Explainable Machine Learning Models for Glioma Subtype Classification and Survival Prediction
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Data Preprocessing
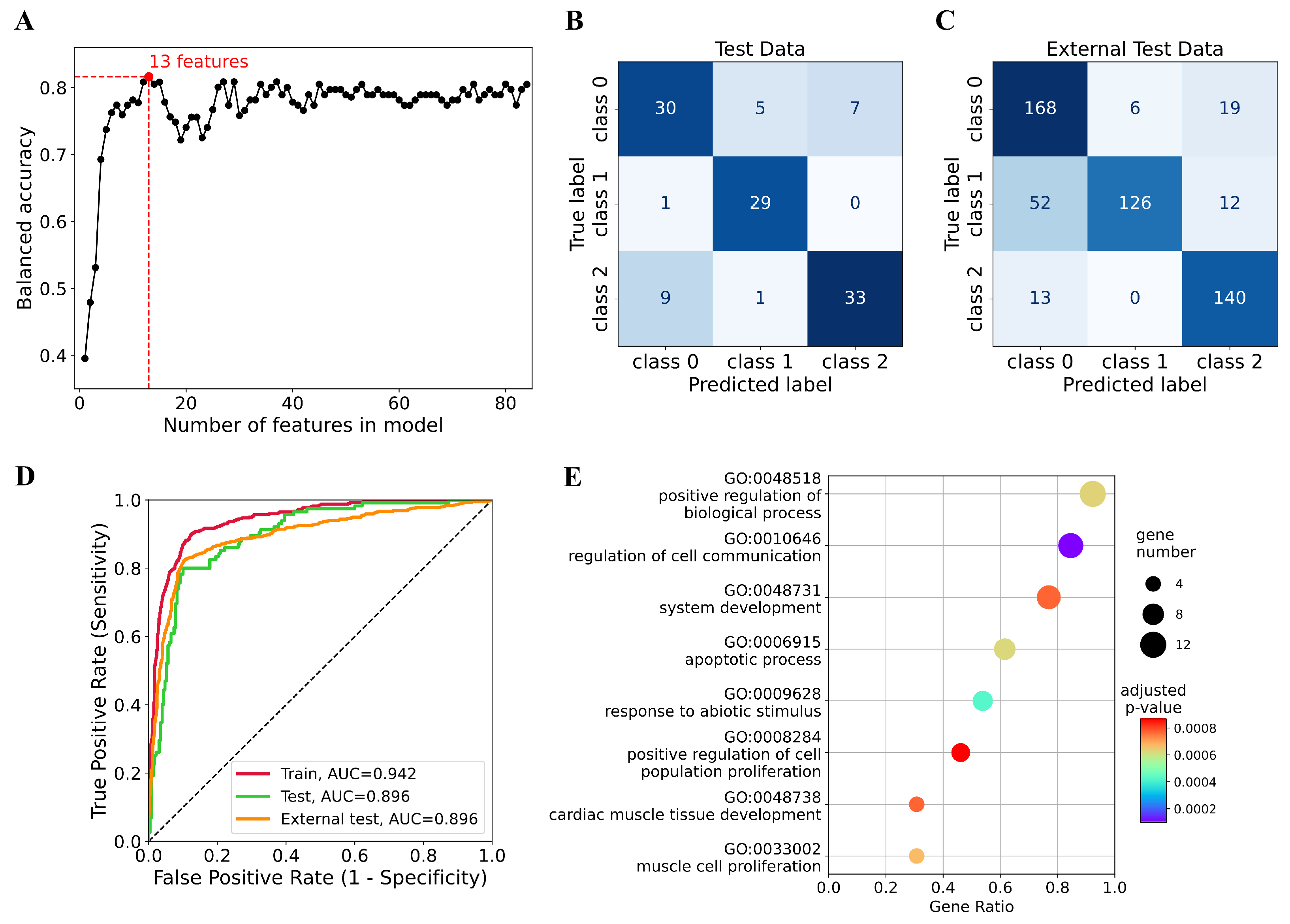
2.3. Feature Selection
2.4. ML Models for Classification
2.5. ML Models for Survival Prediction
2.6. Tuning and Training ML Models
2.7. Explainable Artificial Intelligence
2.8. Functional Enrichment Analysis
2.9. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Selection of Features Associated with Glioma Subtypes
3.3. Classifier for Glioma Subtypes
3.4. Functional Analysis of Prognostic Genes
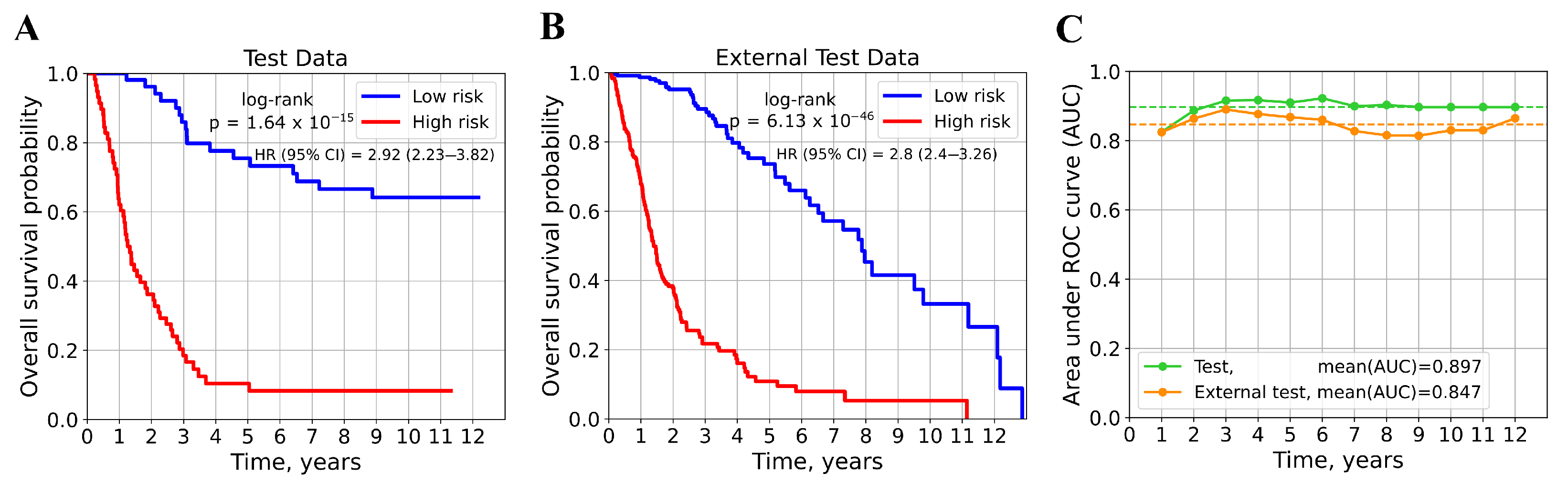
3.5. Model for Survival Prediction
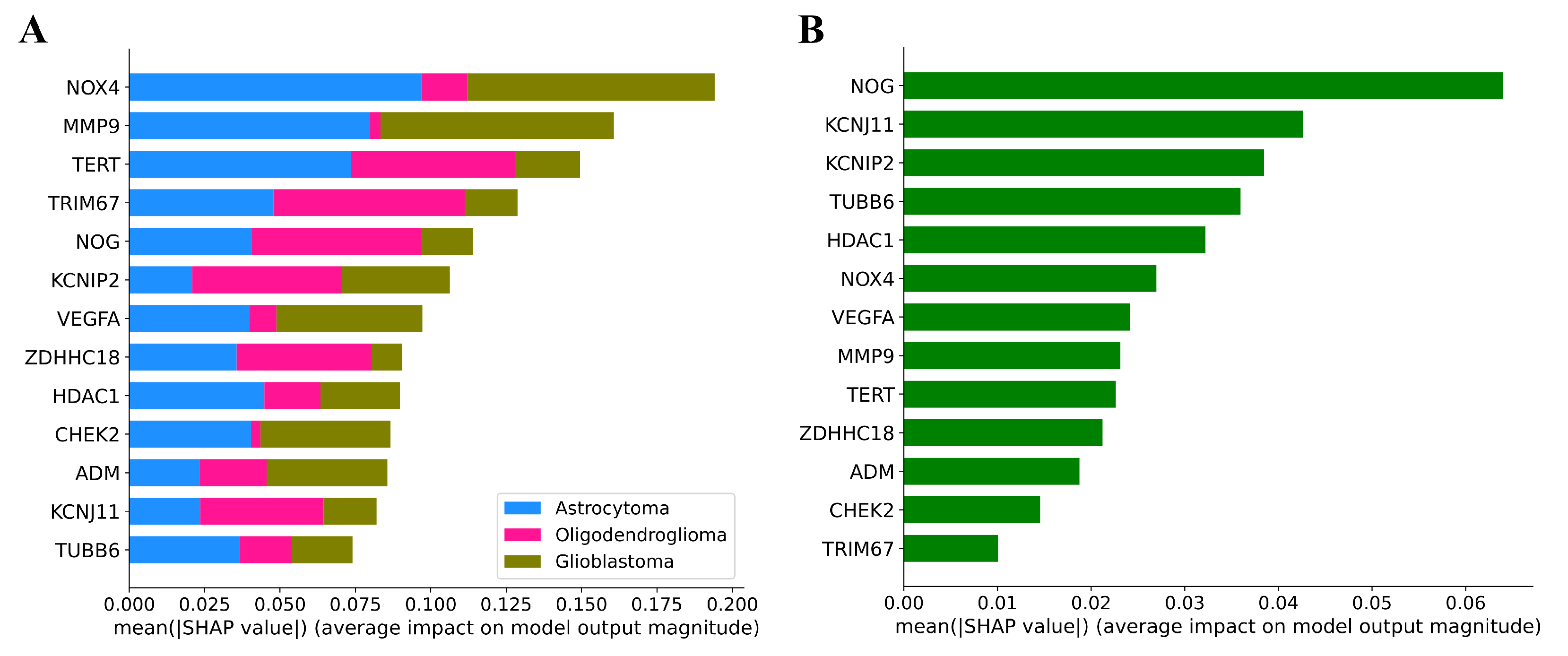
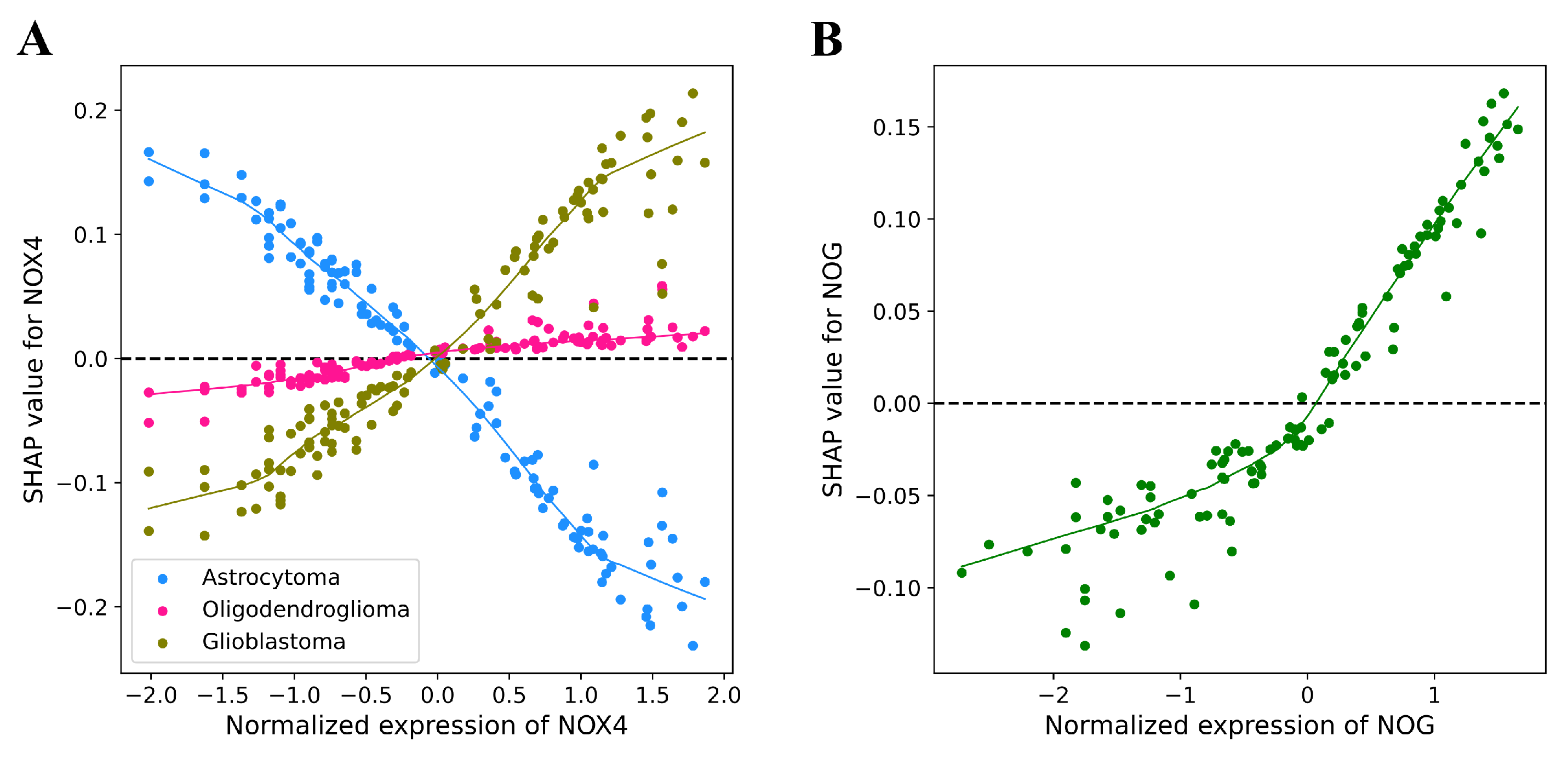
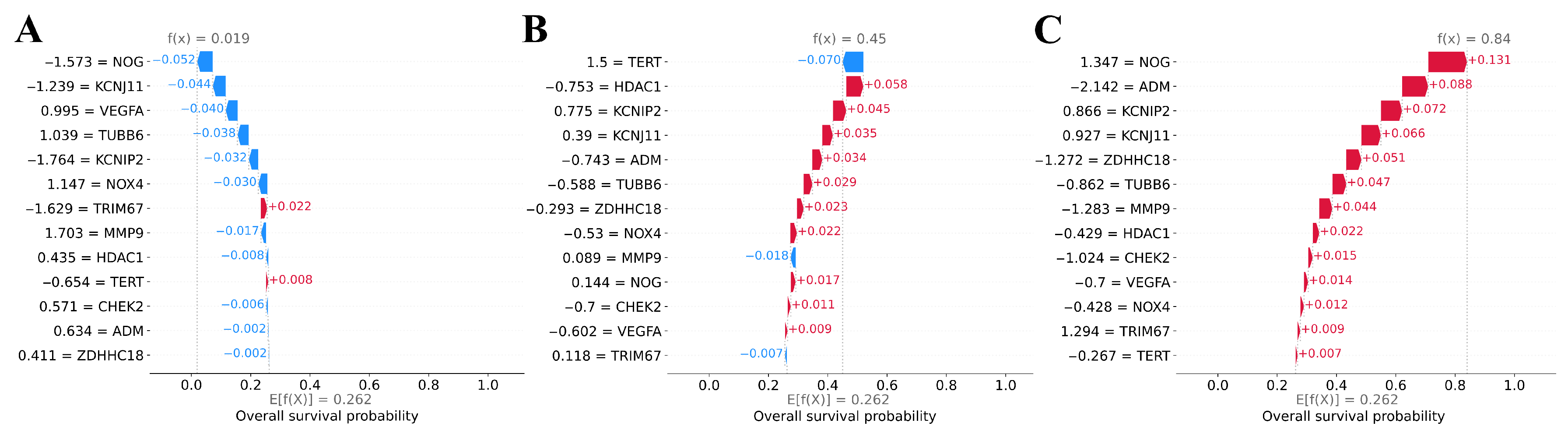
3.6. Explainability of ML Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2015–2019. Neuro-Oncol. 2022, 24, v1–v95. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.Y.; Kesari, S. GMalignant gliomas in adults. N. Engl. J. Med. 2008, 359, 492–507. [Google Scholar] [CrossRef]
- Omuro, A.; DeAngelis, L.M. Glioblastoma and other malignant gliomas: A clinical review. JAMA 2013, 310, 1842–1850. [Google Scholar] [CrossRef] [PubMed]
- Eckel-Passow, J.E.; Lachance, D.H.; Molinaro, A.M.; Walsh, K.M.; Decker, P.A.; Sicotte, H.; Pekmezci, M.; Rice, T.; Kosel, M.L.; Smirnov, I.V.; et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N. Engl. J. Med. 2015, 372, 2499–2508. [Google Scholar] [CrossRef]
- Wijnenga, M.M.J.; French, P.J.; Dubbink, H.J.; Dinjens, W.N.M.; Atmodimedjo, P.N.; Kros, J.M.; Smits, M.; Gahrmann, R.; Rutten, G.-J.; Verheul, J.B.; et al. The impact of surgery in molecularly defined low-grade glioma: An integrated clinical, radiological, and molecular analysis. Neuro-Oncol. 2018, 20, 103–112. [Google Scholar] [CrossRef]
- Patel, S.H.; Bansal, A.G.; Young, E.B.; Batchala, P.P.; Patrie, J.T.; Lopes, M.B.; Jain, R.; Fadul, C.E.; Schiff, D. Extent of Surgical Resection in Lower-Grade Gliomas: Differential Impact Based on Molecular Subtype. Am. J. Neuroradiol. 2019, 40, 1149–1155. [Google Scholar] [CrossRef]
