Unraveling the Clinical Landscape of RNA Modification Regulators with Multi-Omics Insights in Pan-Cancer
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Pan-Cancer Multimodal Data and RNA Modification Genes
2.2. Analysis of Single Nucleotide Variations (SNVs)
2.3. Copy Number Variation Analysis
2.4. Methylation Analysis
2.5. Differential Expression Analysis
2.6. LASSO Regression for Prognostic Key Gene Screening and RMS Construction
2.7. ROC Curve Evaluation of RMS Performance
2.8. Survival Analysis for RMS Validation
2.9. Association Analysis Between TNM Staging and RMS Risk
2.10. Differential Analysis Based on Risk RMS Groups
2.11. Gene Set Enrichment Analysis
2.12. Analysis of TIME and Immunotherapy Response
2.13. Prediction of Drug Sensitivity
2.14. Integration of Single-Cell Transcriptome Data
2.15. Spatial Transcriptomics: Spatial Partitioning and Expression Validation
2.16. Immunohistochemistry and Quantitative Analysis
3. Results
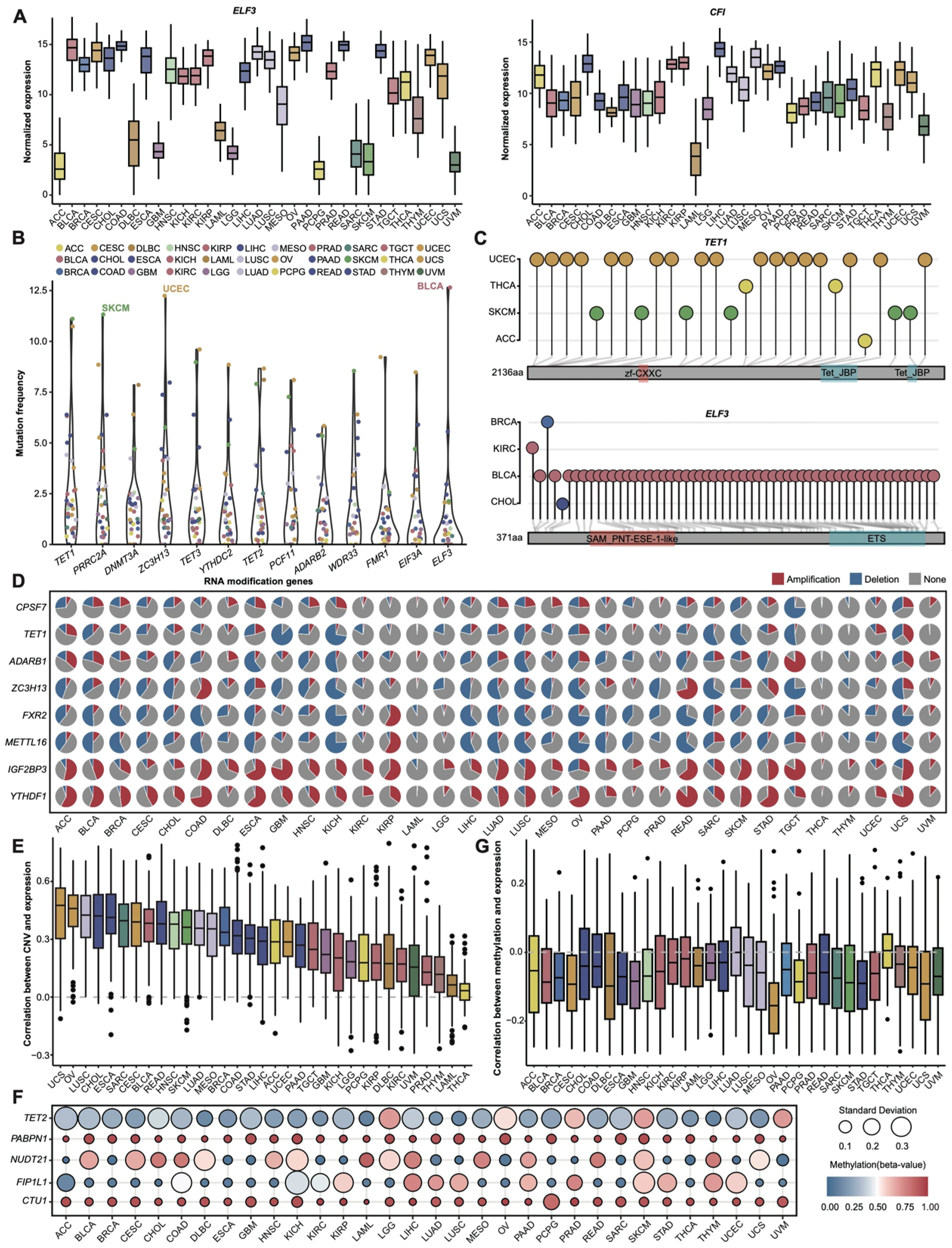
3.1. Pan-Cancer Expression and Genomic Profiles of RNA Modification Genes
3.2. RNA Modification Risk Score Model: Clinical and Biological Associations
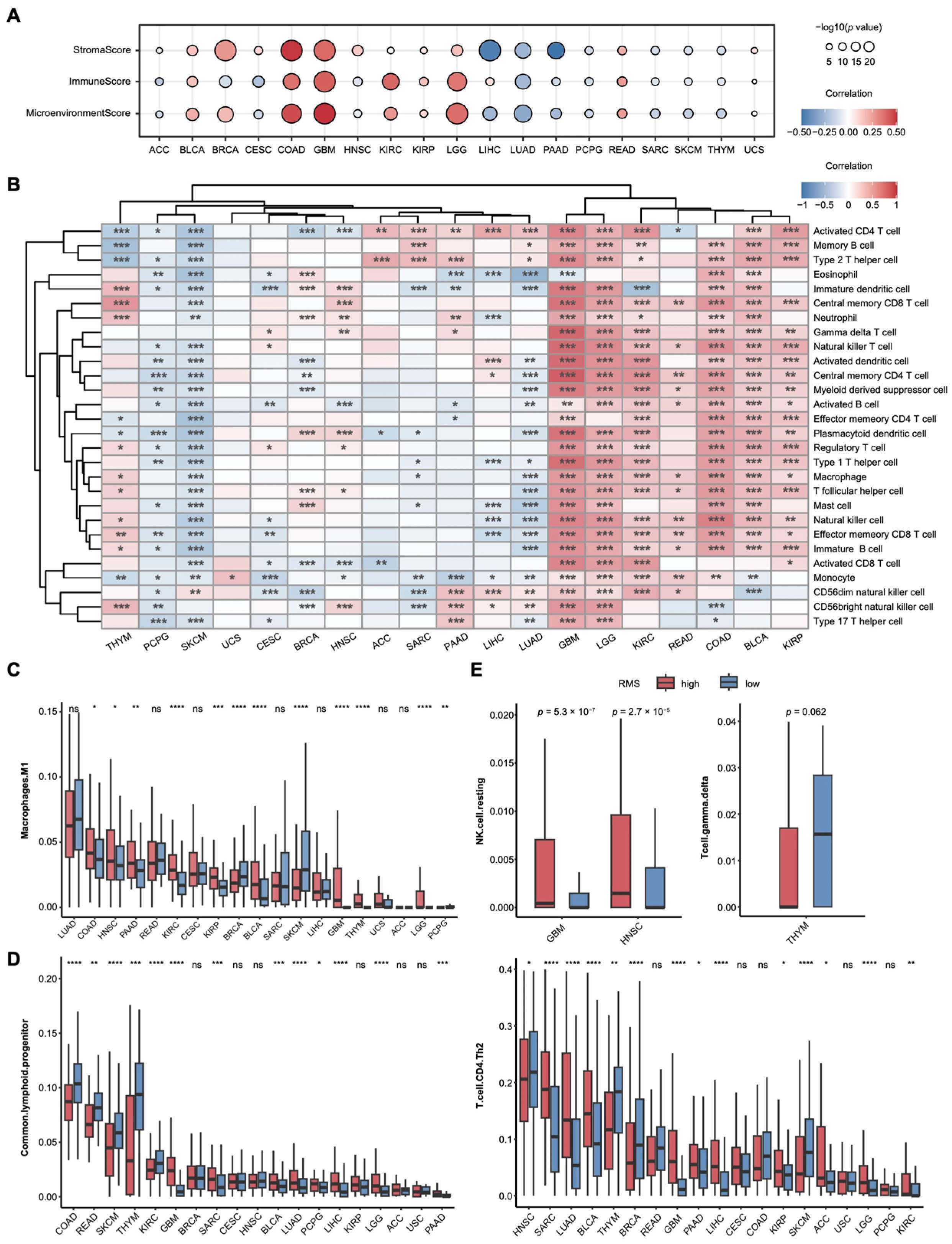
3.3. Regulation of the TIME and Signaling Pathways by RMS
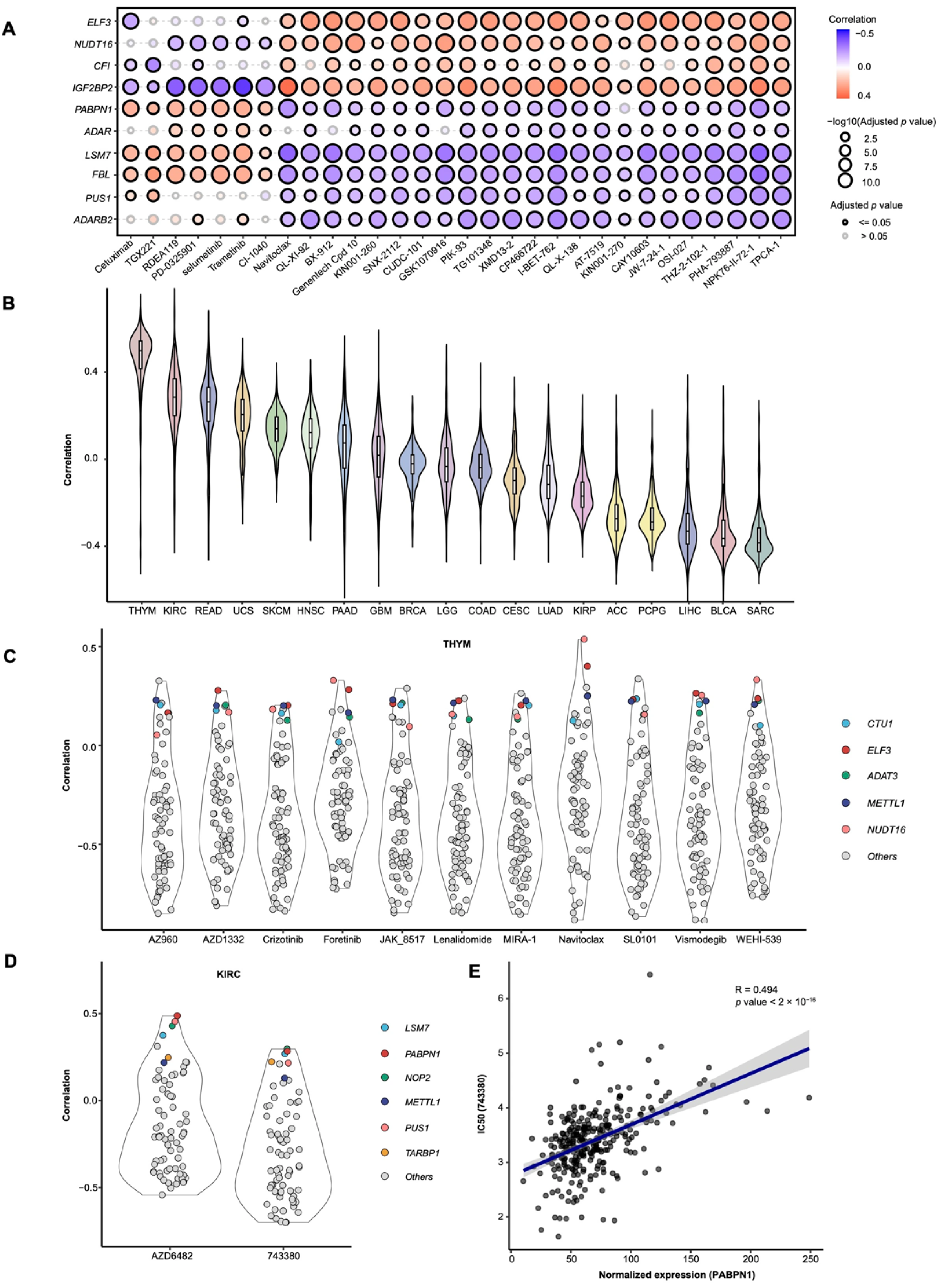
3.4. Drug Sensitivity Analysis Based on RMS and Candidate Drug Screening
3.5. Single-Cell and Spatial Transcriptomic Analyses Reveal Cell-Type-Specific Expression of RMS-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Marx, V. Closing in on cancer heterogeneity with organoids. Nat. Methods 2024, 21, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liu, B.; Zhang, Z. Accelerating the understanding of cancer biology through the lens of genomics. Cell 2023, 186, 1755–1771. [Google Scholar] [CrossRef]
- Xu, H.; Jiao, D.; Liu, A.; Wu, K. Tumor organoids: Applications in cancer modeling and potentials in precision medicine. J. Hematol. Oncol. 2022, 15, 58. [Google Scholar] [CrossRef]
- Yang, D.; Jones, M.G.; Naranjo, S.; Rideout, W.M., 3rd; Min, K.H.J.; Ho, R.; Wu, W.; Replogle, J.M.; Page, J.L.; Quinn, J.J.; et al. Lineage tracing reveals the phylodynamics, plasticity, and paths of tumor evolution. Cell 2022, 185, 1905–1923.e25. [Google Scholar] [CrossRef]
- Ma, X.; Liu, Y.; Liu, Y.; Alexandrov, L.B.; Edmonson, M.N.; Gawad, C.; Zhou, X.; Li, Y.; Rusch, M.C.; Easton, J.; et al. Pan-cancer genome and transcriptome analyses of 1699 paediatric leukaemias and solid tumours. Nature 2018, 555, 371–376. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Stojanov, P.; Mermel, C.H.; Robinson, J.T.; Garraway, L.A.; Golub, T.R.; Meyerson, M.; Gabriel, S.B.; Lander, E.S.; Getz, G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014, 505, 495–501. [Google Scholar] [CrossRef]
