Diagnostic Biomarker Candidates Proposed Using Targeted Lipid Metabolomics Analysis of the Plasma of Patients with PDAC
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
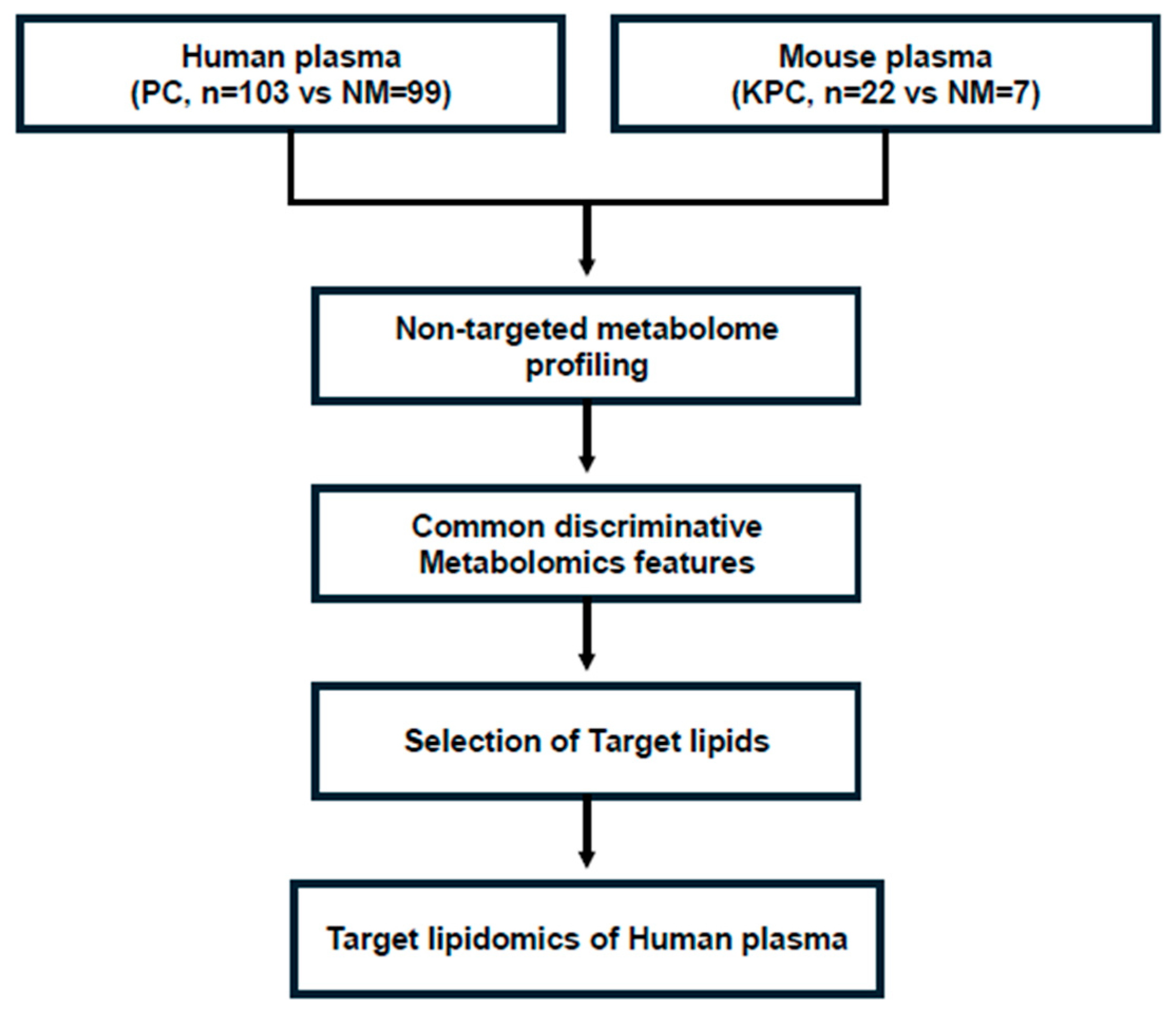
2.1. Non-Targeted Metabolomics Analysis of Human and Mouse Plasma and Selection of a Target Metabolome
2.2. Targeted Lipidomics of Human Plasma
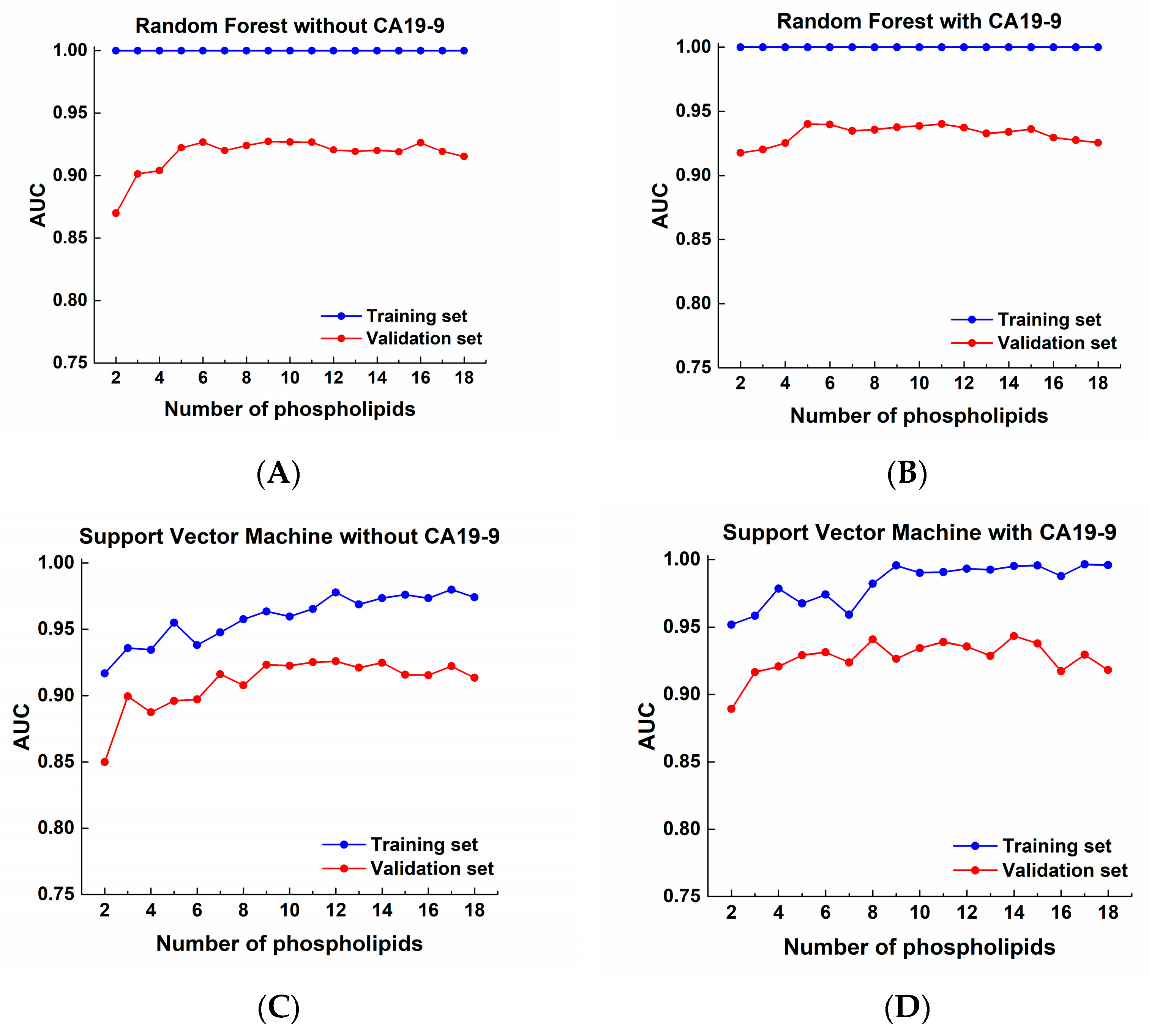
3. Results
3.1. Characteristics of Patients with PDAC
3.2. Non-Targeted Metabolomics Analysis of Human and Mouse Plasma
3.3. Metabolite Identification Process and Selection of Targeted Lipidomics Platforms
3.4. Targeted Lipidomics Analysis of Human Plasma
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PDAC | pancreatic ductal adenocarcinoma |
| LPA | lysophosphatidic acid |
| S1P | sphingosine-1-phosphate |
| GC-MS | gas chromatography–mass spectrometry |
| LC-MS | liquid chromatography–mass spectrometry |
| lysoPC | lysophosphatidylcholine |
Appendix A
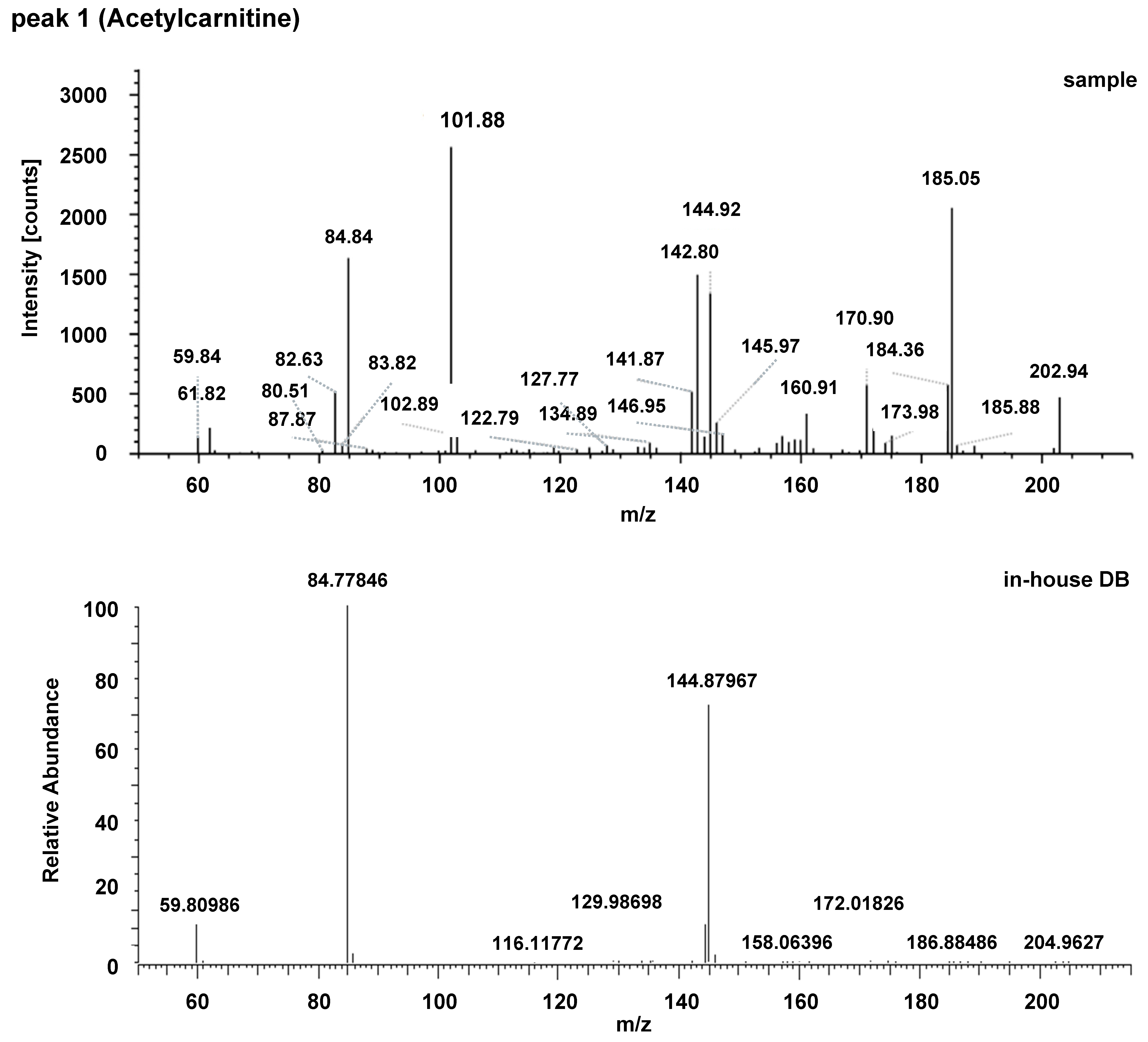

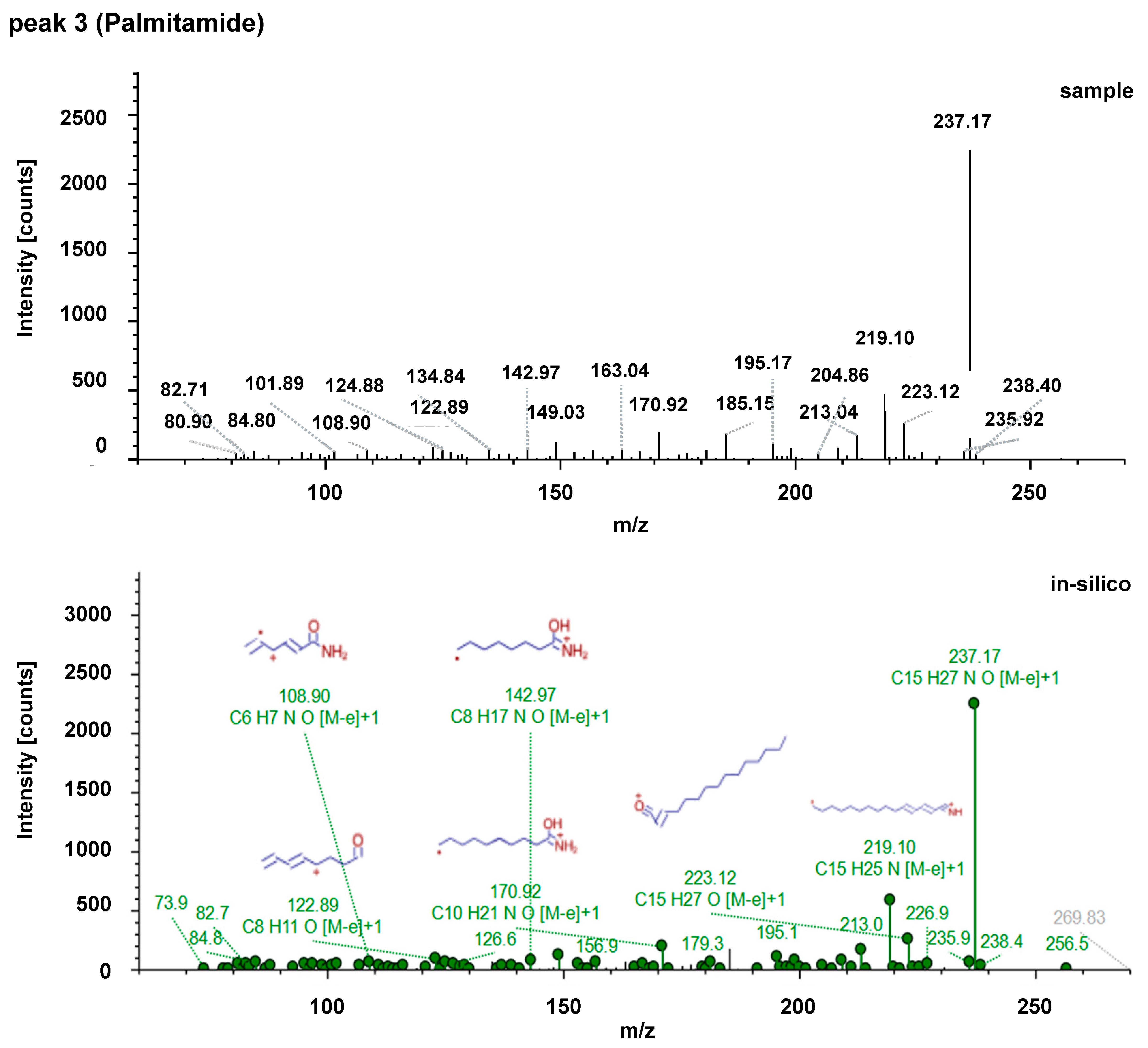



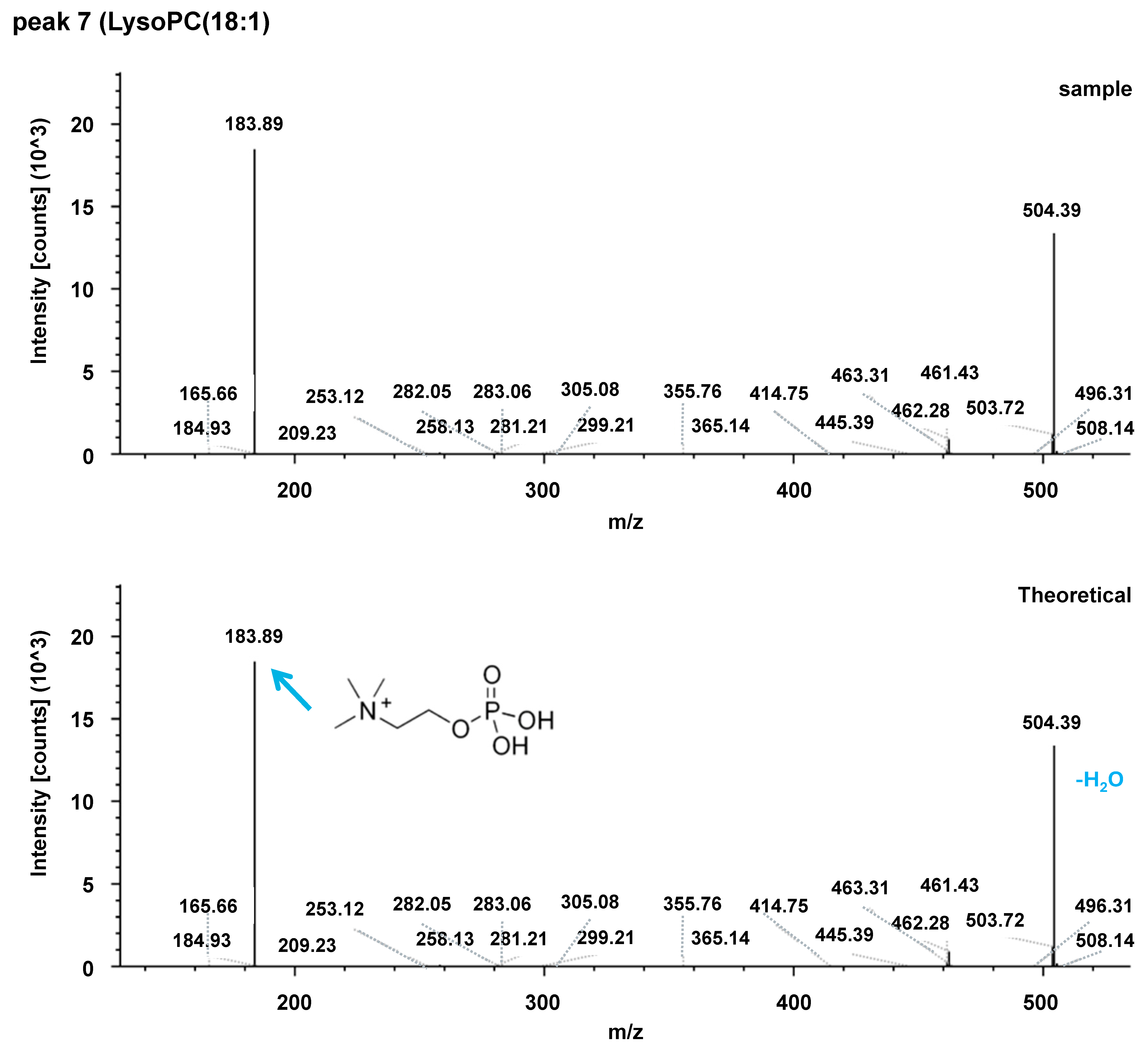
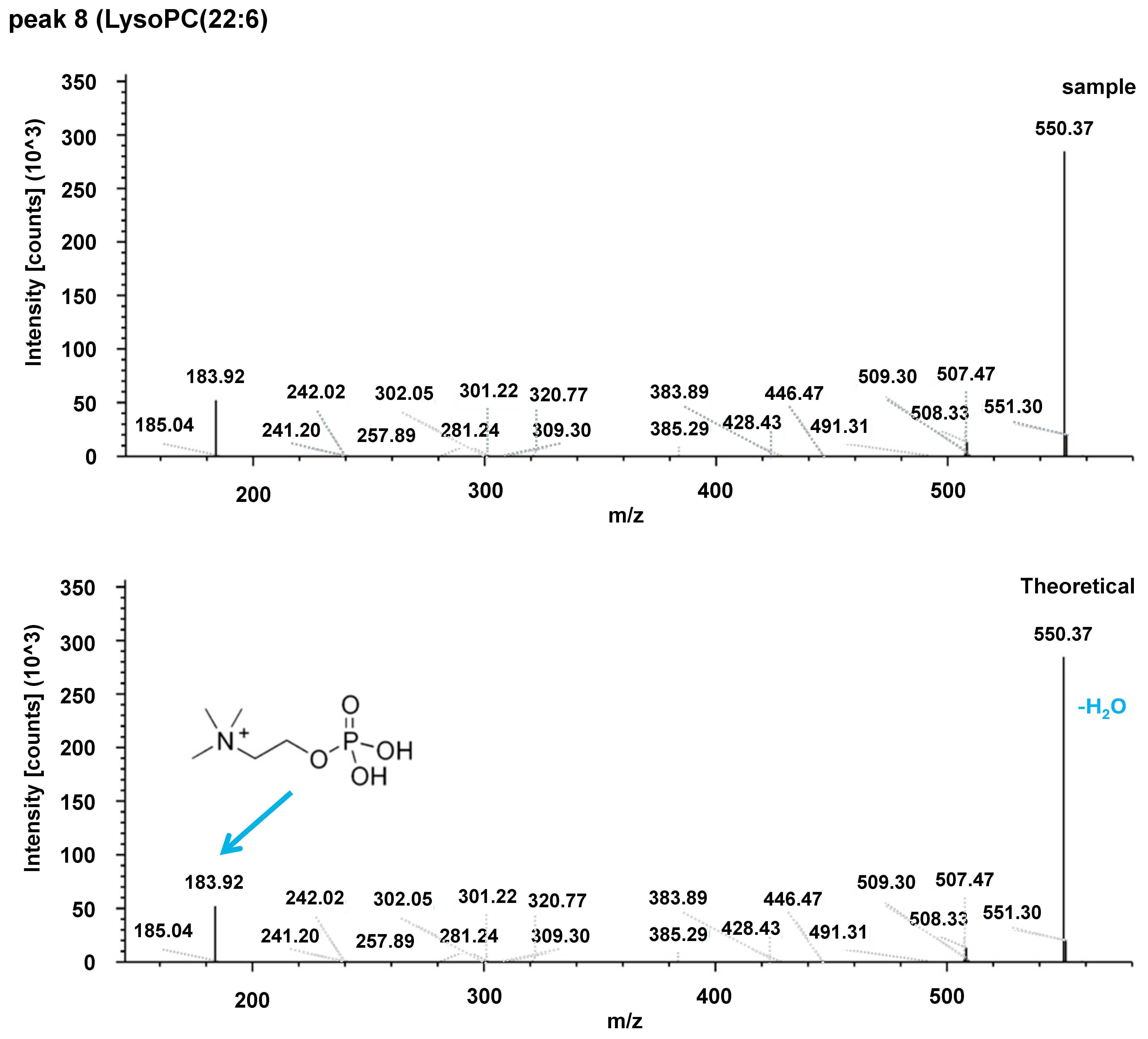
| Acylcarnitines | Fatty Acid Amides | Sphingolipids |
|---|---|---|
| C0-carnitine | Myristamide | C14 Ceramide |
| C2-carnitine | Palmitamide | C16 Ceramide |
| C3-carnitine | Oleamide | C18 Ceramide |
| C4-carnitine | Linoleamide | C18:1 Ceramide |
| C5-carnitine | Stearamide | C20 Ceramide |
| C6-carnitine | Myristoyl EA b,c | C24 Ceramide |
| C8-carnitine | Palmitoyl EA | C24:1 Ceramide |
| C10-carnitine | Oleoyl EA | 16:0 SM |
| C12-carnitine | Linoleoyl EA | 18:0 SM |
| C14-carnitine | Stearoyl EA | 18:1 SM |
| C16-carnitine | Arachidonoyl EA c | 24:0 SM |
| C18-carnitine | EPA EA c | 24:1 SM |
| C18:1-carnitine | DHA EA | |
| Phospholipids | ||
| LysoPC(16:0) | LysoPC(20:2) a | PE(18:0/22:6) |
| LysoPC(18:0) | LysoPC(20:3) a | PE(18:1/18:1) |
| LysoPC(18:1) | LysoPC(22:6) a | PE(18:2/18:2) b,c |
| LysoPC(18:2) | LysoPC(P-16:0) | PE(20:4/20:4) |
| PC(12:0/12:0) b,c | LysoPC(P-18:0) | PE(22:6/22:6) c |
| PC(14:0/14:0) | PC(P-18:0/18:1) | LysoPE(14:0) |
| PC(14:0/16:0) | PC(P-18:0/20:4) | LysoPE(16:0) |
| PC(16:0/16:0) | PC(P-18:0/22:6) | LysoPE(17:1) |
| PC(18:1/14:0) | PE(14:0/14:0) b | LysoPE(18:0) |
