Monte Carlo Gradient Boosted Trees for Cancer Staging: A Machine Learning Approach
Simple Summary
Abstract
1. Introduction
- Introduce both binary and multiclass staging schemas to reflect real-world clinical stratification;
- Apply SMOTE to address class imbalance, comparing strategies applied before vs. after the train–test split;
- Evaluate performance across eight experimental dataflows incorporating full and reduced radiomic sets;
- Use feature importance analysis to identify a reduced subset of 12 biomarkers that maintain high classification accuracy.
2. Data Description
2.1. Radiomic Feature Extraction
- First-order statistics: These features are computed from the distribution of voxel intensities within the region of interest (ROI), without accounting for spatial relationships. They include measures such as the mean, median, skewness, kurtosis, standard deviation, energy, entropy, and various percentile values. These features reflect overall intensity and heterogeneity and may correspond to variations in tumor density or internal necrosis [8];
- Shape features: These geometric descriptors quantify the 3D morphology of the tumor, independent of intensity. They include metrics such as volume, surface area, compactness, sphericity, elongation, and flatness. Shape features are especially valuable for staging, as higher-stage tumors often exhibit greater asymmetry and invasive spread into adjacent tissues [25];
- Texture features: These quantify the spatial relationships between voxel intensities and capture fine-grained heterogeneity patterns:
- ◦
- GLCM (gray-level co-occurrence matrix) features describe the frequency of voxel pairs with specific intensity combinations, emphasizing local contrast and texture [8];
- ◦
- GLSZM (gray-level size zone matrix) features capture the distribution of contiguous zones of uniform intensity, enabling assessment of tumor homogeneity or patchiness;
- ◦
- GLRLM (gray-level run length matrix) features quantify the length of consecutive voxels with the same intensity along different directions, identifying directional textures or streaks;
- ◦
- NGTDM (neighborhood gray tone difference matrix) features compute the contrast between a voxel intensity and the mean gray value of its neighbors, emphasizing local variation in gray tone [8];
- ◦
- GLDM (gray-level dependence matrix) features assess the extent to which groups of voxels depend on a central voxel for their intensity, offering insights into complexity and coarseness [25].
2.2. Clinical Metadata and Feature Engineering
- Sex (male or female);
- Age (in years);
- Number of tumors, derived as a new feature from annotated lesion data.
2.3. Data Cleaning and Filtering
- Records with missing values in radiomic or clinical fields were excluded;
- All features were standardized using z-score normalization to zero-mean and unit-variance across the dataset;
- The final analytic dataset included 398 complete cases, with balanced representation across sex and a wide age distribution.
2.4. Cancer Staging Labels
- Stage I: Localized tumor without lymph node involvement or metastasis;
- Stage II: Larger primary tumor or minor nodal involvement;
- Stage IIIa: Tumor spread to mediastinal lymph nodes on the same side of the chest [26];
- Stage IIIb: Contralateral or subcarinal lymph node involvement, often precluding surgical resection [27].
- Binary class labels (early vs. advanced stage);
- Three-class labels (low, medium, high severity) to better reflect clinical stratification.
3. Methods
3.1. Binary and Multiclass Binning
3.1.1. Binary Classification: Early vs. Advanced Stages
- Low stage: includes patients diagnosed with Stage I and Stage II;
- High stage: includes patients with Stage IIIa and IIIb.
- Simpler decision boundaries in feature space;
- More stable learning under class imbalance;
- Clearer evaluation metrics such as sensitivity and specificity.
3.1.2. Multiclass Classification: Three-Stage Stratification
- Stage I (low): localized tumors with no lymphatic spread;
- Stage II (medium): tumors with greater size or minor lymph node involvement;
- Stages IIIa and IIIb (high): regionally advanced tumors with significant nodal involvement.
- Increased overlap between classes in feature space;
- Greater susceptibility to data imbalance;
- Need for multiclass-capable classifiers (e.g., one-vs.-rest or SoftMax-based models).
