Prospective Evaluation of Cervical Scrapings CDO1 and CELF4 Methylation (epiHERA®) Assay in Detection of Endometrial Cancer
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. The Patients
2.2. Cervical Scrapings Sampling Technique
- Excess mucus was removed from the cervical os if required.
- The central bristles of the broom of the cervical sampler were inserted into the cervical os while the broom was fully in contact with the ectocervix.
- The sampler would be turned clockwise five times.
- The broom of the cervical sampler was put into a storage bottle containing 20 mL of Thinprep preservation solution and gently shaken to allow cervical exfoliated cells to stay in the bottle.
- The specimen was stored at room temperature between 15 °C and 30 °C.
- If the collected specimen contains excess blood (specimen has a red color), it will be discarded and not used for testing.
2.3. Sample Processing (DNA Extraction, Bisulfite Conversion, and Quantitative Methylation-Specific PCR (qMS-PCR))
2.4. Statistical Analysis
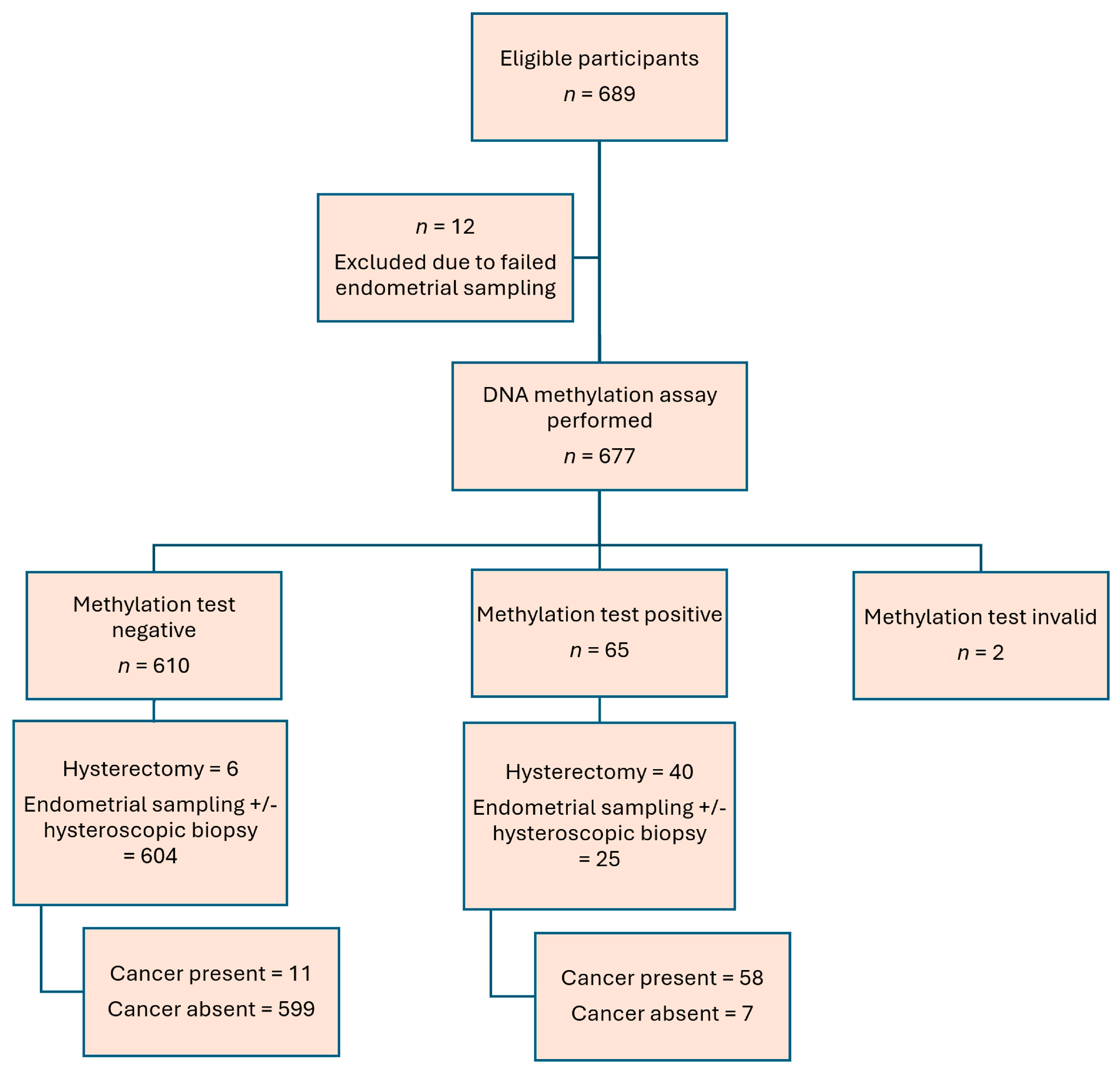
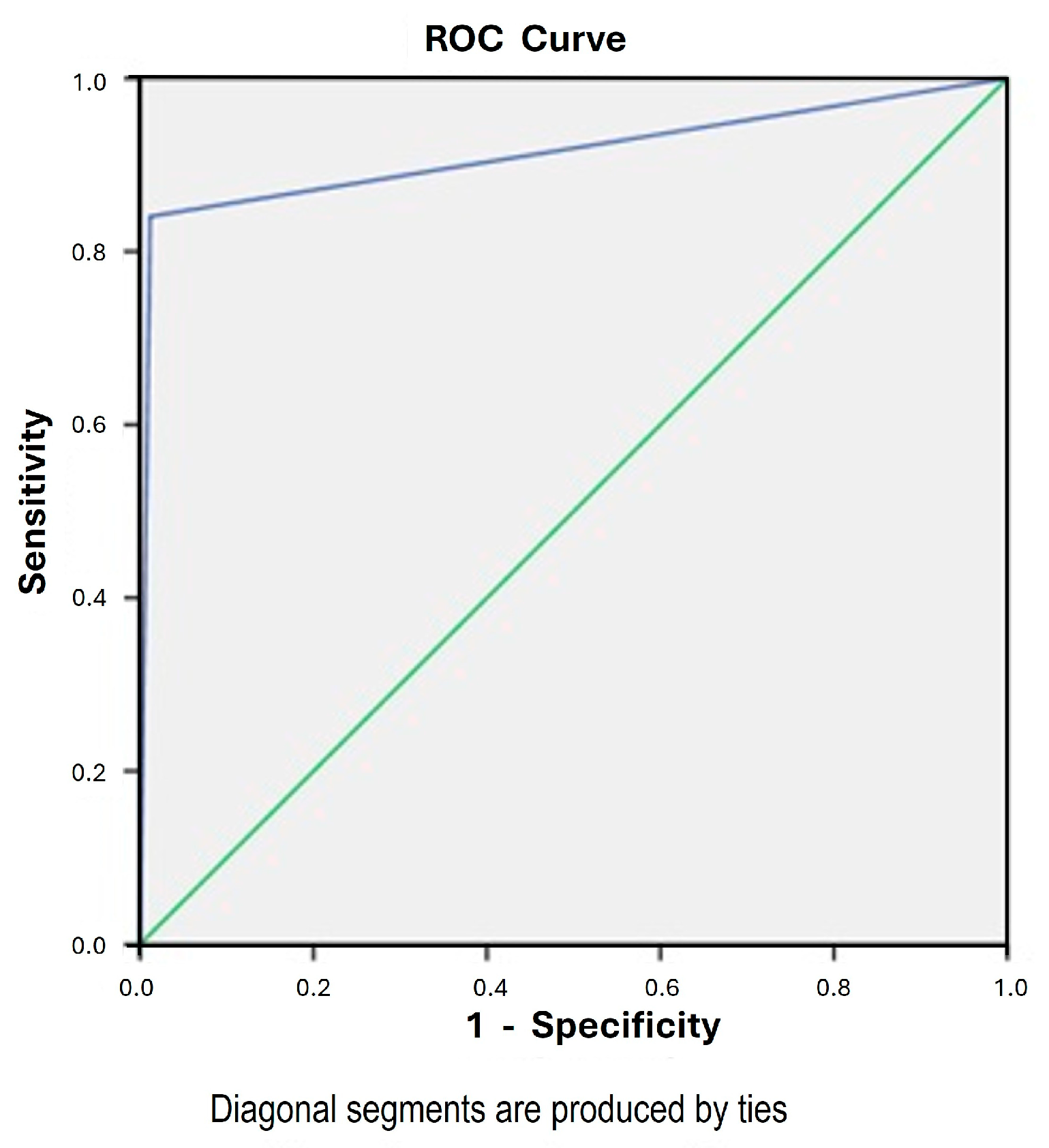
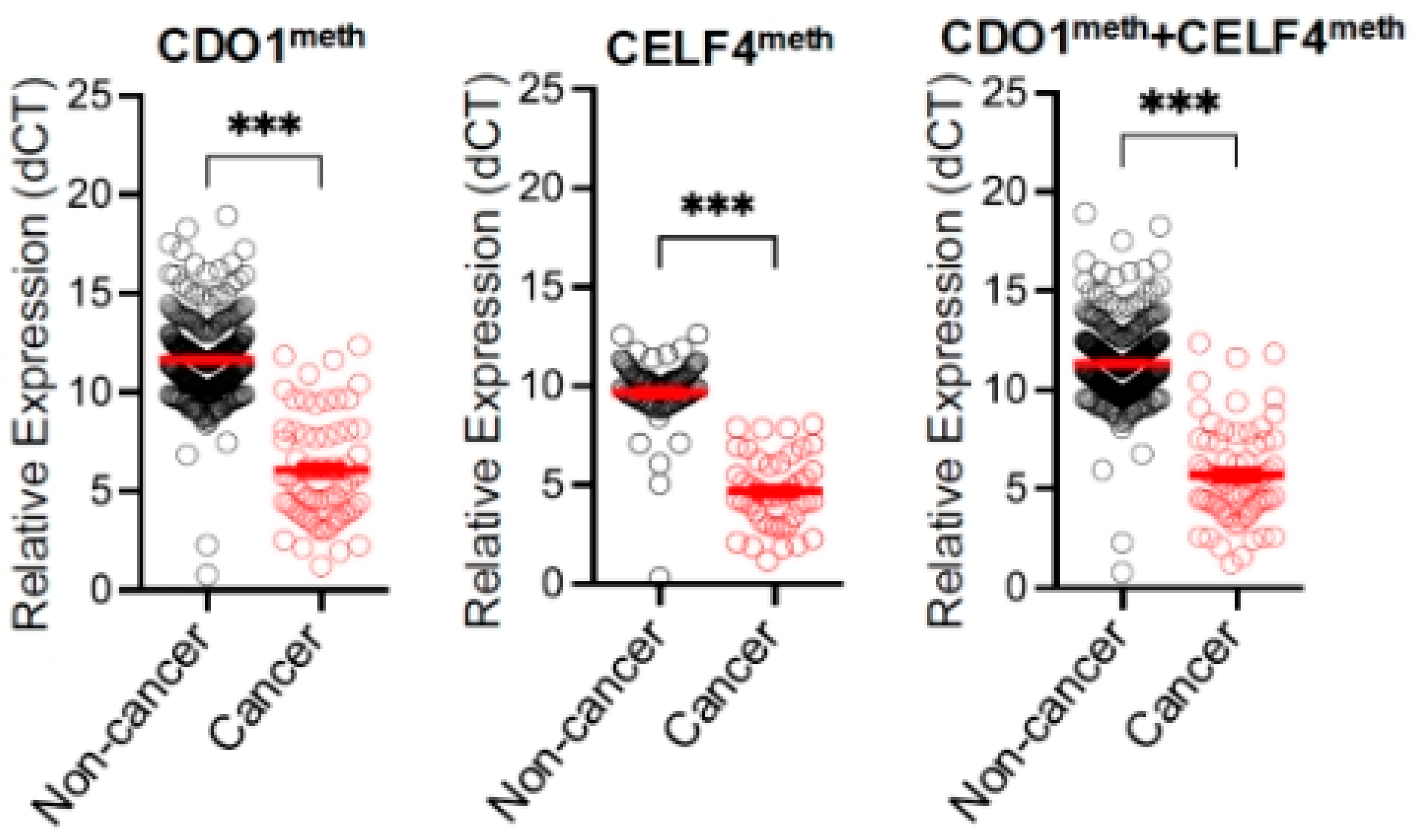
3. Results
4. Discussion
4.1. Performance of DNA Methylation Assay
4.2. Triage for Patients at Risk of Endometrial Cancer
4.3. Implication on Endometrial Hyperplasia Management
4.4. Detection of Cervical Pre-Invasive and Invasive Disease
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PPV | Positive predictive value |
| NPV | Negative predictive value |
| AUC | Area under the curve |
| D&C | Dilatation and curettage |
| BMI | Body mass index |
| PCR | Polymerase chain reaction |
| CIN | Cervical intraepithelial neoplasia |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Cronin, K.A.; Scott, S.; Firth, A.U.; Sung, H.; Henley, S.J.; Sherman, R.L.; Siegel, R.L.; Anderson, R.N.; Kohler, B.A.; Benard, V.B.; et al. Annual report to the nation on the status of cancer, part 1: National cancer statistics. Cancer 2022, 128, 4251–4284. