Machine Learning-Based Gene Expression Analysis to Identify Prognostic Biomarkers in Upper Tract Urothelial Carcinoma
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Samples and RNA Extraction
2.3. Library Preparation, Sequencing, Data Processing, and Analysis
2.4. Gene Expression and Functional Analysis
2.5. Machine Learning Explainability Model
3. Results
3.1. Clinicopathological Features of the Cohort
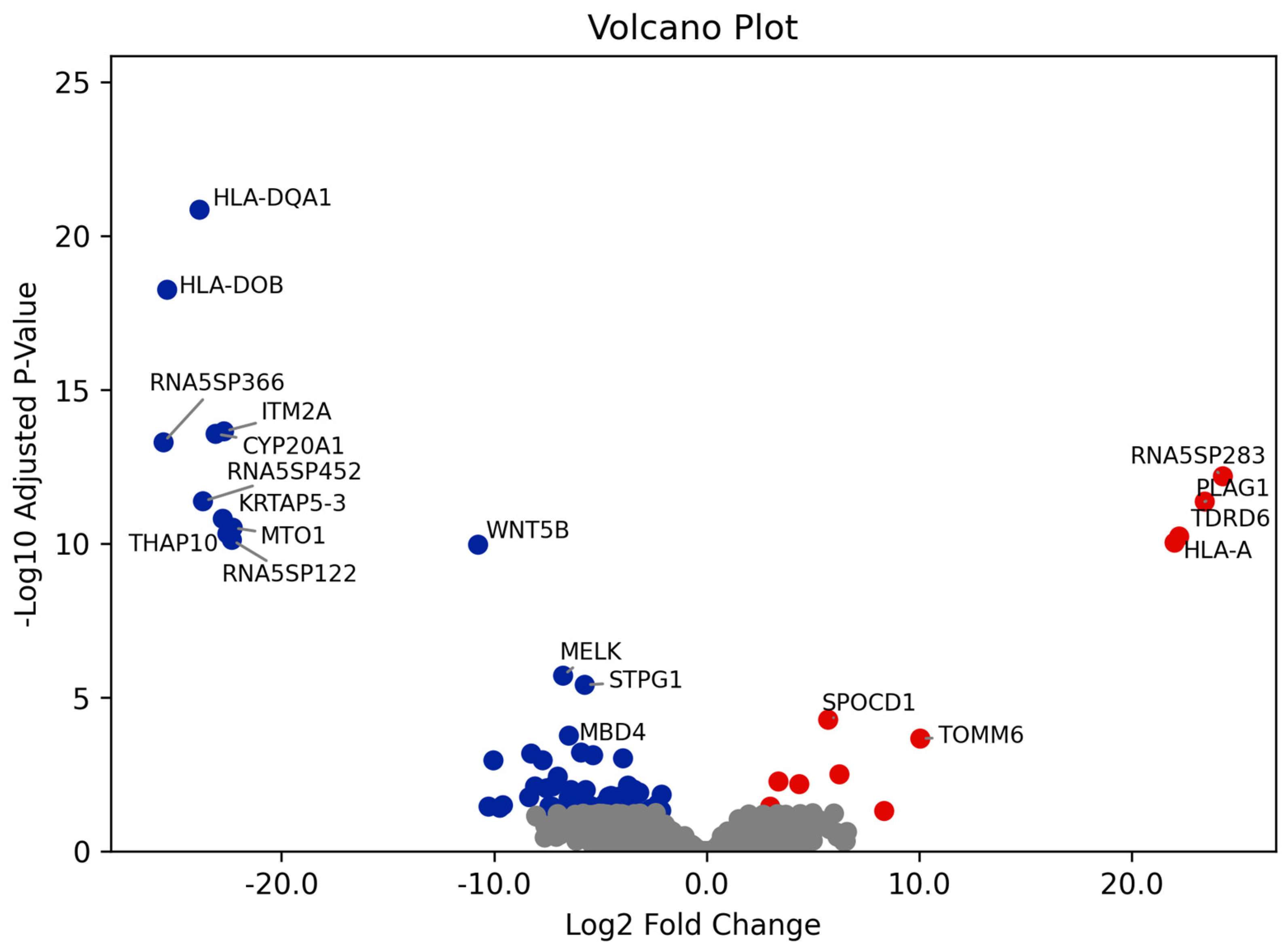
3.2. Differential Gene Expression and GO Enrichment Analyses
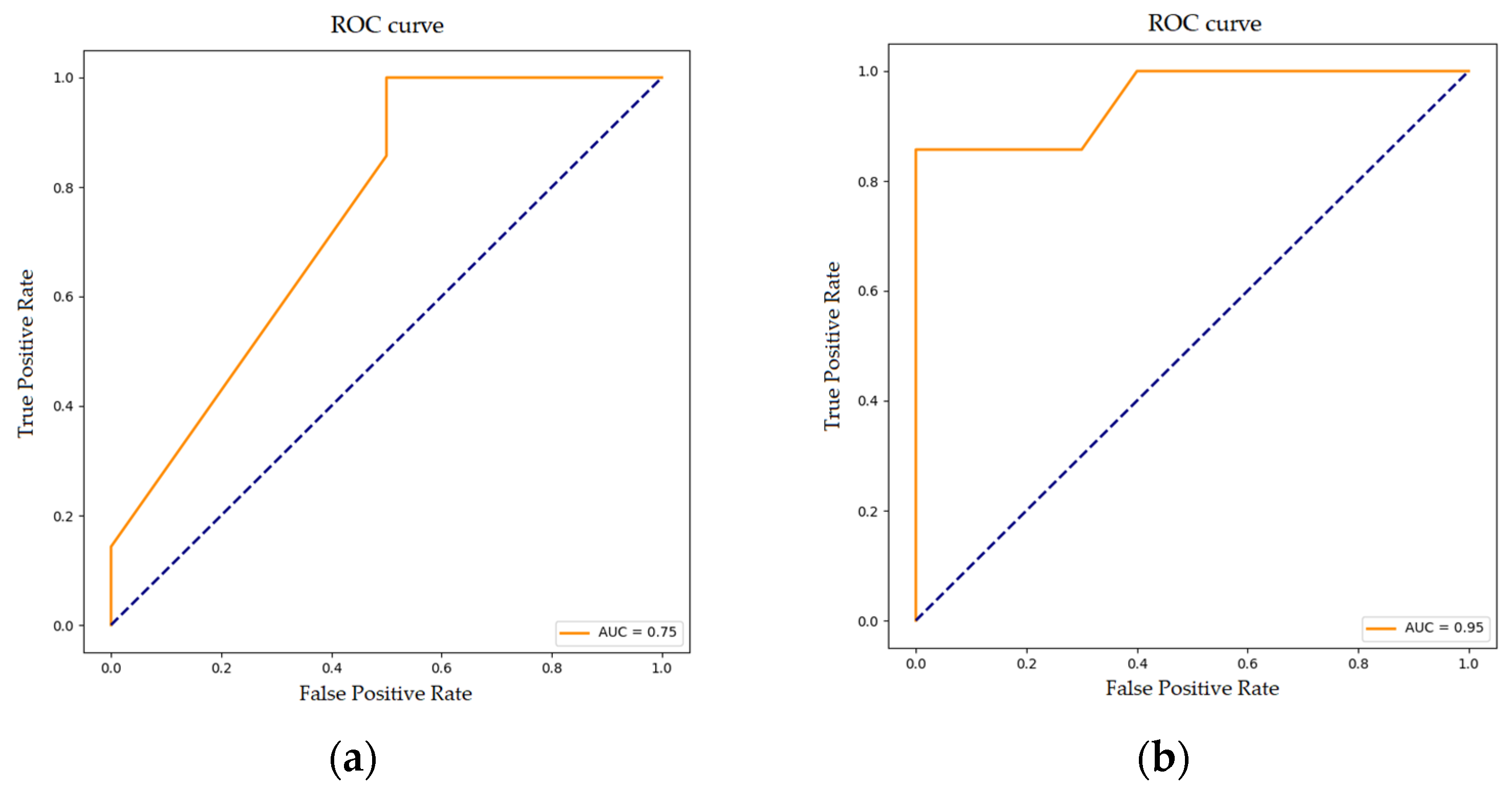
3.3. Random Forest Classification for Disease Progression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soria, F.; Shariat, S.F.; Lerner, S.P.; Fritsche, H.M.; Rink, M.; Kassouf, W.; Spiess, P.E.; Lotan, Y.; Ye, D.; Fernández, M.I.; et al. Epidemiology, Diagnosis, Preoperative Evaluation and Prognostic Assessment of Upper-Tract Urothelial Carcinoma (UTUC). World J. Urol. 2017, 35, 379–387. [Google Scholar] [CrossRef]
- Almås, B.; Halvorsen, O.J.; Johannesen, T.B.; Beisland, C. Higher than expected and significantly increasing incidence of upper tract urothelial carcinoma. A population based study. World J. Urol. 2021, 39, 3385–3391. [Google Scholar] [CrossRef] [PubMed]
- Soualhi, A.; Rammant, E.; George, G.; Russell, B.; Enting, D.; Nair, R.; Van Hemelrijck, M.; Bosco, C. The incidence and prevalence of upper tract urothelial carcinoma: A systematic review. BMC Urol. 2021, 21, 110. [Google Scholar] [CrossRef]
- Rouprêt, M.; Seisen, T.; Birtle, A.J.; Capoun, O.; Compérat, E.M.; Dominguez-Escrig, J.L.; Gürses Andersson, I.; Liedberg, F.; Mariappan, P.; Hugh Mostafid, A.; et al. European Association of Urology Guidelines on Upper Urinary Tract Urothelial Carcinoma: 2023 Update. Eur. Urol. 2023, 84, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Colin, P.; Ouzzane, A.; Pignot, G.; Ravier, E.; Crouzet, S.; Ariane, M.; Audouin, M.; Neuzillet, Y.; Albouy, B.; Hurel, S.; et al. Comparison of oncological outcomes after segmental ureterectomy or radical nephroureterectomy in urothelial carcinomas of the upper urinary tract: Results from a large French multicentre study. BJU Int. 2012, 110, 1134–1141. [Google Scholar] [CrossRef]
- Margulis, V.; Shariat, S.F.; Matin, S.F.; Kamat, A.M.; Zigeuner, R.; Kikuchi, E.; Lotan, Y.; Weizer, A.; Raman, J.D.; Wood, C.G. Outcomes of radical nephroureterectomy: A series from the Upper Tract Urothelial Carcinoma Collaboration. Cancer 2009, 115, 1224–1233. [Google Scholar] [CrossRef]
