Accurate Diagnosis of High-Risk Pulmonary Nodules Using a Non-Invasive Epigenetic Biomarker Test
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
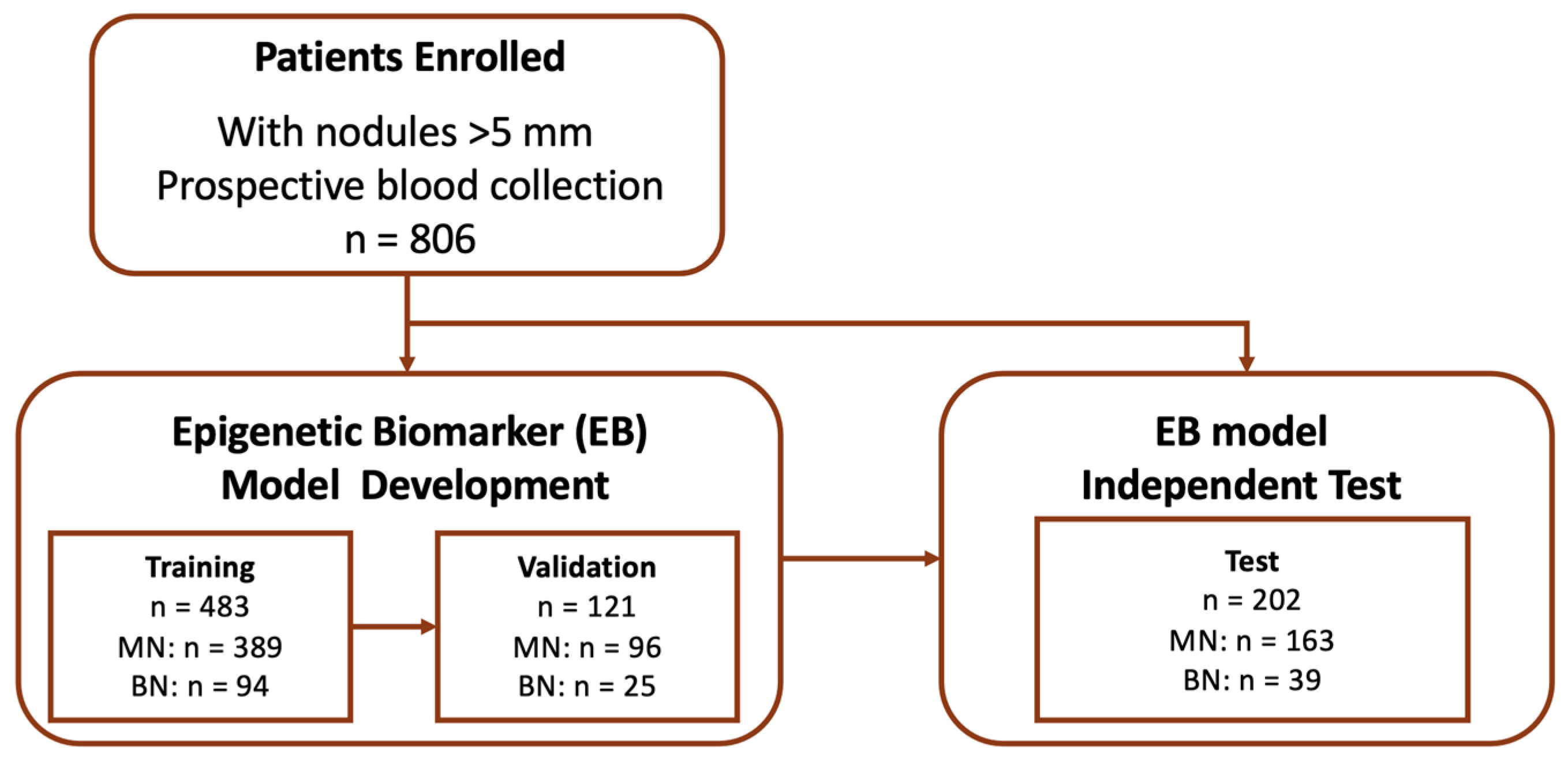
2.1. Study Design and Patients
2.1.1. Blood Sampling
2.1.2. Quantification of Circulating Nucleosomes Using Immunoassays
2.1.3. Chest CT Imaging and Radiological Analysis
2.1.4. Operation Policy and Pathology Evaluation
2.2. Theory/Calculation
2.2.1. EB Model Development for Benign–Malignant Predictions
2.2.2. Comparison with the Mayo Clinic and Veteran Affairs (VA) Models
2.2.3. Statistics
3. Results
3.1. Patient Demographics and Clinicopathologic Features
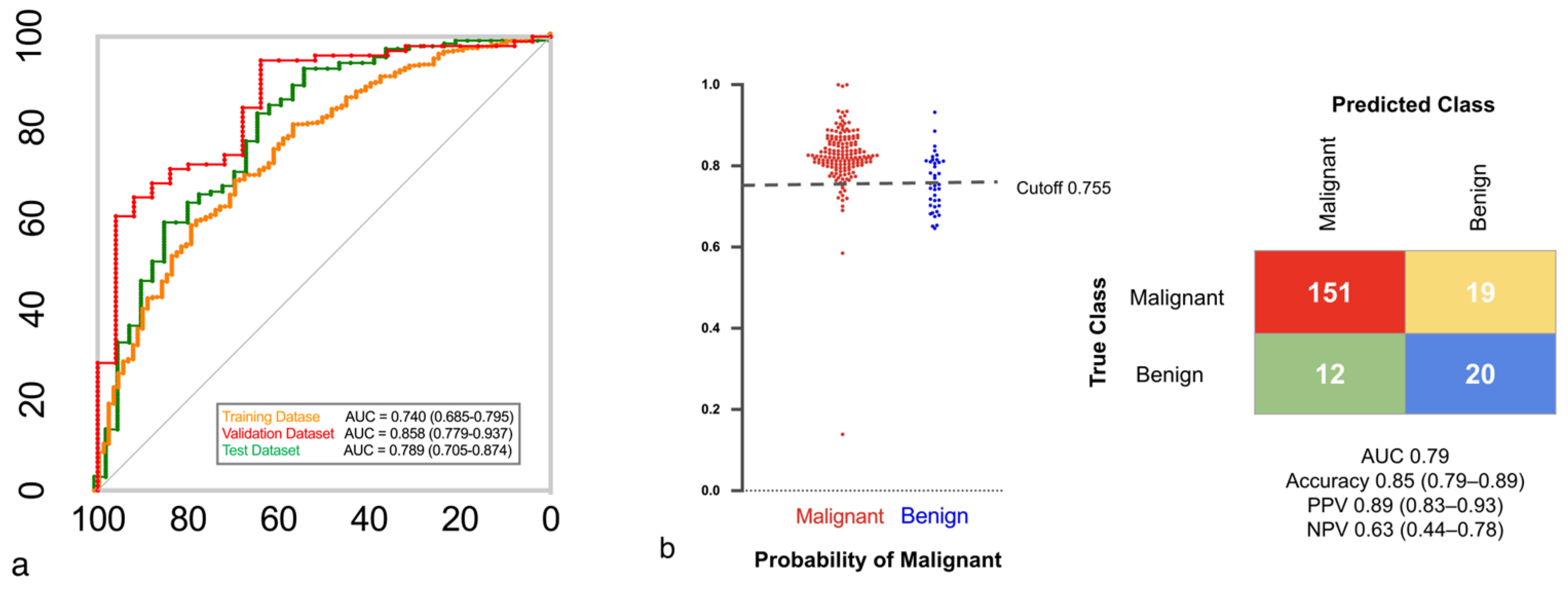
3.2. The EB Model and Lung Cancer Diagnostic Accuracy
Lung-RADS Score Analysis in the Test Dataset
3.3. EB Model Performance in Different Nodule Types
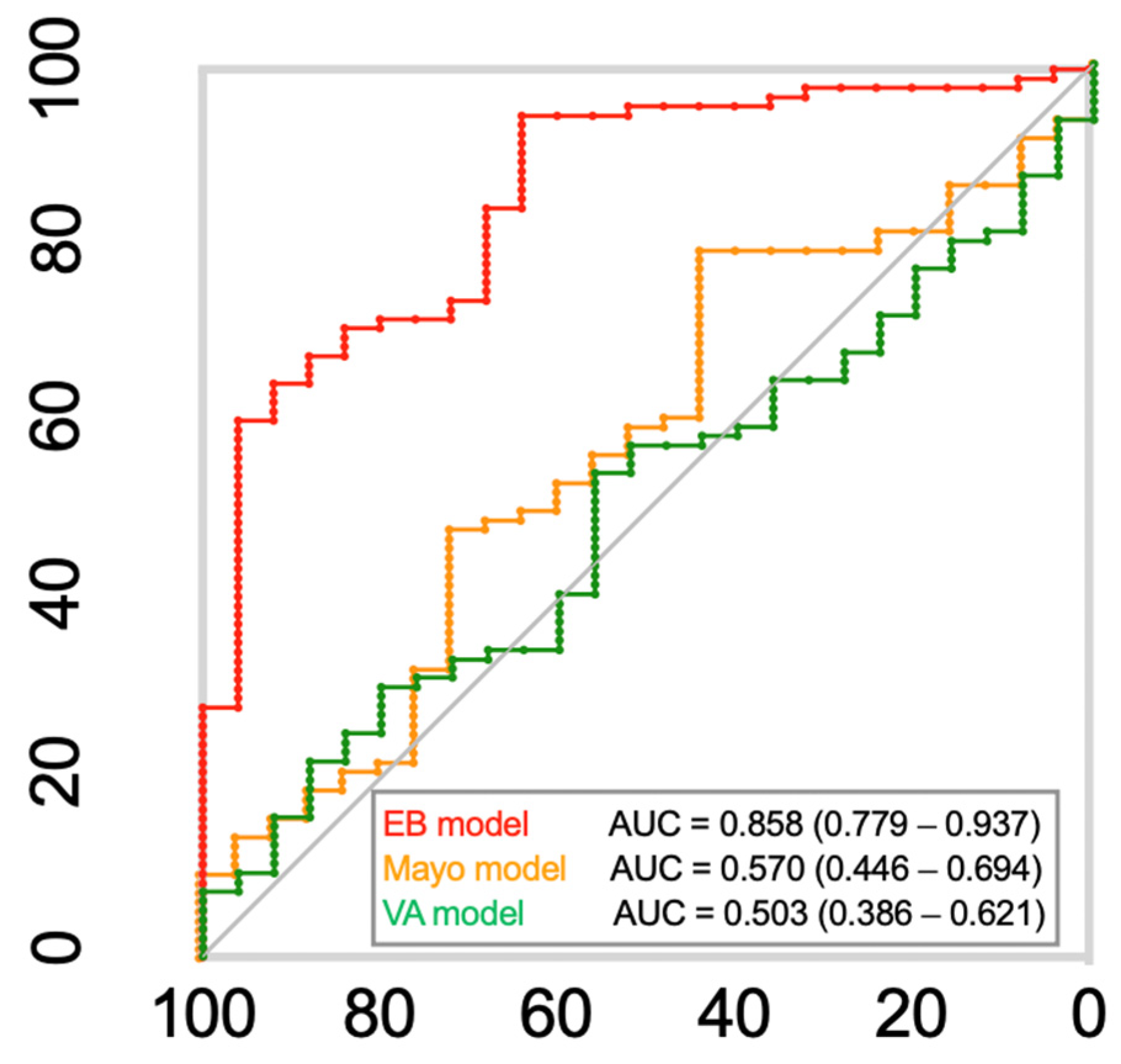
3.4. Conventional Cancer Diagnostic Model Comparison
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef] [PubMed]
- National Lung Screening Trial Research Team; Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef]
- De Koning, H.J.; Van Der Aalst, C.M.; De Jong, P.A.; Scholten, E.T.; Nackaerts, K.; Heuvelmans, M.A.; Lammers, J.J.; Weenink, C.; Yousaf-Khan, U.; Horeweg, N.; et al. Reduced lung-cancer mortality with volume CT screening in a randomized trial. N. Engl. J. Med. 2020, 382, 503–513. [Google Scholar] [CrossRef]
- He, J.; Wang, B.; Tao, J.; Liu, Q.; Peng, M.; Xiong, S.; Li, J.; Cheng, B.; Li, C.; Jiang, S.; et al. Accurate classification of pulmonary nodules by a combined model of clinical, imaging, and cell-free DNA methylation biomarkers: A model development and external validation study. Lancet Digit. Health 2023, 5, e647–e656. [Google Scholar] [CrossRef] [PubMed]
- Vachani, A.; Tanner, N.T.; Aggarwal, J.; Mathews, C.; Kearney, P.; Fang, K.C.; Silvestri, G.; Diette, G.B. Factors that influence physician decision making for indeterminate pulmonary nodules. Ann. Am. Thorac. Soc. 2014, 11, 1586–1591. [Google Scholar] [CrossRef]
- Gierada, D.S.; Pinsky, P.; Nath, H.; Chiles, C.; Duan, F.; Aberle, D.R. Projected outcomes using different nodule sizes to define a positive CT lung cancer screening examination. J. Natl. Cancer Inst. 2014, 106, dju284. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.-C.; Chiu, C.-H.; Yu, C.-J.; Chang, Y.-C.; Chang, Y.-H.; Hsu, K.-H.; Wu, Y.-C.; Chen, C.-Y.; Hsu, H.