| PC(16:1/16:1) | PE(15:0/15:0) | LysoPE(18:1) |
| PC(15:0/18:1) | PE(15:0/18:0) | LysoPE(P-18:0) |
| PC(16:0/18:0) | PE(16:0/16:0) | PE(P-18:0/18:1) |
| PC(18:1/16:0) | PE(16:0/18:1) | PE(P-18:0/20:4) |
| PC(16:0/18:2) | PE(16:0/18:2) | PE(P-18:0/22:6) |
| PC(18:0/18:0) | PE(16:0/20:4) | PE(P-18:0/18:2) a |
| PC(18:0/18:1) | PE(16:0/22:6) | PE(P-16:0/20:4) a |
| PC(18:1/18:1) | PE(16:1/16:1) | LysoPC(O-16:0) |
| PC(16:0/20:4) | PE(17:0/17:0) | PC(O-18:0/2:0) |
| PC(18:0/20:4) | PE(18:0/18:1) | PC(O-18:1/2:0) |
| PC(16:0/22:6) | PE(18:0/18:2) | LysoPC(O-18:0) |
| PC(18:0/22:6) | PE(18:0/20:4) | LysoPC(O-18:1(9z)) |
| LysoPC(18:3) a | ||
| A. Set A (102 PC vs. 68 NM) | B. Set B (78 PC vs. 96 NM) | C. Set C (180 PC vs. 164 NM) | ||||||
|---|---|---|---|---|---|---|---|---|
| Lipid | ↑/↓ | AUC | Lipid | ↑/↓ | AUC | Lipid | ↑/↓ | AUC |
| Palmitamide | ↑ | 0.9591 | ||||||
| PE(22:6/22:6) | ↓ | 0.8502 | ||||||
| Stearamide | ↑ | 0.8297 | ||||||
| LysoPC(O-18:1(9z)) | ↑ | 0.8060 | ||||||
| Linoleoyl EA | ↑ | 0.7733 | ||||||
| C2-carnitine | ↓ | 0.7540 | ||||||
| LysoPC(P-16:0) | ↓ | 0.9028 | ||||||
| LysoPC(18:0) | ↓ | 0.9015 | ||||||
| LysoPC(16:0) | ↓ | 0.8957 | ||||||
| PC(O-18:0/2:0) | ↓ | 0.8906 | ||||||
| LysoPE(P-18:0) | ↓ | 0.8777 | ||||||
| C5-carnitine | ↓ | 0.8677 | ||||||
| PC(18:0/20:4) | ↓ | 0.8656 | ||||||
| LysoPE(18:0) | ↓ | 0.8638 | ||||||
| PC(16:0/20:4) | ↓ | 0.8620 | ||||||
| LysoPE(18:1) | ↓ | 0.8544 | ||||||
| 18:1 SM | ↓ | 0.8479 | ||||||
| PC(O-18:1/2:0) | ↓ | 0.8442 | ||||||
| LysoPC(P-18:0) | ↓ | 0.8431 | ||||||
| 24:0 SM | ↓ | 0.8164 | ||||||
| LysoPE(17:1) | ↓ | 0.8019 | ||||||
| PC(18:1/16:0) | ↓ | 0.7867 | ||||||
| PC(14:0/16:0) | ↓ | 0.7817 | ||||||
| LysoPE(16:0) | ↓ | 0.7693 | ||||||
| C6-carnitine | ↓ | 0.7665 | ||||||
| LysoPC(O-18:0) | ↓ | 0.7539 | ||||||
| PC(16:0/16:0) | ↓ | 0.7539 | ||||||
| 16:0 SM | ↓ | 0.7538 | ||||||
| C4-carnitine | ↓ | 0.8321 | C4-carnitine | ↓ | 0.8762 | |||
| C3-carnitine | ↓ | 0.7537 | C3-carnitine | ↓ | 0.8893 | |||
| C8-carnitine | ↓ | 0.7713 | C8-carnitine | ↓ | 0.8681 | |||
| C12-carnitine | ↓ | 0.8204 | C12-carnitine | ↓ | 0.7872 | |||
| Oleamide | ↑ | 0.9999 | Oleamide | ↑ | 0.8305 | |||
| Linoleamide | ↑ | 1.0000 | Linoleamide | ↑ | 0.7824 | |||