- Mbeutcha, A.; Rouprêt, M.; Kamat, A.M.; Karakiewicz, P.I.; Lawrentschuk, N.; Novara, G.; Raman, J.D.; Seitz, C.; Xylinas, E.; Shariat, S.F. Prognostic factors and predictive tools for upper tract urothelial carcinoma: A systematic review. World J. Urol. 2017, 35, 337–353. [Google Scholar] [CrossRef]
- Huben, R.P.; Mounzer, A.M.; Murphy, G.P. Tumor grade and stage as prognostic variables in upper tract urothelial tumors. Cancer 1988, 62, 2016–2024. [Google Scholar] [CrossRef]
- Hassler, M.R.; Bray, F.; Catto, J.W.F.; Grollman, A.P.; Hartmann, A.; Margulis, V.; Matin, S.F.; Rouprêt, M.; Sfakianos, J.P.; Shariat, S.F.; et al. Molecular Characterization of Upper Tract Urothelial Carcinoma in the Era of Next-generation Sequencing: A Systematic Review of the Current Literature. Eur. Urol. 2020, 78, 209–220. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef]
- Sailer, V.; Eng, K.W.; Zhang, T.; Bareja, R.; Pisapia, D.J.; Sigaras, A.; Bhinder, B.; Romanel, A.; Wilkes, D.; Sticca, E.; et al. Integrative Molecular Analysis of Patients With Advanced and Metastatic Cancer. JCO Precis. Oncol. 2019, 3, 1–12. [Google Scholar] [CrossRef]
- Fujii, Y.; Sato, Y.; Suzuki, H.; Kakiuchi, N.; Yoshizato, T.; Lenis, A.; Maekawa, S.; Yokoyama, A.; Takeuchi, Y.; Inoue, Y.; et al. Molecular classification and diagnostics of upper urinary tract urothelial carcinoma. Cancer Cell 2021, 39, 793–809. [Google Scholar] [CrossRef]
- Mir, M.A.; Pandith, A.A.; Mansoor, S.; Baba, S.M.; Makhdoomi, R.; Ain, Q.U.; Anwar, I.; Para, S.A.; Bhat, A.H.; Koul, A.M.; et al. Differential Expression of SLITRK6 Gene as a Potential Therapeutic Target for Urothelial Carcinoma in Particular Upper Tract Cancer. Gene 2023, 878, 147583. [Google Scholar] [CrossRef]
- Tomiyama, E.; Fujita, K.; Nakano, K.; Kuwahara, K.; Minami, T.; Kato, T.; Hatano, K.; Kawashima, A.; Uemura, M.; Takao, T.; et al. Trop-2 in Upper Tract Urothelial Carcinoma. Curr. Oncol. 2022, 29, 3911–3921. [Google Scholar] [CrossRef]
- Wu, S.; Chen, J.; Dong, P.; Zhang, S.; He, Y.; Sun, L.; Zhu, J.; Cheng, Y.; Li, X.; Tang, A.; et al. Global gene expression profiling identifies ALDH2, CCNE1 and SMAD3 as potential prognostic markers in upper tract urothelial carcinoma. BMC Cancer 2014, 14, 836. [Google Scholar] [CrossRef]