-H.; Wu, M.-T.; et al. Low-dose CT screening among never-smokers with or without a family history of lung cancer in Taiwan: A prospective cohort study. Lancet Respir. Med. 2024, 12, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Gould, M.K.; Donington, J.; Lynch, W.R.; Mazzone, P.J.; Midthun, D.E.; Naidich, D.P.; Wiener, R.S. Evaluation of individuals with pulmonary nodules: When is it lung cancer? Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013, 143, e93S–e120S. [Google Scholar] [CrossRef]
- Kammer, M.N.; Lakhani, D.A.; Balar, A.B.; Antic, S.L.; Kussrow, A.K.; Webster, R.L.; Mahapatra, S.; Barad, U.; Shah, C.; Atwater, T.; et al. Integrated biomarkers for the management of indeterminate pulmonary nodules. Am. J. Respir. Crit. Care Med. 2021, 204, 1306–1316. [Google Scholar] [CrossRef]
- Dawson, S.-J.; Tsui, D.W.Y.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.-F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Casagrande, G.M.S.; Silva, M.O.; Reis, R.M.; Leal, L.F. Liquid biopsy for lung cancer: Up-to-date and perspectives for screening programs. Int. J. Mol. Sci. 2023, 24, 2505. [Google Scholar] [CrossRef] [PubMed]
- Malapelle, U.; Pisapia, P.; Pepe, F.; Russo, G.; Buono, M.; Russo, A.; Gomez, J.; Khorshid, O.; Mack, P.C.; Rolfo, C.; et al. The evolving role of liquid biopsy in lung cancer. Lung Cancer 2022, 172, 53–64. [Google Scholar] [CrossRef]
- Kerr, K.M.; Galler, J.S.; Hagen, J.A.; Laird, P.W.; Laird-Offringa, I.A. The role of DNA methylation in the development and progression of lung adenocarcinoma. Dis. Markers 2007, 23, 5–30. [Google Scholar] [CrossRef] [PubMed]
- Van Den Ackerveken, P.; Lobbens, A.; Pamart, D.; Kotronoulas, A.; Rommelaere, G.; Eccleston, M.; Herzog, M. Epigenetic profiles of elevated cell free circulating H3.1 nucleosomes as potential biomarkers for non-Hodgkin lymphoma. Sci. Rep. 2023, 13, 16335. [Google Scholar] [CrossRef]
- Bajbouj, K.; Al-Ali, A.; Ramakrishnan, R.K.; Saber-Ayad, M.; Hamid, Q. Histone modification in NSCLC: Molecular mechanisms and therapeutic targets. Int. J. Mol. Sci. 2021, 22, 11701. [Google Scholar] [CrossRef]
- Bai, C.; Choi, C.-M.; Chu, C.M.; Anantham, D.; Chung-Man Ho, J.; Khan, A.Z.; Lee, J.-M.; Li, S.Y.; Saenghirunvattana, S.; Yim, A. Evaluation of pulmonary nodules: Clinical Practice Consensus Guidelines for Asia. Chest 2016, 150, 877–893. [Google Scholar] [CrossRef]
