Simple Summary
This study explores the prevalence of BARD1 mutations in breast and ovarian cancer among Chinese patients. BARD1 mutations can vary across different ethnic groups, which is important for assessing cancer risk and developing effective monitoring strategies. This research involved a 30 gene panel and included 2658 patients. It found that BARD1 mutations occurred in 0.45% of breast cancer cases and 0.29% of ovarian cancer cases. Among the 12 patients with BARD1 mutations, eight different mutation types were identified, including three new variants. These mutation carriers were more likely to have family histories of other cancers, such as liver, prostate, and cervical cancers. Most breast tumors in mutation carriers were high-grade invasive ductal carcinoma, with a significant portion being triple-negative. Although BARD1 mutations are rare, the findings suggest that testing for BARD1 should be included in breast cancer panels, and mutation carriers may need closer monitoring due to associated family cancer histories.
Abstract
Background: The prevalence of BARD1 mutations in breast and ovarian cancers varies across different ethnic groups. Evaluating the cancer risk and clinical significance of BARD1 mutations in the local Chinese patients with breast cancer, ovarian cancer, or both is clinically important for designing an appropriate surveillance scheme. Methods: This study used a 30 gene panel to identify BARD1 germline mutations in 2658 breast and ovarian cancer patients. Results: Among this cohort, the BARD1 mutation prevalence was 0.45% for breast cancer and 0.29% for ovarian cancer. In our 12 mutation carriers, we identified eight types of mutation variants, including three novel mutations. BARD1 mutation carriers were more likely to have a family history of liver, prostate, and cervical cancers (p-values = 0.004, 0.018, and 0.037, respectively) than patients who tested negative for mutations. Among the BARD1 mutants, the majority of the breast tumors were invasive ductal carcinoma (NOS type) (10/11, 90.9%) of high-grade disease (9/9, 100%) and half of them were triple-negative breast cancer (5/10, 50%). Conclusions: Although the prevalence of BARD1 mutations is low and the penetrance is incomplete, we recommend including BARD1 in the test panel for breast cancer patients. Our data suggest that more comprehensive surveillance management may be considered in mutation carriers due to the familial aggregation of a relatively wide spectrum of cancers.
1. Introduction
Breast cancer is the most common female cancer and ovarian is the sixth most prevalent cancer in Hong Kong. On average, 1 in 20 women worldwide will be diagnosed with breast cancer in their lifetime, while a woman getting ovarian cancer during her life is about 1 in 78. The risk for breast and ovarian cancers is further enhanced in hereditary breast–ovarian cancer (HBOC). HBOC is a well-studied cancer predisposition syndrome caused by germline loss-of-function mutations and pathogenic variants (PVs) in genes such as BRCA1 or BRCA2. Multigene panel testing has revealed 10–20% HBOC-associated PVs [1,2]. However, further evidence and more conclusive cancer risk assessments are necessary before recommending surveillance management for mutation carriers in genes where PVs confer low or moderate penetrance effects. BRCA1-associated RING domain 1 (BARD1) is one of the genes considered to have low or moderate penetrance [3]. BARD1 consists of a RING-finger domain at its N-terminal region, followed by three intervening ankyrin (ANK) repeat domains and two tandem BRCA1 C-terminal domains (BRCT) [4]. BARD1 shares a high degree of structural and functional homology with BRCA1 within its BRCT and RING-finger domains, and these two proteins form a stable heterodimer [5]. BARD1 has been shown to act as a tumor suppressor in a BRCA1-independent pathway [6] and is involved in the homologous recombination repair pathway [7], stabilizing the p53 tumor suppressor via its ANK and BRCT domains [8]. It interacts with the repeated sequences of the BCL3 ANK domains and modulates the activities of the transcription factor NF-κB in the TP53-dependent apoptotic signaling pathway [9]. Moreover, it also facilitates the ubiquitination of RNA polymerase II, thereby hindering the transcription of damaged DNA and preventing the ubiquitination of ER-alpha and ER-beta, which are involved in cellular proliferation during the development of breast cancer [10]. Additionally, a decrease in BARD1 protein expression has been associated with cellular changes linked to a premalignant phenotype [11]. BARD1 was shown to play a role in maintaining genomic integrity, and the loss of BARD1 leads to chromosomal instability and embryonic death in the early stages [12]. On the other hand, various BARD1 isoforms that lack functional domains, such as the RING-finger and ANK domains due to exon skipping, have been found to be upregulated in different cancers. Abnormal BARD1 isoforms have been detected in non-small cell lung cancer (NSCLC), as well as in breast, colon, and ovarian cancers [13]. These isoforms are thought to have an oncogenic effect by interfering with the function of full-length BARD1 and are believed to contribute to tumorigenesis and cancer progression [13,14].
The BRCA1-BARD1 heterodimers are essential tumor suppressors in breast and ovarian cancers. These heterodimers also have additional functions in regulating the cell cycle, modulating the chromatin structure, and hormone signaling during cancer progression [8]. They are involved in DNA repair, replication fork protection, transcription, and tumor suppression [15]. In cancer cells, mutations that disrupt these heterodimers can lead to the detrimental degradation of both BRCA1 and BARD1 proteins [16].
Another important domain in BARD1 is the BRCT domain. It facilitates the early recruitment of the BRCA1-BARD1 heterodimer to DNA damage sites through a specific interaction with poly (ADP-ribose) polymerase (PARP) [17]. The Food and Drug Administration has approved PARP inhibitors to treat metastatic prostate cancer patients with DNA repair deficiencies due to pathogenic variants in genes involved in homologous recombination repair (HRR), including BARD1. The phase 2 LYNK-002 trial of Olaparib for patients with mutations in HRR or homologous recombination deficiency (HRD)-positive advanced breast carcinoma, malignant solid tumors, and ovarian carcinoma is ongoing [18]. With the widespread use of multiple gene mutation screenings, numerous BARD1 pathogenic variants have been identified in breast and ovarian cancer patients to be considered for PARPi treatment. Moreover, the prevalence and frequency of BARD1 mutations vary across different ethnic groups in breast cancer. Therefore, understanding the cancer risk and phenotypic presentations of BARD1 mutations in Chinese breast cancer patients can contribute to informed clinical management decisions.