- Li, K.; Huang, Z.; Xie, G.; Huang, B.; Song, L.; Zhang, Y.; Yang, J. Transcriptomic insights into UTUC: Role of inflammatory fibrosis and potential for personalized treatment. J. Transl. Med. 2024, 22, 24. [Google Scholar] [CrossRef]
- Ohara, K.; Rendeiro, A.F.; Bhinder, B.; Eng, K.W.; Ravichandran, H.; Nguyen, D.; Pisapia, D.; Vosoughi, A.; Fernandez, E.; Shohdy, K.S.; et al. The evolution of metastatic upper tract urothelial carcinoma through genomic-transcriptomic and single-cell protein markers analysis. Nat. Commun. 2024, 15, 2009. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Fu, J.; Allen, W.; Xia, A.; Ma, Z.; Qi, X. Identification of Biomarkers in Breast Cancer by Gene Expression Profiling Using Human Tissues. Genome Data 2014, 2, 299–301. [Google Scholar] [CrossRef][Green Version]
- Verbakel, J.Y.; Steyerberg, E.W.; Uno, H.; De Cock, B.; Wynants, L.; Collins, G.S.; Van Calster, B. ROC curves for clinical prediction models part 1. ROC plots showed no added value above the AUC when evaluating the performance of clinical prediction models. J. Clin. Epidemiol. 2020, 126, 207–216. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Liu, Y.H.; Jin, J.; Liu, Y.J. Machine learning-based random forest for predicting decreased quality of life in thyroid cancer patients after thyroidectomy. Support. Care Cancer 2022, 30, 2507–2513. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chen, Z.; Fang, Y.; Su, M.; Xu, Y.; Wang, Z.; Gyamfi, M.A.; Zhao, J. Prediction of Prognosis and Recurrence of Bladder Cancer by ECM-Related Genes. J. Immunol. Res. 2022, 2022, 1793005. [Google Scholar] [CrossRef]
- Xu, C.; Coen-Pirani, P.; Jiang, X. Empirical Study of Overfitting in Deep Learning for Predicting Breast Cancer Metastasis. Cancers 2023, 15, 1969. [Google Scholar] [CrossRef]
- Luo, Y.; Deng, X.; Que, J.; Li, Z.; Xie, W.; Dai, G.; Chen, L.; Wang, H. Cell Trajectory-Related Genes of Lung Adenocarcinoma Predict Tumor Immune Microenvironment and Prognosis of Patients. Front. Oncol. 2022, 12, 911401. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.K.; Liao, X.W.; Yang, C.K.; Yu, T.D.; Liu, Z.Q.; Gong, Y.Z.; Huang, K.T.; Zeng, X.M.; Han, C.Y.; Zhu, G.Z.; et al. Diagnostic and prognostic biomarkers of Human Leukocyte Antigen complex for hepatitis B virus-related hepatocellular carcinoma. J. Cancer 2019, 10, 5173–5190. