Integrating Circulating Tumor DNA into Clinical Management of Colorectal Cancer: Practical Implications and Therapeutic Challenges
Simple Summary
Abstract
1. Introduction
2. Methodology
3. Evolution and Current Application of ctDNA in CRC
3.1. Historical Aspects of ctDNA
3.2. Role of ctDNA in Colorectal Cancer
3.3. Clinical Implications and Challenges of Liquid Biopsies in the Management of Colorectal Cancer
- Screening and Diagnostic EvaluationWhile liquid biopsies offer a minimally invasive alternative to traditional tissue biopsies, they generally exhibit lower sensitivity and precision. The detection sensitivity is often limited by the low levels of ctDNA, particularly in early stages of CRC. This can result in false negatives, where ctDNA is not detected despite the presence of residual disease. Dilution of ctDNA by the presence of DNA from healthy non-malignant cells can also lead to inaccurate detection of cancer-specific mutations, generating aberrant results. One strategy to address this limitation is single-cell analysis, which allows detection of CRC mutations using single-cell sequencing that might be missed by bulk sequencing methods. This enhances both the sensitivity and specificity of ctDNA assays, leading to more accurate detection of MRD and early recurrence [81,82]. Using tumor-informed methods to identify patient-specific mutations can also improve ctDNA detection sensitivity, facilitating more exact mutation identification. Standardizing plasma separation and storage protocols to minimize degradation of ctDNA before its analysis can prevent false-positive and false-negative results. Additionally, implementing advanced processing techniques, such as specialized blood collection tubes and double plasma centrifugation, can further improve assay reliability. However, these methodological changes must be carefully weighed against processing complexity and its associated cost for feasibility in routine clinical practice [83,84].
- Low Amount of Target BiomarkersctDNA is released into the bloodstream primarily through apoptosis, necrosis, or secretion from tumor cells. However, in early stages, the tumor burden is minimal, shedding only very small amounts of DNA into circulation. This poses a challenge for the detection of ctDNA in early cancer management [50]. Additionally, with a half-life of only 20–60 min, ctDNA is rapidly removed from circulation, limiting the detection ability even further. This subsequently necessitates highly sensitive analytical methods and techniques [85]. While methylation-based approaches have enhanced ctDNA detection rates, their sensitivity remains low to achieve acceptable detection when used in isolation [86]. To address this, an approach utilizing combination techniques may be required. The integration of genetic and epigenetic alterations, such as combining hotspot mutations with the Screening for the Presence of Tumor by Methylation and Size (SPOT-MAS) assay, developed by Nguyen et al., into a multimodal tool that has previously demonstrated significant improvement in the detection of target biomarkers [87].
- Tumor HeterogeneityTumors exhibit considerable genetic variations across different genetic regions, making it challenging to capture a comprehensive molecular profile of their heterogeneity. Studies indicate that not all tumor mutations are detectable in plasma due to limited shedding and tumor heterogeneity, which can significantly compromise the accuracy of liquid biopsy analyses [88,89]. This limitation affects treatment decisions, as tumors with variations in genetic markers may respond differently to targeted therapies. To overcome this, incorporating additional quality control measures, such as paired whole-blood analysis, can help distinguish tumor-derived mutations from clonal hematopoiesis of indeterminate potential (CHIP) and improve ctDNA assay results [90]. Furthermore, designing personalized ctDNA assays tailored to individual tumor mutation profiles can enhance sensitivity and specificity, allowing superior mutation detection. Hybrid-capture ctDNA sequencing utilizing customized target-enrichment panels has demonstrated high efficacy in detecting MRD, offering a more refined strategy to disease monitoring and treatment optimization [91].
- Lack of StandardizationThe lack of standardized protocols and clinically validated guidelines for ctDNA collection, processing, and analysis can lead to unpredictable results, complicating the implementation of ctDNA assays into clinical practice. Variability in methodologies contributes to inconsistencies and poor reliability of liquid biopsy results. Additionally, the lack of a universally accepted comprehensive ctDNA marker profile imposes an extra layer of complexity for the accurate identification of tumor mutations. Establishing standardized guidelines for variant classification, interpretation, and reporting is necessary to ascertain the clinical utility of ctDNA assays. Collaborative efforts between international societal groups are essential to harmonize the best practices. Differences in blood collection techniques, plasma processing, and DNA extraction methods can significantly impact ctDNA yield and quality, making cross-study comparisons difficult. The American Society of Clinical Oncology (ASCO) and the College of American Pathologists (CAP) have emphasized the need for uniform and reproducible quantification methods to ensure comparable results and interoperability across different laboratories [92,93]. Implementation of internal quality control (IQC) measures and participation in external quality assessment (EQA) programs can improve the reliability and clinical strength of ctDNA assays. Recent data shows that only 45.6% of laboratories engage in EQA programs, underscoring the urgent need for their broader adoption [94].