2. Materials and Methods
2.1. Participants and Selection Criteria
A cohort of 2658 patients with breast and ovarian cancers was recruited by the Hong Kong Hereditary Breast Cancer Family Registry on the following criteria: (1) at least one first- or second-degree relative with BRCA-associated cancer, regardless of age; (2) diagnosis of breast cancer at age 45 or younger; (3) bilateral breast cancer; (4) triple-negative breast cancer; (5) cancers with medullary-type histology; (6) belonging to a BRCA mutation-related family; (7) male breast cancer; (8) ovarian cancer. Medical personnel obtained clinicopathologic characteristics of the patients from their medical records (Table 1). To validate the performance characteristics of next-generation sequencing (NGS) and evaluate its accuracy, known BRCA1/2-positive control and anonymous normal local negative control individuals were included [2]. Patients carrying PVs in our 30 genes panel were excluded from our analysis.

Table 1.
Clinicopathologic characteristics of study cohort.
2.2. Multi-Gene Panel Testing by NGS
Genomic DNA extracted from the peripheral blood underwent multi-gene sequencing analysis utilizing next-generation sequencing (NGS). Library preparation, sequencing, bioinformatics, variant interpretation, annotation, and a statistical analysis were conducted as previously outlined [2]. Paired sequencing reads were aligned to the human reference genome sequence GRCh37/hg19. Variants with a minor allele frequency of at least 1%, as reported by the 1000 Genomes Project [19], were excluded from manual variant curation. The BARD1 reference transcript accession (NM_000465.3) and variant nomenclature adhere to the Human Genome Variation Society (HGVS) guidelines and were verified using LUMC Mutalyzer 3 (http://mutalyzer.nl (30 May 2025)).
2.3. Statistical Analysis
Fisher’s exact test was employed to investigate the association between selection variables and mutation status. The significance threshold for all analyses was established at a p-value of < 0.05. Data analyses were performed using the statistical software R (version 3.4.2) [20].
3. Result
3.1. Patients’ Characteristics of the Cohort
Our testing cohort comprised 2658 breast and ovarian cancer patients. The median age at diagnosis of breast cancer was 43 years (range 18–90), and the median age at diagnosis of ovarian cancer was 47.5 years (range 9–85). Among these patients, 2318 (87.2%) were diagnosed with breast cancer and 199 (7.5%) with ovarian cancer, while 141 (5.3%) were diagnosed with both breast and ovarian cancers. Bilateral breast cancers were observed in 568 patients (23.1%). The majority of breast cancers were classified as ductal carcinoma (NOS type) (2144; 72.7%). A significant proportion of breast cancers were of the luminal A subtype (1263; 56.9%), followed by triple-negative breast cancers (TNBC) (506; 22.8%). Most of the breast tumors were diagnosed at early stages (0, I, or II) (2250; 86.1%), and grading favored grades 2 or 3 (906, 43.8% and 822, 39.7%, respectively). Most ovarian cancers were diagnosed as epithelial cancers (285; 96.3%), and the majority were of high grade (189; 68%). A positive family history of breast cancer (in first- or second-degree relatives) was observed in 1072 patients (40.3%). Family histories of ovarian cancer and prostate cancer were recorded in 189 (7.1%) and 145 (5.5%) of their patient’s relatives, respectively. Comprehensive clinicopathological characteristics are shown in Table 1.
3.2. Characteristics of BARD1 Mutation Carriers
Heterozygous pathogenic mutations in BARD1 were identified in 12 probands, resulting in a mutation frequency rates of 0.45% among high-risk breast cancer patients and 0.29% among ovarian cancer patients, while none of the BARD1 carrier had double heterozygous mutations. Eleven of the mutation carriers had personal breast cancers (91.7%), and only one had ovarian cancer (8.3%). The median age of breast cancer diagnosis was 43 (range 24–69), while the carrier with ovarian cancer was diagnosed at age 31. All of the BARD1 carriers were female. Only one BARD1 carrier had multiple cancers, including ovarian cancer and cancer of the uterus. Unlike BRCA carriers, none of the BARD1 carriers reported having bilateral breast cancer. Most of the breast tumors were diagnosed as invasive ductal carcinoma (NOS type) (10; 90.9%) of grade 3 (9; 100.0%). Half of the breast cancers were found to be triple-negative breast cancers (TNBC). Positive family histories of breast, colorectal, and liver cancers (in first- or second-degree relatives) were observed in four patients (33.3%). In comparison, a family history of ovarian cancer (in first- or second-degree relatives) was noted in only two patients (16.7%). Detailed pathological characteristics and family histories are shown in Table 2 and Table 3.

Table 2.
Characteristics of BARD1 carriers comparing with BRCA1/2 carriers and mutation negative patients.

Table 3.
Germline heterozygous mutations identified in BARD1.