- Leiserson, M.D.; Vandin, F.; Wu, H.T.; Dobson, J.R.; Eldridge, J.V.; Thomas, J.L.; Papoutsaki, A.; Kim, Y.; Niu, B.; McLellan, M.; et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet. 2015, 47, 106–114. [Google Scholar] [CrossRef]
- Zack, T.I.; Schumacher, S.E.; Carter, S.L.; Cherniack, A.D.; Saksena, G.; Tabak, B.; Lawrence, M.S.; Zhsng, C.Z.; Wala, J.; Mermel, C.H.; et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013, 45, 1134–1140. [Google Scholar] [CrossRef]
- Cooper, L.A.; Demicco, E.G.; Saltz, J.H.; Powell, R.T.; Rao, A.; Lazar, A.J. PanCancer insights from The Cancer Genome Atlas: The pathologist’s perspective. J. Pathol. 2018, 244, 512–524. [Google Scholar] [CrossRef] [PubMed]
- Halabi, S. Pan-cancer prognostic models of clinical outcomes: Statistical exercise or clinical tools? Ann. Oncol. 2020, 31, 1427–1429. [Google Scholar] [CrossRef]
- Orsolic, I.; Carrier, A.; Esteller, M. Genetic and epigenetic defects of the RNA modification machinery in cancer. Trends Genet. 2023, 39, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, G.; Heard, E. Advances in epigenetics link genetics to the environment and disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Wang, C.; Hou, X.; Guan, Q.; Zhou, H.; Zhou, L.; Liu, L.; Liu, J.; Li, F.; Li, W.; Liu, H. RNA modification in cardiovascular disease: Implications for therapeutic interventions. Signal Transduct. Target. Ther. 2023, 8, 412. [Google Scholar] [CrossRef]
- Malka-Tunitsky, N.; Sas-Chen, A. Role of RNA modifications in cancer metastasis. Curr. Opin. Genet. Dev. 2024, 87, 102232. [Google Scholar] [CrossRef]
- Cui, L.; Ma, R.; Cai, J.; Guo, C.; Chen, Z.; Yao, L.; Wang, Y.; Fan, R.; Wang, X.; Shi, Y. RNA modifications: Importance in immune cell biology and related diseases. Signal Transduct. Target. Ther. 2022, 7, 334. [Google Scholar] [CrossRef]
- Barbieri, I.; Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 2020, 20, 303–322. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Chen, H.; Pan, Y.; Zhou, Q.; Liang, C.; Wong, C.C.; Zhou, Y.; Huang, D.; Liu, W.; Zhai, J.; Gou, H.; et al. METTL3 Inhibits Antitumor Immunity by Targeting m6A-BHLHE41-CXCL1/CXCR2 Axis to Promote Colorectal Cancer. Gastroenterology 2022, 163, 891–907. [Google Scholar] [CrossRef]