- Kastner, J.; Hossain, R.; Jeudy, J.; Dako, F.; Mehta, V.; Dalal, S.; Dharaiya, E.; White, C. Lung-RADS. version 1.0 versus Lung-RADS. version 1.1: Comparison of Categories Using Nodules from the National Lung Screening Trial. Radiology 2021, 300, 199–206. [Google Scholar] [CrossRef]
- Ye, T.; Deng, L.; Wang, S.; Xiang, J.; Zhang, Y.; Hu, H.; Sun, Y.; Li, Y.; Shen, L.; Xie, L.; et al. Lung adenocarcinomas manifesting as radiological part-solid nodules define a special clinical subtype. J. Thorac. Oncol. 2019, 14, 617–627. [Google Scholar] [CrossRef]
- Hattori, A.; Matsunaga, T.; Hayashi, T.; Takamochi, K.; Oh, S.; Suzuki, K. Prognostic impact of the findings on thin-section computed tomography in patients with subcentimeter non–small cell lung cancer. J. Thorac. Oncol. 2017, 12, 954–962. [Google Scholar] [CrossRef]
- Jacobson, F.L.; Austin, J.H.M.; Field, J.K.; Jett, J.R.; Keshavjee, S.; MacMahon, H.; Mulshine, J.L.; Munden, R.F.; Salgia, R.; Strauss, G.M.; et al. Development of the American Association for Thoracic Surgery guidelines for low-dose computed tomography scans to screen for lung cancer in North America: Recommendations of the American Association for Thoracic Surgery Task Force for Lung Cancer Screening and Surveillance. J. Thorac. Cardiovasc. Surg. 2012, 144, 25–32. [Google Scholar] [CrossRef][Green Version]
- Nicholson, A.G.; Tsao, M.S.; Beasley, M.B.; Borczuk, A.C.; Brambilla, E.; Cooper, W.A.; Dacic, S.; Jain, D.; Kerr, K.M.; Lantuejoul, S.; et al. The 2021 WHO classification of lung tumors: Impact of advances since 2015. J. Thorac. Oncol. 2022, 17, 362–387. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D.; Brambilla, E.; Noguchi, M.; Nicholson, A.G.; Geisinger, K.R.; Yatabe, Y.; Beer, D.G.; Powell, C.A.; Riely, G.J.; Van Schil, P.E.; et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 2011, 6, 244–285. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-S.; Cheng, Y.-J.; Hung, M.-H.; Tseng, Y.-D.; Chen, K.-C.; Lee, Y.-C. Nonintubated thoracoscopic lobectomy for lung cancer. Ann. Surg. 2011, 254, 1038–1043. [Google Scholar] [CrossRef]
- Chen, P.-H.; Hsu, H.-H.; Yang, S.-M.; Tsai, T.-M.; Tsou, K.-C.; Liao, H.-C.; Lin, M.-W.; Chen, J.-S. Preoperative dye localization for thoracoscopic lung surgery: Hybrid versus computed tomography room. Ann. Thorac. Surg. 2018, 106, 1661–1667. [Google Scholar] [CrossRef] [PubMed]