- Korshunov, A.; Sycheva, R.; Golanov, A. Molecular stratification of diagnostically challenging high-grade gliomas composed of small cells: The utility of fluorescence in situ hybridization. Clin. Cancer Res. 2004, 10, 7820–7826. [Google Scholar] [CrossRef]
- Van Den Bent, M.J. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: A clinician’s perspective. Acta Neuropathol. 2010, 120, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Sottoriva, A.; Spiteri, I.; Piccirillo, S.G.; Touloumis, A.; Collins, V.P.; Marioni, J.C.; Curtis, C.; Watts, C.; Tavaré, S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. USA 2013, 110, 4009–4014. [Google Scholar] [CrossRef]
- Kumar, A.; Boyle, E.A.; Tokita, M.; Mikheev, A.M.; Sanger, M.C.; Girard, E.; Silber, J.R.; Gonzalez-Cuyar, L.F.; Hiatt, J.B.; Adey, A.; et al. Deep sequencing of multiple regions of glial tumors reveals spatial heterogeneity for mutations in clinically relevant genes. Genome Biol. 2014, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, C.; Meyer, J.; Balss, J.; Capper, D.; Mueller, W.; Christians, A.; Felsberg, J.; Wolter, M.; Mawrin, C.; Wick, W.; et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: A study of 1010 diffuse gliomas. Acta Neuropathol. 2009, 118, 469–474. [Google Scholar] [CrossRef]
- Puchalski, R.B.; Shah, N.; Miller, J.; Dalley, R.; Nomura, S.R.; Yoon, J.G.; Smith, K.A.; Lankerovich, M.; Bertagnolli, D.; Bickley, K.; et al. An anatomic transcriptional atlas of human glioblastoma. Science 2018, 360, 660–663. [Google Scholar] [CrossRef]
- Barthel, F.P.; Johnson, K.C.; Wesseling, P.; Verhaak, R.G. Evolving insights into the molecular neuropathology of diffuse gliomas in adults. Neurol. Clin. 2018, 36, 421. [Google Scholar] [CrossRef]
- Tang, Y.; Qazi, M.A.; Brown, K.R.; Mikolajewicz, N.; Moffat, J.; Singh, S.K.; McNicholas, P.D. Identification of five important genes to predict glioblastoma subtypes. Neuro-Oncol. Adv. 2021, 3, vdab144. [Google Scholar] [CrossRef]