3.4. Current Progress and Applications in the Management of Colorectal Cancer
3.5. Commercially Available ctDNA Tests in CRC
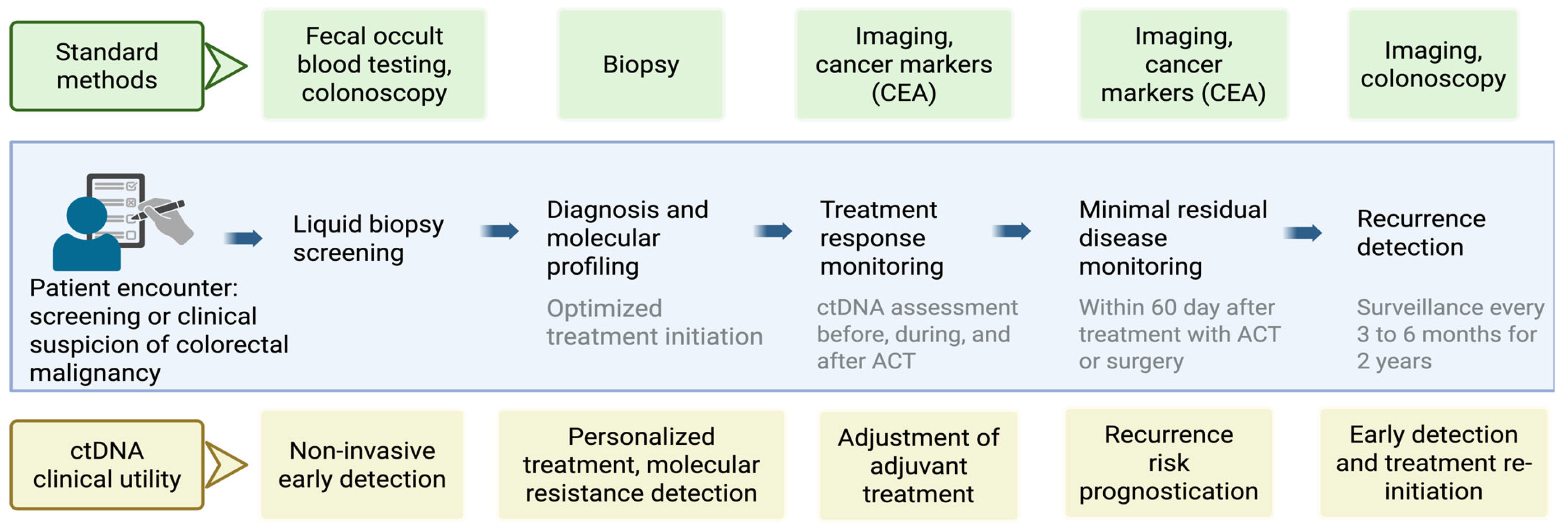
3.6. Current Schematic Strategy and Future Direction
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Saad El, D.K.; Loree, J.M.; Sayre, E.C.; Gill, S.; Brown, C.J.; Dau, H.; Vera De, M.A. Trends in the epidemiology of young-onset colorectal cancer: A worldwide systematic review. BMC Cancer 2020, 20, 288. [Google Scholar] [CrossRef]
- Cheng, L.; Eng, C.; Nieman, L.Z.; Kapadia, A.S.; Du, X.L. Trends in colorectal cancer incidence by anatomic site and disease stage in the United States from 1976 to 2005. Am. J. Clin. Oncol. 2011, 34, 573–580. [Google Scholar] [CrossRef]
- Jonker, D.J.; O’Callaghan, C.J.; Karapetis, C.S.; Zalcberg, J.R.; Tu, D.; Au, H.J.; Berry, R.S.; Krahn, M.; Price, T.; Simes, R.J.; et al. Cetuximab for the treatment of colorectal cancer. N. Engl. J. Med. 2007, 357, 2040–2048. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Peeters, M.; Siena, S.; Humblet, Y.; Hendlisz, A.; Neyns, B.; Canon, J.L.; Van Laetham, J.L.; Maurel, J.; Richardson, G.; et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J. Clin. Oncol. 2007, 25, 1658–1664. [Google Scholar] [CrossRef]
- Su, D.W.; Nieva, J. Biophysical technologies for understanding circulating tumor cell biology and metastasis. Transl. Lung Cancer Res. 2017, 6, 473. [Google Scholar] [CrossRef] [PubMed]
- Nassar, F.J.; Msheik, Z.S.; Nasr, R.R.; Temraz, S.N. Methylated circulating tumor DNA as a biomarker for colorectal cancer diagnosis, prognosis, and prediction. Clin. Epigenet. 2021, 13, 111. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wang, J.; Sun, Y. Circulating Tumor DNA as Biomarkers for Cancer Detection. Genom. Proteom. Bioinform. 2017, 15, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Maheswaran, S.; Sequist, L.V.; Nagrath, S.; Ulkus, L.; Brannigan, B.; Collura, C.V.; Inserra, E.; Diedrichs, S.; Lafrate, A.J.; Bell, D.W.; et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 2008, 359, 366–377. [Google Scholar] [CrossRef]
- Kamat, A.A.; Baldwin, M.; Urbauer, D.; Dang, D.; Han, L.Y.; Godwin, A.; Karlan, B.Y.; Simpson, J.L.; Gershenson, D.M.; Coleman, R.L.; et al. Plasma cell-free DNA in ovarian cancer: An independent prognostic biomarker. Cancer 2010, 116, 1918–1925. [Google Scholar] [CrossRef]
- Lianidou, E.S.; Mavroudis, D.; Sotiropoulou, G.; Agelaki, S.; Pantel, K. What’s new on circulating tumor cells? A meeting report. Breast Cancer Res. 2010, 12, 307. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Circulating tumour cells in cancer patients: Challenges and perspectives. Trends Mol. Med. 2010, 16, 398–406. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Arechederra, M.; Ávila, M.A.; Berasain, C. Liquid biopsy for cancer management: A revolutionary but still limited new tool for precision medicine. Adv. Lab. Med. 2020, 1, 20200009. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.D.; Xiong, W.; Bunker, A.M.; Vaughn, C.P.; Furtado, L.V.; Roberts, W.L.; Fang, J.C.; Samowitz, W.S.; Heichman, K.A. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011, 9, 133. [Google Scholar] [CrossRef] [PubMed]
- Scher, H.I.; Jia, X.; de Bono, J.S.; Fleisher, M.; Pienta, K.J.; Raghavan, D.; Heller, G. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: A reanalysis of IMMC38 trial data. Lancet Oncol. 2009, 10, 233–239. [Google Scholar] [CrossRef]
- Krebs, M.G.; Sloane, R.; Priest, L.; Lancashire, L.; Hou, J.M.; Greystoke, A.; Ward, T.H.; Ferradeschi, R.; Hughes, A.; Glenet, C.; et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. 2011, 29, 1556–1563. [Google Scholar] [CrossRef]
- Bidard, F.C.; Fehm, T.; Ignatiadis, M.; Smerage, J.B.; Alix-Panabières, C.; Janni, W.; Messina, C.; Paoletti, C.; Muller, V.; Hayes, D.F.; et al. Clinical application of circulating tumor cells in breast cancer: Overview of the current interventional trials. Cancer Metastasis Rev. 2013, 32, 179–188. [Google Scholar] [CrossRef]
- Kato, S.; Okamura, R.; Baumgartner, J.M.; Patel, H.; Leichman, L.; Kelly, K.; Sicklick, J.K.; Fanta, P.T.; Lippman, S.M.; Kurzrock, R. Analysis of Circulating Tumor DNA and Clinical Correlates in Patients with Esophageal, Gastroesophageal Junction, and Gastric Adenocarcinoma. Clin. Cancer Res. 2018, 24, 6248–6256. [Google Scholar] [CrossRef]
- Shatsky, R.; Parker, B.A.; Bui, N.Q.; Helsten, T.; Schwab, R.B.; Boles, S.G.; Kurzrock, R. Next-Generation Sequencing of Tissue and Circulating Tumor DNA: The UC San Diego Moores Center for Personalized Cancer Therapy Experience with Breast Malignancies. Mol. Cancer Ther. 2019, 18, 1001–1011. [Google Scholar] [CrossRef]
- Pachmann, K.; Camara, O.; Kroll, T.; Gajda, M.; Gellner, A.K.; Wotschadlo, J.; Runnebaum, I.B. Efficacy control of therapy using circulating epithelial tumor cells (CETC) as “liquid biopsy”: Trastuzumab in HER2/neu-positive breast carcinoma. J. Cancer Res. Clin. Oncol. 2011, 137, 1317–1327. [Google Scholar] [CrossRef]
- Racila, E.; Euhus, D.; Weiss, A.J.; Rao, C.; McConnell, J.; Terstappen, L.W.; Uhr, J.W. Detection and characterization of carcinoma cells in the blood. Proc. Natl. Acad. Sci. USA 1998, 95, 4589–4594. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy and minimal residual disease—Latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Best, M.G.; Sol, N.; In ‘t Veld, S.; Vancura, A.; Muller, M.; Niemeijer, A.N.; Fejes, A.V.; Tjon Kon Fat, L.A.; In ’t Veld, A.H.; Leurs, C.; et al. Swarm Intelligence-Enhanced Detection of Non-Small-Cell Lung Cancer Using Tumor-Educated Platelets. Cancer Cell 2017, 32, 238–252.e9. [Google Scholar] [CrossRef] [PubMed]