- Swensen, S.J.; Silverstein, M.D.; Ilstrup, D.M.; Schleck, C.D.; Edell, E.S. The probability of malignancy in solitary pulmonary nodules: Application to small radiologically indeterminate nodules. Arch. Intern. Med. 1997, 157, 849–855. [Google Scholar] [CrossRef]
- Gould, M.K.; Ananth, L.; Barnett, P.G.; Veterans Affairs SNAP Cooperative Study Group. A clinical model to estimate the pretest probability of lung cancer in patients with solitary pulmonary nodules. Chest 2007, 131, 383–388. [Google Scholar] [CrossRef]
- Ostrow, K.L.; Hoque, M.O.; Loyo, M.; Brait, M.; Greenberg, A.; Siegfried, J.M.; Grandis, J.R.; Gaither Davis, A.; Bigbee, W.L.; Rom, W.; et al. Molecular analysis of plasma DNA for the early detection of lung cancer by quantitative methylation-specific PCR. Clin. Cancer Res. 2010, 16, 3463–3472. [Google Scholar] [CrossRef]
- Belinsky, S.A.; Klinge, D.M.; Dekker, J.D.; Smith, M.W.; Bocklage, T.J.; Gilliland, F.D.; Crowell, R.E.; Karp, D.D.; Stidley, C.A.; Picchi, M.A. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin. Cancer Res. 2005, 11, 6505–6511. [Google Scholar] [CrossRef]
- Li, W.; Liu, J.-B.; Hou, L.-K.; Yu, F.; Zhang, J.; Wu, W.; Tang, X.-M.; Sun, F.; Lu, H.-M.; Deng, J.; et al. Liquid biopsy in lung cancer: Significance in diagnostics, prediction, and treatment monitoring. Mol. Cancer 2022, 21, 25. [Google Scholar] [CrossRef]
- Wei, Y.; Xia, W.; Zhang, Z.; Liu, J.; Wang, H.; Adsay, N.V.; Albarracin, C.; Yu, D.; Abbruzzese, J.L.; Mills, G.B.; et al. Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Mol. Carcinog. 2008, 47, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Mitani, Y.; Oue, N.; Hamai, Y.; Aung, P.P.; Matsumura, S.; Nakayama, H.; Kamata, N.; Yasui, W. Histone H3 acetylation is associated with reduced p21(WAF1/CIP1) expression by gastric carcinomaWAF1/CIP1. J. Pathol. 2005, 205, 65–73. [Google Scholar] [CrossRef]
- Ellinger, J.; Kahl, P.; Von Der Gathen, J.; Heukamp, L.C.; Gütgemann, I.; Walter, B.; Hofstädter, F.; Bastian, P.J.; von Ruecker, A.; Müller, S.C.; et al. Global histone H3K27 methylation levels are different in localized and metastatic prostate cancer. Cancer Investig. 2012, 30, 92–97. [Google Scholar] [CrossRef]