| PC(16:0/18:0) | ↓ | 0.8880 | PC(16:0/18:0) | ↓ | 0.8241 | |||
| LysoPC(18:1) | ↓ | 0.9151 | LysoPC(18:1) | ↓ | 0.8198 | |||
| PC(P-18:0/20:4) | ↓ | 0.8837 | PC(P-18:0/20:4) | ↓ | 0.8167 | |||
| PE(P-18:0/18:1) | ↓ | 0.8786 | PE(P-18:0/18:1) | ↓ | 0.8008 | |||
| PE(18:0/18:2) | ↓ | 0.8156 | PE(18:0/18:2) | ↓ | 0.7803 | |||
| PC(P-18:0/18:1) | ↓ | 0.8435 | PC(P-18:0/18:1) | ↓ | 0.7651 | |||
| PC(18:0/18:1) | ↓ | 0.8789 | PC(18:0/18:1) | ↓ | 0.7637 | |||
| PE(20:4/20:4) | ↓ | 0.8702 | PE(20:4/20:4) | ↓ | 0.8922 | PE(20:4/20:4) | ↓ | 0.8805 |
| LysoPC(18:2) | ↓ | 0.7845 | LysoPC(18:2) | ↓ | 0.9313 | LysoPC(18:2) | ↓ | 0.8680 |
| PC(P-18:0/22:6) | ↓ | 0.8697 | PC(P-18:0/22:6) | ↓ | 0.8544 | PC(P-18:0/22:6) | ↓ | 0.8603 |
| C24 Ceramide | ↓ | 0.8024 | C24 Ceramide | ↓ | 0.9005 | C24 Ceramide | ↓ | 0.8584 |
| PE(P-18:0/22:6) | ↓ | 0.8841 | PE(P-18:0/22:6) | ↓ | 0.9253 | PE(P-18:0/22:6) | ↓ | 0.8537 |
| C10-carnitine | ↓ | 0.8264 | C10-carnitine | ↓ | 0.8564 | C10-carnitine | ↓ | 0.8462 |
| PC(18:0/22:6) | ↓ | 0.8335 | PC(18:0/22:6) | ↓ | 0.8444 | PC(18:0/22:6) | ↓ | 0.8437 |
| PE(P-16:0/20:4) a | ↓ | 0.7809 | PE(P-16:0/20:4) a | ↓ | 0.8558 | PE(P-16:0/20:4) a | ↓ | 0.8252 |
| PC(16:1/16:1) | ↓ | 0.7623 | PC(16:1/16:1) | ↓ | 0.8483 | PC(16:1/16:1) | ↓ | 0.8212 |
| LysoPC(18:3) a | ↓ | 0.8052 | LysoPC(18:3) a | ↓ | 0.8858 | LysoPC(18:3) a | ↓ | 0.8188 |
| LysoPC(22:6) a | ↓ | 0.8599 | LysoPC(22:6) a | ↓ | 0.9152 | LysoPC(22:6) a | ↓ | 0.8092 |
| PC(16:0/22:6) | ↓ | 0.8632 | PC(16:0/22:6) | ↓ | 0.8523 | PC(16:0/22:6) | ↓ | 0.8005 |
| PE(P-18:0/18:2) a | ↓ | 0.8175 | PE(P-18:0/18:2) a | ↓ | 0.9000 | PE(P-18:0/18:2) a | ↓ | 0.8000 |
| PE(P-18:0/20:4) | ↓ | 0.7914 | PE(P-18:0/20:4) | ↓ | 0.9062 | PE(P-18:0/20:4) | ↓ | 0.7989 |
| LysoPC(20:2) a | ↓ | 0.7873 | LysoPC(20:2) a | ↓ | 0.8941 | LysoPC(20:2) a | ↓ | 0.7980 |
| PC(18:1/18:1) | ↓ | 0.7518 | PC(18:1/18:1) | ↓ | 0.9159 | PC(18:1/18:1) | ↓ | 0.7910 |
| PC(14:0/14:0) | ↓ | 0.7504 | PC(14:0/14:0) | ↓ | 0.7865 | PC(14:0/14:0) | ↓ | 0.7793 |
| PC(18:0/18:0) | ↓ | 0.8150 | PC(18:0/18:0) | ↓ | 0.9030 | PC(18:0/18:0) | ↓ | 0.7739 |
| PC(16:0/18:2) | ↓ | 0.7531 | PC(16:0/18:2) | ↓ | 0.8813 | PC(16:0/18:2) | ↓ | 0.7610 |
| LysoPC(20:3) a | ↓ | 0.7539 | LysoPC(20:3)a | ↓ | 0.8567 | LysoPC(20:3) a | ↓ | 0.7563 |
