6314. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Li, Y.; Wang, L.; Yuan, N.; Ma, M.; Chen, Y. LncRNA SNHG8 accelerates proliferation and inhibits apoptosis in HPV-induced cervical cancer through recruiting EZH2 to epigenetically silence RECK expression. J. Cell. Biochem. 2020, 121, 4120–4129. [Google Scholar] [CrossRef] [PubMed]
- Salmerón-Bárcenas, E.G.; Zacapala-Gómez, A.E.; Ortiz-Ortiz, J.; Torres-Rojas, F.I.; Ávila-López, P.A. Integrated bioinformatics analysis reveals that EZH2-rich domains promote transcriptional repression in cervical cancer. EXCLI J. 2022, 21, 852–868. [Google Scholar] [CrossRef]
- Zhang, L.; Tian, S.; Pei, M.; Zhao, M.; Wang, L.; Jiang, Y.; Yang, T.; Zhao, J.; Song, L.; Yang, X. Crosstalk between histone modification and DNA methylation orchestrates the epigenetic regulation of the costimulatory factors, Tim-3 and galectin-9, in cervical cancer. Oncol. Rep. 2019, 42, 2655–2669. [Google Scholar] [CrossRef]
- Azizmohammadi, S.; Azizmohammadi, S.; Safari, A.; Kaghazian, M.; Sadrkhanlo, M.; Behnod, V.; Seifoleslami, M. High-Level Expression of RIPK4 and EZH2 Contributes to Lymph Node Metastasis and Predicts Favorable Prognosis in Patients With Cervical Cancer. Oncol. Res. 2017, 25, 495–501. [Google Scholar] [CrossRef]
- Chen, Q.; Zheng, P.S.; Yang, W.T. EZH2-mediated repression of GSK-3β and TP53 promotes Wnt/β-catenin signaling-dependent cell expansion in cervical carcinoma. Oncotarget 2016, 7, 36115–36129. [Google Scholar] [CrossRef]
- Chen, Z.; Yang, P.; Li, W.; He, F.; Wei, J.; Zhang, T.; Zhong, J.; Chen, H.; Cao, J. Expression of EZH2 is associated with poor outcome in colorectal cancer. Oncol. Lett. 2018, 15, 2953–2961. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Wu, J.; Qiu, X. LINC01116 facilitates colorectal cancer cell proliferation and angiogenesis through targeting EZH2-regulated TPM1. J. Transl. Med. 2021, 19, 45. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, I.; Nosho, K.; Kanno, S.; Igarashi, H.; Kurihara, H.; Ishigami, K.; Ishiguro, K.; Mitsuhashi, K.; Maruyama, R.; Koide, H.; et al. EZH2 expression is a prognostic biomarker in patients with colorectal cancer treated with anti-EGFR therapeutics. Oncotarget 2017, 8, 17810–17818. [Google Scholar] [CrossRef] [PubMed]
- Ghobashi, A.H.; Vuong, T.T.; Kimani, J.W.; Ladaika, C.A.; Hollenhorst, P.C.; O’Hagan, H.M. Activation of AKT induces EZH2-mediated β-catenin trimethylation in colorectal cancer. iScience 2023, 26, 107630. [Google Scholar] [CrossRef]
- Liu, Y.L.; Gao, X.; Jiang, Y.; Zhang, G.; Sun, Z.C.; Cui, B.B.; Yang, Y.M. Expression and clinicopathological significance of EED, SUZ12 and EZH2 mRNA in colorectal cancer. J. Cancer Res. Clin. Oncol. 2015, 141, 661–669. [Google Scholar] [CrossRef]
- Meng, X.; Huang, Z.; Wang, R.; Jiao, Y.; Li, H.; Xu, X.; Feng, R.; Zhu, K.; Jiang, S.; Yan, H.; et al. The prognostic role of EZH2 expression in rectal cancer patients treated with neoadjuvant chemoradiotherapy. Radiat. Oncol. 2014, 9, 188. [Google Scholar] [CrossRef]
- Cheraghi, S.; Asadzadeh, H.; Javadi, G. Dysregulated Expression of Long Non-Coding RNA MINCR and EZH2 in Colorectal Cancer. Iran. Biomed. J. 2022, 26, 64–69. [Google Scholar] [PubMed]
- Abou Gabal, H.; Ahmed, N.; Meckawy, G.; Yassin, R.; Hakim, S. Evaluation of EZH2 and ERRα in colorectal carcinoma: An immunohistochemical study. Pol. J. Pathol. 2021, 72, 200–210. [Google Scholar] [CrossRef]
- Sanches, J.G.P.; Song, B.; Zhang, Q.; Cui, X.; Yabasin, I.B.; Ntim, M.; Li, X.; He, J.; Zhang, Y.; Mao, J.; et al. The Role of KDM2B and EZH2 in Regulating the Stemness in Colorectal Cancer Through the PI3K/AKT Pathway. Front. Oncol. 2021, 11, 637298. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, H.; Maruyama, R.; Ishiguro, K.; Kanno, S.; Yamamoto, I.; Ishigami, K.; Mitsuhashi, K.; Igarashi, H.; Ito, M.; Tanuma, T.; et al. The relationship between EZH2 expression and microRNA-31 in colorectal cancer and the role in evolution of the serrated pathway. Oncotarget 2016, 7, 12704–12717. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Luo, X.; Xiang, L.S.; Li, H.T.; Zha, L.; Li, N.; He, J.M.; Xie, G.F.; Xie, X.; Liang, H.J. EZH2 promotes colorectal cancer stem-like cell expansion by activating p21cip1-Wnt/β-catenin signaling. Oncotarget 2016, 7, 41540–41558. [Google Scholar] [CrossRef]
- Fornaro, L.; Faviana, P.; De Gregorio, V.; Vivaldi, C.; Paolicchi, E.; Masi, G.; Loupakis, F.; Sensi, E.; Lupi, C.; Fontanini, G.; et al. Molecular and pathological characterization of the EZH2 rs3757441 single nucleotide polymorphism in colorectal cancer. BMC Cancer 2015, 15, 874. [Google Scholar] [CrossRef]
- Benard, A.; Goossens-Beumer, I.J.; van Hoesel, A.Q.; Horati, H.; Putter, H.; Zeestraten, E.C.; van de Velde, C.J.; Kuppen, P.J. Prognostic value of polycomb proteins EZH2, BMI1 and SUZ12 and histone modification H3K27me3 in colorectal cancer. PLoS ONE 2014, 9, e108265. [Google Scholar] [CrossRef]
- Wang, C.G.; Ye, Y.J.; Yuan, J.; Liu, F.F.; Zhang, H.; Wang, S. EZH2 and STAT6 expression profiles are correlated with colorectal cancer stage and prognosis. World J. Gastroenterol. 2010, 16, 2421–2427. [Google Scholar] [CrossRef]
- Fluge, Ø.; Gravdal, K.; Carlsen, E.; Vonen, B.; Kjellevold, K.; Refsum, S.; Lilleng, R.; Eide, T.J.; Halvorsen, T.B.; Tveit, K.M.; et al. Expression of EZH2 and Ki-67 in colorectal cancer and associations with treatment response and prognosis. Br. J. Cancer 2009, 101, 1282–1289. [Google Scholar] [CrossRef]
- Mimori, K.; Ogawa, K.; Okamoto, M.; Sudo, T.; Inoue, H.; Mori, M. Clinical significance of enhancer of zeste homolog 2 expression in colorectal cancer cases. Eur. J. Surg. Oncol. (EJSO) 2005, 31, 376–380. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, F.; Zhao, M.; Li, B.; Xing, D.; Wang, J.; Yang, Y. Prognostic significance of EZH2 expression in patients with oesophageal cancer: A meta-analysis. J. Cell. Mol. Med. 2016, 20, 836–841. [Google Scholar] [CrossRef]
- Liu, F.; Gu, L.; Cao, Y.; Fan, X.; Zhang, F.; Sang, M. Aberrant overexpression of EZH2 and H3K27me3 serves as poor prognostic biomarker for esophageal squamous cell carcinoma patients. Biomarkers 2016, 21, 80–90. [Google Scholar] [CrossRef]
- Rehman, A.U.; Iqbal, M.A.; Sattar, R.S.A.; Saikia, S.; Kashif, M.; Ali, W.M.; Medhi, S.; Saluja, S.S.; Husain, S.A. Elevated expression of RUNX3 co-expressing with EZH2 in esophageal cancer patients from India. Cancer Cell Int. 2020, 20, 445. [Google Scholar] [CrossRef]
- Luo, L.; Wang, Z.; Hu, T.; Feng, Z.; Zeng, Q.; Shu, X.; Wu, A.; Huang, P.; Cao, Y.; Tu, Y.; et al. Multiomics characteristics and immunotherapeutic potential of EZH2 in pan-cancer. Biosci. Rep. 2023, 43, BSR20222230. [Google Scholar] [CrossRef]
- Kleer, C.G.; Cao, Q.; Varambally, S.; Shen, R.; Ota, I.; Tomlins, S.A.; Ghosh, D.; Sewalt, R.G.; Otte, A.P.; Hayes, D.F.; et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. USA 2003, 100, 11606–11611. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.J.; Deng, J.; Chen, C.; Hu, W.; Yuan, Y.C.; Xia, Z.K. LncRNA H19 promotes epithelial mesenchymal transition and metastasis of esophageal cancer via STAT3/EZH2 axis. Int. J. Biochem. Cell Biol. 2019, 113, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Nourmohammadi, F.; Forghanifard, M.M.; Abbaszadegan, M.R.; Zarrinpour, V. EZH2 regulates oncomiR-200c and EMT markers in esophageal squamous cell carcinomas. Sci. Rep. 2022, 12, 18290. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, Y.; Li, Z.; Qin, X.; Zhou, X.; Zhang, H.; Li, S. LINC00114 stimulates growth and glycolysis of esophageal cancer cells by recruiting EZH2 to enhance H3K27me3 of DLC1. Clin. Epigenetics 2022, 14, 51. [Google Scholar] [CrossRef]