- Gahlawat, A.W.; Witte, T.; Sinn, P.; Schott, S. Circulating cf-miRNA as a more appropriate surrogate liquid biopsy marker than cfDNA for ovarian cancer. Sci. Rep. 2023, 13, 5503. [Google Scholar] [CrossRef]
- Wang, S.; Lin, Y.; Zhang, Y.; Qiu, X.; Pan, Y.; Yeung, S.-C.J.; Zhang, H. Complex RNA world in small extracellular vesicles for liquid biopsy in cancer management. Extracell. Vesicle 2022, 1, 100015. [Google Scholar] [CrossRef]
- Joosse, S.A.; Pantel, K. Tumor-Educated Platelets as Liquid Biopsy in Cancer Patients. Cancer Cell 2015, 28, 552–554. [Google Scholar] [CrossRef]
- Dell’Olio, F.; Su, J.; Huser, T.; Sottile, V.; Cortés-Hernández, L.E.; Alix-Panabières, C. Photonic technologies for liquid biopsies: Recent advances and open research challenges. Laser Photonics Rev. 2021, 15, 2000255. [Google Scholar] [CrossRef]
- Neumann, M.H.D.; Bender, S.; Krahn, T.; Schlange, T. ctDNA and CTCs in Liquid Biopsy—Current Status and Where We Need to Progress. Comput. Struct. Biotechnol. J. 2018, 16, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Santonja, A.; Cooper, W.N.; Eldridge, M.D.; Edwards, P.A.; Morris, J.A.; Edwards, A.R.; Zhao, H.; Heider, K.; Couturier, D.L.; Vijayaraghavan, A.; et al. Comparison of tumor-informed and tumor-naïve sequencing assays for ctDNA detection in breast cancer. EMBO Mol. Med. 2023, 15, e16505. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.T.; Nagayama, S.; Otaki, M.; Chin, Y.M.; Fukunaga, Y.; Ueno, M.; Nakamura, Y.; Low, S.K. Tumor-informed or tumor-agnostic circulating tumor DNA as a biomarker for risk of recurrence in resected colorectal cancer patients. Front. Oncol. 2023, 12, 1055968. [Google Scholar] [CrossRef]
- Luo, H.; Zhao, Q.; Wei, W.; Zheng, L.; Yi, S.; Li, G.; Wang, W.; Sheng, H.; Pu, H.; Mo, H.; et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci. Transl. Med. 2020, 12, eaax7533. [Google Scholar] [CrossRef]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M. Response to W.C. Taylor, and C. Fiala and E.P. Diamandis. Ann. Oncol. 2020, 31, 1268–1270. [Google Scholar] [CrossRef]
- Chiu, R.W.K.; Heitzer, E.; Lo, Y.M.D.; Mouliere, F.; Tsui, D.W.Y. Cell-Free DNA Fragmentomics: The New “Omics” on the Block. Clin. Chem. 2020, 66, 1480–1484. [Google Scholar] [CrossRef]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef]
- Xu, J.; Wu, W.; Wu, C.; Mao, Y.; Qi, X.; Lu, R.; Xie, S.; Lou, J.; Zhang, Y.; Ding, Y.; et al. A large-scale, multicentered trial evaluating the sensitivity and specificity of digital PCR versus ARMS-PCR for detecting ctDNA-based EGFR p.T790M in non-small-cell lung cancer patients. Transl. Lung Cancer Res. 2021, 10, 3888–3901. [Google Scholar] [CrossRef]
- Sebastião, M.M.; Ho, R.S.; de Carvalho, J.P.V.; Nussbaum, M. Diagnostic Accuracy of Next Generation Sequencing Panel using Circulating Tumor DNA in Patients with Advanced Non-Small Cell Lung Cancer: A Systematic Review and Meta-Analysis. J. Health Econ. Outcomes Res. 2020, 7, 158–163. [Google Scholar] [CrossRef]
- Chen, C.; Douglas, M.P.; Ragavan, M.V.; Phillips, K.A.; Jansen, J.P. Clinical Validity and Utility of Circulating Tumor DNA (ctDNA) Testing in Advanced Non-small Cell Lung Cancer (aNSCLC): A Systematic Literature Review and Meta-analysis. Mol. Diagn. Ther. 2024, 28, 525–536. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef]
- Malkawi, W.; Lutfi, A.; Afghan, M.K.; Shah, L.M.; Costandy, L.; Ramirez, A.B.; George, T.C.; Toor, F.; Salem, A.K.; Kasi, P.M. Circulating tumour cell enumeration, biomarker analyses, and kinetics in patients with colorectal cancer and other GI malignancies. Front. Oncol. 2023, 13, 1305181. [Google Scholar] [CrossRef] [PubMed]
- Molparia, B.; Oliveira, G.; Wagner, J.L.; Spencer, E.G.; Torkamani, A. A feasibility study of colorectal cancer diagnosis via circulating tumor DNA derived CNV detection. PLoS ONE 2018, 13, e0196826. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating tumor DNA as a liquid biopsy for cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Buscail, E.; Alix-Panabières, C.; Quincy, P.; Cauvin, T.; Chauvet, A.; Degrandi, O.; Camount, C.; Verdon, S.; Lamrissi, I.; Moranviller, I.; et al. High Clinical Value of Liquid Biopsy to Detect Circulating Tumor Cells and Tumor Exosomes in Pancreatic Ductal Adenocarcinoma Patients Eligible for Up-Front Surgery. Cancers 2019, 11, 1656. [Google Scholar] [CrossRef]
- Buscail, E.; Maulat, C.; Muscari, F.; Chiche, L.; Cordelier, P.; Dabernat, S.; Panabieres, C.A.; Buscail, L. Liquid Biopsy Approach for Pancreatic Ductal Adenocarcinoma. Cancers 2019, 11, 852. [Google Scholar] [CrossRef]
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.L.; Christie, M.; et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016, 8, 346ra92. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Barlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Diaz, L.A., Jr.; Williams, R.T.; Wu, J.; Kinde, I.; Hecht, J.R.; Berlin, J.; Allen, B.; Bozic, I.; Reiter, J.G.; Nowak, M.A.; et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012, 486, 537–540. [Google Scholar] [CrossRef]
- Liu, W.; Li, Y.; Tang, Y.; Song, Q.; Wang, J.; Li, N.; Chen, S.; Shi, J.; Wang, S.; Li, Y.; et al. Response prediction and risk stratification of patients with rectal cancer after neoadjuvant therapy through an analysis of circulating tumour DNA. EBioMedicine 2022, 78, 103945. [Google Scholar] [CrossRef]
- Parikh, A.R.; Chee, B.H.; Tsai, J.; Rich, T.A.; Price, K.S.; Patel, S.A.; Zhang, L.; Ibrahim, F.; Esquivel, M.; Van Seventer, E.E.; et al. Minimal residual disease using a plasma-only circulating tumor DNA assay to predict recurrence of metastatic colorectal cancer following curative intent treatment. Clin. Cancer Res. 2024, 30, 2964–2973. [Google Scholar] [CrossRef] [PubMed]