| Study No | Data Type | Sample Species | Parts Used in the Paper | URL Link |
|---|---|---|---|---|
| ST004066 | untargeted MS | mouse | Figure 2, Table 2 | https://dev.metabolomicsworkbench.org:22222/data/DRCCMetadata.php?Mode=Study&StudyID=ST004073, accessed on 15 July 2025 |
| ST004067 | untargeted MS | human | Figure A1 | https://dev.metabolomicsworkbench.org:22222/data/DRCCMetadata.php?Mode=Study&StudyID=ST004067, accessed on 15 July 2025 |
| ST004073 | targeted MS | human | Figure 3 and Figure 4, Table 3, Figure A2 Table A2 | https://dev.metabolomicsworkbench.org:22222/data/DRCCMetadata.php?Mode=Study&StudyID=ST004066, accessed on 17 July 2025 |
References
- Bengtsson, A.; Andersson, R.; Ansari, D. The actual 5-year survivors of pancreatic ductal adenocarcinoma based on real-world data. Sci. Rep. 2020, 10, 16425. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Yin, X.; Xu, R.; Song, J.; Ruze, R.; Chen, Y.; Wang, C.; Xu, Q. Lipid metabolism in pancreatic cancer: Emerging roles and potential targets. Cancer Commun. 2022, 42, 1234–1256. [Google Scholar] [CrossRef]
- Su, S.B.; Qin, S.Y.; Chen, W.; Luo, W.; Jiang, H.X. Carbohydrate antigen 19-9 for differential diagnosis of pancreatic carcinoma and chronic pancreatitis. World J. Gastroenterol. 2015, 21, 4323–4333. [Google Scholar] [CrossRef]
- Fahrmann, J.F.; Schmidt, C.M.; Mao, X.; Irajizad, E.; Loftus, M.; Zhang, J.; Patel, N.; Vykoukal, J.; Dennison, J.B.; Long, J.P.; et al. Lead-Time Trajectory of CA19-9 as an Anchor Marker for Pancreatic Cancer Early Detection. Gastroenterology 2021, 160, 1373–1383.e6. [Google Scholar] [CrossRef]
- Luo, G.; Liu, C.; Guo, M.; Cheng, H.; Lu, Y.; Jin, K.; Liu, L.; Long, J.; Xu, J.; Lu, R.; et al. Potential Biomarkers in Lewis Negative Patients With Pancreatic Cancer. Ann. Surg. 2017, 265, 800–805. [Google Scholar] [CrossRef]
- Sefrioui, D.; Blanchard, F.; Toure, E.; Basile, P.; Beaussire, L.; Dolfus, C.; Perdrix, A.; Paresy, M.; Antonietti, M.; Iwanicki-Caron, I.; et al. Diagnostic value of CA19.9, circulating tumour DNA and circulating tumour cells in patients with solid pancreatic tumours. Br. J. Cancer 2017, 117, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Pomare, E.W.; Branch, W.J.; Cummings, J.H. Carbohydrate fermentation in the human colon and its relation to acetate concentrations in venous blood. J. Clin. Investig. 1985, 75, 1448–1454. [Google Scholar] [CrossRef]