- Tzao, C.; Tung, H.-J.; Jin, J.-S.; Sun, G.-H.; Hsu, H.-S.; Chen, B.-H.; Yu, C.-P.; Lee, S.-C. Prognostic significance of global histone modifications in resected squamous cell carcinoma of the esophagus. Mod. Pathol. 2009, 22, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.-Y.; Tong, Z.-T.; Zhu, W.; Wen, Z.-Z.; Rao, H.-L.; Kong, L.-L.; Guan, X.-Y.; Kung, H.-F.; Zeng, Y.-X.; Xie, D. H3K27me3 protein is a promising predictive biomarker of patients’ survival and chemoradioresistance in human nasopharyngeal carcinoma. Mol. Med. 2011, 17, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.-Y.; Hou, J.-H.; Rao, H.-L.; Luo, R.-Z.; Li, M.; Pei, X.-Q.; Lin, M.C.; Guan, X.-Y.; Kung, H.-F.; Zeng, Y.-X.; et al. High expression of H3K27me3 in human hepatocellular carcinomas correlates closely with vascular invasion and predicts worse prognosis in patients. Mol. Med. 2011, 17, 12–20. [Google Scholar] [CrossRef]
- Chervona, Y.; Costa, M. Histone modifications and cancer: Biomarkers of prognosis? Am. J. Cancer Res. 2012, 2, 589–597. [Google Scholar]
- Chen, X.; Song, N.; Matsumoto, K.; Nanashima, A.; Nagayasu, T.; Hayashi, T.; Ying, M.; Endo, D.; Wu, Z.; Koji, T. High expression of trimethylated histone H3 at lysine 27 predicts better prognosis in non-small cell lung cancer. Int. J. Oncol. 2013, 43, 1467–1480. [Google Scholar] [CrossRef]
- Grolleau, E.; Candiracci, J.; Lescuyer, G.; Barthelemy, D.; Benzerdjeb, N.; Haon, C.; Geiguer, F.; Raffin, M.; Hardat, N.; Balandier, J.; et al. Circulating H3K27 methylated nucleosome plasma concentration: Synergistic information with circulating tumor DNA molecular profiling. Biomolecules 2023, 13, 1255. [Google Scholar] [CrossRef]
- Sarno, F.; Benincasa, G.; List, M.; Barabasi, A.-L.; Baumbach, J.; Ciardiello, F.; Filetti, S.; Glass, K.; Loscalzo, J.; Marchese, C.; et al. Clinical epigenetics settings for cancer and cardiovascular diseases: Real-life applications of network medicine at the bedside. Clin. Epigenetics 2021, 13, 66. [Google Scholar] [CrossRef]
- Gandhi, Z.; Gurram, P.; Amgai, B.; Lekkala, S.P.; Lokhandwala, A.; Manne, S.; Mohammed, A.; Koshiya, H.; Dewaswala, N.; Desai, R.; et al. Artificial intelligence and lung cancer: Impact on improving patient outcomes. Cancers 2023, 15, 5236. [Google Scholar] [CrossRef] [PubMed]
- Bahado-Singh, R.; Vlachos, K.T.; Aydas, B.; Gordevicius, J.; Radhakrishna, U.; Vishweswaraiah, S. Precision oncology: Artificial intelligence and DNA methylation analysis of circulating cell-free DNA for lung cancer detection. Front. Oncol. 2022, 12, 790645. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Oh, S.; Hong, S.; Kang, M.; Kang, D.; Ji, Y.-G.; Choi, B.H.; Kang, K.-W.; Jeong, H.; Park, Y.; et al. Early-stage lung cancer diagnosis by deep learning-based spectroscopic analysis of circulating exosomes. ACS Nano 2020, 14, 5435–5444. [Google Scholar] [CrossRef]