- Cai, Y.D.; Zhang, S.; Zhang, Y.H.; Pan, X.; Feng, K.; Chen, L.; Huang, T.; Kong, X. Identification of the gene expression rules that define the subtypes in glioma. J. Clin. Med. 2018, 7, 350. [Google Scholar] [CrossRef]
- Munquad, S.; Si, T.; Mallik, S.; Li, A.; Das, A.B. Subtyping and grading of lower-grade gliomas using integrated feature selection and support vector machine. Briefings Funct. Genom. 2022, 21, 408–421. [Google Scholar] [CrossRef]
- Vieira, F.G.; Bispo, R.; Lopes, M.B. Integration of Multi-Omics Data for the Classification of Glioma Types and Identification of Novel Biomarkers. Bioinform. Biol. Insights 2024, 18, 11779322241249563. [Google Scholar] [CrossRef] [PubMed]
- Munquad, S.; Das, A.B. DeepAutoGlioma: A deep learning autoencoder-based multi-omics data integration and classification tools for glioma subtyping. BioData Min. 2023, 16, 32. [Google Scholar] [CrossRef]
- Wang, Z.; Gao, L.; Guo, X.; Feng, C.; Lian, W.; Deng, K.; Xing, B. Development and validation of a nomogram with an autophagy-related gene signature for predicting survival in patients with glioblastoma. Aging 2019, 11, 12246. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, X.; Chen, X.; Zhang, Q.; Hong, J. Novel immune-related gene signature for risk stratification and prognosis of survival in lower-grade glioma. Front. Genet. 2020, 11, 363. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.; Shu, L.; Li, J.; Wu, L.; Tao, C.; Ye, M.; Zhu, X.; Huang, K. A novel signature constructed by RNA-binding protein coding genes to improve overall survival prediction of glioma patients. Front. Cell Dev. Biol. 2021, 8, 588368. [Google Scholar] [CrossRef]
- Castelvecchi, D. Can we open the black box of AI? Nat. News 2016, 538, 20. [Google Scholar] [CrossRef] [PubMed]
- Adadi, A.; Berrada, M. Peeking inside the black-box: A survey on explainable artificial intelligence (XAI). IEEE Access 2018, 6, 52138–52160. [Google Scholar] [CrossRef]