3.1.3. Label Encoding for Modeling
- For binary classification:
- ◦
- 0: early stage (Stage I and II);
- ◦
- 1: late stage (Stage IIIa and IIIb).
- For multiclass classification:
- ◦
- 0: Stage I;
- ◦
- 1: Stage II;
- ◦
- 2: Stage IIIa or IIIb.
3.2. Addressing Class Imbalance
3.2.1. Nature of the Imbalance
- Stage IIIa and IIIb (advanced stages) accounted for 277 patients (~70%);
- Stages I and II (early stages) were underrepresented, with only 121 patients (~30%).
3.2.2. SMOTE: Synthetic Minority Oversampling Technique
- For each instance in the minority class, a number of K-nearest neighbors (typically K = 5) are identified;
- New instances are created by taking the vector difference between a sample and its neighbors, multiplying by a random scalar between 0 and 1, and adding it to the original vector;
- This results in synthetic examples that lie within the convex hull of existing data points.
3.2.3. Oversampling Strategies
- All class labels were balanced to contain 277 instances each;
- The balanced dataset was then split into 80% training and 20% testing.
- Training and test sets are drawn from an already-balanced distribution;
- The classifier is evaluated on samples from the same (balanced) feature space.
- Synthetic examples may appear in both training and test sets, introducing data leakage;
- Overestimates generalization if the test set is not representative of real-world distributions.
- The training set was oversampled to balance class distributions;
- The test set retained its original imbalanced distribution.
- Preserves the natural distribution in the test set;
- Offers a more realistic estimate of model performance in the real world.
- Training is more difficult due to fewer original examples in minority classes;
- May underperform in accuracy, but with less risk of overfitting.
3.2.4. Integration into the Pipeline
- Binary—Strategy B (SMOTE before): denoted as FDBB and RDBB;
- Binary—Strategy O (SMOTE after): denoted as FDBO and RDBO;
- Multiclass—Strategy B: FDMB and RDMB;
- Multiclass—Strategy O: FDMO and RDMO.
- Oversampling timing;
- Classification granularity;
- Feature-set size (full vs. reduced).
3.3. Gradient Boosted Trees (GBT)
3.3.1. Mathematical Formulation
3.3.2. Feature Handling
- Full-feature set: all 107 radiomic features + age, sex, and tumor count;
- Reduced-feature set: the top 12 radiomic features (Section 3.4) + same clinical variables.
3.3.3. XGBoost for Radiomic Feature Identification
- Automatically handles feature interaction via hierarchical tree splits;
- Provides feature importance scores, enabling selection and interpretation;
- It is robust to outliers;
- Trains efficiently on small to moderate datasets, which is typical in oncology.
3.4. Classification Strategies and Feature Selection
3.4.1. Dataflow Naming and Strategy Design
- Feature set: full (F) vs. reduced (R);
- Classification type: binary (B) vs. multiclass (M);
- Oversampling strategy: SMOTE before (B) vs. SMOTE after (O) train–test split.
- First letter: F = full dataset, R = reduced dataset;
- Second letter: D = dataset context;
- Third letter: B = binary classification, M = multiclass classification;
- Fourth letter: B = SMOTE before split, O = SMOTE after split.
- The dataset was partitioned into 80% training/20% testing;
- SMOTE was applied according to the strategy;
- XGBoost was trained using standardized default hyperparameters (Appendix A.1);
- Predictions on the test set were recorded;
- Accuracy, sensitivity, specificity, F1 score, and confusion matrices were computed.
- Feature reduction effects (full vs. reduced);
- Class structure impact (binary vs. multiclass);
- Oversampling effects on generalization (Strategy B vs. O).
3.4.2. Feature Importance-Based Reduction
- High dimensionality: increases the risk of overfitting;
- Redundancy: many features are redundant or weakly informative;
- Computational cost: scales with feature count.