- Qiu, B.Q.; Lin, X.H.; Ye, X.D.; Huang, W.; Pei, X.; Xiong, D.; Long, X.; Zhu, S.Q.; Lu, F.; Lin, K.; et al. Long non-coding RNA PSMA3-AS1 promotes malignant phenotypes of esophageal cancer by modulating the miR-101/EZH2 axis as a ceRNA. Aging 2020, 12, 1843–1856. [Google Scholar] [CrossRef]
- Zhang, S.; Liao, W.; Wu, Q.; Huang, X.; Pan, Z.; Chen, W.; Gu, S.; Huang, Z.; Wang, Y.; Tang, X.; et al. LINC00152 upregulates ZEB1 expression and enhances epithelial-mesenchymal transition and oxaliplatin resistance in esophageal cancer by interacting with EZH2. Cancer Cell Int. 2020, 20, 569. [Google Scholar] [CrossRef]
- Wang, W.; Zhu, Y.; Li, S.; Chen, X.; Jiang, G.; Shen, Z.; Qiao, Y.; Wang, L.; Zheng, P.; Zhang, Y. Long noncoding RNA MALAT1 promotes malignant development of esophageal squamous cell carcinoma by targeting β-catenin via Ezh2. Oncotarget 2016, 7, 25668–25682. [Google Scholar] [CrossRef]
- Gan, L.; Xu, M.; Hua, R.; Tan, C.; Zhang, J.; Gong, Y.; Wu, Z.; Weng, W.; Sheng, W.; Guo, W. The polycomb group protein EZH2 induces epithelial-mesenchymal transition and pluripotent phenotype of gastric cancer cells by binding to PTEN promoter. J. Hematol. Oncol. 2018, 11, 9. [Google Scholar] [CrossRef]
- Bai, J.; Chen, J.; Ma, M.; Cai, M.; Xu, F.; Wang, G.; Tao, K.; Shuai, X. Inhibiting Enhancer of Zeste Homolog 2 Promotes Cellular Senescence in Gastric Cancer Cells SGC-7901 by Activation of p21 and p16. DNA Cell Biol. 2014, 33, 337–344. [Google Scholar] [CrossRef]
- Song, B.-f.; Xu, L.-z.; Jiang, K.; Cheng, F. MiR-124-3p inhibits tumor progression in prostate cancer by targeting EZH2. Funct. Integr. Genom. 2023, 23, 80. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, L.; Liang, Q.; Qin, H.; Chen, Y.; Wang, T.; Wei, Q.; Luo, Y.; Li, G.; Huang, H. MiR-124-3p inhibits proliferation, migration, and epithelial-mesenchymal transformation in gastric cancer by targeting ITGB1. Am. J. Transl. Res. 2025, 17, 2500–2512. [Google Scholar] [CrossRef]
- Long, H.; Xiang, T.; Luo, J.; Li, F.; Lin, R.; Liu, S.; Jiang, S.; Hu, C.; Chen, G.; Wong, E.; et al. The tumor microenvironment disarms CD8(+) T lymphocyte function via a miR-26a-EZH2 axis. Oncoimmunology 2016, 5, e1245267. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Han, L.; He, N.; Li, R.; He, S. Upregulated Circular RNA KIF4A Promotes Cell Migration and Invasion by Regulating MicroRNA-144-3p/EZH2 Axis in Gastric Cancer. J. Oncol. 2022, 2022, 3985621. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Chen, H.; Li, L.; Yang, F.; Wu, C.; Tao, K. CircGSK3B promotes RORA expression and suppresses gastric cancer progression through the prevention of EZH2 trans-inhibition. J. Exp. Clin. Cancer Res. 2021, 40, 330. [Google Scholar] [CrossRef]
- Ghoreshi, Z.A.; Rezaei Zadeh Rukerd, M.; Askarpour, H.; Kheirkhah Vakilabad, A.A.; Nakhaie, M.; Abbaszadeh Afshar, M.J.; Behboudi, E.; Charostad, J.; Arefinia, N. The Role of Epstein-Barr Virus (EBV) Infected Gastric Cancer in Increasing microRNA124 (miR-124) Promoter Methylation and Enhancer of Zeste Homolog 2 (EZH2) Gene Expression. Medicine 2024, 103, e36534. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, H.C.; Zhang, Y.; Huang, H. MicroRNA-625-3p inhibits gastric cancer metastasis through modulating EZH2. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1177–1185. [Google Scholar] [CrossRef]
- Pan, Y.; Wu, A.; Xu, F.; Chen, C.; Jiang, L.; Jin, R. Lentivirus-mediated overexpression of miR-124 suppresses growth and invasion by targeting JAG1 and EZH2 in gastric cancer. Oncol. Lett. 2018, 15, 7450–7458. [Google Scholar] [CrossRef]
- Deng, M.; Zhang, R.; He, Z.; Qiu, Q.; Lu, X.; Yin, J.; Liu, H.; Jia, X.; He, Z. TET-Mediated Sequestration of miR-26 Drives EZH2 Expression and Gastric Carcinogenesis. Cancer Res. 2017, 77, 6069–6082. [Google Scholar] [CrossRef]
- Sun, M.; Nie, F.; Wang, Y.; Zhang, Z.; Hou, J.; He, D.; Xie, M.; Xu, L.; De, W.; Wang, Z.; et al. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016, 76, 6299–6310. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Min, L.; Qiu, X.; Wu, X.; Liu, C.; Ma, J.; Zhang, D.; Zhu, L. Biological Function of Long Non-coding RNA (LncRNA) Xist. Front. Cell Dev. Biol. 2021, 9, 645647. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Sun, M.; Zhu, Y.-n.; Xia, R.; Liu, Y.-w.; Ding, J.; Ma, H.-w.; He, X.-z.; Zhang, Z.-h.; Liu, Z.-j.; et al. Long noncoding RNA HOXA-AS2 promotes gastric cancer proliferation by epigenetically silencing P21/PLK3/DDIT3 expression. Oncotarget 2015, 6, 33587–33601. [Google Scholar] [CrossRef] [PubMed]
- Kong, R.; Zhang, E.B.; Yin, D.D.; You, L.H.; Xu, T.P.; Chen, W.M.; Xia, R.; Wan, L.; Sun, M.; Wang, Z.X.; et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol. Cancer 2015, 14, 82. [Google Scholar] [CrossRef]
- He, L.J.; Cai, M.Y.; Xu, G.L.; Li, J.J.; Weng, Z.J.; Xu, D.Z.; Luo, G.Y.; Zhu, S.L.; Xie, D. Prognostic significance of overexpression of EZH2 and H3k27me3 proteins in gastric cancer. Asian Pac. J. Cancer Prev. 2012, 13, 3173–3178. [Google Scholar] [CrossRef]
- Choi, J.H.; Song, Y.S.; Yoon, J.S.; Song, K.W.; Lee, Y.Y. Enhancer of zeste homolog 2 expression is associated with tumor cell proliferation and metastasis in gastric cancer. APMIS 2010, 118, 196–202. [Google Scholar] [CrossRef]
- Matsukawa, Y.; Semba, S.; Kato, H.; Ito, A.; Yanagihara, K.; Yokozaki, H. Expression of the enhancer of zeste homolog 2 is correlated with poor prognosis in human gastric cancer. Cancer Sci. 2006, 97, 484–491. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012–2016. Neuro-Oncol. 2019, 21, v1–v100. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, L.; Han, L.; Shi, Z.; Zhang, J.; Pu, P.; Kang, C. EZH2 is a negative prognostic factor and exhibits pro-oncogenic activity in glioblastoma. Cancer Lett. 2015, 356, 929–936. [Google Scholar] [CrossRef]
- Paskeh, M.D.A.; Mehrabi, A.; Gholami, M.H.; Zabolian, A.; Ranjbar, E.; Saleki, H.; Ranjbar, A.; Hashemi, M.; Ertas, Y.N.; Hushmandi, K.; et al. EZH2 as a new therapeutic target in brain tumors: Molecular landscape, therapeutic targeting and future prospects. Biomed. Pharmacother. 2022, 146, 112532. [Google Scholar] [CrossRef]
- Ning, X.; Shi, Z.; Liu, X.; Zhang, A.; Han, L.; Jiang, K.; Kang, C.; Zhang, Q. DNMT1 and EZH2 mediated methylation silences the microRNA-200b/a/429 gene and promotes tumor progression. Cancer Lett. 2015, 359, 198–205. [Google Scholar] [CrossRef]
- Yu, T.; Zhou, F.; Tian, W.; Xu, R.; Wang, B.; Zeng, A.; Zhou, Z.; Li, M.; Wang, Y.; Zhang, J. EZH2 interacts with HP1BP3 to epigenetically activate WNT7B that promotes temozolomide resistance in glioblastoma. Oncogene 2023, 42, 461–470. [Google Scholar] [CrossRef]
- Karlowee, V.; Amatya, V.J.; Takayasu, T.; Takano, M.; Yonezawa, U.; Takeshima, Y.; Sugiyama, K.; Kurisu, K.; Yamasaki, F. Immunostaining of Increased Expression of Enhancer of Zeste Homolog 2 (EZH2) in Diffuse Midline Glioma H3K27M-Mutant Patients with Poor Survival. Pathobiology 2019, 86, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Pang, B.; Gu, G.; Gao, T.; Zhang, R.; Pang, Q.; Liu, Q. Melatonin Inhibits Glioblastoma Stem-like cells through Suppression of EZH2-NOTCH1 Signaling Axis. Int. J. Biol. Sci. 2017, 13, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Pang, B.; Zheng, X.R.; Tian, J.X.; Gao, T.H.; Gu, G.Y.; Zhang, R.; Fu, Y.B.; Pang, Q.; Li, X.G.; Liu, Q. EZH2 promotes metabolic reprogramming in glioblastomas through epigenetic repression of EAF2-HIF1α signaling. Oncotarget 2016, 7, 45134–45143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.X.; Han, L.; Bao, Z.S.; Wang, Y.Y.; Chen, L.Y.; Yan, W.; Yu, S.Z.; Pu, P.Y.; Liu, N.; You, Y.P.; et al. HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro Oncol. 2013, 15, 1595–1603. [Google Scholar] [CrossRef]