There was no significant difference in diagnosis age of breast cancer between BARD1 mutation carriers, BRCA1/2 mutation carriers, and 30 genes non-carriers. However, the histopathology of the breast cancers of the BARD1 mutation carriers and BRCA1/2 mutation carriers showed certain distinguishing characteristics. Interestingly, all BARD1 and BRCA1 mutation carriers were female, whereas 2.8% of BRCA2 mutation carriers were male (Table 2). Unlike BRCA1/2 mutation carriers, bilateral breast cancer was not commonly seen in BARD1 mutation carriers, while 29.8% of the BRCA1 and BRCA2 mutation carriers developed bilateral breast cancers (p-value = 0.039). BARD1 carriers favor the development of high-grade invasion breast cancer, the same as BRCA1 carriers (p-value = 0.204), while BRCA2 carriers and non-carriers develops less aggressive tumors (p-values = 0.004 and <0.001 respectively). BARD1 and BRCA1/2 mutation carriers had a strong family history of breast cancer. A significant increase in the family history of liver cancer was observed in BARD1-mutated families compared to BRCA1-mutated families; 33% of BARD1 mutation carriers had a family history of liver cancer, whereas only 11.6% and 11.3% of BRCA1 mutation carriers and non-carriers did, respectively (p-value = 0.049 and 0.04). There was no significant difference between BARD1 and BRCA2 mutation carriers (p-value = 0.09). A similar observation was made regarding the family history of prostate cancer, which was present in 25% of BARD1 carriers compared to only 2.4% of BRCA1 carriers (p-value = 0.005). No significant difference was found between BARD1 and BRCA2 carriers, confirming the higher chance of having prostate cancer for BRCA2 carriers (p-value = 0.216). BARD1 carriers had significant family histories of liver, prostate, and cervical cancers compared to non-carriers (p-values = 0.04, 0.018, and 0.037, respectively). Additionally, BARD1 mutations were associated with a higher grade of disease compared to BRCA2 carriers and mutation-free non-carriers (p-values = 0.004 and <0.001, respectively; Table 2). The associations of age at ovarian cancer diagnosis and histology between BARD1 mutation carriers and BRCA1/2 mutation carriers were not calculated due to limited case numbers. Details of family histories of BARD1 mutation carriers are shown in Figure 1 and Table 3.
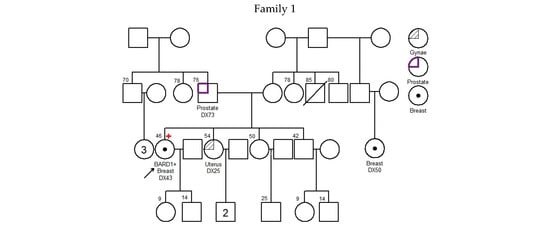
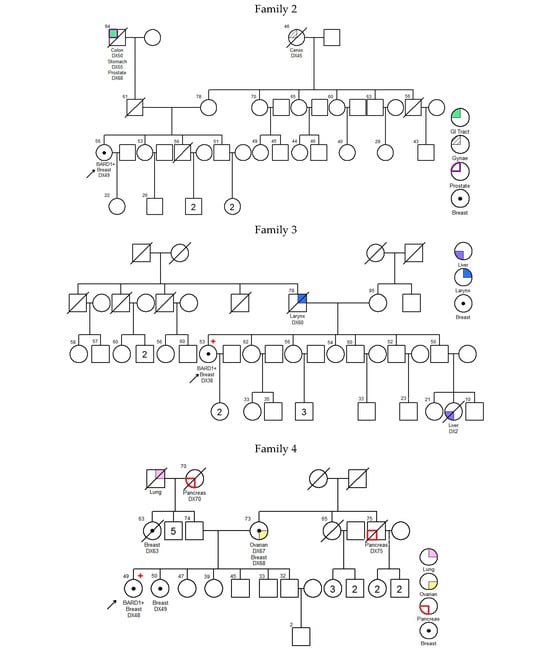
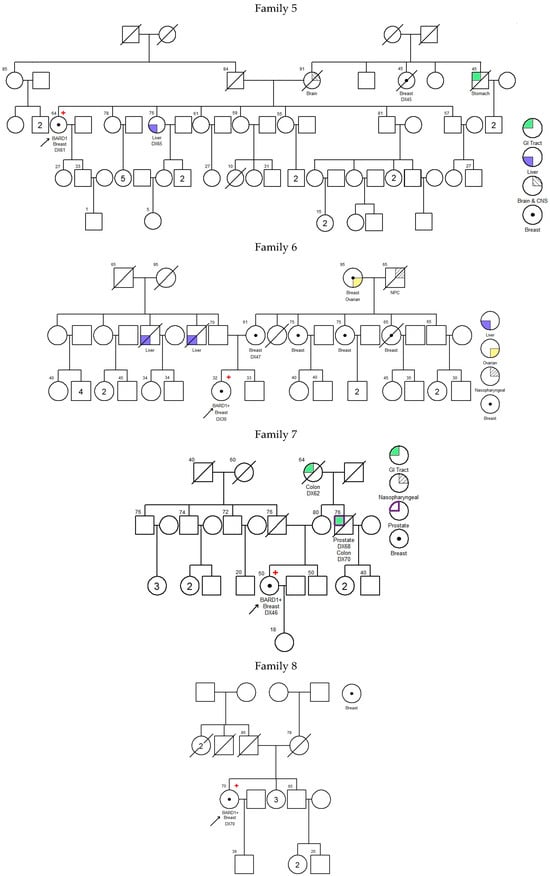
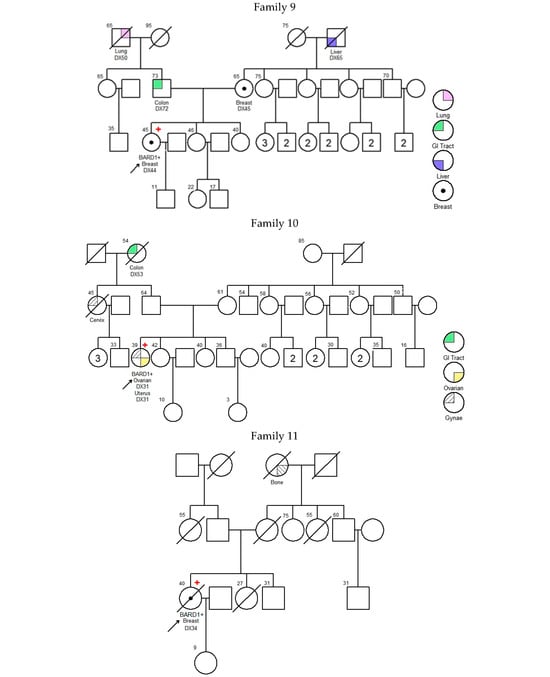
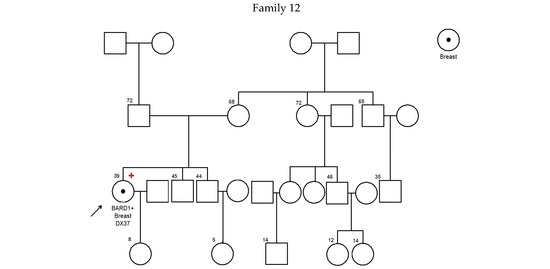
Figure 1.