- Slater, S.; Bryant, A.; Aresu, M.; Begum, R.; Chen, H.-C.; Peckitt, C.; Alcausi, R.L.; Carter, P.; Anandappa, G.; Khakoo, S.; et al. Tissue-free liquid biopsies combining genomic and methylation signals for minimal residual disease detection in patients with early colorectal cancer from the UK TRACC part B study. Clin. Cancer Res. 2024, 30, 3459–3469. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Chen, J.; Yu, M.; Liu, D. Using circulating tumor DNA as a novel biomarker to screen and diagnose colorectal cancer: A meta-analysis. J. Clin. Med. 2023, 12, 408. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Watanabe, J.; Akazawa, N.; Hirata, K.; Kataoka, K.; Yokota, M.; Kato, K.; Kotaka, M.; Kagawa, Y.; Yeh, K.H.; et al. ctDNA-based molecular residual disease and survival in resectable colorectal cancer. Nat. Med. 2024, 30, 3272–3283. [Google Scholar] [CrossRef]
- Henriksen, T.; Demuth, C.; Frydendahl, A.; Nors, J.; Nesic, M.; Rasmussen, M.; Reinert, T.; Lansen, O.H.; Jaensch, C.; Leve, U.S.; et al. Unraveling the potential clinical utility of circulating tumor DNA detection in colorectal cancer—Evaluation in a nationwide Danish cohort. Ann. Oncol. 2024, 35, 229–239. [Google Scholar] [CrossRef]
- Nassar, A.; Aly, N.E.; Jin, Z.; Aly, E.H. ctDNA as a predictor of outcome after curative resection for locally advanced rectal cancer: Systematic review and meta-analysis. Color. Dis. 2024, 26, 1346–1358. [Google Scholar] [CrossRef]
- Reinert, T.; Henriksen, T.V.; Christensen, E.; Sharma, S.; Salari, R.; Sethi, H.; Knudsen, M.; Nordentoft, I.; Wu, H.T.; Tin, A.S.; et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients with Stages I to III Colorectal Cancer. JAMA Oncol. 2019, 5, 1124–1131. [Google Scholar] [CrossRef]
- Tarazona, N.; Gimeno-Valiente, F.; Gambardella, V.; Zuñiga, S.; Rentero-Garrido, P.; Huerta, M.; Rosello, S.; Ciarpaglini, C.M.; Asins, J.A.C.; Carrasco, F.; et al. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann. Oncol. 2019, 30, 1804–1812. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Wang, Y.; Christie, M.; Simons, K.; Lee, M.; Wong, R.; Kosmider, S.; Ananda, S.; McKendrick, J.; et al. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol. 2019, 5, 1710–1717. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Lo, S.N.; Wang, Y.; Li, L.; Christie, M.; Lee, M.; Wong, R.; Kosminder, S.; Skinner, L.; et al. Prognostic significance of postsurgery circulating tumor DNA in nonmetastatic colorectal cancer: Individual patient pooled analysis of three cohort studies. Int. J. Cancer 2021, 148, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, C.; Lin, G.; Xiao, Y.; Jia, W.; Xiao, G.; Liu, Q.; Wu, B.; Wu, A.; Qiu, H.; et al. Serial Circulating Tumor DNA in Predicting and Monitoring the Effect of Neoadjuvant Chemoradiotherapy in Patients with Rectal Cancer: A Prospective Multicenter Study. Clin. Cancer Res. 2021, 27, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Henriksen, T.V.; Tarazona, N.; Frydendahl, A.; Reinert, T.; Gimeno-Valiente, F.; Carbonell-Asins, J.A.; Sharma, S.; Renner, D.; Hafez, D.; Roda, D.; et al. Circulating Tumor DNA in Stage III Colorectal Cancer, beyond Minimal Residual Disease Detection, toward Assessment of Adjuvant Therapy Efficacy and Clinical Behavior of Recurrences. Clin. Cancer Res. 2022, 28, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Taieb, J.; Taly, V.; Henriques, J.; Bourreau, C.; Mineur, L.; Bennouna, J.; Desrame, J.; Louvet, C.; Lepere, C.; Mabro, M.; et al. Prognostic Value and Relation with Adjuvant Treatment Duration of ctDNA in Stage III Colon Cancer: A Post Hoc Analysis of the PRODIGE-GERCOR IDEA-France Trial. Clin. Cancer Res. 2021, 27, 5638–5646. [Google Scholar] [CrossRef]
- Parikh, A.R.; Van Seventer, E.E.; Siravegna, G.; Hartwig, A.V.; Jaimovich, A.; He, Y.; Kanter, K.; Fish, M.G.; Fosbenner, K.D.; Miao, B.; et al. Minimal Residual Disease Detection using a Plasma-only Circulating Tumor DNA Assay in Patients with Colorectal Cancer. Clin. Cancer Res. 2021, 27, 5586–5594. [Google Scholar] [CrossRef]
- Li, Y.; Mo, S.; Zhang, L.; Ma, X.; Hu, X.; Huang, D.; Lu, B.; Luo, C.; Peng, H.; Cai, S.; et al. Postoperative circulating tumor DNA combined with consensus molecular subtypes can better predict outcomes in stage III colon cancers: A prospective cohort study. Eur. J. Cancer 2022, 169, 198–209. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Wang, Y.; Christie, M.; Simons, K.; Lee, M.; Wong, R.; Kosminder, S.; Ananda, S.; McKendrick, J.; et al. Circulating tumor DNA analyses as markers of recurrence risk and adjuvant benefit in stage II colon cancer. N. Engl. J. Med. 2022, 386, 226–236. [Google Scholar] [CrossRef]
- Kotani, D.; Oki, E.; Nakamura, Y.; Yukami, H.; Mishima, S.; Bando, H.; Shirasu, H.; Yamazaki, K.; Watanabe, J.; Kotaka, M.; et al. Molecular residual disease and efficacy of adjuvant chemotherapy in patients with colorectal cancer. Nat. Med. 2023, 29, 127–134. [Google Scholar] [CrossRef]
- Lonardi, S.; Pietrantonio, F.; Tarazona Llavero, N.; Montagut Viladot, C.; Sartore Bianchi, A.; Zampino, M.G.; Elex Fernandez, M.E.; Santos Vivas, C.; Mandala, M.; Tamberi, S.; et al. LBA28 The PEGASUS trial: Post-surgical liquid biopsy-guided treatment of stage III and high-risk stage II colon cancer patients. Ann. Oncol. 2023, 34, S1268–S1269. [Google Scholar] [CrossRef]
- Dhiman, A.; Kothary, V.; Witmer, H.D.D.; Bregio, C.; Sood, D.; Ong, C.T.; Blasé, P.; Eng, O.S.; Shergill, A.; Turaga, K.K.; et al. Role of Tumor-informed Personalized Circulating Tumor DNA Assay in Informing Recurrence in Patients with Peritoneal Metastases From Colorectal and High-grade Appendix Cancer Undergoing Curative-intent Surgery. Ann. Surg. 2023, 278, 925–931. [Google Scholar] [CrossRef]