- Scimia, M.; Du, J.; Pepe, F.; Bianco, M.A.; Russo Spena, S.; Patell-Socha, F.; Sun, Q.; Powell, M.J.; Malapelle, U.; Troncone, G. Evaluation of a novel liquid biopsy-based ColoScape assay for mutational analysis of colorectal neoplasia and triage of FIT+ patients: A pilot study. J. Clin. Pathol. 2018, 71, 1123–1126. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Javed, A.A.; Thoburn, C.; Wong, F.; Tie, J.; Gibbs, P.; Schmidt, C.M.; Yip-Schneider, M.T.; Allen, P.J.; Schattner, M.; et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc. Natl. Acad. Sci. USA 2017, 114, 10202–10207. [Google Scholar] [CrossRef]
- Klein, E.A.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, B.; Yu, Y.; Lu, J.; Lou, Y.; Qian, F.; Chen, T.; Zhang, L.; Yang, J.; Zhong, H.; et al. Multimodal fusion of liquid biopsy and CT enhances differential diagnosis of early-stage lung adenocarcinoma. npj Precis. Oncol. 2024, 8, 50. [Google Scholar] [CrossRef]
- Henschke, C.I.; Yankelevitz, D.F.; Mirtcheva, R.; McGuinness, G.; McCauley, D.; Miettinen, O.S. CT screening for lung cancer. AJR Am. J. Roentgenol. 2002, 178, 1053–1057. [Google Scholar] [CrossRef]
- Kuo, S.W.; Chen, P.H.; Lu, T.P.; Chen, K.C.; Liao, H.C.; Tsou, K.C.; Tsai, T.M.; Lin, M.W.; Hsu, H.H.; Chen, J.S. Primary tumor resection for stage IV non-small-cell lung cancer without progression after first-line epidermal growth factor receptor-tyrosine kinase inhibitor treatment: A retrospective case-control study. Ann. Surg. Oncol. 2022, 29, 4873–4884. [Google Scholar] [CrossRef]
| Patient Characteristics | Whole Cohort (n = 806) | Training Dataset (n = 483) n (%) | Validation Dataset (n =121) n (%) | Test Set (n = 202) n (%) | p-Value |
|---|---|---|---|---|---|
| Mean age (years) (range) | 59.44 ± 11.75 (23–89) | 59.04 ± 11.45 (26–88) | 60.03 ± 12.41 (33–89) | 60.02 ± 11.66 (23–85) | 0.51 |
| Female | 511 (63.40%) | 306 (63.35%) | 79 (65.29%) | 126 (62.38%) | 0.87 |
| Non-smoker | 630 (78.16%) | 379 (78.47%) | 92 (76.03%) | 159 (78.71%) | 0.88 |
| History of alcohol consumption | 87 (10.79%) | 53 (10.97%) | 13 (10.74%) | 21 (10.40%) | 0.97 |
| Lung cancer family history | 280 (34.74%) | 164 (33.95%) | 41 (33.88%) | 75 (37.13%) | 0.75 |
| Nodule type | 0.28 | ||||