- Yang, S.; Wei, J.; Cui, Y.H.; Park, G.; Shah, P.; Deng, Y.; Aplin, A.E.; Lu, Z.; Hwang, S.; He, C.; et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019, 10, 2782. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Huang, W.; Li, Y.; Weng, H. Roles of METTL3 in cancer: Mechanisms and therapeutic targeting. J. Hematol. Oncol. 2020, 13, 117. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, J.; Chen, W.; Xu, Y.; Shen, Y.; Xu, X. Targeting IGF2BP3 in Cancer. Int. J. Mol. Sci. 2023, 24, 9423. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Li, M.; Chang, H.; Wang, R.; Zhang, Z.; Zhang, J.; He, Y.; Ma, H. The m6A demethylase ALKBH5 promotes tumor progression by inhibiting RIG-I expression and interferon alpha production through the IKKε/TBK1/IRF3 pathway in head and neck squamous cell carcinoma. Mol. Cancer 2022, 21, 97. [Google Scholar] [CrossRef]
- Xu, X.; Cui, J.; Wang, H.; Ma, L.; Zhang, X.; Guo, W.; Xue, X.; Wang, Y.; Qiu, S.; Tian, X.; et al. IGF2BP3 is an essential N6-methyladenosine biotarget for suppressing ferroptosis in lung adenocarcinoma cells. Mater. Today Bio. 2022, 17, 100503. [Google Scholar] [CrossRef]
- Du, M.; Peng, Y.; Li, Y.; Sun, W.; Zhu, H.; Wu, J.; Zong, D.; Wu, L.; He, X. MYC-activated RNA N6-methyladenosine reader IGF2BP3 promotes cell proliferation and metastasis in nasopharyngeal carcinoma. Cell Death Discov. 2022, 8, 53. [Google Scholar] [CrossRef]
- He, J.; Zhou, M.; Yin, J.; Wan, J.; Chu, J.; Jia, J.; Sheng, J.; Wang, C.; Yin, H.; He, F. METTL3 restrains papillary thyroid cancer progression via m6A/c-Rel/IL-8-mediated neutrophil infiltration. Mol. Ther. 2021, 29, 1821–1837. [Google Scholar] [CrossRef]
- Nombela, P.; Miguel-Lopez, B.; Blanco, S. The role of m6A, m5C and Ψ RNA modifications in cancer: Novel therapeutic opportunities. Mol. Cancer 2021, 20, 18. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Seliger, B.; Marincola, F.M.; Ferrone, S.; Abken, H. The complex role of B7 molecules in tumor immunology. Trends Mol. Med. 2008, 14, 550–559. [Google Scholar] [CrossRef]
- Schoenfeld, A.J.; Hellmann, M.D. Acquired Resistance to Immune Checkpoint Inhibitors. Cancer Cell 2020, 37, 443–455. [Google Scholar] [CrossRef]
- Darmanis, S.; Sloan, S.A.; Croote, D.; Mignardi, M.; Chernikova, S.; Samghababi, P.; Zhang, Y.; Neff, N.; Kowarsky, M.; Caneda, C.; et al. Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma. Cell Rep. 2017, 21, 1399–1410. [Google Scholar] [CrossRef]
- Lv, X.; Wang, B.; Liu, K.; Li, M.J.; Yi, X.; Wu, X. Decoding heterogeneous and coordinated tissue architecture in glioblastoma using spatial transcriptomics. iScience 2024, 27, 110064. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.L.; Li, S.Y.; Dang, S.S.; Cheng, Y.A.; Zhang, X.; Wang, W.J.; Hughes, C.E.; Caterson, B. Increased expression of chondroitin sulphate proteoglycans in rat hepatocellular carcinoma tissues. World J. Gastroenterol. 2012, 18, 3962–3976. [Google Scholar] [CrossRef]