Pedigree of BARD1 + families. Arrow refers to proband.
3.3. Mutation Spectrum in BARD1
In our cohort, 8 unique mutations were identified from 12 probands. Four (50%) of the mutations were nonsense mutations, two (25%) resulted in frameshifts and early termination, one (12.5%) occurred at splice sites, and one (12.5%) was a large deletion of exons 1–11. Three novel mutations were identified: c.540T>A; p.(Tyr180*), c.2167_2174delCATGCGAG; p.(His723Thrfs*4), and deletion of exons 1–11. There was no specific genomic regional clustering for these mutations in BARD1. The most frequent mutation, c.1338C>A; p.(Tyr446*) was seen in 5 (41.7%) unrelated families. This variant is located in exon 5, within the repeated domains of ANK. Details of family histories and distribution on the functional domains of these mutation variants are shown in Figure 2 and Table 3.
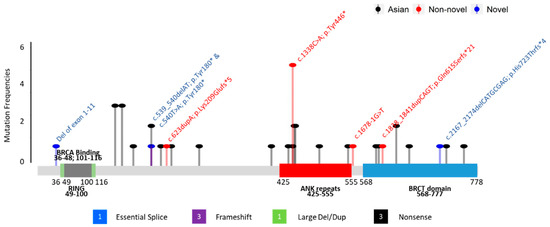
Figure 2.
Mutations found in BARD1. The protein domains RING (Grey), BRCA binding (green), ankyrin (ANK, red), and BRCA1 carboxy-terminal (BRCT, blue) are indicated;  —Asian mutation;
—Asian mutation;  —novel mutation;
—novel mutation;  —recurrent mutation.
—recurrent mutation.
 —Asian mutation;
—Asian mutation;  —novel mutation;
—novel mutation;  —recurrent mutation.
—recurrent mutation.
4. Discussion
Case–control studies have found a low to moderate association between breast cancer and pathogenic and likely pathogenic (P/LP) variants in the BARD1 gene, with a prevalence range of 0.1% to 0.51% in patients with breast cancer [1,3,28]. Recently, the absolute risk range for breast cancer in BARD1 carriers has been revised from 15–40% to 20–40% in the NCCN Genetic/Familial High-Risk Assessment 2023.3 Guidelines. BARD1 PVs in breast cancer patients of European ancestry had an odds ratio (OR) of 2.2 (p-value = 0.002; n = 28,536) [1]. Another study found an OR of 3.2 (p-value = 0.012; n = 2127) for breast cancer patients with a family history of breast cancer, while a large-scale case–control study indicated an OR of 2.3 (p-value = 0.04; n = 13,935) [3,29], showing that BARD1 is associated with low to moderate risk for breast cancer. In a retrospective study of approximately 48,700 breast cancer cases and 20,800 ovarian cancer cases, BARD1 was identified as a moderate-risk gene for breast cancer (OR = 2.90, 95% CIs: 2.25–3.75, p-value < 0.0001) but not for ovarian cancer (OR = 1.36, 95% CIs: 0.87–2.11, p-value = 0.1733) [30]. Moreover, a stronger association (OR = 5.4; p-value < 0.00001; n = 4469) between BARD1 PVs and familial breast cancer patients was reported. The risk was further enhanced (OR = 12.0; p-value < 0.00001; n = 782) for breast cancer patients who diagnosed under 40 years of age, suggesting that BARD1 may be a risk gene for early-onset familial breast cancer [28]. However, the association of BARD1 with ovarian cancer has not been convincingly established [1,22,30]. In a meta-analysis, the mutation frequency rates in the BARD1 gene among breast cancer and ovarian cancer patients from mixed populations were 0.25% and 0.12%, respectively [30]. In another study including data from Australia, USA, and UK, the prevalence of PVs in BARD1 in the breast cancer group was only 0.12% [22]. In our Asian cohort, the prevalence rates of PVs in BARD1 in the breast cancer group (0.45%) and the ovarian cancer group (0.29%) were significantly higher than the reported mutation frequencies. However, the breast cancer risk estimates of BARD1 PVs for Caucasians and Asians show no substantial difference and the frequency of BARD1 mutations in general population controls (from mixed populations) is 0.09% [30].
Individuals with BARD1 and BRCA1 germline pathogenic mutations have been found to have a higher incidence rates of aggressive breast cancer phenotypes, such as TNBCs, which are associated with higher rates of recurrence, progression, and mortality [31,32,33]. At the molecular and protein levels, BARD1 shows a significant degree of structural and functional similarity to BRCA1, and breast cancers occurring in individuals with BARD1 germline PVs exhibit a similar somatic gene expression profile to those with BRCA1 pathogenic variants. An example of this is when a patient with breast cancer had a germline BARD1 deletion or loss of heterozygosity in the tumor, resulting in a basal-like gene expression profile similar to those observed in cancers associated with BRCA1 germline PVs [31,34]. No significant differences in clinicopathological characteristics were observed in our Chinese cohort between BARD1 and BRCA1 mutation carriers, except that carriers of the BRCA1 mutation had a higher incidence of bilateral breast cancer. However, BARD1 mutation carriers with bilateral breast cancer have been reported in Polish and Belarusian populations [29] but not in Asian populations. Significant differences in molecular subtypes and grading were observed between BARD1 and BRCA2 mutation carriers. Among BRCA2 mutation carriers, 84.5% had a preference for being ER-positive with a molecular subtype of luminal A (71%), while only 54.5% of BARD1 mutation carriers developed ER-positive breast cancer (p-value = 0.022), with 30% in the molecular subtype of luminal A. In our BARD1 mutation carriers, we observed that 50% of them harbored TNBC, a frequency similar to that of BRCA1 mutation carriers (58.2%). In comparison, only 11.6% of BRCA2 mutation carriers were TNBC (p-value = 0.005), and 18.6% of mutation-negative patients were TNBC (p-value = 0.025). This association aligns with studies in the Spanish population and other European studies [35,36]. The Breast Cancer Association Consortium and the CARRIERS case–control studies also found associations between BARD1 PVs and an increased risk of triple-negative breast cancer [37,38]. All of our BARD1 mutation carriers developed high-grade breast cancer, while only 44.7% of BRCA2 mutation carriers (p-value = 0.004) and 39.7% of mutation-negative patients (p-value < 0.001) developed high-grade breast cancer. Another Asian study from Singapore also found that patients with BARD1 PVs developed more aggressive triple-negative breast cancer and high-grade breast cancers [39].