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Costas, L.; Frias-Gomez, J.; Guardiola, M.; Benavente, Y.; Pineda, M.; Pavón, M.Á.; Martínez, J.M.; Climent, M.; Barahona, M.; Canet, J.; et al. New perspectives on screening and early detection of endometrial cancer. Int. J. Cancer 2019, 145, 3194–3206. [Google Scholar] [CrossRef]
- Wen, K.-C.; Huang, R.-L.; Chen, L.-Y.; Wu, T.-I.; Lu, C.-H.; Chu, T.-Y.; Ou, Y.-C.; Wu, C.-H.; Hsu, S.-T.; Ding, D.-C.; et al. Endometrial Cancer Detection Using a Cervical DNA Methylation Assay (MPap) in Women with Abnormal Uterine Bleeding: A Multicenter Hospital-Based Validation Study. Cancers 2022, 14, 4343. [Google Scholar] [CrossRef]
- Wong, A.; Lao, T.; Cheung, C.; Yeung, S.; Fan, H.; Ng, P.; Yuen, P.; Sahota, D. Reappraisal of endometrial thickness for the detection of endometrial cancer in postmenopausal bleeding: A retrospective cohort study. BJOG Int. J. Obstet. Gynaecol. 2015, 123, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Smith-Bindman, R.; Kerlikowske, K.; Feldstein, V.A.; Subak, L.; Scheidler, J.; Segal, M.; Brand, R.; Grady, D. Endovaginal Ultrasound to Exclude Endometrial Cancer and Other Endometrial Abnormalities. JAMA 1998, 280, 1510–1517. [Google Scholar] [CrossRef] [PubMed]
- Dijkhuizen, F.P.; Mol, B.W.; Brölmann, H.A.; Heintz, A.P. The accuracy of endometrial sampling in the diagnosis of patients with endometrial carcinoma and hyperplasia: A meta-analysis. Cancer 2000, 89, 1765–1772. [Google Scholar] [CrossRef]
- van Hanegem, N.; Prins, M.M.; Bongers, M.Y.; Opmeer, B.C.; Sahota, D.S.; Mol, B.W.J.; Timmermans, A. The accuracy of endometrial sampling in women with postmenopausal bleeding: A systematic review and meta-analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2016, 197, 147–155. [Google Scholar] [CrossRef]
- Hong Kong cancer registry. Corpus Uteri Cancer in 2022 [Internet]. Hong Kong. Hospital Authority; Oct 2022. Available online: https://www3.ha.org.hk/cancereg/pdf/factsheet/2022/corpus_2022.pdf (accessed on 21 April 2025).