- Wu, H.X.; Chen, Y.X.; Wang, Z.X.; Zhao, Q.; He, M.M.; Wang, Y.N.; Wang, F.; Xu, R.H. Alteration in TET1 as potential biomarker for immune checkpoint blockade in multiple cancers. J. Immunother. Cancer 2019, 7, 264. [Google Scholar] [CrossRef]
- Stasik, S.; Juratli, T.A.; Petzold, A.; Richter, S.; Zolal, A.; Schackert, G.; Dahl, A.; Krex, D.; Thiede, C. Exome sequencing identifies frequent genomic loss of TET1 in IDH-wild-type glioblastoma. Neoplasia 2020, 22, 800–808. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Su, Y.; Zhang, Z. Characterizing m6A modification factors and their interactions in colorectal cancer: Implications for tumor subtypes and clinical outcomes. Discov. Oncol. 2024, 15, 457. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Lv, N.; Liao, J.; Long, J.; Xue, R.; Ai, N.; Xu, D.; Fan, X. Copy number variation is highly correlated with differential gene expression: A pan-cancer study. BMC Med. Genet. 2019, 20, 175. [Google Scholar] [CrossRef]
- Kamdar, S.; Isserlin, R.; Van der Kwast, T.; Zlotta, A.R.; Bader, G.D.; Fleshner, N.E.; Bapat, B. Exploring targets of TET2-mediated methylation reprogramming as potential discriminators of prostate cancer progression. Clin. Epigenetics 2019, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Xu, J.; Cui, Z.; Wu, S.; Xie, T.; Zhang, X. Multi-omics analysis of N6-methyladenosine reader IGF2BP3 as a promising biomarker in pan-cancer. Front. Immunol. 2023, 14, 1071675. [Google Scholar] [CrossRef]
- Chae, Y.K.; Davis, A.A.; Agte, S.; Pan, A.; Simon, N.I.; Iams, W.T.; Cruz, M.R.; Tamragouri, K.; Rhee, K.; Mohindra, N.; et al. Clinical Implications of Circulating Tumor DNA Tumor Mutational Burden (ctDNA TMB) in Non-Small Cell Lung Cancer. Oncologist 2019, 24, 820–828. [Google Scholar] [CrossRef]
- Li, L.; Liu, Z. SRF Facilitates Transcriptional Inhibition of Gem Expression by m6A Methyltransferase METTL3 to Suppress Neuronal Damage in Epilepsy. Mol. Neurobiol. 2025, 62, 2903–2925. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Meng, J.; Song, X.; An, Q. m6A mRNA Methylation Analysis Provides Novel Insights into Pigmentation in Sheep Skin. Epigenetics 2023, 18, 2230662. [Google Scholar] [CrossRef]
- Wei, P.; Tang, H.; Li, D. Insights into pancreatic cancer etiology from pathway analysis of genome-wide association study data. PLoS ONE 2012, 7, e46887. [Google Scholar] [CrossRef]
- Yang, Y.; Li, J.; Jing, C.; Zhai, Y.; Bai, Z.; Yang, Y.; Deng, W. Inhibition of neuroactive ligand-receptor interaction pathway can enhance immunotherapy response in colon cancer: An in silico study. Expert Rev. Anticancer Ther. 2023, 23, 1205–1215. [Google Scholar] [CrossRef]