- Zakrzewska, M.; Fendler, W.; Zakrzewski, K.; Sikorska, B.; Grajkowska, W.; Dembowska-Bagińska, B.; Filipek, I.; Stefańczyk, Ł.; Liberski, P.P. Altered MicroRNA Expression Is Associated with Tumor Grade, Molecular Background and Outcome in Childhood Infratentorial Ependymoma. PLoS ONE 2016, 11, e0158464. [Google Scholar] [CrossRef]
- Mochizuki, D.; Misawa, Y.; Kawasaki, H.; Imai, A.; Endo, S.; Mima, M.; Yamada, S.; Nakagawa, T.; Kanazawa, T.; Misawa, K. Aberrant Epigenetic Regulation in Head and Neck Cancer Due to Distinct EZH2 Overexpression and DNA Hypermethylation. Int. J. Mol. Sci. 2018, 19, 3707. [Google Scholar] [CrossRef]
- Nienstedt, J.C.; Schroeder, C.; Clauditz, T.; Simon, R.; Sauter, G.; Muenscher, A.; Blessmann, M.; Hanken, H.; Pflug, C. EZH2 overexpression in head and neck cancer is related to lymph node metastasis. J. Oral Pathol. Med. 2018, 47, 240–245. [Google Scholar] [CrossRef]
- Révész, M.; Oberna, F.; Slezák, A.; Tóth, E.; Ferenczi, Ö.; Kenessey, I.; Takácsi-Nagy, Z. EZH2 Expression in Head-and-Neck Squamous Cell Cancer in Young Patients. Int. J. Mol. Sci. 2024, 25, 5250. [Google Scholar] [CrossRef]
- Chang, J.W.; Gwak, S.Y.; Shim, G.A.; Liu, L.; Lim, Y.C.; Kim, J.M.; Jung, M.G.; Koo, B.S. EZH2 is associated with poor prognosis in head-and-neck squamous cell carcinoma via regulating the epithelial-to-mesenchymal transition and chemosensitivity. Oral Oncol. 2016, 52, 66–74. [Google Scholar] [CrossRef]
- Eichenauer, T.; Simmendinger, L.; Fraune, C.; Mandelkow, T.; Blessin, N.C.; Kluth, M.; Hube-Magg, C.; Möller, K.; Clauditz, T.; Weidemann, S.; et al. High level of EZH2 expression is linked to high density of CD8-positive T-lymphocytes and an aggressive phenotype in renal cell carcinoma. World J. Urol. 2021, 39, 481–490. [Google Scholar] [CrossRef]
- Yuan, J.B.; Yang, L.Y.; Tang, Z.Y.; Zu, X.B.; Qi, L. Down-regulation of EZH2 by RNA interference inhibits proliferation and invasion of ACHN cells via the Wnt/β-catenin pathway. Asian Pac. J. Cancer Prev. 2012, 13, 6197–6201. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, M.; Shi, H.; Hu, J.; Wang, Y.; Sun, Z.; Xu, S. Reduced E-cadherin facilitates renal cell carcinoma progression by WNT/β-catenin signaling activation. Oncotarget 2017, 8, 19566–19576. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.Q.; Zhang, L.; Gao, B.S.; Wan, Y.G.; Zhang, X.H.; Chen, B.; Wang, Y.T.; Sun, N.; Fu, Y.W. EZH2 promotes tumor progression by increasing VEGF expression in clear cell renal cell carcinoma. Clin. Transl. Oncol. 2015, 17, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Qin, W.; Zheng, J.; Wei, M.; Zhou, X.; Wang, H.; Wen, W. [Expressions of EZH2 and RUNX3 in renal cell carcinoma and their clinical significance]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2013, 29, 82–84, 88. (In Chinese) [Google Scholar] [PubMed]
- Sakurai, T.; Bilim, V.N.; Ugolkov, A.V.; Yuuki, K.; Tsukigi, M.; Motoyama, T.; Tomita, Y. The enhancer of zeste homolog 2 (EZH2), a potential therapeutic target, is regulated by miR-101 in renal cancer cells. Biochem. Biophys. Res. Commun. 2012, 422, 607–614. [Google Scholar] [CrossRef]
- Lyu, C.; Wang, L.; Stadlbauer, B.; Noessner, E.; Buchner, A.; Pohla, H. Identification of EZH2 as Cancer Stem Cell Marker in Clear Cell Renal Cell Carcinoma and the Anti-Tumor Effect of Epigallocatechin-3-Gallate (EGCG). Cancers 2022, 14, 4200. [Google Scholar] [CrossRef]
- Wu, L.; Jiang, X.; Qi, C.; Zhang, C.; Qu, B.; Shen, N. EZH2 Inhibition Interferes With the Activation of Type I Interferon Signaling Pathway and Ameliorates Lupus Nephritis in NZB/NZW F1 Mice. Front. Immunol. 2021, 12, 653989. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Zhao, C.; Li, S.; Wang, J.; Zhou, Q.; Sun, J.; Ding, Q.; Liu, M.; Ding, G. EZH2 Expression is increased in BAP1-mutant renal clear cell carcinoma and is related to poor prognosis. J. Cancer 2018, 9, 3787–3796. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.H.; Kapur, P.; Eckel-Passow, J.E.; Christie, A.; Joseph, R.W.; Serie, D.J.; Cheville, J.C.; Thompson, R.H.; Homayoun, F.; Panwar, V.; et al. Multicenter Validation of Enhancer of Zeste Homolog 2 Expression as an Independent Prognostic Marker in Localized Clear Cell Renal Cell Carcinoma. J. Clin. Oncol. 2017, 35, 3706–3713. [Google Scholar] [CrossRef]
- Liu, L.; Xu, Z.; Zhong, L.; Wang, H.; Jiang, S.; Long, Q.; Xu, J.; Guo, J. Prognostic Value of EZH2 Expression and Activity in Renal Cell Carcinoma: A Prospective Study. PLoS ONE 2013, 8, e81484. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, J.; Valind, A.; Jansson, C.; O’Sullivan, M.J.; Holmquist Mengelbier, L.; Gisselsson, D. Aberrant epigenetic regulation in clear cell sarcoma of the kidney featuring distinct DNA hypermethylation and EZH2 overexpression. Oncotarget 2016, 7, 11127–11136. [Google Scholar] [CrossRef]
- Glaser, K.; Schepers, E.J.; Zwolshen, H.M.; Lake, C.M.; Timchenko, N.A.; Karns, R.A.; Cairo, S.; Geller, J.I.; Tiao, G.M.; Bondoc, A.J. EZH2 is a key component of hepatoblastoma tumor cell growth. Pediatr. Blood Cancer 2024, 71, e30774. [Google Scholar] [CrossRef]
- Zhou, J.; Che, J.; Xu, L.; Yang, W.; Li, Y.; Zhou, W.; Zou, S. Enhancer of zeste homolog 2 promotes hepatocellular cancer progression and chemoresistance by enhancing protein kinase B activation through microRNA-381-mediated SET domain bifurcated 1. Bioengineered 2022, 13, 5737–5755. [Google Scholar] [CrossRef]
- Xiao, G.; Jin, L.L.; Liu, C.Q.; Wang, Y.C.; Meng, Y.M.; Zhou, Z.G.; Chen, J.; Yu, X.J.; Zhang, Y.J.; Xu, J.; et al. EZH2 negatively regulates PD-L1 expression in hepatocellular carcinoma. J. Immunother. Cancer 2019, 7, 300. [Google Scholar] [CrossRef]
- Wang, B.; Liu, Y.; Liao, Z.; Wu, H.; Zhang, B.; Zhang, L. EZH2 in hepatocellular carcinoma: Progression, immunity, and potential targeting therapies. Exp. Hematol. Oncol. 2023, 12, 52. [Google Scholar] [CrossRef]
- Gao, S.-B.; Zheng, Q.-F.; Xu, B.; Pan, C.-B.; Li, K.-L.; Zhao, Y.; Zheng, Q.-L.; Lin, X.; Xue, L.-X.; Jin, G.-H. EZH2 Represses Target Genes through H3K27-Dependent and H3K27-Independent Mechanisms in Hepatocellular Carcinoma. Mol. Cancer Res. 2014, 12, 1388–1397. [Google Scholar] [CrossRef]
- You, Z.; Peng, D.; Cao, Y.; Zhu, Y.; Yin, J.; Zhang, G.; Peng, X. P53 suppresses the progression of hepatocellular carcinoma via miR-15a by decreasing OGT expression and EZH2 stabilization. J. Cell. Mol. Med. 2021, 25, 9168–9182. [Google Scholar] [CrossRef]
- Vella, S.; Pomella, S.; Leoncini, P.P.; Colletti, M.; Conti, B.; Marquez, V.E.; Strillacci, A.; Roma, J.; Gallego, S.; Milano, G.M.; et al. MicroRNA-101 is repressed by EZH2 and its restoration inhibits tumorigenic features in embryonal rhabdomyosarcoma. Clin. Epigenetics 2015, 7, 82. [Google Scholar] [CrossRef]
- Chi, X.Z.; Yang, J.O.; Lee, K.Y.; Ito, K.; Sakakura, C.; Li, Q.L.; Kim, H.R.; Cha, E.J.; Lee, Y.H.; Kaneda, A.; et al. RUNX3 suppresses gastric epithelial cell growth by inducing p21(WAF1/Cip1) expression in cooperation with transforming growth factor {beta}-activated SMAD. Mol. Cell. Biol. 2005, 25, 8097–8107. [Google Scholar] [CrossRef]
- Wu, C.S.; Chien, Y.C.; Yen, C.J.; Wu, J.Y.; Bai, L.Y.; Yu, Y.L. EZH2-mediated epigenetic silencing of tumor-suppressive let-7c/miR-99a cluster by hepatitis B virus X antigen enhances hepatocellular carcinoma progression and metastasis. Cancer Cell Int. 2023, 23, 199. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Li, H.; Zhan, H.; Han, T.; Dong, Y.; Tian, C.; Guo, Y.; Yan, F.; Dai, D.; Liu, P. Identification of Potential Biomarkers for Liver Cancer Through Gene Mutation and Clinical Characteristics. Front. Oncol. 2021, 11, 733478. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiao, Y.; Chen, K.; Chen, S.; Zhang, M.; Wu, Z.; Wu, Y. Enhancer of zeste homolog 2 depletion arrests the proliferation of hepatoblastoma cells. Mol. Med. Rep. 2016, 13, 2724–2728. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zheng, F.; Liao, Y.J.; Cai, M.Y.; Liu, T.H.; Chen, S.P.; Wu, P.H.; Wu, L.; Bian, X.W.; Guan, X.Y.; Zeng, Y.X.; et al. Systemic delivery of microRNA-101 potently inhibits hepatocellular carcinoma in vivo by repressing multiple targets. PLoS Genet. 2015, 11, e1004873. [Google Scholar] [CrossRef]