- Schwenk, R.W.; Holloway, G.P.; Luiken, J.J.; Bonen, A.; Glatz, J.F. Fatty acid transport across the cell membrane: Regulation by fatty acid transporters. Prostaglandins Leukot. Essent. Fat. Acids 2010, 82, 149–154. [Google Scholar] [CrossRef]
- Lust, C.A.C.; Bi, X.; Henry, C.J.; Ma, D.W.L. Development of Fatty Acid Reference Ranges and Relationship with Lipid Biomarkers in Middle-Aged Healthy Singaporean Men and Women. Nutrients 2021, 13, 435. [Google Scholar] [CrossRef]
- Kim, S.Y. Fatty acid addiction in cancer: A high-fat diet directly promotes tumor growth through fatty acid oxidation. Biochim. Biophys. Acta Rev. Cancer 2025, 1880, 189428. [Google Scholar] [CrossRef] [PubMed]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J. Am. Soc. Mass. Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef]
- Mahajan, U.M.; Oehrle, B.; Goni, E.; Strobel, O.; Kaiser, J.; Grutzmann, R.; Werner, J.; Friess, H.; Gress, T.M.; Seufferlein, T.W.; et al. Validation of two plasma multimetabolite signatures for patients at risk of or with suspected pancreatic ductal adenocarcinoma (METAPAC): A prospective, multicentre, investigator-masked, enrichment design, phase 4 diagnostic study. Lancet Gastroenterol. Hepatol. 2025, 10, 634–647. [Google Scholar] [CrossRef]
- Woo, S.M.; Lee, H.; Kang, J.H.; Kang, M.; Choi, W.; Sim, S.H.; Chun, J.W.; Han, N.; Kim, K.H.; Ham, W.; et al. Loss of SLC25A20 in Pancreatic Adenocarcinoma Reversed the Tumor-Promoting Effects of a High-Fat Diet. Theranostics 2025, 15, 6516–6533. [Google Scholar] [CrossRef]
- Deken, M.A.; Niewola-Staszkowska, K.; Peyruchaud, O.; Mikulčić, N.; Antolić, M.; Shah, P.; Cheasty, A.; Tagliavini, A.; Nizzardo, A.; Pergher, M.; et al. Characterization and translational development of IOA-289, a novel autotaxin inhibitor for the treatment of solid tumors. Immuno-Oncol. Technol. 2023, 18, 100384. [Google Scholar] [CrossRef]
- Komachi, M.; Sato, K.; Tobo, M.; Mogi, C.; Yamada, T.; Ohta, H.; Tomura, H.; Kimura, T.; Im, D.S.; Yanagida, K.; et al. Orally active lysophosphatidic acid receptor antagonist attenuates pancreatic cancer invasion and metastasis in vivo. Cancer Sci. 2012, 103, 1099–1104. [Google Scholar] [CrossRef]