- Ponnusamy, L.; Natarajan, S.R.; Thangaraj, K.; Manoharan, R. Therapeutic aspects of AMPK in breast cancer: Progress, challenges, and future directions. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188379. [Google Scholar] [CrossRef]
- Liu, W.W.; Zheng, S.Q.; Li, T.; Fei, Y.F.; Wang, C.; Zhang, S.; Wang, F.; Jiang, G.M.; Wang, H. RNA modifications in cellular metabolism: Implications for metabolism-targeted therapy and immunotherapy. Signal Transduct. Target. Ther. 2024, 9, 70. [Google Scholar] [CrossRef]
- Du, R.; Bai, Y.; Li, L. Biological networks in gestational diabetes mellitus: Insights into the mechanism of crosstalk between long non-coding RNA and N6-methyladenine modification. BMC Pregnancy Childbirth 2022, 22, 384. [Google Scholar] [CrossRef]
- Szymanski, L.; Matak, D.; Bartnik, E.; Szczylik, C.; Czarnecka, A.M. Thyroid Hormones as Renal Cell Cancer Regulators. J. Signal Transduct. 2016, 2016, 1362407. [Google Scholar] [CrossRef]
- Li, J.; Zhang, W.; Ma, X.; Wei, Y.; Zhou, F.; Li, J.; Zhang, C.; Yang, Z. Cuproptosis/ferroptosis-related gene signature is correlated with immune infiltration and predict the prognosis for patients with breast cancer. Front. Pharmacol. 2023, 14, 1192434. [Google Scholar] [CrossRef]
- Zhang, L.; Yan, X.; Wang, Y.; Wang, Q.; Yan, H.; Yan, Y. Identification and validation of a novel robust glioblastoma prognosis model based on bioinformatics. Heliyon 2024, 10, e37374. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Tian, C.; Li, Q.; Xu, Q. TET1 Knockdown Inhibits Porphyromonas gingivalis LPS/IFN-γ-Induced M1 Macrophage Polarization through the NF-κB Pathway in THP-1 Cells. Int. J. Mol. Sci. 2019, 20, 2023. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Chen, C.; Tang, Z.; Yuan, W.; Yue, K.; Cui, P.; Qiu, X.; Zhang, H.; Li, T.; Zhu, X.; et al. TREM2 deficiency aggravates renal injury by promoting macrophage apoptosis and polarization via the JAK-STAT pathway in mice. Cell Death Dis. 2024, 15, 401. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, X.; Wan, C.; Liu, Y.; Wang, Y.; Meng, C.; Zhang, Y.; Jiang, C. NLRP3 inflammasome mediates M1 macrophage polarization and IL-1β production in inflammatory root resorption. J. Clin. Periodontol. 2020, 47, 451–460. [Google Scholar] [CrossRef]
- Hsieh, S.L.; Yang, S.Y.; Lin, C.Y.; He, X.Y.; Tsai, C.H.; Fong, Y.C.; Lo, Y.S.; Tang, C.H. MCP-1 controls IL-17-promoted monocyte migration and M1 polarization in osteoarthritis. Int. Immunopharmacol. 2024, 132, 112016. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Yu, H.; Peng, T.; Yang, K.; Xu, X.; Gu, W. Glycolytic enzyme PGK1 promotes M1 macrophage polarization and induces pyroptosis of acute lung injury via regulation of NLRP3. Respir. Res. 2024, 25, 291. [Google Scholar] [CrossRef]