- Gao, S.B.; Sun, S.L.; Zheng, Q.L.; Zhang, L.; Zhu, Y.; Jin, G.H.; Xue, L.X. Genetic alteration and misexpression of Polycomb group genes in hepatocellular carcinoma. Am. J. Cancer Res. 2015, 5, 2969–2979. [Google Scholar]
- Xu, L.; Beckebaum, S.; Iacob, S.; Wu, G.; Kaiser, G.M.; Radtke, A.; Liu, C.; Kabar, I.; Schmidt, H.H.; Zhang, X.; et al. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. J. Hepatol. 2014, 60, 590–598. [Google Scholar] [CrossRef]
- Nakagawa, S.; Okabe, H.; Sakamoto, Y.; Hayashi, H.; Hashimoto, D.; Yokoyama, N.; Sakamoto, K.; Kuroki, H.; Mima, K.; Nitta, H.; et al. Enhancer of zeste homolog 2 (EZH2) promotes progression of cholangiocarcinoma cells by regulating cell cycle and apoptosis. Ann. Surg. Oncol. 2013, 20 (Suppl. S3), S667–S675. [Google Scholar] [CrossRef]
- Cai, M.Y.; Hou, J.H.; Rao, H.L.; Luo, R.Z.; Li, M.; Pei, X.Q.; Lin, M.C.; Guan, X.Y.; Kung, H.F.; Zeng, Y.X.; et al. High expression of H3K27me3 in human hepatocellular carcinomas correlates closely with vascular invasion and predicts worse prognosis in patients. Mol. Med. 2011, 17, 12–20. [Google Scholar] [CrossRef]
- Yonemitsu, Y.; Imazeki, F.; Chiba, T.; Fukai, K.; Nagai, Y.; Miyagi, S.; Arai, M.; Aoki, R.; Miyazaki, M.; Nakatani, Y.; et al. Distinct expression of polycomb group proteins EZH2 and BMI1 in hepatocellular carcinoma. Hum. Pathol. 2009, 40, 1304–1311. [Google Scholar] [CrossRef] [PubMed]
- Toyokawa, G.; Takada, K.; Tagawa, T.; Hamamoto, R.; Yamada, Y.; Shimokawa, M.; Oda, Y.; Maehara, Y. A Positive Correlation Between the EZH2 and PD-L1 Expression in Resected Lung Adenocarcinomas. Ann. Thorac. Surg. 2019, 107, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, J.S.; Alnoor, F.; Sadiq, Q.; Solares, J.; Gradowski, J.F. SMARCA4 (BRG1) and SMARCB1 (INI1) expression in TTF-1 negative neuroendocrine carcinomas including merkel cell carcinoma. Pathol. Res. Pract. 2021, 219, 153341. [Google Scholar] [CrossRef] [PubMed]
- Behrens, C.; Solis, L.M.; Lin, H.; Yuan, P.; Tang, X.; Kadara, H.; Riquelme, E.; Galindo, H.; Moran, C.A.; Kalhor, N.; et al. EZH2 protein expression associates with the early pathogenesis, tumor progression, and prognosis of non-small cell lung carcinoma. Clin. Cancer Res. 2013, 19, 6556–6565. [Google Scholar] [CrossRef]
- Geng, J.; Li, X.; Zhou, Z.; Wu, C.-L.; Bai, X.; Dai, M. EZH2 promotes tumor progression via regulating VEGF-A/AKT signaling in non-small cell lung cancer. Cancer Lett. 2015, 359, 275–287. [Google Scholar] [CrossRef]
- LaFave, L.M.; Béguelin, W.; Koche, R.; Teater, M.; Spitzer, B.; Chramiec, A.; Papalexi, E.; Keller, M.D.; Hricik, T.; Konstantinoff, K.; et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat. Med. 2015, 21, 1344–1349. [Google Scholar] [CrossRef]
- Fan, K.; Zhang, C.L.; Qi, Y.F.; Dai, X.; Birling, Y.; Tan, Z.F.; Cao, F. Prognostic Value of EZH2 in Non-Small-Cell Lung Cancers: A Meta-Analysis and Bioinformatics Analysis. BioMed Res. Int. 2020, 2020, 2380124. [Google Scholar] [CrossRef]
- Matsubara, T.; Toyokawa, G.; Takada, K.; Kinoshita, F.; Kozuma, Y.; Akamine, T.; Shimokawa, M.; Haro, A.; Osoegawa, A.; Tagawa, T.; et al. The association and prognostic impact of enhancer of zeste homologue 2 expression and epithelial–mesenchymal transition in resected lung adenocarcinoma. PLoS ONE 2019, 14, e0215103. [Google Scholar] [CrossRef]
- Toyokawa, G.; Takada, K.; Tagawa, T.; Kinoshita, F.; Kozuma, Y.; Matsubara, T.; Haratake, N.; Takamori, S.; Akamine, T.; Hirai, F.; et al. Prevalence of Enhancer of Zeste Homolog 2 in Patients with Resected Small Cell Lung Cancer. Anticancer Res. 2018, 38, 3707–3711. [Google Scholar] [CrossRef]
- Shinozaki-Ushiku, A.; Ushiku, T.; Morita, S.; Anraku, M.; Nakajima, J.; Fukayama, M. Diagnostic utility of BAP1 and EZH2 expression in malignant mesothelioma. Histopathology 2017, 70, 722–733. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, H.; Lv, L.; Bao, L.; Wang, X.; Han, S. Prognostic Significance of EZH2 Expression in Non-Small Cell Lung Cancer: A Meta-analysis. Sci. Rep. 2016, 6, 19239. [Google Scholar] [CrossRef]
- Liu, H.; Li, W.; Yu, X.; Gao, F.; Duan, Z.; Ma, X.; Tan, S.; Yuan, Y.; Liu, L.; Wang, J.; et al. EZH2-mediated Puma gene repression regulates non-small cell lung cancer cell proliferation and cisplatin-induced apoptosis. Oncotarget 2016, 7, 56338–56354. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Hao, K.; Hu, H.; Sheng, Z.; Yan, J.; Wang, Q.; Yu, L. Expression of the enhancer of zeste homolog 2 in biopsy specimen predicts chemoresistance and survival in advanced non-small cell lung cancer receiving first-line platinum-based chemotherapy. Lung Cancer 2014, 86, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Li, X.; Shen, H.; Bai, X. Quantitative analysis of EZH2 expression and its correlations with lung cancer patients’ clinical pathological characteristics. Clin. Transl. Oncol. 2013, 15, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Yuan, C.; Xiao, X.; Wang, X.; Ji, X.; Yu, H.; Wu, Z.; Zhang, J. The expression and significance of the enhancer of zeste homolog 2 in lung adenocarcinoma. Oncol. Rep. 2012, 28, 147–154. [Google Scholar] [CrossRef]
- Huqun; Ishikawa, R.; Zhang, J.; Miyazawa, H.; Goto, Y.; Shimizu, Y.; Hagiwara, K.; Koyama, N. Enhancer of zeste homolog 2 is a novel prognostic biomarker in nonsmall cell lung cancer. Cancer 2012, 118, 1599–1606. [Google Scholar] [CrossRef]
- Lu, J.; Zhao, F.P.; Peng, Z.; Zhang, M.W.; Lin, S.X.; Liang, B.J.; Zhang, B.; Liu, X.; Wang, L.; Li, G.; et al. EZH2 promotes angiogenesis through inhibition of miR-1/Endothelin-1 axis in nasopharyngeal carcinoma. Oncotarget 2014, 5, 11319–11332. [Google Scholar] [CrossRef][Green Version]
- Balinth, S.; Fisher, M.L.; Hwangbo, Y.; Wu, C.; Ballon, C.; Sun, X.; Mills, A.A. EZH2 regulates a SETDB1/ΔNp63α axis via RUNX3 to drive a cancer stem cell phenotype in squamous cell carcinoma. Oncogene 2022, 41, 4130–4144. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, X.; Niu, X.; Wang, X.; Jiang, R.; Xu, T.; Liu, Y.; Liang, L.; Ou, X.; Xing, X.; et al. EZH2 suppresses the nucleotide excision repair in nasopharyngeal carcinoma by silencing XPA gene. Mol. Carcinog. 2017, 56, 447–463. [Google Scholar] [CrossRef]
- Alajez, N.M.; Shi, W.; Hui, A.B.; Bruce, J.; Lenarduzzi, M.; Ito, E.; Yue, S.; O’Sullivan, B.; Liu, F.F. Enhancer of Zeste homolog 2 (EZH2) is overexpressed in recurrent nasopharyngeal carcinoma and is regulated by miR-26a, miR-101, and miR-98. Cell Death Dis. 2010, 1, e85. [Google Scholar] [CrossRef]
- Li, X.; Lin, Y.; Yang, X.; Wu, X.; He, X. Long noncoding RNA H19 regulates EZH2 expression by interacting with miR-630 and promotes cell invasion in nasopharyngeal carcinoma. Biochem. Biophys. Res. Commun. 2016, 473, 913–919. [Google Scholar] [CrossRef]
- Fan, D.C.; Zhao, Y.R.; Qi, H.; Hou, J.X.; Zhang, T.H. MiRNA-506 presents multiple tumor suppressor activities by targeting EZH2 in nasopharyngeal carcinoma. Auris Nasus Larynx 2020, 47, 632–642. [Google Scholar] [CrossRef]
- Chen, J.; Tang, S.; Zheng, Q.; Li, J.; Jiang, H.; Lu, H.; Liao, G.; Li, K.; Liang, Y. The competitive mechanism of EZH1 and EZH2 in promoting oral squamous cell carcinoma. Exp. Cell Res. 2024, 436, 113957. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, D.; Dafar, A.; Niklasson, J.; Sandberg, I.; Braz-Silva, P.; Sapkota, D.; Öhman, J.; Giglio, D.; Hasséus, B. EZH2 Expression Correlates With T-Cell Infiltration in Oral Leukoplakia and Predicts Cancer Transformation. Anticancer Res. 2023, 43, 1533–1542. [Google Scholar] [CrossRef] [PubMed]