- Zhou, J.; Tomashefski, J.F.; Khiyami, A. ThinPrep Pap tests in patients with endometrial cancer: A histo-cytological correlation. Diagn. Cytopathol. 2007, 35, 448–453. [Google Scholar] [CrossRef]
- Wang, L.; Dong, L.; Xu, J.; Guo, L.; Wang, Y.; Wan, K.; Jing, W.; Zhao, L.; Feng, X.; Zhang, K.; et al. Hypermethylated CDO1 and ZNF454 in Cytological Specimens as Screening Biomarkers for Endometrial Cancer. Front. Oncol. 2022, 12, 714663. [Google Scholar] [CrossRef]
- Huang, R.-L.; Su, P.-H.; Liao, Y.-P.; Wu, T.-I.; Hsu, Y.-T.; Lin, W.-Y.; Wang, H.-C.; Weng, Y.-C.; Ou, Y.-C.; Huang, T.H.-M.; et al. Integrated Epigenomics Analysis Reveals a DNA Methylation Panel for Endometrial Cancer Detection Using Cervical Scrapings. Clin. Cancer Res. 2017, 23, 263–272. [Google Scholar] [CrossRef]
- Locke, W.J.; Guanzon, D.; Ma, C.; Liew, Y.J.; Duesing, K.R.; Fung, K.Y.; Ross, J.P. DNA Methylation Cancer Biomarkers: Translation to the Clinic. Front. Genet. 2019, 10, 1150. [Google Scholar] [CrossRef]
- Bakkum-Gamez, J.N.; Wentzensen, N.; Maurer, M.J.; Hawthorne, K.M.; Voss, J.S.; Kroneman, T.N.; Famuyide, A.O.; Clayton, A.C.; Halling, K.C.; Kerr, S.E.; et al. Detection of endometrial cancer via molecular analysis of DNA collected with vaginal tampons. Gynecol. Oncol. 2015, 137, 14–22. [Google Scholar] [CrossRef]
- Evans, I.; Reisel, D.; Jones, A.; Bajrami, A.; Nijjar, S.; Solangon, S.A.; Arora, R.; Redl, E.; Schreiberhuber, L.; Ishaq-Parveen, I.; et al. Performance of the WID-qEC test versus sonography to detect uterine cancers in women with abnormal uterine bleeding (EPI-SURE): A prospective, consecutive observational cohort study in the UK. Lancet Oncol. 2023, 24, 1375–1386. [Google Scholar] [CrossRef]
- Tao, M.H.; Freudenheim, J.L. DNA methylation in endometrial cancer. Epigenetics 2010, 5, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Nieminen, T.T.; Gylling, A.; Abdel-Rahman, W.M.; Nuorva, K.; Aarnio, M.; Renkonen-Sinisalo, L.; Järvinen, H.J.; Mecklin, J.-P.; Bützow, R.; Peltomäki, P. Molecular Analysis of Endometrial Tumorigenesis: Importance of Complex Hyperplasia Regardless of Atypia. Clin. Cancer Res. 2009, 15, 5772–5783. [Google Scholar] [CrossRef]
- Pan, Y.; Liu, G.; Zhou, F.; Su, B.; Li, Y. DNA methylation profiles in cancer diagnosis and therapeutics. Clin. Exp. Med. 2017, 18, 1–14. [Google Scholar] [CrossRef]
- Hao, X.; Luo, H.; Krawczyk, M.; Wei, W.; Wang, W.; Wang, J.; Flagg, K.; Hou, J.; Zhang, H.; Yi, S.; et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc. Natl. Acad. Sci. USA 2017, 114, 7414–7419. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-C.; Wang, H.-C.; Liao, Y.-P.; Chen, Y.-C.; Weng, Y.-C.; Yu, M.-H.; Lai, H.-C. The feasibility of detecting endometrial and ovarian cancer using DNA methylation biomarkers in cervical scrapings. J. Gynecol. Oncol. 2018, 29, e17. [Google Scholar] [CrossRef] [PubMed]