- Lygre, K.B.; Forthun, R.B.; Høysæter, T.; Hjelle, S.M.; Eide, G.E.; Gjertsen, B.T.; Pfeffer, F.; Hovland, R. Assessment of postoperative circulating tumour DNA to predict early recurrence in patients with stage I-III right-sided colon cancer: Prospective observational study. BJS Open 2024, 8, zrad146. [Google Scholar] [CrossRef] [PubMed]
- Morris, V.K.; Yothers, G.; Kopetz, S.; Puhalla, S.L.; Lucas, P.C.; Iqbal, A.; Boland, P.M.; Deming, D.A.; Scott, A.J.; Lim, H.J.; et al. Phase II results of circulating tumor DNA as a predictive biomarker in adjuvant chemotherapy in patients with stage II colon cancer: NRG-GI005 (COBRA) phase II/III study. J. Clin. Oncol. 2024, 42 (Suppl. S3), 5. [Google Scholar] [CrossRef]
- Kasi, P.M.; Aushev, V.N.; Ensor, J.; Langer, N.; Wang, C.G.; Cannon, T.L.; Berim, L.D.; Feinstein, T.; Grothey, A.; McCollom, J.W.; et al. Circulating tumor DNA (ctDNA) for informing adjuvant chemotherapy (ACT) in stage II/III colorectal cancer (CRC): Interim analysis of BESPOKE CRC study. J. Clin. Oncol. 2024, 42 (Suppl. S3), 9. [Google Scholar] [CrossRef]
- Yukami, H.; Nakamura, Y.; Mishima, S.; Ando, K.; Bando, H.; Watanabe, J.; Hirata, K.; Akazawa, N.; Ikeda, M.; Yokota, M.; et al. Circulating tumor DNA (ctDNA) dynamics in patients with colorectal cancer (CRC) with molecular residual disease: Updated analysis from GALAXY study in the CIRCULATE-JAPAN. J. Clin. Oncol. 2024, 42 (Suppl. S3), 6. [Google Scholar] [CrossRef]
- Jones, R.P.; Pugh, S.A.; Graham, J.; Primrose, J.N.; Barriuso, J. Circulating tumour DNA as a biomarker in resectable and irresectable stage IV colorectal cancer; a systematic review and meta-analysis. Eur. J. Cancer 2021, 144, 368–381. [Google Scholar] [CrossRef]
- Callesen, L.B.; Hamfjord, J.; Boysen, A.K.; Pallisgaard, N.; Guren, T.K.; Kure, E.H.; Spindler, H.L.G. Circulating tumour DNA and its clinical utility in predicting treatment response or survival in patients with metastatic colorectal cancer: A systematic review and meta-analysis. Br. J. Cancer 2022, 127, 500–513. [Google Scholar] [CrossRef]
- Faulkner, L.G.; Howells, L.M.; Pepper, C.; Shaw, J.A.; Thomas, A.L. The utility of ctDNA in detecting minimal residual disease following curative surgery in colorectal cancer: A systematic review and meta-analysis. Br. J. Cancer 2023, 128, 297–309. [Google Scholar] [CrossRef]
- Do, Q.; Katiyar, V.; Malloy, M.; Sharma, V.R. Circulating tumor DNA detection as a prognostic tool in patients with localized colon cancer: A systematic review and meta-analysis. J. Clin. Oncol. 2023, 41 (Suppl. S16), e15629. [Google Scholar] [CrossRef]
- Chang, L.; Zhang, X.; He, L.; Ma, Q.; Fang, T.; Jiang, C.; Ma, Z.; Li, Q.; Wu, C. Prognostic Value of ctDNA Detection in Patients with Locally Advanced Rectal Cancer Undergoing Neoadjuvant Chemoradiotherapy: A Systematic Review and Meta-analysis. Oncologist 2023, 28, e1198–e1208. [Google Scholar] [CrossRef]
- Kojima, M.; Harada, T.; Fukazawa, T.; Kurihara, S.; Touge, R.; Saeki, I.; Takahashi, S.; Hiyama, E. Single-cell next-generation sequencing of circulating tumor cells in patients with neuroblastoma. Cancer Sci. 2023, 114, 1616–1624. [Google Scholar] [CrossRef]
- Nagasawa, S.; Kashima, Y.; Suzuki, A.; Suzuki, Y. Single-cell and spatial analyses of cancer cells: Toward elucidating the molecular mechanisms of clonal evolution and drug resistance acquisition. Inflamm. Regen. 2021, 41, 22. [Google Scholar] [CrossRef]
- Sherwood, J.L.; Corcoran, C.; Brown, H.; Sharpe, A.D.; Musilova, M.; Kohlmann, A. Optimised pre-analytical methods improve KRAS mutation detection in circulating tumour DNA (ctDNA) from patients with non-small cell lung cancer (NSCLC). PLoS ONE 2016, 11, e0150197. [Google Scholar] [CrossRef] [PubMed]
- Medina Diaz, I.; Nocon, A.; Mehnert, D.H.; Fredebohm, J.; Diehl, F.; Holtrup, F. Performance of Streck cfDNA blood collection tubes for liquid biopsy testing. PLoS ONE 2016, 11, e0166354. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoli, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Toth, K.; Sipos, F.; Kalmar, A.; Patai, A.V.; Wichmann, B.; Stoehr, R.; Golcher, H.; Scheller, V.; Tulassay, Z.; Molnar, B. Detection of methylated SEPT9 in plasma is a reliable screening method for both left-and right-sided colon cancers. PLoS ONE 2012, 7, e46000. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.H.; Vu, G.H.; Nguyen, T.T.; Nguyen, T.A.; Tran, V.U.; Vu, L.T.; Huong Nguyen, G.T.; Duy Nguyen, N.; Tran, T.H.; Chi Nguyen, V.T.; et al. Combination of Hotspot Mutations with Methylation and Fragmentomic Profiles to Enhance Multi-Cancer Early Detection. Cancer Med. 2025, 14, e70575. [Google Scholar] [CrossRef]
- De Mattos-Arruda, L.; Weigelt, B.; Cortes, J.; Won, H.; Ng, C.; Nuciforo, P.; Bidard, F.C.; Aura, C.; Saura, C.; Peg, V.; et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: A proof-of-principle. Ann. Oncol. 2014, 25, 1729–1735. [Google Scholar] [CrossRef]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–992. [Google Scholar] [CrossRef]
- Perdigones, N.; Murtaza, M. Capturing tumor heterogeneity and clonal evolution in solid cancers using circulating tumor DNA analysis. Pharmacol. Ther. 2017, 174, 22–26. [Google Scholar] [CrossRef]
- Ryoo, S.-B.; Heo, S.; Lim, Y.; Lee, W.; Cho, S.H.; Ahn, J.; Kang, J.K.; Kim, S.Y.; Kim, H.P.; Bang, D.; et al. Personalised circulating tumour DNA assay with large-scale mutation coverage for sensitive minimal residual disease detection in colorectal cancer. Br. J. Cancer 2023, 129, 374–381. [Google Scholar] [CrossRef]
- Allan, Z.; Liu, D.S.; Lee, M.M.; Tie, J.; Clemons, N.J. A practical approach to interpreting circulating tumor DNA in the management of gastrointestinal cancers. Clin. Chem. 2024, 70, 49–59. [Google Scholar] [CrossRef]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. Arch. Pathol. Lab. Med. 2018, 142, 1242–1253. [Google Scholar] [CrossRef] [PubMed]