Germline copy number variants (CNVs) in the BRCA1 and BRCA2 genes account for less than 5% of known pathogenic variants in these genes [40]. CNVs in the BARD1 gene have also been observed. Deletions of exon 1 [41] and exon 2 [42] have been reported in breast cancer patients, while deletions of exons 8 to 11 and the entire gene were identified in ovarian cancer patients [43]. Additionally, a deletion of exons 8 to 11 in the BARD1 gene has also been identified in a family with hereditary colorectal cancer syndrome [44]. Our cohort identified a deletion of the entire BARD1 gene from exons 1 to 11. Two CNVs (deletion of exons 4 to 11 and duplication of exons 1 to 9) in the BARD1 gene were also identified among non-cancer controls [23]. CNVs in BARD1 are not rare events; they accounted for at least 8.3% of known PVs in our cohort.
Two regions in the BARD1 gene have been reported to have an increased density of pathogenic variants [30]. The first overlaps with the RING-finger domain, extending from exon 2 to around 230 amino acids in exon 4. The second “hotspot” region extends from exon 5 to exon 10, covering the ANK repeat and the BRCT domains. However, unlike the BRCA1 and BRCA2 genes, which have clustered regions associated with breast and ovarian cancers, no clear hotspot could be identified in BARD1. All mutations identified in our cohort, except for the CNV mentioned earlier, were located in ANK repeat and BRCT domains (see Figure 2). Among the 12 BARD1 mutation carriers we identified, five carried a c.1338C>A; p.(Tyr446*) mutation in exon 5, which is located in the repeated domains of ANK. This variant has also been reported in unselected breast and colorectal cancer patients in the Chinese population [23,24], and there were two submissions from Ambry Genetics and Invitae in ClinVar. Excluding the above-mentioned patients from China, this c.1338C>A; p.(Tyr446*) mutation was not reported in a large cumulative summary of the BARD1 PVs mutation spectrum identified from breast and ovarian cancer patients, with the entire BARD1 coding sequence sequenced [30]. In the gnomAD control, this variant was also seen once in the East Asian and European (non-Finnish) populations. These findings demonstrate that although the mutation is shared across ethnicities, it is not a common recurrent mutation; however, the frequency of its identification in the Chinese population is relatively higher (Table 2 and Supplementary Table S1).
Many studies have widely discussed the association of the BARD1 missense mutation c.1670G>C; p.(Cys557Ser) to breast cancer risk. Extensive studies in Iceland, Finland, Latin or South America, and Italy showed this germline variant was associated with an increase of two- to four-fold in breast cancer risk [26,27]. In contrast, other reports from Yoruba, Chinese, Japanese, Australian, and African American individuals did not show similar findings [45]. A meta-analysis also found no evidence to support this association, except in women with a strong family history, where these carriers had a 3.4-fold increase in breast cancer risk [46]. The association between familial breast cancer susceptibility on this missense variant remains controversial. However, none of our high-risk patients carried this missense variant, further impeding our understanding of the clinical relevance of BARD1.
BARD1 mutation carriers in our study were more likely to have a family history of liver, prostate, and cervical cancers than patients who tested negative for the 30 gene panel (p-values = 0.04, 0.018, and 0.037, respectively). In our study, no significant difference was found in the family histories of breast cancer between BARD1 mutated carriers and non-carriers, largely due to selection bias, as ‘family history of breast cancer’ is one of our recruiting criteria. Current NCCN surveillance management guidelines recommend only annual breast screening starting at age 40 for BARD1 mutation carriers but no surveillance management for other related cancers. However, BARD1 PVs have been identified in patients with not only breast cancer but also in patients with neuroblastoma, colon cancer, liver cancer, lung cancer, and acute myeloid leukemia [21]. BARD1 has also be found as prognosis-related genes of liver cancer and used for predicting the survival of liver cancer patients [47]. For prostate cancer, a study from Poland confirmed BARD1 mutation carriers were not at elevated risk of prostate cancer [48]; however, another study of 9185 men with aggressive prostate cancer from 18 international studies provided evidence of greater risk (OR ≥ 2) but the carrier frequency differences between aggressive and non-aggressive prostate cancer were not statistically significant [49]. These variants may confer low to moderate penetrance effects, which still require more evidence and convincing risk assessments for recommendations on surveillance for carriers of BARD1 pathogenic variants concerning other cancers.
5. Conclusions
We demonstrated that the mutation frequency rates of BARD1 were 0.45% among high-risk breast cancer patients and 0.29% among ovarian cancer patients. We identified three novel mutations and a recurrent mutation in the BARD1 gene. Half of the BARD1 mutation carriers were found to have TNBC and were likely to have familial aggregation of liver, prostate, and cervical cancers compared to patients who tested negative for mutations in the 30 gene panel. Mutation screening for BARD1 should be included in the test panel for breast cancer patients. However, more comprehensive surveillance management may be considered, even given the low penetrance of BARD1, especially for Asian patients. More clinical evidence is needed to demonstrate the effectiveness of PARP inhibitors in patients with BARD1 mutations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers17152524/s1, Table S1. BARD1 pathogenic variants identified from breast or ovarian cancer patients from Asian populations.
Author Contributions
A.K., C.H.A., and E.S.K.M. designed the study. A.K. coordinated the prospective data collection for the Hong Kong Hereditary Breast Cancer Family Registry. C.Y.S.H. retrieved and collected data for this study, interpreted the results, and drafted the manuscript. A.K., C.H.A., and E.S.K.M. reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by Dr. Ellen Li Charitable Foundation, Kerry Kuok Foundation, Health and Medical Research Fund (03143406), Asian Fund for Cancer Research, and Hong Kong Hereditary Breast Cancer Family Registry.