- Sangtani, A.; Wang, C.; Weaver, A.; Hoppman, N.L.; Kerr, S.E.; Abyzov, A.; Shridhar, V.; Staub, J.; Kocher, J.-P.A.; Voss, J.S.; et al. Combining copy number, methylation markers, and mutations as a panel for endometrial cancer detection via intravaginal tampon collection. Gynecol. Oncol. 2020, 156, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Maritschnegg, E.; Wang, Y.; Pecha, N.; Horvat, R.; Van Nieuwenhuysen, E.; Vergote, I.; Heitz, F.; Sehouli, J.; Kinde, I.; Diaz, L.A.; et al. Lavage of the Uterine Cavity for Molecular Detection of Müllerian Duct Carcinomas: A Proof-of-Concept Study. J. Clin. Oncol. 2015, 33, 4293–4300. [Google Scholar] [CrossRef]
- A De Strooper, L.M.; van Zummeren, M.; Steenbergen, R.D.M.; Bleeker, M.C.G.; Hesselink, A.T.; A Wisman, G.B.; Snijders, P.J.F.; Heideman, D.A.M.; Meijer, C.J.L.M. CADM1, MAL and miR124-2 methylation analysis in cervical scrapes to detect cervical and endometrial cancer. J. Clin. Pathol. 2014, 67, 1067–1071. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Du, J.; Wang, Y.; Liu, Z.; Wang, Y.; Li, L.; Liu, P.; Wang, L.; Liu, Q.; Meng, Z. The endometrial cancer detection using non-invasive hypermethylation of CDO1 and CELF4 genes in women with postmenopausal bleeding in Northwest China. Cytojournal 2024, 21, 15. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, Y.; Fu, Y.; Lv, W.; Xu, D. DNA methylation detection is a significant biomarker for screening endometrial cancer in premenopausal women with abnormal uterine bleeding. Int. J. Gynecol. Cancer 2024, 34, 1165–1171. [Google Scholar] [CrossRef] [PubMed]
- Qi, B.; Sun, Y.; Lv, Y.; Hu, P.; Ma, Y.; Gao, W.; Li, S.; Zhang, X.; Jin, X.; Liou, Y.; et al. Hypermethylated CDO1 and CELF4 in cytological specimens as triage strategy biomarkers in endometrial malignant lesions. Front. Oncol. 2023, 13, 1289366. [Google Scholar] [CrossRef]
- Chen, M.; Zhu, J.-Y.; Mu, W.-J.; Guo, L. Cysteine dioxygenase type 1 (CDO1): Its functional role in physiological and pathophysiological processes. Genes Dis. 2023, 10, 877–890. [Google Scholar] [CrossRef]
- Nasiri-Aghdam, M.; Garcia-Garduño, T.C.; Jave-Suárez, L.F. CELF Family Proteins in Cancer: Highlights on the RNA-Binding Protein/Noncoding RNA Regulatory Axis. Int. J. Mol. Sci. 2021, 22, 11056. [Google Scholar] [CrossRef]
- Daud, S.; Jalil, S.S.; Griffin, M.; Ewies, A.A. Endometrial hyperplasia – the dilemma of management remains: A retrospective observational study of 280 women. Eur. J. Obstet. Gynecol. Reprod. Biol. 2011, 159, 172–175. [Google Scholar] [CrossRef]
- Giannella, L.; Piva, F.; Carpini, G.D.; Di Giuseppe, J.; Grelloni, C.; Giulietti, M.; Sopracordevole, F.; Giorda, G.; Del Fabro, A.; Clemente, N.; et al. Concurrent Endometrial Cancer in Women with Atypical Endometrial Hyperplasia: What Is the Predictive Value of Patient Characteristics? Cancers 2023, 16, 172. [Google Scholar] [CrossRef]
- Sadek, K.H.; Cagampang, F.R.; Bruce, K.D.; Shreeve, N.; Macklon, N.; Cheong, Y. Variation in stability of housekeeping genes in endometrium of healthy and polycystic ovarian syndrome women. Hum. Reprod. 2011, 27, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Ayakannu, T.; Taylor, A.H.; Konje, J.C. Selection of Endogenous Control Reference Genes for Studies on Type 1 or Type 2 Endometrial Cancer. Sci. Rep. 2020, 10, 8468. [Google Scholar] [CrossRef] [PubMed]