- Rixe, O.; Villano, J.L.; Wesolowski, R.; Noonan, A.M.; Puduvalli, V.K.; Wise-Draper, T.M.; Curry, R., 3rd; Yilmaz, E.; Cruze, C.; Ogretmen, B.; et al. A First-in-Human Phase I Study of BXQ-350, a First-in-Class Sphingolipid Metabolism Regulator, in Patients with Advanced/Recurrent Solid Tumors or High-Grade Gliomas. Clin. Cancer Res. 2024, 30, 5053–5060. [Google Scholar] [CrossRef]
- Guillermet-Guibert, J.; Davenne, L.; Pchejetski, D.; Saint-Laurent, N.; Brizuela, L.; Guilbeau-Frugier, C.; Delisle, M.B.; Cuvillier, O.; Susini, C.; Bousquet, C. Targeting the sphingolipid metabolism to defeat pancreatic cancer cell resistance to the chemotherapeutic gemcitabine drug. Mol. Cancer Ther. 2009, 8, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Wolrab, D.; Jirasko, R.; Cifkova, E.; Horing, M.; Mei, D.; Chocholouskova, M.; Peterka, O.; Idkowiak, J.; Hrnciarova, T.; Kuchar, L.; et al. Lipidomic profiling of human serum enables detection of pancreatic cancer. Nat. Commun. 2022, 13, 124. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Gemma, A.; Tatsumi, K.; Hattori, N.; Ushiki, A.; Tsushima, K.; Saito, Y.; Abe, M.; Horimasu, Y.; Kashiwada, T.; et al. Identification and characterization of lysophosphatidylcholine 14:0 as a biomarker for drug-induced lung disease. Sci. Rep. 2022, 12, 19819. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, X.; Liu, Y.; Yan, F.; Zeng, Y.; Song, Y.; Fang, H.; Song, D.; Wang, X. Lysophosphatidylcholine inhibits lung cancer cell proliferation by regulating fatty acid metabolism enzyme long-chain acyl-coenzyme A synthase 5. Clin. Transl. Med. 2023, 13, e1180. [Google Scholar] [CrossRef]
- Kalia, V.; Reyes-Dumeyer, D.; Dubey, S.; Nandakumar, R.; Lee, A.J.; Lantigua, R.; Medrano, M.; Rivera, D.; Honig, L.S.; Mayeux, R.; et al. Lysophosphatidylcholines are associated with P-tau181 levels in early stages of Alzheimer’s Disease. medRxiv 2023. [Google Scholar] [CrossRef]
- Chen, Y.; Ma, Z.; Zhong, J.; Li, L.; Min, L.; Xu, L.; Li, H.; Zhang, J.; Wu, W.; Dai, L. Simultaneous quantification of serum monounsaturated and polyunsaturated phosphatidylcholines as potential biomarkers for diagnosing non-small cell lung cancer. Sci. Rep. 2018, 8, 7137. [Google Scholar] [CrossRef] [PubMed]