Institutional Review Board Statement
The study was performed in accordance with the Declaration of Helsinki. This study was approved by the Institutional Review Board of the University of Hong Kong/Hospital Authority West Cluster and respective authorities of other contributing hospitals in Hong Kong (UW06-274T-1299).
Informed Consent Statement
Written informed consent was obtained from all participants recruited in this study.
Data Availability Statement
The dataset supporting the conclusions of this article is included within the article.
Acknowledgments
We thank all researchers, clinicians, technicians, and nursing staff who enabled this work to be carried out.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Couch, F.J.; Shimelis, H.; Hu, C.; Hart, S.N.; Polley, E.C.; Na, J.; Hallberg, E.; Moore, R.; Thomas, A.; Lilyquist, J.; et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. 2017, 3, 1190–1196. [Google Scholar] [CrossRef]
- Kwong, A.; Shin, V.Y.; Chen, J.; Cheuk, I.W.Y.; Ho, C.Y.S.; Au, C.H.; Chan, K.K.L.; Ngan, H.Y.S.; Chan, T.L.; Ford, J.M.; et al. Germline Mutation in 1338 BRCA-Negative Chinese Hereditary Breast and/or Ovarian Cancer Patients: Clinical Testing with a Multigene Test Panel. J. Mol. Diagn. 2020, 22, 544–554. [Google Scholar] [CrossRef]
- Slavin, T.P.; Maxwell, K.N.; Lilyquist, J.; Vijai, J.; Neuhausen, S.L.; Hart, S.N.; Ravichandran, V.; Thomas, T.; Maria, A.; Villano, D.; et al. The contribution of pathogenic variants in breast cancer susceptibility genes to familial breast cancer risk. NPJ Breast Cancer 2017, 3, 22, Erratum in NPJ Breast Cancer 2017, 3, 44. [Google Scholar] [CrossRef]
- Fox, D., 3rd; Le Trong, I.; Rajagopal, P.; Brzovic, P.S.; Stenkamp, R.E.; Klevit, R.E. Crystal structure of the BARD1 ankyrin repeat domain and its functional consequences. J. Biol. Chem. 2008, 283, 21179–21186. [Google Scholar] [CrossRef]
- Brzovic, P.S.; Rajagopal, P.; Hoyt, D.W.; King, M.C.; Klevit, R.E. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 2001, 8, 833–837. [Google Scholar] [CrossRef] [PubMed]
- Hawsawi, Y.; Faris, F. The Fundamental Role of BARD1 Mutations and Their Applications as a Prognostic Biomarker for Cancer Treatment. In BRCA1 and BRCA2 Mutations-Diagnostic and Therapeutic Implications; IntechOpen: London, UK, 2023. [Google Scholar] [CrossRef]
- Stewart, M.D.; Zelin, E.; Dhall, A.; Walsh, T.; Upadhyay, E.; Corn, J.E.; Chatterjee, C.; King, M.C.; Klevit, R.E. BARD1 is necessary for ubiquitylation of nucleosomal histone H2A and for transcriptional regulation of estrogen metabolism genes. Proc. Natl. Acad. Sci. USA 2018, 115, 1316–1321. [Google Scholar] [CrossRef]
- Irminger-Finger, I.; Ratajska, M.; Pilyugin, M. New concepts on BARD1: Regulator of BRCA pathways and beyond. Int. J. Biochem. Cell Biol. 2016, 72, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Irminger-Finger, I.; Leung, W.C.; Li, J.; Dubois-Dauphin, M.; Harb, J.; Feki, A.; Jefford, C.E.; Soriano, J.V.; Jaconi, M.; Montesano, R.; et al. Identification of BARD1 as mediator between pro-apoptotic stress and p53-dependent apoptosis. Mol. Cell 2001, 8, 1255–1266. [Google Scholar] [CrossRef] [PubMed]
- Dizin, E.; Irminger-Finger, I. Negative feedback loop of BRCA1-BARD1 ubiquitin ligase on estrogen receptor alpha stability and activity antagonized by cancer-associated isoform of BARD1. Int. J. Biochem. Cell Biol. 2010, 42, 693–700. [Google Scholar] [CrossRef]
- Irminger-Finger, I.; Soriano, J.V.; Vaudan, G.; Montesano, R.; Sappino, A.P. In vitro repression of BRCA1-associated RING domain gene, BARD1, induces phenotypic changes in mammary epithelial cells. J. Cell Biol. 1998, 143, 1329–1339. [Google Scholar] [CrossRef]
- McCarthy, E.E.; Celebi, J.T.; Baer, R.; Ludwig, T. Loss of BARD1, the heterodimeric partner of the BRCA1 tumor suppressor, results in early embryonic lethality and chromosomal instability. Mol. Cell Biol. 2003, 23, 5056–5063. [Google Scholar] [CrossRef] [PubMed]
- Hawsawi, Y.M.; Shams, A.; Theyab, A.; Abdali, W.A.; Hussien, N.A.; Alatwi, H.E.; Alzahrani, O.R.; Oyouni, A.A.A.; Babalghith, A.O.; Alreshidi, M. BARD1 mystery: Tumor suppressors are cancer susceptibility genes. BMC Cancer 2022, 22, 599. [Google Scholar] [CrossRef]
- Li, L.; Ryser, S.; Dizin, E.; Pils, D.; Krainer, M.; Jefford, C.E.; Bertoni, F.; Zeillinger, R.; Irminger-Finger, I. Oncogenic BARD1 isoforms expressed in gynecological cancers. Cancer Res. 2007, 67, 11876–11885. [Google Scholar] [CrossRef] [PubMed]
- Tarsounas, M.; Sung, P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell Biol. 2020, 21, 284–299. [Google Scholar] [CrossRef]
- Alzahrani, F.A.; Ahmed, F.; Sharma, M.; Rehan, M.; Mahfuz, M.; Baeshen, M.N.; Hawsawi, Y.; Almatrafi, A.; Alsagaby, S.A.; Kamal, M.A.; et al. Investigating the pathogenic SNPs in BLM helicase and their biological consequences by computational approach. Sci. Rep. 2020, 10, 12377. [Google Scholar] [CrossRef]
- Li, M.; Yu, X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell 2013, 23, 693–704. [Google Scholar] [CrossRef]
- Hyman, D.M.; Hendifar, A.; Chung, H.C.; Maio, M.; Leary, A.; Spanggaard, I.; Rhee, J.; Marton, M.; Chen, M.; Krishnan, S.; et al. Phase II study of olaparib in previously treated advanced solid tumors with homologous recombination repair mutation (HRRm) or homologous recombination repair deficiency (HRD): LYNK-002. Ann. Oncol. 2019, 30 (Suppl. 5), v53–v54. [Google Scholar] [CrossRef]
- 1000 Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 28 February 2025).