- Linder, M.W.; Huggett, J.F.; Baluchova, K.; Capoluongo, E.D.; Payne, D.A.; Salinas, A.V.; Haselmann, V.; Ashavaid, T.; Pan, S.; Ahmad-Nejad, P.; et al. Results from an IFCC global survey on laboratory practices for the analysis of circulating tumor DNA. Clin. Chim. Acta 2023, 547, 117398. [Google Scholar] [CrossRef] [PubMed]
- Malla, M.; Loree, J.M.; Kasi, P.M.; Parikh, A.R. Using Circulating Tumor DNA in Colorectal Cancer: Current and Evolving Practices. J. Clin. Oncol. 2022, 40, 2846–2857. [Google Scholar] [CrossRef] [PubMed]
- Boysen, A.K.; Pallisgaard, N.; Andersen, C.S.A.; Spindler, K.G. Circulating tumor DNA as a marker of minimal residual disease following local treatment of metastases from colorectal cancer. Acta Oncol. 2020, 59, 1424–1429. [Google Scholar] [CrossRef]
- Lecomte, T.; Berger, A.; Zinzindohoué, F.; Micard, S.; Landi, B.; Blons, H.; Beaune, P.; Cugenc, P.H.; Puig, P.L. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int. J. Cancer. 2002, 100, 542–548. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Verheul, H.M.; Flamen, P.; Rougier, P.; Beets-Tan, R.; Glynne-Jones, R.; Seufferlein, T. Imaging in colorectal cancer: Progress and challenges for the clinicians. Cancers 2016, 8, 81. [Google Scholar] [CrossRef]
- Schøler, L.V.; Reinert, T.; Ørntoft, M.W.; Kassentoft, C.G.; Árnadóttir, S.S.; Vang, S.; Nordentoft, I.; Kundsen, M.; Lamy, P.; Andreasen, D.; et al. Clinical Implications of Monitoring Circulating Tumor DNA in Patients with Colorectal Cancer. Clin. Cancer Res. 2017, 23, 5437–5445. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Lahouel, K.; Lo, S.N.; Wang, Y.; Kosmider, S.; Wong, R.; Shapiro, J.; Lee, M.; Harris, S.; et al. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N. Engl. J. Med. 2022, 386, 2261–2272. [Google Scholar] [CrossRef]
- Khan, K.; Rata, M.; Cunningham, D.; Koh, D.M.; Tunariu, N.; Hahne, J.C.; Vlachogiannis, G.; Hedayat, S.; Marchetti, S.; Lampis, A.; et al. Functional imaging and circulating biomarkers of response to regorafenib in treatment-refractory metastatic colorectal cancer patients in a prospective phase II study. Gut 2018, 67, 1484–1492. [Google Scholar] [CrossRef]
- Pappas, L.; Yaeger, R.; Kasi, P.M.; Shah, M.A.; Azad, N.S.; Horick, N.K.; Corcoran, R.B.; Diaz, L.A.; Parikh, A.R. Early identification and treatment of occult metastatic disease in stage III colon cancer (SU2C ACT3 clinical trial). Am. Soc. Clin. Oncol. 2024, 42 (Suppl. S3), 148. [Google Scholar] [CrossRef]
- Taieb, J.; Souglakos, J.; Boukovinas, I.; Falcoz, A.; Pages, F.; Messaritakis, I.; Bennouna, J.; Artru, P.; Louvet, C.; Lepere, C.; et al. Combined Analyses of Circulating Tumor DNA and Immunoscore in Patients with Stage III Colon Cancer: A Post Hoc Analysis of the PRODIGE-GERCOR IDEA-France/HORG-IDEA-Greece Trials. J. Clin. Oncol. 2025, 43, 1564–1577. [Google Scholar] [CrossRef]
- Siri, G.; Alesaeidi, S.; Dizghandi, S.E.; Alani, B.; Mosallaei, M.; Soosanabadi, M. Analysis of SDC2 gene promoter methylation in whole blood for noninvasive early detection of colorectal cancer. J. Cancer Res. Ther. 2022, 18 (Suppl. S2), S354–S358. [Google Scholar] [CrossRef] [PubMed]
- Sidaway, P. Diagnosis using methylation. Nat. Rev. Clin. Oncol. 2020, 17, 196. [Google Scholar] [CrossRef] [PubMed]
- Shweikeh, F.; Zeng, Y.; Jabir, A.R.; Whittenberger, E.; Kadatane, S.P.; Huang, Y.; Mouchli, M.; Castillo, D.R. The emerging role of blood-based biomarkers in early detection of colorectal cancer: A systematic review. Cancer Treat. Res. Commun. 2024, 42, 100872. [Google Scholar] [CrossRef] [PubMed]
- Wade, R.; Nevitt, S.; Liu, Y.; Harden, M.; Khouja, C.; Raine, G.; Churchill, R.; Dias, S. Multi-cancer early detection tests for general population screening: A systematic literature review. Health Technol. Assess. 2025, 29, 1–105. [Google Scholar] [CrossRef]
- Schraa, S.; Van Rooijen, K.; Van Der Kruijssen, D.; Rubio Alarcón, C.; Phallen, J.; Sausen, M.; Simmons, J.; Coupe, V.M.H.; Van Grevenstein, W.M.U.; Elias, S.; et al. Circulating tumor DNA guided adjuvant chemotherapy in stage II colon cancer (MEDOCC-CrEATE): Study protocol for a trial within a cohort study. BMC Cancer 2020, 20, 790. [Google Scholar] [CrossRef]
- Taniguchi, H.; Nakamura, Y.; Kotani, D.; Yukami, H.; Mishima, S.; Sawada, K.; Shirasu, H.; Ebi, H.; Yamanaka, T.; Aleshin, A.; et al. CIRCULATE-Japan: Circulating tumor DNA–guided adaptive platform trials to refine adjuvant therapy for colorectal cancer. Cancer Sci. 2021, 112, 2915–2920. [Google Scholar] [CrossRef]
- Hashimoto, T.; Tsukada, Y.; Ito, M.; Kanato, K.; Mizusawa, J.; Fukuda, H.; Tsukamoto, S.; Takashima, A.; Kanemitsu, Y. Utility of circulating tumour DNA for prognosis and prediction of therapeutic effect in locally recurrent rectal cancer: Study protocol for a multi-institutional, prospective observational study (JCOG1801A1, CAP-LR study). BMJ Open 2023, 13, e073217. [Google Scholar] [CrossRef]
- Campani, C.; Imbeaud, S.; Couchy, G.; Ziol, M.; Hirsch, T.Z.; Rebouissou, S.; Noblet, B.; Nahon, P.; Hormigos, K.; Sidali, S.; et al. Circulating tumour DNA in patients with hepatocellular carcinoma across tumour stages and treatments. Gut 2024, 73, 1870–1982. [Google Scholar] [CrossRef]
- Sato, S.; Nakamura, Y.; Oki, E.; Yoshino, T. Molecular Residual Disease-guided Adjuvant Treatment in Resected Colorectal Cancer: Focus on CIRCULATE-Japan. Clin. Color. Cancer 2023, 22, 53–58. [Google Scholar] [CrossRef]
- Guardant Health. Guardant Reveal Product Sheet. Available online: https://www.guardantcomplete.com/hcp/solutions/guardant-reveal (accessed on 27 July 2025).