| Variable | Set A (n = 102) | Set B (n = 78) |
|---|---|---|
| Sex, n (%) | ||
| Male | 47 (46.1%) | 44 (56.4%) |
| Female | 55 (53.9%) | 34 (43.6%) |
| Median age, years | 68.5 | 64 |
| BMI, mean ± SD | 23.3 ± 3.0 | 23.7 ± 3.1 |
| Smoking history, n (%) | 40 (39.2%) | 38 (48.7%) |
| Diabetes mellitus, n (%) | 72 (70.6%) | 49 (62.8%) |
| Tumor stage, n (%) | ||
| Stage I | 84 (82.4%) | 54 (69.2%) |
| Stage II | 6 (5.9%) | 9 (11.5%) |
| Stage III | 6 (5.9%) | 10 (12.8%) |
| Stage IV | 6 (5.9%) | 5 (6.4%) |
| Tumor location, n (%) | ||
| Body and tail | 53 (52.0%) | 29 (37.2%) |
| Body and head | 1 (1.0%) | - |
| Head and neck | 44 (43.1%) | 45 (57.7%) |
| Uncinate process | 4 (3.9%) | - |
| AoV | - | 1 (1.3%) |
| N/A | - | 3 (3.8%) |
| No | Metabolite Identification | Pattern (↑/↓) | Human Plasma | Mouse Plasma | ||||
|---|---|---|---|---|---|---|---|---|
| m/z | rt (min) | AUC | m/z | rt (min) | AUC | |||
| 1 | Acetylcarnitine | ↓ | 204.12197 | 1.161 | 0.7956 | 204.1219 | 1.138 | 0.9286 |
| 2 | Mystamide | ↑ | 228.2311 | 3.756 | 0.6715 | 228.23091 | 3.75 | 0.9578 |
| 3 | Palmitamide | ↑ | 256.2623 | 4.258 | 0.7247 | 256.26229 | 4.25 | 0.7857 |
| 4 | Palmitoylcarnitine | ↓ | 400.34047 | 5.558 | 0.6103 | 400.34014 | 5.514 | 0.9058 |
| 5 | LysoPC(18:3) | ↓ | 518.32194 | 3.596 | 0.7287 | 518.32159 | 3.605 | 1.0000 |
| 6 | LysoPC(18:2) | ↓ | 520.3374 | 3.863 | 0.6689 | 520.33689 | 3.844 | 0.7597 |
| 7 | LysoPC(18:1) | ↓ | 522.35298 | 4.153 | 0.6081 | 522.35272 | 4.147 | 0.8474 |
| 8 | LysoPC(22:6) | ↓ | 568.3375 | 3.8 | 0.6431 | 568.33677 | 3.785 | 0.7890 |
| Lipid | MRM Transition | rt (min) | AUC | |||
|---|---|---|---|---|---|---|
| Q1 | Q3 | Set A | Set B | Set C | ||
| PE(20:4/20:4) | 788.5 | 647.5 | 2.32 | 0.8702 | 0.8922 | 0.8805 |
| LysoPC(18:2) | 520.3 | 184.1 | 0.62 | 0.7845 | 0.9313 | 0.8680 |
| PC(P-18:0/22:6) | 818.3 | 184.1 | 4.76 | 0.8697 | 0.8544 | 0.8603 |
| C24 Ceramide | 650.5 | 632.5 | 10.50 | 0.8024 | 0.9005 | 0.8584 |
| PE(P-18:0/22:6) | 776.7 | 385.3 | 4.64 | 0.8841 | 0.9253 | 0.8537 |
| C10-carnitine | 316.4 | 84.9 | 4.16 | 0.8264 | 0.8564 | 0.8462 |
| PC(18:0/22:6) | 834.6 | 184.1 | 4.14 | 0.8335 | 0.8444 | 0.8437 |