- Yan, S.; Ding, J.; Wang, Z.; Zhang, F.; Li, J.; Zhang, Y.; Wu, S.; Yang, L.; Pang, X.; Zhang, Y.; et al. CTRP6 regulates M1 macrophage polarization via the PPAR-γ/NF-κB pathway and reprogramming glycolysis in recurrent spontaneous abortion. Int. Immunopharmacol. 2023, 124 Pt A, 110840. [Google Scholar] [CrossRef]
- Pang, X.; Wang, Y.; Liu, M. M1-macrophage polarization is upregulated in deep vein thrombosis and contributes to the upregulation of adhesion molecules. Hum. Immunol. 2019, 80, 883–889. [Google Scholar] [CrossRef]
- Shanley, M.; Daher, M.; Dou, J.; Li, S.; Basar, R.; Rafei, H.; Dede, M.; Gumin, J.; Pantaleomicronn Garciotaa, J.; Nunez Cortes, A.K.; et al. Interleukin-21 engineering enhances NK cell activity against glioblastoma via CEBPD. Cancer Cell 2024, 42, 1450–1466.e11. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, S.; Wang, Z. Identification of genetic mechanisms underlying lipid metabolism-mediated tumor immunity in head and neck squamous cell carcinoma. BMC Med. Genom. 2023, 16, 110. [Google Scholar] [CrossRef] [PubMed]
- Mensurado, S.; Blanco-Dominguez, R.; Silva-Santos, B. The emerging roles of γδ T cells in cancer immunotherapy. Nat. Rev. Clin. Oncol. 2023, 20, 178–191. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Yu, J.; Ge, S.; Fan, X. Novel insight into RNA modifications in tumor immunity: Promising targets to prevent tumor immune escape. Innovation 2023, 4, 100452. [Google Scholar] [CrossRef]
- Kerdivel, G.; Amrouche, F.; Calmejane, M.A.; Carallis, F.; Hamroune, J.; Hantel, C.; Bertherat, J.; Assie, G.; Boeva, V. DNA hypermethylation driven by DNMT1 and DNMT3A favors tumor immune escape contributing to the aggressiveness of adrenocortical carcinoma. Clin. Epigenetics 2023, 15, 121. [Google Scholar] [CrossRef]
- Li, R.; Chen, H.; Li, C.; Qi, Y.; Zhao, K.; Wang, J.; You, C.; Huang, H. The prognostic value and immune landscaps of m6A/m5C-related lncRNAs signature in the low grade glioma. BMC Bioinform. 2023, 24, 274. [Google Scholar] [CrossRef]
- Otto, T.; Sicinski, P. Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer 2017, 17, 93–115. [Google Scholar] [CrossRef]
- Vu, T.; Datta, P.K. Regulation of EMT in Colorectal Cancer: A Culprit in Metastasis. Cancers 2017, 9, 171. [Google Scholar] [CrossRef]
- Xiong, M.; Liu, C.; Li, W.; Jiang, H.; Long, W.; Zhou, M.; Yang, C.; Kazobinka, G.; Sun, Y.; Zhao, J.; et al. PABPN1 promotes clear cell renal cell carcinoma progression by suppressing the alternative polyadenylation of SGPL1 and CREG1. Carcinogenesis 2023, 44, 576–586. [Google Scholar] [CrossRef]