- Randall, M.P.; Egolf, L.E.; Vaksman, Z.; Samanta, M.; Tsang, M.; Groff, D.; Evans, J.P.; Rokita, J.L.; Layeghifard, M.; Shlien, A.; et al. BARD1 germline variants induce haploinsufficiency and DNA repair defects in neuroblastoma. J. Natl. Cancer Inst. 2024, 116, 138–148. [Google Scholar] [CrossRef]
- Ramus, S.J.; Song, H.; Dicks, E.; Tyrer, J.P.; Rosenthal, A.N.; Intermaggio, M.P.; Fraser, L.; Gentry-Maharaj, A.; Hayward, J.; Philpott, S.; et al. Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J. Natl. Cancer Inst. 2015, 107, djv214. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Meng, H.; Yao, L.; Lv, M.; Bai, J.; Zhang, J.; Wang, L.; Ouyang, T.; Li, J.; Wang, T.; et al. Germline Mutations in Cancer Susceptibility Genes in a Large Series of Unselected Breast Cancer Patients. Clin. Cancer Res. 2017, 23, 6113–6119. [Google Scholar] [CrossRef]
- Gong, R.; He, Y.; Liu, X.Y.; Wang, H.Y.; Sun, L.Y.; Yang, X.H.; Li, B.; Cao, X.K.; Ye, Z.L.; Kong, L.H.; et al. Mutation spectrum of germline cancer susceptibility genes among unselected Chinese colorectal cancer patients. Cancer Manag. Res. 2019, 11, 3721–3739. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Hart, S.N.; Polley, E.C.; Gnanaolivu, R.; Shimelis, H.; Lee, K.Y.; Lilyquist, J.; Na, J.; Moore, R.; Antwi, S.O.; et al. Association Between Inherited Germline Mutations in Cancer Predisposition Genes and Risk of Pancreatic Cancer. JAMA 2018, 319, 2401–2409. [Google Scholar] [CrossRef] [PubMed]
- Ratajska, M.; Antoszewska, E.; Piskorz, A.; Brozek, I.; Borg, Å.; Kusmierek, H.; Biernat, W.; Limon, J. Cancer predisposing BARD1 mutations in breast-ovarian cancer families. Breast Cancer Res. Treat. 2012, 131, 89–97. [Google Scholar] [CrossRef]
- De Brakeleer, S.; De Grève, J.; Loris, R.; Janin, N.; Lissens, W.; Sermijn, E.; Teugels, E. Cancer predisposing missense and protein truncating BARD1 mutations in non-BRCA1 or BRCA2 breast cancer families. Hum. Mutat. 2010, 31, E1175–E1185. [Google Scholar] [CrossRef] [PubMed]
- Weber-Lassalle, N.; Borde, J.; Weber-Lassalle, K.; Horváth, J.; Niederacher, D.; Arnold, N.; Kaulfuß, S.; Ernst, C.; Paul, V.G.; Honisch, E.; et al. Germline loss-of-function variants in the BARD1 gene are associated with early-onset familial breast cancer but not ovarian cancer. Breast Cancer Res. 2019, 21, 55. [Google Scholar] [CrossRef]
- Suszynska, M.; Kluzniak, W.; Wokolorczyk, D.; Jakubowska, A.; Huzarski, T.; Gronwald, J.; Debniak, T.; Szwiec, M.; Ratajska, M.; Klonowska, K.; et al. BARD1 is a low/moderate breast cancer risk gene: Evidence based on an association study of the Central European p.Q564X recurrent mutation. Cancers 2019, 11, 740. [Google Scholar] [CrossRef]
- Suszynska, M.; Kozlowski, P. Summary of BARD1 mutations and precise estimation of breast and ovarian cancer risks associated with the mutations. Genes 2020, 11, 798. [Google Scholar] [CrossRef]
- Atchley, D.P.; Albarracin, C.T.; Lopez, A.; Valero, V.; Amos, C.I.; Gonzalez-Angulo, A.M.; Hortobagyi, G.N.; Arun, B.K. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J. Clin. Oncol. 2008, 26, 4282–4288. [Google Scholar] [CrossRef]
- Couch, F.J.; Hart, S.N.; Sharma, P.; Toland, A.E.; Wang, X.; Miron, P.; Olson, J.E.; Godwin, A.K.; Pankratz, V.S.; Olswold, C.; et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J. Clin. Oncol. 2015, 33, 304–311. [Google Scholar] [CrossRef]
- Buys, S.S.; Sandbach, J.F.; Gammon, A.; Patel, G.; Kidd, J.; Brown, K.L.; Sharma, L.; Saam, J.; Lancaster, J.; Daly, M.B. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer 2017, 123, 1721–1730. [Google Scholar] [CrossRef]
- Sabatier, R.; Adélaïde, J.; Finetti, P.; Ferrari, A.; Huiart, L.; Sobol, H.; Chaffanet, M.; Birnbaum, D.; Bertucci, F. BARD1 homozygous deletion, a possible alternative to BRCA1 mutation in basal breast cancer. Genes Chromosomes Cancer 2010, 49, 1143–1151. [Google Scholar] [CrossRef]
- Rofes, P.; Del Valle, J.; Torres-Esquius, S.; Feliubadaló, L.; Stradella, A.; Moreno-Cabrera, J.M.; López-Doriga, A.; Munté, E.; De Cid, R.; Campos, O.; et al. BARD1 pathogenic variants are associated with triple-negative breast cancer in a Spanish hereditary breast and ovarian cancer cohort. Genes 2021, 12, 150. [Google Scholar] [CrossRef] [PubMed]