- Foundation Medicine. FoundationOne Tracker Technical Overview. Available online: https://www.foundationmedicine.com/test/foundationone-liquid-cdx?utm_source=google&utm_medium=cpc&utm_campaign=liquid-cdx&utm_content=general-ub&utm_term=ctdna%20t (accessed on 27 July 2025).
- Kinde, I.; Wu, J.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 9530–9535. [Google Scholar] [CrossRef]
- Sullivan, R.; Alatise, O.I.; Anderson, B.O.; Audisio, R.; Autier, P.; Aggarwal, A.; Balch, C.; Brennan, M.F.; Dare, A.; Cruz, A.D.; et al. Global cancer surgery: Delivering safe, affordable, and timely cancer surgery. Lancet Oncol. 2015, 16, 1193–1224. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef]
- Tan, D.S.; Mok, T.S.; Rebbeck, T.R. Cancer Genomics: Diversity and Disparity Across Ethnicity and Geography. J. Clin. Oncol. 2016, 34, 91–101. [Google Scholar] [CrossRef]



| Author | Year | Design | Timing of ctDNA Assessment | Primary Endpoint | Findings |
|---|---|---|---|---|---|
| Reinert et al. [59] | 2019 | Multicenter prospective study | Before surgery, 30 days after, and every third month for 3 years | Recurrence rates | 70% recurrence in ctDNA-positive patients (95% CI, 34.2–93.1%); recurrence in the ctDNA negative group was 11.9% (95% CI, 6.3–20.1%) |
| Tarazona et al. [60] | 2019 | Prospective cohort study | 6–8 weeks after surgery, and every 4 months for up to 5 years | DFS | 57.1% recurrence in the ctDNA-positive after surgery and 85% after ACT |
| Tie et al. [61] | 2019 | Multicenter cohort study | 4–10 weeks after surgery and after ACT | 3-year RFI | ctDNA-positive patients had RFI of 30% (95% CI, 9–55%) at 3 years, and ctDNA-negative patients had 77% (95% CI, 60–87%) |
| Tie et al. [62] | 2020 | Multicenter cohort study | 4–6, 6–8, and 8–10 weeks after surgery | RFS | RFS was inferior for the post-surgery ctDNA-positive group at 5 years (38.6% vs. 85.5%; HR, 7.56; 95% CI, 4.85–11.79%) |
| Zhou et al. [63] | 2021 | Multicenter prospective study | Before nCRT, one cycle after nCT, 7 weeks after nCRT, before surgery, and within 1 month after | Metastasis-free survival | ctDNA is an independent predictor of MFS (HR, 1.267; p < 0.001) |
| Henriksen et al. [64] | 2021 | Prospective cohort study | 2–4 weeks after surgery, before initiation of ACT | RFS | 18% recurrence rate in ctDNA negative group, 80% in ctDNA positive group |
| Taieb et al. [65] | 2021 | Randomized controlled trial | 6–8 weeks after surgery | DFS and OS | 3-year DFS rate 66.39% in ctDNA positive and 76.71% for ctDNA negative group (p = 0.015) |
| Parikh et al. [66] | 2021 | Prospective study | Before surgery, 4 weeks after surgery, 4 weeks after ACT | Detection of ctDNA and RFS | 100% recurrence in the CtDNA-positive group and 24.5% recurrence in the CtDNA-negative group |
| Liu et al. [52] | 2022 | Multicenter randomized trial | During and after NAT and before TME | RFS at 3 years | ctDNA predictor of recurrence in high risk (HR = 21.27; 95% CI, 5.15–87.92%); or low risk (HR = 16.39; 95% CI, 1.46–184.30%) |
| Li et al. [67] | 2022 | Prospective cohort study | 1 week before ACT and 2–4 weeks after ACT | RFS | 3-year RFS in the ctDNA-positive group was 45.5% and in the ctDNA negative group was 72.7%; 24.8% recurrence ctDNA negative group and 54.5% recurrence in the ctDNA positive patients after ACT |
| Tie et al. [68] | 2022 | Multicenter randomized controlled trial | 4 or 7 weeks after surgery | RFS at 2 years | 2-year recurrence-free survival was 93.2% in the ctDNA-guided and 92.4% in standard management |
| Kotani et al. [69] | 2023 | Multicenter prospective study | 4–12 weeks after surgery | DFS | 9.5% recurrence in the ctDNA negative group and 61.4% in the ctDNA positive patients |
| Leonardi et al. [70] | 2023 | Randomized controlled trial | After surgery and after ACT | Post-surgery false negative cases | 34% relapse in the CTDNA-positive group, 9% relapse in the CTDNA-negative group |
| Dhiman et al. [71] | 2023 | Prospective cohort study | 4–6 weeks after surgery, 4–6 weeks, and every 3 months for 1 year | RFS | 90% recurrence in the rising ctDNA levels group vs. 21% in the stable ctDNA group |
| Lygre et al. [72] | 2024 | Prospective observational study | 1 month after surgery, 3 months, and then every 6 months | RFS | Higher recurrence in ctDNA-positive patients vs. ctDNA-negative (HR: 172.91; 95%CI: 8.70 to 3437.24%) |
| Morris et al. [73] | 2024 | Multicenter prospective study | At 6 months after ACT | Clearance of ctDNA, RFS | Clearance of ctDNA was observed at 43% (95% CI 10–82%) in the control arm and 11% (95% CI 0.3–48%) in the experimental arm (p = 0.98) |
| Henriksen et al. [57] | 2024 | Multicenter prospective study | Within 60 days after the operation and every 3–4 months for up to 36 months | RFS | ctDNA detection was prognostic of recurrence (HR 11.3, 95% CI 7.8–16.4%) |