- Loh, H.W.; Ooi, C.P.; Seoni, S.; Barua, P.D.; Molinari, F.; Acharya, U.R. Application of explainable artificial intelligence for healthcare: A systematic review of the last decade (2011–2022). Comput. Methods Programs Biomed. 2022, 226, 107161. [Google Scholar] [CrossRef]
- Ma, L.; Xiao, Z.; Li, K.; Li, S.; Li, J.; Yi, X. Game theoretic interpretability for learning based preoperative gliomas grading. Future Generation Computer Systems. Future Gener. Comput. Syst. 2020, 112, 1–10. [Google Scholar] [CrossRef]
- Chakraborty, S.; Sudhakar, S.; Swaminathan, R. An Explainable Machine Learning Model for Differentiation of Glioma Sub-types using MR Image Texture Analysis of Cerebral Edema. IFAC-PapersOnLine 2024, 58, 227–232. [Google Scholar] [CrossRef]
- Ayaz, H.; Oladimeji, O.; McLoughlin, I.; Tormey, D.; Booth, T.C.; Unnikrishnan, S. An eXplainable Deep Learning Model for Multi-Modal MRI Grading of IDH-Mutant Astrocytomas. Results Eng. 2024, 24, 103353. [Google Scholar] [CrossRef]
- Alahakoon, A.M.H.H.; Walgampaya, C.K.; Walgampaya, S.; Ekanayake, I.U.; Alawatugoda, J. Overall Survival predictions of GBM Patients using Radiomics: An Explainable AI Approach using SHAP. IEEE Access 2024, 12, 145234–145253. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, K.-N.; Wang, Q.; Li, G.; Zeng, F.; Zhang, Y.; Wu, F.; Chai, R.; Wang, Z.; Zhang, C.; et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genom. Proteom. Bioinform. 2021, 19, 1–12. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jenkins, D.F.; Manimaran, S.; Johnson, W.E. Alternative empirical Bayes models for adjusting for batch effects in genomic studies. BMC Bioinform. 2018, 19, 262. [Google Scholar] [CrossRef] [PubMed]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- Han, H.; Men, K. How does normalization impact RNA-seq disease diagnosis? J. Biomed. Inform. 2018, 85, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Ross, B.C. Mutual Information between Discrete and Continuous Data Sets. PLoS ONE 2014, 9, e87357. [Google Scholar] [CrossRef]