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Xie, T.; Shan, H.; Cheng, G. HLA-DQA1 expression is associated with prognosis and predictable with radiomics in breast cancer. Radiat. Oncol. 2023, 18, 117. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.Y.; Choi, K.U.; Kim, A.; Lee, S.J.; Kim, K.; Kim, J.Y.; Lee, I.S.; Chung, S.H.; Kim, J.I. Evaluation of immune-biomarker expression in high-grade soft-tissue sarcoma: HLA-DQA1 expression as a prognostic marker. Exp. Ther. Med. 2020, 20, 107. [Google Scholar] [CrossRef]
- Li, Y.; Li, H.; Yang, B.; Wei, J.; Zhen, C.; Feng, L. Clinical significance of PI3 and HLA-DOB as potential prognostic predicators for ovarian cancer. Transl. Cancer Res. 2020, 9, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xu, T.; Xia, Y.; Wang, Z.; Li, X.; Chen, W. ITM2A as a tumor suppressor and its correlation with PD-L1 in breast cancer. Front. Oncol. 2021, 10, 581733. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Wang, M.; Yang, J.; Xiong, H.; Wang, Y.; Tang, J. Integral membrane protein 2A inhibits cell growth in human breast cancer via enhancing autophagy induction. Cell Commun. Signal. 2019, 17, 105. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Shin, I.W.; Lee, T.J.; Park, J.; Kim, J.H.; Park, M.S.; Lee, E.J. Loss of ITM2A, a novel tumor suppressor of ovarian cancer through G2/M cell cycle arrest, is a poor prognostic factor of epithelial ovarian cancer. Gynecol. Oncol. 2016, 140, 545–553. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Gao, C.; Hu, Q.; Mao, X. Integral membrane protein 2A enhances sensitivity to chemotherapy via notch signaling pathway in cervical cancer. Bioengineered 2021, 12, 10183–10193. [Google Scholar] [CrossRef]
- Jiang, J.; Xu, J.; Ou, L.; Yin, C.; Wang, Y.; Shi, B. ITM2A inhibits the progression of bladder cancer by downregulating the phosphorylation of STAT3. Am. J. Cancer Res. 2024, 14, 2202–2215. [Google Scholar] [CrossRef]
- Zhu, M.; Yan, C.; Ren, C.; Huang, X.; Zhu, X.; Gu, H.; Wang, M.; Wang, S.; Gao, Y.; Ji, Y.; et al. Exome Array Analysis Identifies Variants in SPOCD1 and BTN3A2 That Affect Risk for Gastric Cancer. Gastroenterology 2017, 152, 2011–2021. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, X.Y.; Qin, Y.Y.; Yan, X.L.; Chen, H.M.; Huang, Q.D.; Chen, J.K.; Zheng, J.M. SPOCD1 promotes the proliferation and metastasis of glioma cells by up-regulating PTX3. Am. J. Cancer Res. 2018, 8, 624–635. [Google Scholar] [PubMed]