- Sihavong, P.; Kitkumthorn, N.; Srimaneekarn, N.; Bumalee, D.; Lapthanasupkul, P. Differential Expression of EZH2 and H3K27me3 in Oral Verrucous Carcinoma and Oral Verrucous Hyperplasia. Head Neck Pathol. 2021, 15, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Chen, L.; Tang, J.; Zhang, C.; Wen, Y.; Wen, W. Targeting EZH2 depletes LMP1-induced activated regulatory T cells enhancing antitumor immunity in nasopharyngeal carcinoma. J. Cancer Res. Ther. 2020, 16, 309–319. [Google Scholar] [CrossRef]
- Zheng, M.; Cao, M.X.; Luo, X.J.; Li, L.; Wang, K.; Wang, S.S.; Wang, H.F.; Tang, Y.J.; Tang, Y.L.; Liang, X.H. EZH2 promotes invasion and tumour glycolysis by regulating STAT3 and FoxO1 signalling in human OSCC cells. J. Cell. Mol. Med. 2019, 23, 6942–6954. [Google Scholar] [CrossRef]
- Zhao, L.; Yu, Y.; Wu, J.; Bai, J.; Zhao, Y.; Li, C.; Sun, W.; Wang, X. Role of EZH2 in oral squamous cell carcinoma carcinogenesis. Gene 2014, 537, 197–202. [Google Scholar] [CrossRef]
- Zhai, L.; Tai, W.L.; Pan, Y.Q.; Luo, J.B.; Ma, L.; Zheng, Y.T.; Guo, M.Y.; Zhang, X. Expression of EZH2 and P53 and their correlation in ovarian cancer tissues. Int. J. Clin. Exp. Pathol. 2020, 13, 456–464. [Google Scholar]
- Sun, S.; Zhao, S.; Yang, Q.; Wang, W.; Cai, E.; Wen, Y.; Yu, L.; Wang, Z.; Cai, J. Enhancer of zeste homolog 2 promotes cisplatin resistance by reducing cellular platinum accumulation. Cancer Sci. 2018, 109, 1853–1864. [Google Scholar] [CrossRef]
- Cardenas, H.; Zhao, J.; Vieth, E.; Nephew, K.P.; Matei, D. EZH2 inhibition promotes epithelial-to-mesenchymal transition in ovarian cancer cells. Oncotarget 2016, 7, 84453–84467. [Google Scholar] [CrossRef]
- Lai, Y.; Han, X.; Xie, B.; Xu, Y.; Yang, Z.; Wang, D.; Li, W.; Xie, Y.; Song, W.; Zhang, X.; et al. EZH2 suppresses ferroptosis in hepatocellular carcinoma and reduces sorafenib sensitivity through epigenetic regulation of TFR2. Cancer Sci. 2024, 115, 2220–2234. [Google Scholar] [CrossRef]
- Li, R.; Zhou, T.; Chen, S.; Li, N.; Cai, Z.; Ling, Y.; Feng, Z. Small cell carcinoma of the ovary, hypercalcemic type (SCCOHT): A challenge for clinicopathological diagnosis. Int. J. Clin. Exp. Pathol. 2019, 12, 2166–2172. [Google Scholar]
- Luo, Y.; Li, L.; Hu, Q.; Zhang, Z.; Liu, F.; Peng, Y.; Zou, Y.; Chen, L. Iron overload increases the sensitivity of endometriosis stromal cells to ferroptosis via a PRC2-independent function of EZH2. Int. J. Biochem. Cell Biol. 2024, 169, 106553. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hong, J.H.; Huang, Y.; Liu, S.; Yin, J.; Deng, P.; Sun, Y.; Yu, Z.; Zeng, X.; Xiao, R.; et al. EZH2 mediated metabolic rewiring promotes tumor growth independently of histone methyltransferase activity in ovarian cancer. Mol. Cancer 2023, 22, 85. [Google Scholar] [CrossRef] [PubMed]
- Reid, B.M.; Vyas, S.; Chen, Z.; Chen, A.; Kanetsky, P.A.; Permuth, J.B.; Sellers, T.A.; Saglam, O. Morphologic and molecular correlates of EZH2 as a predictor of platinum resistance in high-grade ovarian serous carcinoma. BMC Cancer 2021, 21, 714. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Yang, Q.; Cai, E.; Huang, B.; Ying, F.; Wen, Y.; Cai, J.; Yang, P. EZH2/H3K27Me3 and phosphorylated EZH2 predict chemotherapy response and prognosis in ovarian cancer. PeerJ 2020, 8, e9052. [Google Scholar] [CrossRef]
- Huo, X.; Sun, H.; Qian, Q.; Ma, X.; Peng, P.; Yu, M.; Zhang, Y.; Yang, J.; Cao, D.; Gui, T.; et al. CYP27B1 Downregulation: A New Molecular Mechanism Regulating EZH2 in Ovarian Cancer Tumorigenicity. Front. Cell Dev. Biol. 2020, 8, 561804. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, S.Y.; Karnezis, A.N.; Colborne, S.; Santos, N.D.; Lang, J.D.; Hendricks, W.P.; Orlando, K.A.; Yap, D.; Kommoss, F.; et al. The histone methyltransferase EZH2 is a therapeutic target in small cell carcinoma of the ovary, hypercalcaemic type. J. Pathol. 2017, 242, 371–383. [Google Scholar] [CrossRef]
- Xu, Y.; Li, X.; Wang, H.; Xie, P.; Yan, X.; Bai, Y.; Zhang, T. Hypermethylation of CDH13, DKK3 and FOXL2 promoters and the expression of EZH2 in ovary granulosa cell tumors. Mol. Med. Rep. 2016, 14, 2739–2745. [Google Scholar] [CrossRef]
- Tsang, D.P.; Cheng, A.S. Epigenetic regulation of signaling pathways in cancer: Role of the histone methyltransferase EZH2. J. Gastroenterol. Hepatol. 2011, 26, 19–27. [Google Scholar] [CrossRef] [PubMed]
- McCleary-Wheeler, A.L.; Lomberk, G.A.; Weiss, F.U.; Schneider, G.; Fabbri, M.; Poshusta, T.L.; Dusetti, N.J.; Baumgart, S.; Iovanna, J.L.; Ellenrieder, V.; et al. Insights into the epigenetic mechanisms controlling pancreatic carcinogenesis. Cancer Lett. 2013, 328, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Dai, H.; Gong, Y.; Zhang, C.; Shu, J.; Luo, Y.; Jiang, Y.; Liu, W.; Bie, P. ANLN-induced EZH2 upregulation promotes pancreatic cancer progression by mediating miR-218-5p/LASP1 signaling axis. J. Exp. Clin. Cancer Res. 2019, 38, 347. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; To, K.F.; Tong, J.H.; Xiao, Z.; Xia, T.; Lai, P.B.; Chow, S.C.; Zhu, Y.X.; Chan, S.L.; Marquez, V.E.; et al. Enhancer of zeste homolog 2 silences microRNA-218 in human pancreatic ductal adenocarcinoma cells by inducing formation of heterochromatin. Gastroenterology 2013, 144, 1086–1097.e1089. [Google Scholar] [CrossRef]
- Patil, S.; Steuber, B.; Kopp, W.; Kari, V.; Urbach, L.; Wang, X.; Küffer, S.; Bohnenberger, H.; Spyropoulou, D.; Zhang, Z.; et al. EZH2 Regulates Pancreatic Cancer Subtype Identity and Tumor Progression via Transcriptional Repression of GATA6. Cancer Res. 2020, 80, 4620–4632. [Google Scholar] [CrossRef]
- Chibaya, L.; Murphy, K.C.; DeMarco, K.D.; Gopalan, S.; Liu, H.; Parikh, C.N.; Lopez-Diaz, Y.; Faulkner, M.; Li, J.; Morris, J.P.; et al. EZH2 inhibition remodels the inflammatory senescence-associated secretory phenotype to potentiate pancreatic cancer immune surveillance. Nat. Cancer 2023, 4, 872–892. [Google Scholar] [CrossRef]
- Li, H.; Wang, H.; Cui, Y.; Jiang, W.; Zhan, H.; Feng, L.; Gao, M.; Zhao, K.; Zhang, L.; Xie, X.; et al. EZH2 regulates pancreatic cancer cells through E2F1, GLI1, CDK3, and Mcm4. Hereditas 2023, 160, 23. [Google Scholar] [CrossRef]
- Zhou, X.; Gao, W.; Hua, H.; Ji, Z. LncRNA-BLACAT1 Facilitates Proliferation, Migration and Aerobic Glycolysis of Pancreatic Cancer Cells by Repressing CDKN1C via EZH2-Induced H3K27me3. Front. Oncol. 2020, 10, 539805. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, J.; Weng, Y.C.; Wang, J.C. EZH2-Mediated microRNA-139-5p Regulates Epithelial-Mesenchymal Transition and Lymph Node Metastasis of Pancreatic Cancer. Mol. Cells 2018, 41, 868–880. [Google Scholar] [CrossRef]
- Han, T.; Jiao, F.; Hu, H.; Yuan, C.; Wang, L.; Jin, Z.L.; Song, W.F.; Wang, L.W. EZH2 promotes cell migration and invasion but not alters cell proliferation by suppressing E-cadherin, partly through association with MALAT-1 in pancreatic cancer. Oncotarget 2016, 7, 11194–11207. [Google Scholar] [CrossRef]
- Chen, J.; Xu, H.; Zou, X.; Wang, J.; Zhu, Y.; Chen, H.; Shen, B.; Deng, X.; Zhou, A.; Chin, Y.E.; et al. Snail recruits Ring1B to mediate transcriptional repression and cell migration in pancreatic cancer cells. Cancer Res. 2014, 74, 4353–4363. [Google Scholar] [CrossRef]
- Yamamoto, K.; Tateishi, K.; Kudo, Y.; Sato, T.; Yamamoto, S.; Miyabayashi, K.; Matsusaka, K.; Asaoka, Y.; Ijichi, H.; Hirata, Y.; et al. Loss of histone demethylase KDM6B enhances aggressiveness of pancreatic cancer through downregulation of C/EBPα. Carcinogenesis 2014, 35, 2404–2414. [Google Scholar] [CrossRef]