| PE(P-16:0/20:4) a | 724.5 | 361.2 | 3.42 | 0.7809 | 0.8558 | 0.8252 |
| PC(16:1/16:1) | 730.5 | 184.1 | 2.22 | 0.7623 | 0.8483 | 0.8212 |
| LysoPC(18:3) a | 518.3 | 184.0 | 0.54 | 0.8052 | 0.8858 | 0.8188 |
| LysoPC(22:6) a | 568.3 | 184.0 | 0.58 | 0.8599 | 0.9152 | 0.8092 |
| PC(16:0/22:6) | 806.6 | 184.1 | 2.88 | 0.8632 | 0.8523 | 0.8005 |
| PE(P-18:0/18:2) a | 728.6 | 337.3 | 5.20 | 0.8175 | 0.9000 | 0.8000 |
| PE(P-18:0/20:4) | 752.7 | 361.1 | 4.94 | 0.7914 | 0.9062 | 0.7989 |
| LysoPC(20:2) a | 548.4 | 184.0 | 0.71 | 0.7873 | 0.8941 | 0.7980 |
| PC(18:1/18:1) | 786.6 | 184.1 | 4.38 | 0.7518 | 0.9159 | 0.7910 |
| PC(14:0/14:0) | 678.5 | 184.1 | 1.94 | 0.7504 | 0.7865 | 0.7793 |
| PC(18:0/18:0) | 790.6 | 184.1 | 7.91 | 0.8150 | 0.9030 | 0.7739 |
| PC(16:0/18:2) | 758.6 | 184.1 | 3.20 | 0.7531 | 0.8813 | 0.7610 |
| LysoPC(20:3) a | 546.4 | 184.0 | 0.64 | 0.7539 | 0.8567 | 0.7563 |
| CA19-9 | 0.7571 | 0.7356 | 0.7489 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, S.-S.; Woo, S.M.; Lee, J.H.; Kang, J.H.; Park, S.-J.; Lee, W.J.; Park, H.M.; Chun, J.W.; Kim, S.J.; Yoo, H.J.; et al. Diagnostic Biomarker Candidates Proposed Using Targeted Lipid Metabolomics Analysis of the Plasma of Patients with PDAC. Cancers 2025, 17, 2988. https://doi.org/10.3390/cancers17182988
Han S-S, Woo SM, Lee JH, Kang JH, Park S-J, Lee WJ, Park HM, Chun JW, Kim SJ, Yoo HJ, et al. Diagnostic Biomarker Candidates Proposed Using Targeted Lipid Metabolomics Analysis of the Plasma of Patients with PDAC. Cancers. 2025; 17(18):2988. https://doi.org/10.3390/cancers17182988
Chicago/Turabian StyleHan, Sung-Sik, Sang Myung Woo, Jun Hwa Lee, Joon Hee Kang, Sang-Jae Park, Woo Jin Lee, Hyeong Min Park, Jung Won Chun, Su Jung Kim, Hyun Ju Yoo, and et al. 2025. "Diagnostic Biomarker Candidates Proposed Using Targeted Lipid Metabolomics Analysis of the Plasma of Patients with PDAC" Cancers 17, no. 18: 2988. https://doi.org/10.3390/cancers17182988
APA StyleHan, S.-S., Woo, S. M., Lee, J. H., Kang, J. H., Park, S.-J., Lee, W. J., Park, H. M., Chun, J. W., Kim, S. J., Yoo, H. J., Kim, K.-H., & Kim, S.-Y. (2025). Diagnostic Biomarker Candidates Proposed Using Targeted Lipid Metabolomics Analysis of the Plasma of Patients with PDAC. Cancers, 17(18), 2988. https://doi.org/10.3390/cancers17182988