- Liang, J.; Zhao, H.; Hu, J.; Liu, Y.; Li, Z. SPOCD1 promotes cell proliferation and inhibits cell apoptosis in human osteosarcoma. Mol. Med. Rep. 2018, 17, 3218–3225. [Google Scholar] [CrossRef]
- van der Heijden, A.G.; Mengual, L.; Lozano, J.J.; Ingelmo-Torres, M.; Ribal, M.J.; Fernández, P.L.; Oosterwijk, E.; Schalken, J.A.; Alcaraz, A.; Witjes, J.A. A five-gene expression signature to predict progression in T1G3 bladder cancer. Eur. J. Cancer 2016, 64, 127–136. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, C.; Qin, M.; Ye, Y.; Mo, Y.; Meng, Q.; Yang, G.; Feng, G.; Lin, R.; Xian, S.; et al. Investigating cellular similarities and differences between upper tract urothelial carcinoma and bladder urothelial carcinoma using single-cell sequencing. Front. Immunol. 2024, 15, 1298087. [Google Scholar] [CrossRef] [PubMed]
- De Souza Santos, E.; De Bessa, S.A.; Netto, M.M.; Nagai, M.A. Silencing of LRRC49 and THAP10 genes by bidirectional promoter hypermethylation is a frequent event in breast cancer. Int. J. Oncol. 2008, 33, 25–31. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Q.; Guo, Z.; Wang, Y.; Wang, L.; Liu, X.; Lu, M.; Ju, L.; Xiao, Y.; Wang, X. Inhibition of MELK produces potential anti-tumour effects in bladder cancer by inducing G1/S cell cycle arrest via the ATM/CHK2/p53 pathway. J. Cell Mol. Med. 2020, 24, 1804–1821. [Google Scholar] [CrossRef]
- Maggisano, V.; Capriglione, F.; Mio, C.; Bulotta, S.; Damante, G.; Russo, D.; Celano, M. RNA Profile of Cell Bodies and Exosomes Released by Tumorigenic and Non-Tumorigenic Thyroid Cells. Int. J. Mol. Sci. 2024, 2, 1407. [Google Scholar] [CrossRef] [PubMed]
- Perkins, R.S.; Murray, G.; Suthon, S.; Davis, L.; Perkins, N.B., III; Fletcher, L.; Bozzi, A.; Schreiber, S.L.; Lin, J.; Laxton, S.; et al. WNT5B drives osteosarcoma stemness, chemoresistance and metastasis. Clin. Transl. Med. 2024, 5, 1670. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, A.; He, R.; Dang, W.; Liu, X.; Yang, T.; Shi, P.; Bu, X.; Gao, D.; Zhang, N.; et al. Gene polymorphism of cytochrome P450 significantly affects lung cancer susceptibility. Cancer Med. 2019, 10, 4892–4905. [Google Scholar] [CrossRef]