| Solid | 357 (44.29%) | 217 (44.93%) | 51 (42.15%) | 89 (44.06%) | |
| Part-solid | 183 (22.71%) | 114 (23.60%) | 25 (20.66%) | 44 (21.78%) | |
| GGO | 266 (33.00%) | 152 (31.47%) | 45 (37.19%) | 69 (34.16%) | |
| Lung-RADS | 0.30 | ||||
| 2 | 284 (35.24%) | 164 (33.96%) | 47 (38.84%) | 73 (36.14%) | |
| 3 | 69 (8.56%) | 40 (8.28%) | 10 (8.27%) | 19 (9.41%) | |
| 4A | 107 (13.28%) | 71 (14.70%) | 19 (15.70%) | 17 (8.41%) | |
| 4B, 4X | 346 (42.92%) | 208 (43.06%) | 45 (37.19%) | 93 (46.04%) | |
| Nodule size (cm) | 0.29 | ||||
| <1 cm | 236 (29.28%) | 136 (28.16%) | 38 (31.41%) | 62 (30.69%) | |
| 1–2 cm | 274 (34.00%) | 173 (35.82%) | 44 (36.36%) | 57 (28.22%) | |
| >2 cm | 296 (36.72%) | 174 (36.02%) | 39 (32.23%) | 83 (41.09%) | |
| Mean tumor size: cm (range) | 2.05 ± 1.70 (0.3–10.2) | 2.00 ± 1.64 (0.3–10.1) | 1.92 ± 1.63 (0.5–9.6) | 2.24 ± 1.87 (0.4–10.2) | 0.15 |
| Nodule location | 0.16 | ||||
| Right upper lobe | 211 (26.18%) | 125 (25.88%) | 35 (28.92%) | 51 (25.25%) | |
| Right middle lobe | 67 (8.31%) | 49 (10.15%) | 9 (7.44%) | 9 (4.45%) | |
| Right lower lobe | 171 (21.22%) | 98 (20.29%) | 28 (23.14%) | 45 (22.28%) | |
| Left upper lobe | 231 (28.66%) | 137 (28.36%) | 31 (25.62%) | 63 (31.19%) | |
| Left lower lobe | 113 (14.12%) | 68 (14.08%) | 18 (14.88%) | 27 (13.37%) | |
| Others * | 13 (1.61%) | 6 (1.24%) | 0 (0.00%) | 7 (3.46%) | |
| Malignancy | 648 (80.40%) | 389 (80.54%) | 96 (79.34%) | 163 (80.69%) | 0.95 |
| Lung-RADS | ||||
|---|---|---|---|---|
| 2 | 3 | 4A + 4B | 4X | |
| (n = 73) | (n = 19) | (n = 81) | (n = 29) | |
| AUC | 0.75 | 0.81 | 0.78 | 0.98 |
| Accuracy | 0.84 (0.73–0.91) | 0.84 (0.60–0.97) | 0.82 (0.71–0.89) | 0.97 (0.82–1.00) |
| Sensitivity | 0.90 (0.79–0.96) | 1.00 (0.73–1.00) | 0.92 (0.82–0.97) | 0.96 (0.79–1.00) |
| Specificity | 0.57 (0.30–0.81) | 0.40 (0.07–0.83) | 0.44 (0.22–0.69) | 1.00 (1.00–1.00) |
| PPV | 0.90 (0.79–0.96) | 0.82 (0.56–0.95) | 0.85 (0.74–0.92) | 1.00 (0.84–1.00) |
| NPV | 0.57 (0.30–0.81) | 1.000 (0.20–1.00) | 0.62 (0.32–0.85) | 0.67 (0.13–0.98) |
| Component | |||
|---|---|---|---|
| Solid | Part-Solid | GGO | |
| All Nodule Sizes | (n = 89) | (n = 44) | (n = 69) |
| Accuracy | 0.84 (0.75–0.91) | 0.84 (0.70–0.93) | 0.86 (0.75–0.93) |
| Sensitivity | 0.96 (0.87–0.99) | 0.89 (0.73–0.96) | 0.91 (0.80–0.97) |
| Specificity | 0.45 (0.24–0.68) | 0.63 (0.26–0.90) | 0.55 (0.25–0.82) |
| PPV | 0.86 (0.76–0.92) | 0.91 (0.76–0.98) | 0.91 (0.80–0.97) |