- Moore, J.H.; White, B.C. Tuning ReliefF for genome-wide genetic analysis. In Proceedings of the European Conference on Evolutionary Computation, Machine Learning and Data Mining in Bioinformatics, Valencia, Spain, 11–13 April 2007; Springer: Berlin/Heidelberg, Germany, 2007; pp. 166–175. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Urbanowicz, R.J.; Olson, R.S.; Schmitt, P.; Meeker, M.; Moore, J.H. Benchmarking Relief-Based Feature Selection Methods for Bioinformatics Data Mining. arXiv 2017, arXiv:1711.08477. [Google Scholar] [CrossRef]
- Peterson, L.E. K-nearest neighbor. Scholarpedia 2009, 4, 1883. [Google Scholar] [CrossRef]
- Vapnik, V.N. Statistical Learning Theory; Wiley: New York, NY, USA, 2008; 1780p. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. LightGBM: A Highly Efficient Gradient Boosting Decision Tree. In Proceedings of the 31st International Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; p. 30. [Google Scholar]
- Prokhorenkova, L.; Gusev, G.; Vorobev, A.; Dorogush, A.V.; Gulin, A. CatBoost: Unbiased boosting with categorical features. arXiv 2017, arXiv:1706.09516. [Google Scholar]
- Arik, S.O.; Pfister, T. TabNet: Attentive Interpretable Tabular Learning. arXiv 2019, arXiv:1908.07442. [Google Scholar] [CrossRef]
- Joseph, M.; Raj, H. GANDALF: Gated Adaptive Network for Deep Automated Learning of Features. arXiv 2022, arXiv:2207.08548. [Google Scholar]
- Joseph, M. PyTorch Tabular: A Framework for Deep Learning with Tabular Data. arXiv 2021, arXiv:2104.13638. [Google Scholar] [CrossRef]
- Mosley, L. A Balanced Approach to the Multi-Class Imbalance Problem. Ph.D. Thesis, Iowa State University, Ames, IA, USA, 2013. [Google Scholar]
- Cox, D.R. Analysis of Survival Data, 1st ed.; Chapman & Hall/CRC: New York, NY, USA, 1984; p. 212. [Google Scholar]
- Ishwaran, H.; Kogalur, U.B.; Blackstone, E.H.; Lauer, M.S. Random survival forests. Ann. Appl. Stat. 2008, 2, 841–860. [Google Scholar] [CrossRef]
- XGBoost Survival Embeddings. Available online: https://loft-br.github.io/xgboost-survival-embeddings/index.html (accessed on 30 January 2025).