- Kuroki, H.; Hayashi, H.; Okabe, H.; Hashimoto, D.; Takamori, H.; Nakahara, O.; Nakagawa, S.; Fukushima, Y.; Chikamoto, A.; Beppu, T.; et al. EZH2 is associated with malignant behavior in pancreatic IPMN via p27Kip1 downregulation. PLoS ONE 2014, 9, e100904. [Google Scholar] [CrossRef][Green Version]
- Maftouh, M.; Avan, A.; Funel, N.; Paolicchi, E.; Vasile, E.; Pacetti, P.; Vaccaro, V.; Faviana, P.; Campani, D.; Caponi, S.; et al. A polymorphism in the promoter is associated with EZH2 expression but not with outcome in advanced pancreatic cancer patients. Pharmacogenomics 2014, 15, 609–618. [Google Scholar] [CrossRef]
- Xin, L. EZH2 accompanies prostate cancer progression. Nat. Cell Biol. 2021, 23, 934–936. [Google Scholar] [CrossRef]
- Xu, K.; Wu, Z.J.; Groner, A.C.; He, H.H.; Cai, C.; Lis, R.T.; Wu, X.; Stack, E.C.; Loda, M.; Liu, T.; et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 2012, 338, 1465–1469. [Google Scholar] [CrossRef]
- Wee, Z.N.; Li, Z.; Lee, P.L.; Lee, S.T.; Lim, Y.P.; Yu, Q. EZH2-Mediated Inactivation of IFN-γ-JAK-STAT1 Signaling Is an Effective Therapeutic Target in MYC-Driven Prostate Cancer. Cell Rep. 2014, 8, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, Y.; Lu, X.; Song, B.; Fong, K.W.; Cao, Q.; Licht, J.D.; Zhao, J.C.; Yu, J. Polycomb- and Methylation-Independent Roles of EZH2 as a Transcription Activator. Cell Rep. 2018, 25, 2808–2820.e4. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Fong, K.W.; Kim, J.; Wang, F.; Lu, X.; Lee, Y.; Brea, L.T.; Wadosky, K.; Guo, C.; Abdulkadir, S.A.; et al. Posttranslational regulation of FOXA1 by Polycomb and BUB3/USP7 deubiquitin complex in prostate cancer. Sci. Adv. 2021, 7, eabe2261. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.J.; Engers, R.; Florl, A.R.; Otte, A.P.; Muller, M.; Schulz, W.A. Expression changes in EZH2, but not in BMI-1, SIRT1, DNMT1 or DNMT3B are associated with DNA methylation changes in prostate cancer. Cancer Biol. Ther. 2007, 6, 1403–1412. [Google Scholar] [CrossRef]
- Spetsieris, N.; Boukovala, M.; Patsakis, G.; Alafis, I.; Efstathiou, E. Neuroendocrine and Aggressive-Variant Prostate Cancer. Cancers 2020, 12, 3792. [Google Scholar] [CrossRef]
- Yamada, Y.; Beltran, H. Clinical and Biological Features of Neuroendocrine Prostate Cancer. Curr. Oncol. Rep. 2021, 23, 15. [Google Scholar] [CrossRef]
- Wang, J.; Park, K.S.; Yu, X.; Gong, W.; Earp, H.S.; Wang, G.G.; Jin, J.; Cai, L. A cryptic transactivation domain of EZH2 binds AR and AR’s splice variant, promoting oncogene activation and tumorous transformation. Nucleic Acids Res. 2022, 50, 10929–10946. [Google Scholar] [CrossRef] [PubMed]
- Dardenne, E.; Beltran, H.; Benelli, M.; Gayvert, K.; Berger, A.; Puca, L.; Cyrta, J.; Sboner, A.; Noorzad, Z.; MacDonald, T.; et al. N-Myc Induces an EZH2-Mediated Transcriptional Program Driving Neuroendocrine Prostate Cancer. Cancer Cell 2016, 30, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Fischetti, I.; Botti, L.; Sulsenti, R.; Cancila, V.; Enriquez, C.; Ferri, R.; Bregni, M.; Crivelli, F.; Tripodo, C.; Colombo, M.P.; et al. Combined therapy targeting AR and EZH2 curbs castration-resistant prostate cancer enhancing anti-tumor T-cell response. Epigenomics 2024, 16, 653–670. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Nie, Q.; Zhang, X.; Huang, L.; Pang, G.; Chu, J.; Yuan, X. miR-26a-5p restoration via EZH2 silencing blocks the IL-6/STAT3 axis to repress the growth of prostate cancer. Expert Opin. Ther. Targets 2023, 27, 1285–1297. [Google Scholar] [CrossRef]
- Su, Z.; Zhang, M.; Luo, H.; Zhong, J.; Tan, J.; Xu, Y.; Pan, X.; Zeng, H.; Nie, L.; Xu, M.; et al. circEZH2(E2) (/E3) is a dual suppressor of miR363/miR708 to promote EZH2 expression and prostate cancer progression. Cancer Sci. 2023, 114, 1378–1395. [Google Scholar] [CrossRef]
- Hansen, A.F.; Høiem, T.S.; Selnaes, K.M.; Bofin, A.M.; Størkersen, Ø.; Bertilsson, H.; Wright, A.J.; Giskeødegård, G.F.; Bathen, T.F.; Rye, M.B.; et al. Prediction of recurrence from metabolites and expression of TOP2A and EZH2 in prostate cancer patients treated with radiotherapy. NMR Biomed. 2023, 36, e4694. [Google Scholar] [CrossRef]
- Huang, K.; Tang, Y. SChLAP1 promotes prostate cancer development through interacting with EZH2 to mediate promoter methylation modification of multiple miRNAs of chromosome 5 with a DNMT3a-feedback loop. Cell Death Dis. 2021, 12, 188. [Google Scholar] [CrossRef]
- Ma, L.; Yan, Y.; Bai, Y.; Yang, Y.; Pan, Y.; Gang, X.; Karnes, R.J.; Zhang, J.; Lv, Q.; Wu, Q.; et al. Overcoming EZH2 Inhibitor Resistance by Taxane in PTEN-Mutated Cancer. Theranostics 2019, 9, 5020–5034. [Google Scholar] [CrossRef]
- Xu, S.G.; Yu, J.J.; Shi, Q.; Niu, Q.; Guo, Z.; Guo, B.Y.; Zhou, G.C.; Gu, X.; Wu, Y.X. Conditionally replicative adenovirus carrying shRNA targeting EZH2 inhibits prostate cancer growth and invasion. Oncol. Rep. 2019, 42, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Scott, H.; Carlsson, S.V.; Sjoberg, D.D.; Cerundolo, L.; Lilja, H.; Prevo, R.; Rieunier, G.; Macaulay, V.; Higgins, G.S.; et al. Increased EZH2 expression in prostate cancer is associated with metastatic recurrence following external beam radiotherapy. Prostate 2019, 79, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, V.; Yadav, A.; Tiwari, R.; Nigam, S.; Goel, S.; Carskadon, S.; Gupta, N.; Goel, A.; Palanisamy, N.; Ateeq, B. Epigenetic Silencing of miRNA-338-5p and miRNA-421 Drives SPINK1-Positive Prostate Cancer. Clin. Cancer Res. 2019, 25, 2755–2768. [Google Scholar] [CrossRef] [PubMed]
- Patil, P.A.; McKenney, J.K.; Reynolds, J.P.; Przybycin, C.G.; Magi-Galluzzi, C. Clinical significance and EZH2, ERG and SPINK1 protein expression in pure and mixed ductal adenocarcinoma of the prostate. Histol. Histopathol. 2019, 34, 381–390. [Google Scholar] [CrossRef]
- Lobo, J.; Rodrigues, Â.; Antunes, L.; Graça, I.; Ramalho-Carvalho, J.; Vieira, F.Q.; Martins, A.T.; Oliveira, J.; Jerónimo, C.; Henrique, R. High immunoexpression of Ki67, EZH2, and SMYD3 in diagnostic prostate biopsies independently predicts outcome in patients with prostate cancer. Urol. Oncol. Semin. Orig. Investig. 2018, 36, e167. [Google Scholar] [CrossRef]
- Labbé, D.P.; Sweeney, C.J.; Brown, M.; Galbo, P.; Rosario, S.; Wadosky, K.M.; Ku, S.Y.; Sjöström, M.; Alshalalfa, M.; Erho, N.; et al. TOP2A and EZH2 Provide Early Detection of an Aggressive Prostate Cancer Subgroup. Clin. Cancer Res. 2017, 23, 7072–7083. [Google Scholar] [CrossRef]
- Abdelrahman, A.E.; Arafa, S.A.; Ahmed, R.A. Prognostic Value of Twist-1, E-cadherin and EZH2 in Prostate Cancer: An Immunohistochemical Study. Turk Patoloji Derg. 2017, 1, 198–210. [Google Scholar] [CrossRef]
- Melling, N.; Thomsen, E.; Tsourlakis, M.C.; Kluth, M.; Hube-Magg, C.; Minner, S.; Koop, C.; Graefen, M.; Heinzer, H.; Wittmer, C.; et al. Overexpression of enhancer of zeste homolog 2 (EZH2) characterizes an aggressive subset of prostate cancers and predicts patient prognosis independently from pre- and postoperatively assessed clinicopathological parameters. Carcinogenesis 2015, 36, 1333–1340. [Google Scholar] [CrossRef]
- Matsika, A.; Srinivasan, B.; Day, C.; Mader, S.A.; Kiernan, D.M.; Broomfield, A.; Fu, J.; Hooper, J.D.; Kench, J.G.; Samaratunga, H. Cancer stem cell markers in prostate cancer: An immunohistochemical study of ALDH1, SOX2 and EZH2. Pathology 2015, 47, 622–628. [Google Scholar] [CrossRef]
- Jacobs, C.; Tumati, V.; Kapur, P.; Yan, J.; Hong, D.; Bhuiyan, M.; Xie, X.J.; Pistenmaa, D.; Yu, L.; Hsieh, J.T.; et al. DOC-2/DAB2 interacting protein status in high-risk prostate cancer correlates with outcome for patients treated with radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 2014, 89, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Changchien, Y.C.; Tátrai, P.; Papp, G.; Sápi, J.; Fónyad, L.; Szendrői, M.; Pápai, Z.; Sápi, Z. Poorly differentiated synovial sarcoma is associated with high expression of enhancer of zeste homologue 2 (EZH2). J. Transl. Med. 2012, 10, 216. [Google Scholar] [CrossRef]