| Number of Patients | Percentage | Number of Patients | Percentage | ||
|---|---|---|---|---|---|
| Age (n = 675) | Pap smear cytology (n = 218) | ||||
| - <30–39 | 11 | 1.6% | Adenocarcinoma | 1 | 0.5% |
| - 40–49 | 106 | 15.7% | Abnormal glandular cell | 2 | 1% |
| - 50–59 | 240 | 35.6% | AGC not otherwise specified | 6 | 2.8% |
| - 60–69 | 234 | 34.7% | ASCH | 1 | 0.5% |
| - 70–79 | 74 | 11% | LSIL | 2 | 1% |
| - ≥80 | 10 | 1.5% | ASCUS | 12 | 5.5% |
| Mean age: 58.7, Median 59 | Negative for intra-epithelial neoplasia | 194 | 89% | ||
| BMI (n = 672) | Endometrial assessment pathology (n = 675) | ||||
| - <18.5 | 22 | 3.3% | Insufficient | 317 (menopause n = 301, pre-menopause n = 16) | 47% |
| 203 | 30.1% | ||||
| 70 | 10.4% | ||||
| 5 | 0.7% | ||||
| - 18.5–22.9 | 189 | 28.1% | Endometrial hyperplasia | ||
| - 23–24.9 | 148 | 22% | - With atypia | 7 | 1% |
| - 25–29.9 | 212 | 31.5% | - Without atypia | 4 | 0.6% |
| - 30–34.9 | 83 | 12.4% | Malignant | ||
| - ≥35 | 18 | 2.7% | Endometrioid adenocarcinoma grade 1 | 40 | 5.9% |
| Mean BMI: 24.8, Median 24.1 | Endometrioid adenocarcinoma grade 2 | 12 | 1.8% | ||
| Missing data = 3 | Endometrioid adenocarcinoma grade 3 | 11 | 1.6% | ||
| Menopaused | 538/675 | 79.7% | Carcinosarcoma | 3 | 0.4% |
| Family history of gynecological cancer | 46/674 | 6.8% | Clear cell carcinoma | 2 | 0.3% |
| Missing data = 1 | SCC | 1 | 0.1% | ||
| Hypertension present (n = 675) | 205/675 | 30.4% | Stage of endometrial cancer (n = 68) | ||
| Diabetes present (n = 675) | 89/675 | 13.2% | - 1A | 27 | 39.7% |
| Hyperlipidaemia present (n = 675) | 92/675 | 13.6% | - 1B | 11 | 16.2% |
| - II | 14 | 20.6% | |||
| Presentation symptoms (n = 674) | - IIIA | 1 | 1.5% | ||
| - Postmenopausal bleeding | 444 | 65.8% | - IIIB | 2 | 2.9% |
| - Menorrhagia | 82 | 12.2% | - IIIC | 12 | 17.6% |
| - Irregular menstruation | 25 | 3.7% | - IVB | 1 | 1.5% |
| - Intermenstrual bleeding | 21 | 3.1% | Missing data = 1 | ||
| - Hypermetabolic activity in endometrium on PET | 1 | 0.1% | |||
| - History of endometrial hyperplasia | 3 | 0.4% | |||
| - Asymptomatic thickened endometrial thickness or hydrometra | 46 | 6.8% | |||
| - Abnormal cervical smear | 3 | 0.4% | |||
| - Post-coital bleeding | 8 | 1.2% | |||
| - Foul-smelling vaginal discharge | 2 | 0.3% | |||
| - Others | 39 | 5.8% | |||
| Missing data = 1 | |||||
| Combine CDO1 and CELF4 Methylation Assay | Negative | Positive | Accuracy | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) | PLR (95% CI) | NLR (95% CI) | AUC (95% CI) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Endometrial assessment pathology | Pathology: benign/insufficient | N = 599 | N = 7 | 97.3% (657/675) | 84.1% (73.3% to 91.8%) | 98.8% (97.6% to 99.5%) | 89.2% (79.8% to 94.6%) | 98.2% (96.9% to 98.9%) | 72.8 (34.6 to 153.1) | 0.16 (0.09 to 0.28) | 0.92 (0.86 to 0.97) |
| Pathology: malignant | N = 11 | N = 58 |
| Accurate | Inaccurate | p Value | |
|---|---|---|---|
| Age (n = 675) (Mean) | 58.7 (SD 9.53) | 59.3 (SD 10.1) | 0.78 |