- Katzman, J.L.; Shaham, U.; Cloninger, A.; Bates, J.; Jiang, T.; Kluger, Y. Deepsurv: Personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med. Res. Methodol. 2018, 18, 24. [Google Scholar] [CrossRef] [PubMed]
- Kvamme, H.; Borgan, Ø.; Scheel, I. Time-to-Event Prediction with Neural Networks and Cox Regression. J. Mach. Learn. Res. 2019, 20, 1–30. [Google Scholar]
- Kvamme, H.; Borgan, Ø. Continuous and discrete-time survival prediction with neural networks. arXiv 2019, arXiv:1910.06724. [Google Scholar] [CrossRef]
- Fotso, S. Deep neural networks for survival analysis based on a multi-task framework. arXiv 2018, arXiv:1801.05512. [Google Scholar] [CrossRef]
- Pölsterl, S. scikit-survival: A Library for Time-to-Event Analysis Built on Top of scikit-learn. J. Mach. Learn. Res. 2020, 21, 1–6. [Google Scholar]
- Harrell, F.E., Jr.; Lee, K.L.; Mark, D.B. Multivariable prognostic models: Issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat. Med. 1996, 15, 361–387. [Google Scholar] [CrossRef]
- Heagerty, P.J.; Lumley, T.; Pepe, M.S. Time-dependent ROC Curves for Censored Survival Data and a Diagnostic Marker. Biometrics 2000, 56, 337–344. [Google Scholar] [CrossRef]
- Graf, E.; Schmoor, C.; Sauerbrei, W.; Schumacher, M. Assessment and comparison of prognostic classification schemes for survival data. Stat. Med. 2018, 56, 2529–2545. [Google Scholar] [CrossRef]
- Bergstra, J.; Bardenet, R.; Bengio, Y.; Kégl, B. Algorithms for hyper-parameter optimization. In Proceedings of the 25th International Conference on Neural Information Processing Systems (NIPS 2011), Granada, Spain, 12–14 December 2011; p. 24. [Google Scholar]
- Watanabe, S. Tree-Structured Parzen Estimator: Understanding Its Algorithm Components and Their Roles for Better Empirical Performance. arXiv 2023, arXiv:2304.11127. [Google Scholar] [CrossRef]
- Akiba, T.; Sano, S.; Yanase, T.; Ohta, T.; Koyama, M. Optuna: A Next-generation Hyperparameter Optimization Framework. arXiv 2019, arXiv:1907.10902. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; pp. 4768–4777. [Google Scholar]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. g:Profiler—Interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef] [PubMed]
- Davidson-Pilon, C. Lifelines: Survival analysis in Python. J. Open Source Softw. 2019, 4, 1317. [Google Scholar] [CrossRef]
- Chen, S.C.-C.; Lo, C.-M.; Wang, S.-H.; Su, E.C.-Y. RNA editing-based classification of diffuse gliomas: Predicting isocitrate dehydrogenase mutation and chromosome 1p/19q codeletion. BMC Bioinform. 2019, 20, 659. [Google Scholar] [CrossRef]
- Park, Y.W.; Eom, J.; Kim, D.; Ahn, S.S.; Kim, E.H.; Kang, S.-G.; Chang, J.H.; Kim, S.H.; Lee, S.-K. A fully automatic multiparametric radiomics model for differentiation of adult pilocytic astrocytomas from high-grade gliomas. Eur. Radiol. 2022, 32, 4500–4509. [Google Scholar] [CrossRef]
- Gashi, M.; Vuković, M.; Jekic, N.; Thalmann, S.; Holzinger, A.; Jean-Quartier, C.; Jeanquartier, F. State-of-the-Art Explainability Methods with Focus on Visual Analytics Showcased by Glioma Classification. BioMedInformatics 2022, 2, 139–158. [Google Scholar] [CrossRef]
- Hu, X.; Martinez-Ledesma, E.; Zheng, S.; Kim, H.; Barthel, F.; Jiang, T.; Hess, K.R.; Verhaak, R.G.W. Multigene signature for predicting prognosis of patients with 1p19q co-deletion diffuse glioma. Neuro-Oncol. 2017, 19, 786–795. [Google Scholar] [CrossRef]
- Liu, B.; Liu, J.; Liu, K.; Huang, H.; Li, Y.; Hu, X.; Wang, K.; Cao, H.; Cheng, Q. A prognostic signature of five pseudogenes for predicting lower-grade gliomas. Biomed. Pharmacother. 2019, 117, 109116. [Google Scholar] [CrossRef]
- Song, L.R.; Weng, J.C.; Li, C.B.; Huo, X.L.; Li, H.; Hao, S.Y.; Wu, Z.; Wang, L.; Li, D.; Zhang, J.T. Prognostic and predictive value of an immune infiltration signature in diffuse lower-grade gliomas. JCI Insight 2020, 5, e133811. [Google Scholar] [CrossRef]
- Shono, T.; Yokoyama, N.; Uesaka, T.; Kuroda, J.; Takeya, R.; Yamasaki, T.; Amano, T.; Mizoguchi, M.; Suzuki, S.O.; Niiro, H.; et al. Enhanced expression of NADPH oxidase Nox4 in human gliomas and its roles in cell proliferation and survival. Cancer Cell Biol. 2008, 123, 787–792. [Google Scholar] [CrossRef]
- Lin, L.; Li, X.; Zhu, S.; Long, Q.; Hu, Y.; Zhang, L.; Liu, Z.; Li, B.; Li, X. Ferroptosis-related NFE2L2 and NOX4 Genes are Potential Risk Prognostic Biomarkers and Correlated with Immunogenic Features in Glioma. Cell Biochem. Biophys. 2023, 81, 7–17. [Google Scholar] [CrossRef]
- Xue, Q.; Cao, L.; Chen, X.-Y.; Zhao, J.; Gao, L.; Li, S.-Z.; Fei, Z. High expression of MMP9 in glioma affects cell proliferation and is associated with patient survival rates. Oncol. Lett. 2017, 13, 1325–1330. [Google Scholar] [CrossRef]