- Simeone, N.; Frezza, A.M.; Zaffaroni, N.; Stacchiotti, S. Tazemetostat for advanced epithelioid sarcoma: Current status and future perspectives. Future Oncol. 2021, 17, 1253–1263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Peng, H. LncRNA-ANCR regulates the cell growth of osteosarcoma by interacting with EZH2 and affecting the expression of p21 and p27. J. Orthop. Surg. Res. 2017, 12, 103. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, M.; Liaño-Pons, J.; Wang, J.; Alzrigat, M.; Yuan, Y.; Ruiz-Pérez, M.V.; Chen, Y.; Kashuba, E.; de Flon, F.H.; Brodin, B.; et al. EZH2 inhibition sensitizes retinoic acid-driven senescence in synovial sarcoma. Cell Death Dis. 2024, 15, 836. [Google Scholar] [CrossRef]
- Yalçınkaya, U.; Uğraş, N.; Özgün, G.; Ocakoğlu, G.; Deligönül, A.; Çetintaş, S.K.; Bilgen, M.S. Enhancer of zeste homologue 2 (EZH2) expression in synovial sarcomas as a promising indicator of prognosis. Bosn. J. Basic Med. Sci. 2017, 17, 302–308. [Google Scholar] [CrossRef][Green Version]
- Ramaglia, M.; D’Angelo, V.; Iannotta, A.; Di Pinto, D.; Pota, E.; Affinita, M.C.; Donofrio, V.; Errico, M.E.; Lombardi, A.; Indolfi, C.; et al. High EZH2 expression is correlated to metastatic disease in pediatric soft tissue sarcomas. Cancer Cell Int. 2016, 16, 59. [Google Scholar] [CrossRef]
- Sun, R.; Shen, J.; Gao, Y.; Zhou, Y.; Yu, Z.; Hornicek, F.; Kan, Q.; Duan, Z. Overexpression of EZH2 is associated with the poor prognosis in osteosarcoma and function analysis indicates a therapeutic potential. Oncotarget 2016, 7, 38333–38346. [Google Scholar] [CrossRef]
- Béguelin, W.; Popovic, R.; Teater, M.; Jiang, Y.; Bunting, K.L.; Rosen, M.; Shen, H.; Yang, S.N.; Wang, L.; Ezponda, T.; et al. EZH2 Is Required for Germinal Center Formation and Somatic EZH2 Mutations Promote Lymphoid Transformation. Cancer Cell 2013, 23, 677–692. [Google Scholar] [CrossRef]
- Gartin, A.K.; Frost, T.C.; Cushman, C.H.; Leeper, B.A.; Gokhale, P.C.; DeCaprio, J.A. Merkel Cell Carcinoma Sensitivity to EZH2 Inhibition Is Mediated by SIX1 Derepression. J. Investig. Dermatol. 2022, 142, 2783–2792.e15. [Google Scholar] [CrossRef]
- Durand, M.-A.; Drouin, A.; Mouchard, A.; Durand, L.; Esnault, C.; Berthon, P.; Tallet, A.; Le Corre, Y.; Hainaut-Wierzbicka, E.; Blom, A.; et al. Distinct Regulation of EZH2 and its Repressive H3K27me3 Mark in Polyomavirus-Positive and -Negative Merkel Cell Carcinoma. J. Investig. Dermatol. 2023, 143, 1937–1946.e7. [Google Scholar] [CrossRef]
- Huang, X.M.; Shi, S.S.; Jian, T.M.; Tang, D.R.; Wu, T.; Sun, F.Y. LncRNA PVT1 knockdown affects proliferation and apoptosis of uveal melanoma cells by inhibiting EZH2. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2880–2887. [Google Scholar] [CrossRef]
- Uebel, A.; Kewitz-Hempel, S.; Willscher, E.; Gebhardt, K.; Sunderkötter, C.; Gerloff, D. Resistance to BRAF Inhibitors: EZH2 and Its Downstream Targets as Potential Therapeutic Options in Melanoma. Int. J. Mol. Sci. 2023, 24, 1963. [Google Scholar] [CrossRef] [PubMed]
- Acikalin, A.; Bagir, E.; Paydas, S. Prognostic and predictive value of EZH2 expression and the tumor immune microenvironment in Merkel cell carcinoma. Pol. J. Pathol. 2021, 72, 140–147. [Google Scholar] [PubMed]
- Hoffmann, F.; Niebel, D.; Aymans, P.; Ferring-Schmitt, S.; Dietrich, D.; Landsberg, J. H3K27me3 and EZH2 expression in melanoma: Relevance for melanoma progression and response to immune checkpoint blockade. Clin. Epigenetics 2020, 12, 24. [Google Scholar] [CrossRef]
- Cao, J.; Pontes, K.C.; Heijkants, R.C.; Brouwer, N.J.; Groenewoud, A.; Jordanova, E.S.; Marinkovic, M.; van Duinen, S.; Teunisse, A.F.; Verdijk, R.M.; et al. Overexpression of EZH2 in conjunctival melanoma offers a new therapeutic target. J. Pathol. 2018, 245, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Harms, K.L.; Chubb, H.; Zhao, L.; Fullen, D.R.; Bichakjian, C.K.; Johnson, T.M.; Carskadon, S.; Palanisamy, N.; Harms, P.W. Increased expression of EZH2 in Merkel cell carcinoma is associated with disease progression and poorer prognosis. Hum. Pathol. 2017, 67, 78–84. [Google Scholar] [CrossRef]
- Yu, H.; Ma, M.; Yan, J.; Xu, L.; Yu, J.; Dai, J.; Xu, T.; Tang, H.; Wu, X.; Li, S.; et al. Identification of coexistence of BRAF V600E mutation and EZH2 gain specifically in melanoma as a promising target for combination therapy. J. Transl. Med. 2017, 15, 243. [Google Scholar] [CrossRef]
- Veija, T.; Koljonen, V.; Bohling, T.; Kero, M.; Knuutila, S.; Sarhadi, V.K. Aberrant expression of ALK and EZH2 in Merkel cell carcinoma. BMC Cancer 2017, 17, 236. [Google Scholar] [CrossRef]
- Tiffen, J.; Gallagher, S.J.; Filipp, F.; Gunatilake, D.; Emran, A.A.; Cullinane, C.; Dutton-Register, K.; Aoude, L.; Hayward, N.; Chatterjee, A.; et al. EZH2 Cooperates with DNA Methylation to Downregulate Key Tumor Suppressors and IFN Gene Signatures in Melanoma. J. Investig. Dermatol. 2020, 140, 2442–2454.e5. [Google Scholar] [CrossRef]
- Rao, R.C.; Chan, M.P.; Andrews, C.A.; Kahana, A. EZH2, Proliferation Rate, and Aggressive Tumor Subtypes in Cutaneous Basal Cell Carcinoma. JAMA Oncol. 2016, 2, 962–963. [Google Scholar] [CrossRef] [PubMed]
- Montagnani, V.; Benelli, M.; Apollo, A.; Pescucci, C.; Licastro, D.; Urso, C.; Gerlini, G.; Borgognoni, L.; Luzzatto, L.; Stecca, B. Thin and thick primary cutaneous melanomas reveal distinct patterns of somatic copy number alterations. Oncotarget 2016, 7, 30365–30378. [Google Scholar] [CrossRef] [PubMed]
- Emadali, A.; Hoghoughi, N.; Duley, S.; Hajmirza, A.; Verhoeyen, E.; Cosset, F.L.; Bertrand, P.; Roumier, C.; Roggy, A.; Suchaud-Martin, C.; et al. Haploinsufficiency for NR3C1, the gene encoding the glucocorticoid receptor, in blastic plasmacytoid dendritic cell neoplasms. Blood 2016, 127, 3040–3053. [Google Scholar] [CrossRef] [PubMed]
- Harms, P.W.; Collie, A.M.; Hovelson, D.H.; Cani, A.K.; Verhaegen, M.E.; Patel, R.M.; Fullen, D.R.; Omata, K.; Dlugosz, A.A.; Tomlins, S.A.; et al. Next generation sequencing of Cytokeratin 20-negative Merkel cell carcinoma reveals ultraviolet-signature mutations and recurrent TP53 and RB1 inactivation. Mod. Pathol. 2016, 29, 240–248. [Google Scholar] [CrossRef]
- Stacchiotti, S.; Astolfi, A.; Gronchi, A.; Fontana, A.; Pantaleo, M.A.; Negri, T.; Brenca, M.; Tazzari, M.; Urbini, M.; Indio, V.; et al. Evolution of Dermatofibrosarcoma Protuberans to DFSP-Derived Fibrosarcoma: An Event Marked by Epithelial-Mesenchymal Transition-like Process and 22q Loss. Mol. Cancer Res. MCR 2016, 14, 820–829. [Google Scholar] [CrossRef]
- Masudo, K.; Suganuma, N.; Nakayama, H.; Oshima, T.; Rino, Y.; Iwasaki, H.; Matsuzu, K.; Sugino, K.; Ito, K.; Kondo, T.; et al. EZH2 Overexpression as a Useful Prognostic Marker for Aggressive Behaviour in Thyroid Cancer. In Vivo 2018, 32, 25–31. [Google Scholar] [CrossRef]
- Borbone, E.; Troncone, G.; Ferraro, A.; Jasencakova, Z.; Stojic, L.; Esposito, F.; Hornig, N.; Fusco, A.; Orlando, V. Enhancer of zeste homolog 2 overexpression has a role in the development of anaplastic thyroid carcinomas. J. Clin. Endocrinol. Metab. 2011, 96, 1029–1038. [Google Scholar] [CrossRef]
- Sawicka-Gutaj, N.; Shawkat, S.; Andrusiewicz, M.; Ziółkowska, P.; Czarnywojtek, A.; Gut, P.; Ruchała, M. EZH2 and SMYD3 expression in papillary thyroid cancer. Oncol. Lett. 2021, 21, 342. [Google Scholar] [CrossRef]
- Xue, L.; Yan, H.; Chen, Y.; Zhang, Q.; Xie, X.; Ding, X.; Wang, X.; Qian, Z.; Xiao, F.; Song, Z.; et al. EZH2 upregulation by ERα induces proliferation and migration of papillary thyroid carcinoma. BMC Cancer 2019, 19, 1094. [Google Scholar] [CrossRef]