| BMI (n = 672) (Mean) | 24.8 (SD 4.45) | 25.5 (SD 4.98) | 0.51 |
| Menopausal status (n = 675) | |||
| Menopause | 525 (97.6%) | 13 (2.4%) | 0.38 |
| Not menopause | 132 (96.4%) | 5 (3.6%) | |
| Hypertension (n = 675) | |||
| Present | 200 (97.6%) | 5 (2.4%) | 1.0 |
| Absent | 457 (97.2%) | 13 (2.8%) | |
| Diabetes (n = 675) | |||
| - Present | 85 (95.5%) | 4 (4.5%) | 0.28 |
| - Absent | 572 (97.6%) | 14 (2.4%) | |
| Hyperlipidaemia (n = 675) | |||
| Present | 91 (98.9%) | 1 (1.1%) | 0.49 |
| Absent | 566 (97.1%) | 17 (2.9%) | |
| Fibroid (n = 674) | |||
| - Present | 152 (97.4%) | 4 (2.6%) | 1.0 |
| - Absent | 505 (97.5%) | 13 (2.5%) | |
| Adenomyosis (n = 674) | |||
| - Present | 10 (100%) | 0 (0%) | 1.0 |
| - Absent | 647 (97.4%) | 17 (2.6%) | |
| Time from LMP (n = 122) | |||
| - ≤ 14 days | 40 (97.6%) | 1 (2.4%) | 1.0 |
| - > 14 days | 80 (98.8%) | 1 (1.2%) | |
| Endometrial thickness (n = 533) | |||
| - ≤ 4 mm | 281 (99.6%) | 1 (0.4%) | <0.001 * |
| - > 4 mm | 240 (95.6%) | 11 (4.4%) | |
| High-grade/Glandular abnormality smear (n = 218) | |||
| - Yes | 8 (80%) | 2 (20%) | 0.02 * |
| - No | 205 (98.6%) | 3 (1.4%) | |
| Endometrial hyperplasia (n = 675) | |||
| - Present | 6 (54.5%) | 5 (45%) | <0.001 * |
| - Absent | 651 (98%) | 13 (92%) | |
| Stage of tumor (n = 68) | |||
| - Stage 1 | 31 (79.5%) | 8 (20.5%) | 0.33 |
| - > Stage 1 | 27 (90%) | 3 (10%) | |
| Histology of tumor (n = 69) | |||
| - Endometrioid | 53 (84.1%) | 10 (15.9%) | 1.0 |
| - Non-endometrioid | 5 (83.3%) | 1 (16.7%) | |
| Grade of tumor (n = 63) | |||
| - Grade 1 | 32 (82.1%) | 7 (17.9%) | 0.18 |
| - Grade 2 | 12 (100%) | 0 (0%) | |
| - Grade 3 | 8 (72.7%) | 3 (27.3%) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.-S.J.; Wu, S.; Yeung, S.-Y.; Cheung, C.-W.; Fung, W.-Y.L.; Kwok, P.-K.S.; Yung, K.-K.; Wong, T.-K.S.; Kanneganti, A.; Lau, T.-S. Prospective Evaluation of Cervical Scrapings CDO1 and CELF4 Methylation (epiHERA®) Assay in Detection of Endometrial Cancer. Cancers 2025, 17, 3010. https://doi.org/10.3390/cancers17183010
Lee H-SJ, Wu S, Yeung S-Y, Cheung C-W, Fung W-YL, Kwok P-KS, Yung K-K, Wong T-KS, Kanneganti A, Lau T-S. Prospective Evaluation of Cervical Scrapings CDO1 and CELF4 Methylation (epiHERA®) Assay in Detection of Endometrial Cancer. Cancers. 2025; 17(18):3010. https://doi.org/10.3390/cancers17183010
Chicago/Turabian StyleLee, Ho-Sze Jacqueline, Shiye Wu, Suet-Ying Yeung, Chun-Wai Cheung, Wen-Ying Linda Fung, Pui-Kei Sonia Kwok, Kar-Kei Yung, Tsz-Kei Sani Wong, Abhiram Kanneganti, and Tat-San Lau. 2025. "Prospective Evaluation of Cervical Scrapings CDO1 and CELF4 Methylation (epiHERA®) Assay in Detection of Endometrial Cancer" Cancers 17, no. 18: 3010. https://doi.org/10.3390/cancers17183010
APA StyleLee, H.-S. J., Wu, S., Yeung, S.-Y., Cheung, C.-W., Fung, W.-Y. L., Kwok, P.-K. S., Yung, K.-K., Wong, T.-K. S., Kanneganti, A., & Lau, T.-S. (2025). Prospective Evaluation of Cervical Scrapings CDO1 and CELF4 Methylation (epiHERA®) Assay in Detection of Endometrial Cancer. Cancers, 17(18), 3010. https://doi.org/10.3390/cancers17183010