- Zhou, W.; Yu, X.; Sun, S.; Zhang, X.; Yang, W.; Zhang, J.; Zhang, X.; Zheng Jiang, Z. Increased expression of MMP-2 and MMP-9 indicates poor prognosis in glioma recurrence. Biomed. Pharmacother. 2019, 118, 109369. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, B.; Cai, J.; Sun, Y.; Wang, G.; Li, Y.; Li, R.; Feng, Y.; Han, B.; Li, J.; et al. Comparative Analysis of Matrix Metalloproteinase Family Members Reveals That MMP9 Predicts Survival and Response to Temozolomide in Patients with Primary Glioblastoma. PLoS ONE 2016, 11, e0151815. [Google Scholar] [CrossRef]
- Sun, F.; Liao, M.; Tao, Z.; Hu, R.; Qin, J.; Tao, W.; Liu, W.; Wang, Y.; Pi, G.; Lei, J.; et al. Identification of PANoptosis-related predictors for prognosis and tumor microenvironment by multiomics analysis in glioma. J. Cancer 2024, 15, 2486–2504. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K. 474P Potassium inward rectifier channel subfamily J member 11 mRNA expression in glioma and its significance in predicting prognosis and chemotherapy sensitivity. Ann. Oncol. 2024, 35, S418. [Google Scholar] [CrossRef]
- Luo, X.; Xu, S.; Zhong, Y.; Tu, T.; Xu, Y.; Li, X.; Wang, B.; Yang, F. High gene expression levels of VEGFA and CXCL8 in the peritumoral brain zone are associated with the recurrence of glioblastoma: A bioinformatics analysis. Oncol. Lett. 2019, 18, 6171–6179. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, C.; Wang, Z.; Guo, Y.; Wang, Y.; Fan, X.; Jiang, T. Expression changes in ion channel and immunity genes are associated with glioma-related epilepsy in patients with diffuse gliomas. J. Cancer Res. Clin. Oncol. 2022, 148, 2793–2802. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Quan, Q.; Jiang, J.; Peng, R. An immune and epithelial–mesenchymal transition-related risk model and immunotherapy strategy for grade II and III gliomas. Front. Genet. 2023, 13, 1070630. [Google Scholar] [CrossRef]
- Pan, P.; Guo, A.; Peng, L. Establishment of glioma prognosis nomogram based on the function of meox1 in promoting the progression of cancer. Heliyon 2024, 10, e29827. [Google Scholar] [CrossRef]
- Demirdizen, E.; Al-Ali, R.; Narayanan, A.; Sun, X.; Varga, J.P.; Steffl, B.; Brom, M.; Krunic, D.; Schmidt, C.; Schmidt, G.; et al. TRIM67 drives tumorigenesis in oligodendrogliomas through Rho GTPase-dependent membrane blebbing. Neuro-Oncol. 2023, 25, 1031–1043. [Google Scholar] [CrossRef]
- Lu, F.; Shen, S.H.; Wu, S.; Zheng, P.; Lin, K.; Liao, J.; Jiang, X.; Zeng, G.; Wei, D. Hypomethylation-induced prognostic marker zinc finger DHHC-type palmitoyltransferase 12 contributes to glioblastoma progression. Ann. Transl. Med. 2022, 10, 334. [Google Scholar] [CrossRef]
- Ying, T.; Lai, Y.; Lu, S.; E, S. Identification and validation of a glycolysis-related taxonomy for improving outcomes in glioma. CNS Neurosci. Ther. 2024, 30, e14601. [Google Scholar] [CrossRef]
- Li, J.; Yan, X.; Liang, C.; Chen, H.; Liu, M.; Wu, Z.; Zheng, J.; Dang, J.; La, X.; Liu, Q. Comprehensive Analysis of the Differential Expression and Prognostic Value of Histone Deacetylases in Glioma. Front. Cell Dev. Biol. 2022, 10, 840759. [Google Scholar] [CrossRef]
- Jiang, L.; Zhu, X.; Yang, H.; Chen, T.; Lv, K. Bioinformatics Analysis Discovers Microtubular Tubulin Beta 6 Class V (TUBB6) as a Potential Therapeutic Target in Glioblastoma. Front. Genet. 2020, 11, 566579. [Google Scholar] [CrossRef] [PubMed]
- La Torre, D.; Aguennouz, M.; Conti, A.; Giusa, M.; Raffa, G.; Abbritti, R.V.; Germano, A.; Angileri, F.F. Potential clinical role of telomere length in human glioblastoma. Transl. Med. UniSa 2011, 1, 243–270. [Google Scholar]
- Sampl, S.; Pramhas, S.; Stern, C.; Preusser, M.; Marosi, C.; Holzmann, K. Expression of telomeres in astrocytoma WHO grade 2 to 4: TERRA level correlates with telomere length, telomerase activity, and advanced clinical grade. Transl. Oncol. 2012, 5, 56-IN4. [Google Scholar] [CrossRef]
- Sallinen, S.-L.; Konen, T.; Haapasalo, H.; Schleutker, J. CHEK2 mutations in primary glioblastomas. J.-Neuro-Oncol. 2005, 74, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.-S.; Hou, W.-G.; Li, X.-L.; Jin, T.-B.; Li, Y.; Feng, D.-Y.; Liu, D.-B.; Gao, G.-D.; Yin, Z.-M.; Qin, H.-Z. Genetic association of CHEK2, GSTP1, and ERCC1 with glioblastoma in the Han Chinese population. Tumor Biol. 2014, 35, 4937–4941. [Google Scholar] [CrossRef]
- Tlais, D.; Barros Guinle, M.I.; Wheeler, J.R.; Prolo, L.M.; Vogel, H.; Partap, S. CHEK2 mutations in pediatric brain tumors. Neuro-Oncol. Adv. 2023, 5, vdad038. [Google Scholar] [CrossRef]
- Wilson, T.J.; Zamler, D.B.; Doherty, R.; Castro, M.G.; Lowenstein, P.R. Reversibility of glioma stem cells’ phenotypes explains their complex in vitro and in vivo behavior: Discovery of a novel neurosphere-specific enzyme, cGMP-dependent protein kinase 1, using the genomic landscape of human glioma stem cells as a discovery tool. Oncotarget 2016, 7, 63020–63041. [Google Scholar] [PubMed]