- Guo, W.; Wu, S.; Wang, L.; Wei, X.; Liu, X.; Wang, J.; Lu, Z.; Hollingshead, M.; Fang, B. Antitumor activity of a novel oncrasin analogue is mediated by JNK activation and STAT3 inhibition. PLoS ONE 2011, 6, e28487. [Google Scholar] [CrossRef]
- Suvasini, R.; Shruti, B.; Thota, B.; Shinde, S.V.; Friedmann-Morvinski, D.; Nawaz, Z.; Prasanna, K.V.; Thennarasu, K.; Hegde, A.S.; Arivazhagan, A.; et al. Insulin growth factor-2 binding protein 3 (IGF2BP3) is a glioblastoma-specific marker that activates phosphatidylinositol 3-kinase/mitogen-activated protein kinase (PI3K/MAPK) pathways by modulating IGF-2. J. Biol. Chem. 2011, 286, 25882–25890. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Dou, X.; Zhao, Y.; Zhang, L.; Zhang, L.; Dai, Q.; Liu, J.; Wu, T.; Xiao, Y.; He, C. IGF2BP3 promotes mRNA degradation through internal m(7)G modification. Nat. Commun. 2024, 15, 7421. [Google Scholar] [CrossRef] [PubMed]
- Meri, S. Complement activation in diseases presenting with thrombotic microangiopathy. Eur. J. Intern. Med. 2013, 24, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Shen, L.; Cai, Y.; Wu, J.; Chen, K.; Xu, D.; Lei, Y.; Chai, S.; Xiong, N. The Role of Coagulation-Related Genes in Glioblastoma: A Comprehensive Analysis of the Tumor Microenvironment, Prognosis, and Treatment. Biochem. Genet. 2025. ahead of print. [Google Scholar] [CrossRef]
- Feng, Z.Y.; Wang, T.; Su, X.; Guo, S. Identification of the m6A RNA Methylation Regulators WTAP as a Novel Prognostic Biomarker and Genomic Alterations in Cutaneous Melanoma. Front. Mol. Biosci. 2021, 8, 665222. [Google Scholar] [CrossRef]
- Yu, H.; Yang, X.; Tang, J.; Si, S.; Zhou, Z.; Lu, J.; Han, J.; Yuan, B.; Wu, Q.; Lu, Q.; et al. ALKBH5 Inhibited Cell Proliferation and Sensitized Bladder Cancer Cells to Cisplatin by m6A-CK2α-Mediated Glycolysis. Mol. Ther. Nucleic Acids 2021, 23, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Panneerdoss, S.; Eedunuri, V.K.; Yadav, P.; Timilsina, S.; Rajamanickam, S.; Viswanadhapalli, S.; Abdelfattah, N.; Onyeagucha, B.C.; Cui, X.; Lai, Z.; et al. Cross-talk among writers, readers, and erasers of m6A regulates cancer growth and progression. Sci. Adv. 2018, 4, eaar8263. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Kang, Y.; Wang, L.; Huff, S.; Tang, R.; Hui, H.; Agrawal, K.; Gonzalez, G.M.; Wang, Y.; Patel, S.P.; et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc. Natl. Acad. Sci. USA 2020, 117, 20159–20170. [Google Scholar] [CrossRef]