- Sabour-Takanlou, M.; Sabour-Takanlou, L.; Biray-Avci, C. EZH2-associated tumor malignancy: A prominent target for cancer treatment. Clin. Genet. 2024, 106, 377–385. [Google Scholar] [CrossRef]
- Wang, Z.; Dai, J.; Yan, J.; Zhang, Y.; Yin, Z. Targeting EZH2 as a novel therapeutic strategy for sorafenib-resistant thyroid carcinoma. J. Cell. Mol. Med. 2019, 23, 4770–4778. [Google Scholar] [CrossRef]
- Ma, X.; Li, Y.; Song, Y.; Xu, G. Long Noncoding RNA CCDC26 Promotes Thyroid Cancer Malignant Progression via miR-422a/EZH2/Sirt6 Axis. OncoTargets Ther. 2021, 14, 3083–3094. [Google Scholar] [CrossRef]
- Rinke, J.; Müller, J.P.; Blaess, M.F.; Chase, A.; Meggendorfer, M.; Schäfer, V.; Winkelmann, N.; Haferlach, C.; Cross, N.C.P.; Hochhaus, A.; et al. Molecular characterization of EZH2 mutant patients with myelodysplastic/myeloproliferative neoplasms. Leukemia 2017, 31, 1936–1943. [Google Scholar] [CrossRef] [PubMed]
- Stomper, J.; Meier, R.; Ma, T.; Pfeifer, D.; Ihorst, G.; Blagitko-Dorfs, N.; Greve, G.; Zimmer, D.; Platzbecker, U.; Hagemeijer, A.; et al. Integrative study of EZH2 mutational status, copy number, protein expression and H3K27 trimethylation in AML/MDS patients. Clin. Epigenetics 2021, 13, 77. [Google Scholar] [CrossRef] [PubMed]
- Lasho, T.L.; Mudireddy, M.; Finke, C.M.; Hanson, C.A.; Ketterling, R.P.; Szuber, N.; Begna, K.H.; Patnaik, M.M.; Gangat, N.; Pardanani, A.; et al. Targeted next-generation sequencing in blast phase myeloproliferative neoplasms. Blood Adv. 2018, 2, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Venton, G.; Courtier, F.; Charbonnier, A.; D’Incan, E.; Saillard, C.; Mohty, B.; Mozziconacci, M.J.; Birnbaum, D.; Murati, A.; Vey, N.; et al. Impact of gene mutations on treatment response and prognosis of acute myeloid leukemia secondary to myeloproliferative neoplasms. Am. J. Hematol. 2018, 93, 330–338. [Google Scholar] [CrossRef]
- Kosalai, S.T.; Morsy, M.H.A.; Papakonstantinou, N.; Mansouri, L.; Stavroyianni, N.; Kanduri, C.; Stamatopoulos, K.; Rosenquist, R.; Kanduri, M. EZH2 upregulates the PI3K/AKT pathway through IGF1R and MYC in clinically aggressive chronic lymphocytic leukaemia. Epigenetics 2019, 14, 1125–1140. [Google Scholar] [CrossRef]
- Zheng, Z.; Li, L.; Li, G.; Zhang, Y.; Dong, C.; Ren, F.; Chen, W.; Ma, Y. EZH2/EHMT2 Histone Methyltransferases Inhibit the Transcription of DLX5 and Promote the Transformation of Myelodysplastic Syndrome to Acute Myeloid Leukemia. Front. Cell Dev. Biol. 2021, 9, 619795. [Google Scholar] [CrossRef]
- Chai, J.; Choudhuri, J.; Gong, J.Z.; Wang, Y.; Tian, X. Upregulation of Enhancer of Zeste Homolog 2 (EZH2) with Associated pERK Co-Expression and PRC2 Complex Protein SUZ12 Correlation in Adult T-Cell Leukemia/Lymphoma. Cancers 2024, 16, 646. [Google Scholar] [CrossRef]
- Rinke, J.; Chase, A.; Cross, N.C.P.; Hochhaus, A.; Ernst, T. EZH2 in Myeloid Malignancies. Cells 2020, 9, 1639. [Google Scholar] [CrossRef]
- Stasik, S.; Middeke, J.M.; Kramer, M.; Röllig, C.; Krämer, A.; Scholl, S.; Hochhaus, A.; Crysandt, M.; Brümmendorf, T.H.; Naumann, R.; et al. EZH2 mutations and impact on clinical outcome: An analysis in 1,604 patients with newly diagnosed acute myeloid leukemia. Haematologica 2020, 105, e228–e231. [Google Scholar] [CrossRef]
- Shen, J.K.; Cote, G.M.; Gao, Y.; Choy, E.; Mankin, H.J.; Hornicek, F.J.; Duan, Z. Targeting EZH2-mediated methylation of H3K27 inhibits proliferation and migration of Synovial Sarcoma in vitro. Sci. Rep. 2016, 6, 25239. [Google Scholar] [CrossRef]
- Li, B.; Chng, W.J. EZH2 abnormalities in lymphoid malignancies: Underlying mechanisms and therapeutic implications. J. Hematol. Oncol. 2019, 12, 118. [Google Scholar] [CrossRef]
- Kim, D.H.; Siddiqui, S.; Jain, P.; Wang, M.; Thakral, B.; Li, S.; Miranda, R.; Vega, F.; Medeiros, L.J.; Ok, C.Y. TP53 mutation is frequent in mantle cell lymphoma with EZH2 expression and have dismal outcome when both are present. Hum. Pathol. 2024, 146, 1–7. [Google Scholar] [CrossRef]
- Zhao, Y.; Ding, L.; Wang, D.; Ye, Z.; He, Y.; Ma, L.; Zhu, R.; Pan, Y.; Wu, Q.; Pang, K.; et al. EZH2 cooperates with gain-of-function p53 mutants to promote cancer growth and metastasis. EMBO J. 2019, 38, e99599. [Google Scholar] [CrossRef]
- Profitós-Pelejà, N.; Santos, J.C.; Marín-Niebla, A.; Roué, G.; Ribeiro, M.L. Regulation of B-Cell Receptor Signaling and Its Therapeutic Relevance in Aggressive B-Cell Lymphomas. Cancers 2022, 14, 860. [Google Scholar] [CrossRef]
- Martínez-Laperche, C.; Sanz-Villanueva, L.; Díaz Crespo, F.J.; Muñiz, P.; Martín Rojas, R.; Carbonell, D.; Chicano, M.; Suárez-González, J.; Menárguez, J.; Kwon, M.; et al. EZH2 mutations at diagnosis in follicular lymphoma: A promising biomarker to guide frontline treatment. BMC Cancer 2022, 22, 982. [Google Scholar] [CrossRef] [PubMed]
- Groß, E.; Hilger, R.-A.; Schümann, F.L.; Bauer, M.; Bouska, A.; Rohde, C.; Willscher, E.; Lützkendorf, J.; Müller, L.P.; Edemir, B.; et al. SAM-Competitive EZH2-Inhibitors Induce Platinum Resistance by EZH2-Independent Induction of ABC-Transporters. Cancers 2023, 15, 3043. [Google Scholar] [CrossRef] [PubMed]
- Schümann, F.L.; Groß, E.; Bauer, M.; Rohde, C.; Sandmann, S.; Terziev, D.; Müller, L.P.; Posern, G.; Wienke, A.; Fend, F.; et al. Divergent Effects of EZH1 and EZH2 Protein Expression on the Prognosis of Patients with T-Cell Lymphomas. Biomedicines 2021, 9, 1842. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Baquero, D.; Sakhdari, A.; Mo, H.; Kim, D.H.; Kanagal-Shamanna, R.; Li, S.; Young, K.H.; O’Malley, D.P.; Dogan, A.; Jain, P.; et al. EZH2 expression is associated with inferior overall survival in mantle cell lymphoma. Mod. Pathol. 2021, 34, 2183–2191. [Google Scholar] [CrossRef]
- Wu, S.-y.; Xie, Z.-y.; Yan, L.-y.; Liu, X.-f.; Zhang, Y.; Wang, D.-a.; Dong, J.; Sun, H.-t. The correlation of EZH2 expression with the progression and prognosis of hepatocellular carcinoma. BMC Immunol. 2022, 23, 28. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lv, H.; Jia, X.; Hu, G.; Kong, L.; Zhang, T.; Li, L.; Pan, Y.; Zhai, Q.; Meng, B.; et al. Clinical significance of enhancer of zeste homolog 2 and histone deacetylases 1 and 2 expression in peripheral T-cell lymphoma. Oncol. Lett. 2019, 18, 1415–1423. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Chen, X.; Huang, C.; Chen, G.; Chen, F.; Lu, J.; Shi, X.; He, C.; Zeng, Z.; Qiu, Y.; et al. EZH2/Bcl-2 Coexpression Predicts Worse Survival in Diffuse Large B-cell Lymphomas and Demonstrates Poor Efficacy to Rituximab in Localized Lesions. J. Cancer 2019, 10, 2006–2017. [Google Scholar] [CrossRef] [PubMed]
- Huet, S.; Xerri, L.; Tesson, B.; Mareschal, S.; Taix, S.; Mescam-Mancini, L.; Sohier, E.; Carrère, M.; Lazarovici, J.; Casasnovas, O.; et al. EZH2 alterations in follicular lymphoma: Biological and clinical correlations. Blood Cancer J. 2017, 7, e555. [Google Scholar] [CrossRef]
- Wang, X.; Sehgal, L.; Jain, N.; Khashab, T.; Mathur, R.; Samaniego, F. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J. Transl. Med. 2016, 14, 346. [Google Scholar] [CrossRef]
- Oh, E.J.; Kim, S.H.; Yang, W.I.; Ko, Y.H.; Yoon, S.O. Long Non-coding RNA HOTAIR Expression in Diffuse Large B-Cell Lymphoma: In Relation to Polycomb Repressive Complex Pathway Proteins and H3K27 Trimethylation. J. Pathol. Transl. Med. 2016, 50, 369–376. [Google Scholar] [CrossRef]
- Tian, X.; Pelton, A.; Shahsafaei, A.; Dorfman, D.M. Differential expression of enhancer of zeste homolog 2 (EZH2) protein in small cell and aggressive B-cell non-Hodgkin lymphomas and differential regulation of EZH2 expression by p-ERK1/2 and MYC in aggressive B-cell lymphomas. Mod. Pathol. 2016, 29, 1050–1057. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).