- Shimelis, H.; LaDuca, H.; Hu, C.; Hart, S.N.; Na, J.; Thomas, A.; Akinhanmi, M.; Moore, R.M.; Brauch, H.; Cox, A.; et al. Triple-negative breast cancer risk genes identified by multigene hereditary cancer panel testing. J. Natl. Cancer Inst. 2018, 110, 855–862. [Google Scholar] [CrossRef]
- Breast Cancer Association Consortium; Dorling, L.; Carvalho, S.; Allen, J.; González-Neira, A.; Luccarini, C.; Wahlström, C.; Pooley, K.A.; Parsons, M.T.; Fortuno, C.; et al. Breast cancer risk genes-association analysis in more than 113,000 Women. N. Engl. J. Med. 2021, 384, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Hart, S.N.; Gnanaolivu, R.; Huang, H.; Lee, K.Y.; Na, J.; Gao, C.; Lilyquist, J.; Yadav, S.; Boddicker, N.J.; et al. A population-based study of genes previously implicated in breast cancer. N. Engl. J. Med. 2021, 384, 440–451. [Google Scholar] [CrossRef]
- Toh, M.R.; Chong, S.T.; Chan, S.H.; Low, C.E.; Ishak, N.D.B.; Lim, J.Q.; Courtney, E.; Ngeow, J. Functional analysis of clinical BARD1 germline variants. Cold Spring Harb. Mol. Case Stud. 2019, 5, a004093. [Google Scholar] [CrossRef] [PubMed]
- Kwong, A.; Chen, J.; Shin, V.Y.; Ho, J.C.; Law, F.B.; Au, C.H.; Chan, T.L.; Ma, E.S.; Ford, J.M. The importance of analysis of long-range rearrangement of BRCA1 and BRCA2 in genetic diagnosis of familial breast cancer. Cancer Genet. 2015, 208, 448–454. [Google Scholar] [CrossRef]
- Adedokun, B.; Zheng, Y.; Ndom, P.; Gakwaya, A.; Makumbi, T.; Zhou, A.Y.; Yoshimatsu, T.F.; Rodriguez, A.; Madduri, R.K.; Foster, I.T.; et al. Prevalence of inherited mutations in breast cancer predisposition genes among women in Uganda and Cameroon. Cancer Epidemiol. Biomark. Prev. 2020, 29, 359–367. [Google Scholar] [CrossRef]
- Tung, N.; Battelli, C.; Allen, B.; Kaldate, R.; Bhatnagar, S.; Bowles, K.; Timms, K.; Garber, J.E.; Herold, C.; Ellisen, L.; et al. Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer 2015, 121, 25–33. [Google Scholar] [CrossRef]
- Carter, N.J.; Marshall, M.L.; Susswein, L.R.; Zorn, K.K.; Hiraki, S.; Arvai, K.J.; Torene, R.I.; McGill, A.K.; Yackowski, L.; Murphy, P.D.; et al. Germline pathogenic variants identified in women with ovarian tumors. Gynecol. Oncol. 2018, 151, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Carrera, S.; Rodríguez-Martínez, A.B.; Garin, I.; Sarasola, E.; Martínez, C.; Maortua, H.; Callejo, A.; Ruiz de Lobera, A.; Muñoz, A.; Miñambres, N.; et al. Germline heterozygous exons 8–11 pathogenic BARD1 gene deletion reported for the first time in a family with suspicion of a hereditary colorectal cancer syndrome: More than an incidental finding? Hered. Cancer Clin. Pract. 2023, 21, 2. [Google Scholar] [CrossRef] [PubMed]
- Stacey, S.N.; Sulem, P.; Johannsson, O.T.; Helgason, A.; Gudmundsson, J.; Kostic, J.P.; Kristjansson, K.; Jonsdottir, T.; Sigurdsson, H.; Hrafnkelsson, J.; et al. The BARD1 Cys557Ser variant and breast cancer risk in Iceland. PLoS Med. 2006, 3, e217. [Google Scholar] [CrossRef]
- Gonzalez-Hormazabal, P.; Reyes, J.M.; Blanco, R.; Bravo, T.; Carrera, I.; Peralta, O.; Gomez, F.; Waugh, E.; Margarit, S.; Ibañez, G.; et al. The BARD1 Cys557Ser variant and risk of familial breast cancer in a South-American population. Mol. Biol. Rep. 2012, 39, 8091–8098. [Google Scholar] [CrossRef]
- Apizi, A.; Wang, L.; Wusiman, L.; Song, E.; Han, Y.; Jia, T.; Zhang, W. Establishment and verification of a prognostic model of liver cancer by RNA-binding proteins based on the TCGA database. Transl. Cancer Res. 2022, 11, 1925–1937. [Google Scholar] [CrossRef]
- Stempa, K.; Wokołorczyk, D.; Kluźniak, W.; Rogoża-Janiszewska, E.; Malińska, K.; Rudnicka, H.; Huzarski, T.; Gronwald, J.; Gliniewicz, K.; Dębniak, T.; et al. Do BARD1 Mutations Confer an Elevated Risk of Prostate Cancer? Cancers 2021, 13, 5464. [Google Scholar] [CrossRef]
- Darst, B.F.; Saunders, E.; Dadaev, T.; Sheng, X.; Wan, P.; Pooler, L.; Xia, L.Y.; Chanock, S.; Berndt, S.I.; Wang, Y.; et al. Germline Sequencing Analysis to Inform Clinical Gene Panel Testing for Aggressive Prostate Cancer. JAMA Oncol. 2023, 9, 1514–1524. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).