- Wang, P.; Zheng, H.; Zhang, J.; Wang, Y.; Liu, P.; Xuan, X.; Li, Q.; Du, Y. Identification of key gene modules and genes in colorectal cancer by co-expression analysis weighted gene co-expression network analysis. Biosci. Rep. 2020, 40, 2020–2044. [Google Scholar] [CrossRef] [PubMed]



| Progressive UTUC (n = 7) | Non-Progressive UTUC (n = 10) | Total UTUC (n = 17) | |
|---|---|---|---|
| Gender, n (%) | |||
| Male | 4 (57) | 7 (70) | 11 (65) |
| Female | 3 (43) | 3 (30) | 6 (35) |
| Tumor location, n (%) | |||
| Pelvis | 1 (14) | 8 (80) | 9 (53) |
| Ureter | 4 (57) | 1 (10) | 5 (29) |
| Both | 2 (29) | 1 (10) | 3 (18) |
| Pathological Stage, n (%) | |||
| pT2 | 3 (43) | 4 (40) | 7 (41) |
| pT3 | 4 (57) | 6 (60) | 10 (59) |
| Histological Grade, n (%) | |||
| Low | - | - | - |
| High | 7 (100) | 10 (100) | 17 (100) |
| Metastasis, n (%) | |||
| Local | - | - | - |
| Distant | 5 (71) | - | 5 (29) |
| Local + distant | 2 (29) | - | 2 (12) |
| Nodes, n (%) | 1 (14) | - | 1 (6) |
| Adjuvant Chemotherapy, n (%) | 3 (43) | 3 (30) | 6 (35) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padullés, B.; López-Aladid, R.; Ingelmo-Torres, M.; Roldán, F.L.; Martínez, C.; Juez, J.; Izquierdo, L.; Mengual, L.; Alcaraz, A. Machine Learning-Based Gene Expression Analysis to Identify Prognostic Biomarkers in Upper Tract Urothelial Carcinoma. Cancers 2025, 17, 2619. https://doi.org/10.3390/cancers17162619
Padullés B, López-Aladid R, Ingelmo-Torres M, Roldán FL, Martínez C, Juez J, Izquierdo L, Mengual L, Alcaraz A. Machine Learning-Based Gene Expression Analysis to Identify Prognostic Biomarkers in Upper Tract Urothelial Carcinoma. Cancers. 2025; 17(16):2619. https://doi.org/10.3390/cancers17162619
Chicago/Turabian StylePadullés, Bernat, Ruben López-Aladid, Mercedes Ingelmo-Torres, Fiorella L. Roldán, Carmen Martínez, Judith Juez, Laura Izquierdo, Lourdes Mengual, and Antonio Alcaraz. 2025. "Machine Learning-Based Gene Expression Analysis to Identify Prognostic Biomarkers in Upper Tract Urothelial Carcinoma" Cancers 17, no. 16: 2619. https://doi.org/10.3390/cancers17162619
APA StylePadullés, B., López-Aladid, R., Ingelmo-Torres, M., Roldán, F. L., Martínez, C., Juez, J., Izquierdo, L., Mengual, L., & Alcaraz, A. (2025). Machine Learning-Based Gene Expression Analysis to Identify Prognostic Biomarkers in Upper Tract Urothelial Carcinoma. Cancers, 17(16), 2619. https://doi.org/10.3390/cancers17162619