- Liu, M.; Xu, Z.; Du, Z.; Wu, B.; Jin, T.; Xu, K.; Xu, L.; Li, E.; Xu, H. The identification of key genes and pathways in glioma by bioinformatics analysis. J. Immunol. Res. 2017, 1, 1278081. [Google Scholar] [CrossRef]
- Ngo, M.T.; Harley, B.A. Perivascular signals alter global gene expression profile of glioblastoma and response to temozolomide in a gelatin hydrogel. Biomaterials 2019, 198, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Alhajala, H.S.; Nguyen, H.S.; Shabani, S.; Best, B.; Kaushal, M.; Al-Gizawiy, M.M.; Ahn, E.Y.; Knipstein, J.A.; Mirza, S.; Schmainda, K.M.; et al. Irradiation of pediatric glioblastoma cells promotes radioresistance and enhances glioma malignancy via genome-wide transcriptome changes. Oncotarget 2018, 9, 34122. [Google Scholar] [CrossRef]
- Kabir, F.; Apu, M.N.H. Multi-omics analysis predicts fibronectin 1 as a prognostic biomarker in glioblastoma multiforme. Genomics 2022, 114, 110378. [Google Scholar] [CrossRef]
- Pan, X.; Zeng, T.; Yuan, F.; Zhang, Y.H.; Chen, L.; Zhu, L.; Wan, S.; Huang, T.; Cai, Y.D. Screening of methylation signature and gene functions associated with the subtypes of isocitrate dehydrogenase-mutation gliomas. Front. Bioeng. Biotechnol. 2019, 7, 339. [Google Scholar] [CrossRef] [PubMed]

| Characteristic | CGGA, mRNAseq_693 (Training Set, n = 398) | CGGA, mRNAseq_325 (Validation/Test Set, n = 229) | TCGA, TCGA-LGG and TCGA-GBM (External Test Set, n = 536) | p Adjusted (Validation/Test Set vs. Training Set) | p Adjusted (External Test Set vs. Training Set) |
|---|---|---|---|---|---|
| Histology | 0.590 | <0.001 | |||
| Astrocytoma | 167 | 84 | 193 | ||
| Oligodendroglioma | 91 | 60 | 190 | ||
| Glioblastoma | 140 | 85 | 153 | ||
| Grade | 0.058 | 0.213 | |||
| WHO II (G2) | 130 | 94 | 154 | ||
| WHO III (G3) | 128 | 50 | 187 | ||
| WHO IV (G4) | 140 | 85 | 153 | ||
| Unknown | 0 | 0 | 42 | ||
| IDH mutation status | 0.704 | 0.125 | |||
| Wildtype | 175 | 112 | 216 | ||
| Mutant | 200 | 116 | 313 | ||
| Unknown | 23 | 1 | 7 | ||
| MGMT promoter methylation | 0.058 | <0.001 | |||
| Methylated | 185 | 99 | 365 | ||
| Unmethylated | 136 | 116 | 140 | ||
| Unknown | 77 | 14 | 31 | ||
| Age, years | 0.823 | <0.001 | |||
| Range | 11–76 | 10–79 | 17–89 | ||
| Median | 43 | 43 | 48 | ||
| Gender | 0.590 | 0.926 | |||
| Male | 233 | 142 | 311 | ||
| Female | 165 | 87 | 225 | ||
| Survival status | 0.115 | <0.001 | |||
| Alive | 179 | 85 | 313 | ||
| Dead | 204 | 139 | 221 | ||
| Unknown | 15 | 5 | 2 | ||
| OS, days | 0.315 | <0.001 | |||
| Range | 27–4725 | 19–4809 | 1–6423 | ||
| Median | 1283 | 1118 | 544 |
| Model | BA, Train | BA, CV | BA, Test | BA, External Test |
|---|---|---|---|---|
| GANDALF | 0.793 | 0.824 | 0.808 | 0.785 |
| kNN | 0.802 | 0.805 | 0.805 | 0.794 |
| SVM | 0.843 | 0.823 | 0.805 | 0.821 |
| LightGBM | 0.852 | 0.833 | 0.778 | 0.781 |
| TabNet | 0.830 | 0.826 | 0.777 | 0.784 |
| XGBoost | 0.843 | 0.826 | 0.775 | 0.775 |
| RF | 0.934 | 0.821 | 0.772 | 0.780 |
| ERT | 0.917 | 0.827 | 0.767 | 0.788 |
| CatBoost | 0.783 | 0.842 | 0.714 | 0.728 |
| Model | C-Index, Train | C-Index, CV | C-Index, Test | C-Index, External Test |
|---|---|---|---|---|
| RSF | 0.851 | 0.781 | 0.815 | 0.816 |
| PCHazard | 0.811 | 0.787 | 0.814 | 0.783 |
| EST | 0.810 | 0.779 | 0.813 | 0.815 |
| CoxPH | 0.783 | 0.765 | 0.812 | 0.803 |
| CoxCC | 0.802 | 0.790 | 0.809 | 0.800 |
| XGBSE-DBCE | 0.919 | 0.778 | 0.804 | 0.799 |
| XGBSE-KN | 0.858 | 0.784 | 0.800 | 0.812 |
| N-MTLR | 0.808 | 0.787 | 0.796 | 0.784 |
| DeepSurv | 0.800 | 0.787 | 0.787 | 0.786 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vershinina, O.; Turubanova, V.; Krivonosov, M.; Trukhanov, A.; Ivanchenko, M. Explainable Machine Learning Models for Glioma Subtype Classification and Survival Prediction. Cancers 2025, 17, 2614. https://doi.org/10.3390/cancers17162614
Vershinina O, Turubanova V, Krivonosov M, Trukhanov A, Ivanchenko M. Explainable Machine Learning Models for Glioma Subtype Classification and Survival Prediction. Cancers. 2025; 17(16):2614. https://doi.org/10.3390/cancers17162614
Chicago/Turabian StyleVershinina, Olga, Victoria Turubanova, Mikhail Krivonosov, Arseniy Trukhanov, and Mikhail Ivanchenko. 2025. "Explainable Machine Learning Models for Glioma Subtype Classification and Survival Prediction" Cancers 17, no. 16: 2614. https://doi.org/10.3390/cancers17162614
APA StyleVershinina, O., Turubanova, V., Krivonosov, M., Trukhanov, A., & Ivanchenko, M. (2025). Explainable Machine Learning Models for Glioma Subtype Classification and Survival Prediction. Cancers, 17(16), 2614. https://doi.org/10.3390/cancers17162614