- Wang, L.; Hui, H.; Agrawal, K.; Kang, Y.; Li, N.; Tang, R.; Yuan, J.; Rana, T.M. m6 A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J. 2020, 39, e104514. [Google Scholar] [CrossRef]
- Huang, Q.; Mo, J.; Liao, Z.; Chen, X.; Zhang, B. The RNA m6A writer WTAP in diseases: Structure, roles, and mechanisms. Cell Death Dis. 2022, 13, 852. [Google Scholar] [CrossRef]
- Zhou, B.; Bie, F.; Zang, R.; Zhang, M.; Song, P.; Liu, L.; Peng, Y.; Bai, G.; Zhao, J.; Gao, S. RNA modification writer expression profiles predict clinical outcomes and guide neoadjuvant immunotherapy in non-small cell lung cancer. EBioMedicine 2022, 84, 104268. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Mei, W.; Qu, C.; Lu, J.; Shang, L.; Cao, F.; Li, F. Role of m6A writers, erasers and readers in cancer. Exp. Hematol. Oncol. 2022, 11, 45. [Google Scholar] [CrossRef]
- Liu, X.; Guo, W.; Wu, S.; Wang, L.; Wang, J.; Dai, B.; Kim, E.S.; Heymach, J.V.; Wang, M.; Girard, L.; et al. Antitumor activity of a novel STAT3 inhibitor and redox modulator in non-small cell lung cancer cells. Biochem. Pharmacol. 2012, 83, 1456–1464. [Google Scholar] [CrossRef]
- Qi, Y.J.; Su, G.H.; You, C.; Zhang, X.; Xiao, Y.; Jiang, Y.Z.; Shao, Z.M. Radiomics in breast cancer: Current advances and future directions. Cell Rep. Med. 2024, 5, 101719. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Manfrin, A.; Lutolf, M.P. Progress and potential in organoid research. Nat. Rev. Genet. 2018, 19, 671–687. [Google Scholar] [CrossRef]
- Wang, S.W.; Gao, C.; Zheng, Y.M.; Yi, L.; Lu, J.C.; Huang, X.Y.; Cai, J.B.; Zhang, P.F.; Cui, Y.H.; Ke, A.W. Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Mol. Cancer 2022, 21, 57. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Zhang, J.; Cao, Z.; Wang, J.; Song, J.; Yi, X. Unraveling the Clinical Landscape of RNA Modification Regulators with Multi-Omics Insights in Pan-Cancer. Cancers 2025, 17, 2695. https://doi.org/10.3390/cancers17162695
Li Q, Zhang J, Cao Z, Wang J, Song J, Yi X. Unraveling the Clinical Landscape of RNA Modification Regulators with Multi-Omics Insights in Pan-Cancer. Cancers. 2025; 17(16):2695. https://doi.org/10.3390/cancers17162695
Chicago/Turabian StyleLi, Qingman, Jingjing Zhang, Zuyi Cao, Jiale Wang, Jiaxing Song, and Xianfu Yi. 2025. "Unraveling the Clinical Landscape of RNA Modification Regulators with Multi-Omics Insights in Pan-Cancer" Cancers 17, no. 16: 2695. https://doi.org/10.3390/cancers17162695
APA StyleLi, Q., Zhang, J., Cao, Z., Wang, J., Song, J., & Yi, X. (2025). Unraveling the Clinical Landscape of RNA Modification Regulators with Multi-Omics Insights in Pan-Cancer. Cancers, 17(16), 2695. https://doi.org/10.3390/cancers17162695