| NPV | 0.75 (0.43–0.93) | 0.56 (0.23–0.85) | 0.55 (0.25–0.82) |
| Epigenetic Simple Regression Model | |||
|---|---|---|---|
| Training Dataset | Validation Dataset | Test Dataset | |
| All Nodule Sizes | (n = 483) | (n = 121) | (n = 202) |
| AUC | 0.74 | 0.86 | 0.79 |
| Accuracy | 0.80 (0.77–0.84) | 0.88 (0.81–0.94) | 0.85 (0.79–0.89) |
| Sensitivity | 0.91 (0.87–0.93) | 0.95 (0.88–0.98) | 0.93 (0.87–0.96) |
| Specificity | 0.37 (0.28–0.48) | 0.64 (0.43–0.81) | 0.51 (0.35–0.67) |
| PPV A | 0.86 (0.82–0.89) | 0.91 (0.83–0.96) | 0.89 (0.83–0.93) |
| NPV A | 0.49 (0.37–0.61) | 0.76 (0.53–0.91) | 0.63 (0.44–0.78) |
| Nodules sized 5–10 mm | (n = 142) | (n = 43) | (n = 61) |
| AUC | 0.70 | 0.89 | 0.80 |
| Accuracy (95% CI) | 0.76 (0.68–0.83) | 0.88 (0.75–0.96) | 0.85 (0.74–0.93) |
| Sensitivity (95% CI) | 0.91 (0.83–0.95) | 1.000 (0.86–1.000) | 0.94 (0.82–0.98) |
| Specificity (95% CI) | 0.27 (0.14–0.46) | 0.62 (0.32–0.85) | 0.54 (0.26–0.80) |
| PPV (95% CI) B | 0.81 (0.72–0.87) | 0.86 (0.69–0.95) | 0.88 (0.75–0.95) |
| NPV (95% CI) B | 0.47 (0.25–0.71) | 1.000 (0.60–1.000) | 0.70 (0.35–0.92) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, P.-H.; Tsai, T.-M.; Lu, T.-P.; Lu, H.-H.; Pamart, D.; Kotronoulas, A.; Herzog, M.; Micallef, J.V.; Hsu, H.-H.; Chen, J.-S. Accurate Diagnosis of High-Risk Pulmonary Nodules Using a Non-Invasive Epigenetic Biomarker Test. Cancers 2025, 17, 916. https://doi.org/10.3390/cancers17060916
Chen P-H, Tsai T-M, Lu T-P, Lu H-H, Pamart D, Kotronoulas A, Herzog M, Micallef JV, Hsu H-H, Chen J-S. Accurate Diagnosis of High-Risk Pulmonary Nodules Using a Non-Invasive Epigenetic Biomarker Test. Cancers. 2025; 17(6):916. https://doi.org/10.3390/cancers17060916
Chicago/Turabian StyleChen, Pei-Hsing, Tung-Ming Tsai, Tzu-Pin Lu, Hsiao-Hung Lu, Dorian Pamart, Aristotelis Kotronoulas, Marielle Herzog, Jacob Vincent Micallef, Hsao-Hsun Hsu, and Jin-Shing Chen. 2025. "Accurate Diagnosis of High-Risk Pulmonary Nodules Using a Non-Invasive Epigenetic Biomarker Test" Cancers 17, no. 6: 916. https://doi.org/10.3390/cancers17060916
APA StyleChen, P.-H., Tsai, T.-M., Lu, T.-P., Lu, H.-H., Pamart, D., Kotronoulas, A., Herzog, M., Micallef, J. V., Hsu, H.-H., & Chen, J.-S. (2025). Accurate Diagnosis of High-Risk Pulmonary Nodules Using a Non-Invasive Epigenetic Biomarker Test. Cancers, 17(6), 916. https://doi.org/10.3390/cancers17060916