| Kasi et al. [74] | 2024 | Multicenter prospective study | 4–12 weeks after surgery | DFS | 44.2% with ctDNA positive had recurrence; ctDNA positive had significantly worse DFS compared to ctDNA negative (HR = 124.3, 95% CI: 29.8–518.7%) |
| Yu Kami et al. [75] | 2024 | Multicenter prospective study | 1, 3, 6, 9, 12, 18, and 24 months post-surgery until recurrence | DFS | ctDNA positive patients were 5 times more likely to recur vs. ctDNA negative patients (HR: 5.4, 95%CI: 3.58–7.67%) |
| Slater et al. [54] | 2024 | Multicenter prospective study | Before and after surgery, after ACT, every 3 months for year 1 and every 6 months for 2 years after | RFS at 2 years | RFS in ctDNA positive patients was 50.4% and 91.1% in the ctDNA negative group (95% CI, 84.1–95.1%) |
| Parikh et al. [53] | 2024 | Multicenter prospective study | Before surgery, 3 and 10 weeks after surgery, and every 12 to 24 weeks for up to 5 years | 3-week ctDNA detection rate, RFS and OS | 94.7% recurrence in the ctDNA positive and 43.5% recurrence in the ctDNA negative group; sensitivity (40.8–73.6%), specificity (62.3–99.5%) |
| Nakamura et al. [56] | 2024 | Multicenter prospective study | 4, 12, 24, 36, 48, 72, and 96 weeks after surgery until recurrence | DFS and OS | 78.27% recurrence in the ctDNA positive group and 13.14% in the ctDNA negative group; 24-month OS 83.20% in the ctDNA positive group vs. 99.30% in the ctDNA negative group. |
| Author | Year | Population | Findings |
|---|---|---|---|
| Jones et al. [76] | 2021 | 2823 | Poor OS (HR 2.2, 95% CI 1.79–2.69%) and PFS (HR 3.15, 95% CI 2.10–4.73%) in the ctDNA-positive groups after treatment. |
| Callesen et al. [77] | 2022 | 6930 | High baseline ctDNA is associated with short PFS (HR = 2.2; 95% CI 1.8–2.8%; n = 509) and OS (HR = 2.4; 95% CI 1.9–3.1%; n = 1336) |
| Faulkner et al. [78] | 2022 | 3002 | Worse PFS with ctDNA positive at the first liquid biopsy post-surgery [HR: 6.92, 95% CI: 4.49–10.64%] |
| Do et al. [79] | 2023 | 3311 | ctDNA positive groups had a higher risk of recurrence vs. the ctDNA negative group (RR = 7.73, 95% CI: 5.73–10.42%) |
| Min et al. [55] | 2023 | 8076 | Combined sensitivity of 0.723, specificity of 0.920, and diagnostic OR 23.30 (95%CI: 9.3–57.9%) with an AUC of 0.860 |
| Chang et al. [80] | 2023 | 475 | ctDNA positive after nCRT had worse RFS (HR = 9.16, 95% CI, 5.48–15.32%), worse OS (HR = 8.49, 95% CI, 2.20–32.72%), and worse pCR results (OR = 0.40, 95% CI, 0.18–0.89%) |
| Nassar et al. [58] | 2024 | 1022 | Preoperative ctDNA + 5x risk of distant metastasis (RR [95% CI] 5.03 [3.31–7.65%], p < 0.001), postoperative ctDNA + 6x risk of distant metastasis (RR [95% CI] 6.17 [2.38–15.95%], p < 0.001) |
| Assay Name | Company | Assay Type | Technology | Approximate Cost (USD) | MRD Use | Turnaround Time | Notes |
|---|---|---|---|---|---|---|---|
| Signatera™ | Natera | Tumor-informed, bespoke | NGS-based multiplex PCR | ~$3500 per timepoint | Yes | 10–14 days | Requires initial tumor tissue for personalized assay design. Widely used in MRD trials (e.g., DYNAMIC, CIRCULATE-US) [68,112] |
| Guardant Reveal ™ [113] | Guardant Health | Tumor-naïve | Targeted hybrid-capture NGS | ~$2000–$2500 | Yes | 7–10 days | Does not require tumor tissue. Includes methylation and genomic alterations [113]. |
| FoundationOne® Tracker [112] | Foundation Medicine | Tumor-informed | Hybrid-capture NGS | ~$3000–$3500 | Yes | 10–14 days | Requires FFPE tissue; used for longitudinal monitoring [114]. |
| Safe-SeqS | Johns Hopkins/PGDx | Tumor-informed | NGS with molecular barcoding | Research use only | Yes | Variable | Used in academic settings; basis for some platforms [115]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vojjala, N.; Gibatova, V.; Shah, R.N.; Singal, S.; Prabhu, R.; Krishnamoorthy, G.; Riggins, K.; Moka, N. Integrating Circulating Tumor DNA into Clinical Management of Colorectal Cancer: Practical Implications and Therapeutic Challenges. Cancers 2025, 17, 2520. https://doi.org/10.3390/cancers17152520
Vojjala N, Gibatova V, Shah RN, Singal S, Prabhu R, Krishnamoorthy G, Riggins K, Moka N. Integrating Circulating Tumor DNA into Clinical Management of Colorectal Cancer: Practical Implications and Therapeutic Challenges. Cancers. 2025; 17(15):2520. https://doi.org/10.3390/cancers17152520
Chicago/Turabian StyleVojjala, Nikhil, Viktoriya Gibatova, Raj N. Shah, Sakshi Singal, Rishab Prabhu, Geetha Krishnamoorthy, Karen Riggins, and Nagaishwarya Moka. 2025. "Integrating Circulating Tumor DNA into Clinical Management of Colorectal Cancer: Practical Implications and Therapeutic Challenges" Cancers 17, no. 15: 2520. https://doi.org/10.3390/cancers17152520
APA StyleVojjala, N., Gibatova, V., Shah, R. N., Singal, S., Prabhu, R., Krishnamoorthy, G., Riggins, K., & Moka, N. (2025). Integrating Circulating Tumor DNA into Clinical Management of Colorectal Cancer: Practical Implications and Therapeutic Challenges. Cancers, 17(15), 2520. https://doi.org/10.3390/cancers17152520