- Gain: The average reduction in loss (e.g., log-loss) when a feature is used for splitting;
- Weight: The number of times a feature is used in all splits;
- Cover: The number of samples affected by the splits using this feature.
- A Monte Carlo approach was used to assess the impact of randomness by running the FDBB model (full, binary, SMOTE before) multiple times (100 times in this study);
- In each run, the gain-based importance of each feature was recorded;
- Importance scores for each feature were averaged across multiple runs;
- Features were ranked by their mean importance.
3.4.3. Validation of Feature Reduction
- Faster training and evaluation;
- Improved model interpretability;
- Potential for hardware-efficient deployment in clinical environments.
4. Results
4.1. Feature Importance and Interpretability
- Sphericity consistently ranked number one, suggesting that tumors with more irregular or elongated shapes (lower sphericity) were strongly associated with advanced stages. These matches established clinical observations that advanced-stage lung tumors are more invasive and spatially heterogeneous [8,9];
- Elongation and flatness, both geometric descriptors, also ranked in the top five, indicating that shape complexity is a dominant signal in staging classification;
- First-order features such as skewness, median, and maximum 3D diameter ranked high as well. These features likely capture aspects of tumor density, asymmetry in intensity distribution (e.g., due to necrosis), and size, all known correlates of disease progression;
- Texture features (e.g., GLDM dependence entropy, GLCM contrast) appeared lower in the ranking but still contributed to performance. Their value may be more nuanced, distinguishing between medium and high stages, or capturing subtle heterogeneity not seen in geometry alone.
4.2. Redundancy and Complementarity in Radiomic Features
- High correlation among shape features, particularly between sphericity, elongation, and flatness. While this may suggest overlap, each metric captures a different aspect of morphology (e.g., sphericity measures roundness, elongation measures axis distortion), justifying their inclusion;
- First-order features such as median and skewness showed moderate to low correlation, indicating they offer orthogonal information about tumor intensity distribution;
- Texture features were weakly correlated with both shape and statistical features, confirming their complementary value. For instance, GLDM dependence entropy was uncorrelated with geometric features but contributed to distinguishing among mid-stage cases (e.g., Stage II).
4.3. Decision Tree Interpretability
- The root node splits on “Number of Rows”, a clinical feature derived from patient metadata, and represents the number of tumors a patient has. This reflects the fact that patients with multiple nodules are more likely to be at advanced stages due to intrapulmonary metastasis or synchronous tumors;
- Subsequent splits use skewness, maximum 3D diameter, and sphericity, highlighting a hierarchy where the model first evaluates tumor burden and size, then examines shape irregularity;
- Leaf nodes assign probabilities to stage labels, allowing soft decision-making.

- The number of tumors is the root node similar to panel (a);
- The first split is based on the shape feature, “maximum 2D diameter column”;
- Then, “age” is a major consideration, along with “zone entropy” and “dependence variance”;
- The remaining splits are based on shape features and first-order statistics.
- The first split is based on the “median”, a first-order statistics feature, that corresponds to the median density of cancerous tissue. It was identified as the top 12 impactful radiomics by the proposed feature selection procedure;
- The next set of splits is entirely based on the value of the “sphericity”, a shape feature;
- The next set of node splits is based on the shape category of radiomics along with the clinical features;
4.4. Classification Accuracy and Comparative Pipeline Performance
- Overall accuracy;
- Class-specific metrics (TPR, FPR, TNR, FNR);
- Effect of order when balancing the dataset (using SMOTE);
- Full- vs. reduced-feature set.
4.4.1. Binary vs. Multiclass Confusion Matrices
- Most confusion occurred between Stage II and Stage IIIa, likely due to biological overlap in tumor aggressiveness and nodal involvement;
- Stage I predictions were very accurate, perhaps because early-stage tumors have distinctive geometric and intensity features.


4.4.2. Error Decomposition
- True positive rate (sensitivity): The proportion of correctly identified high-stage cases;
- True negative rate (specificity): The proportion of correctly identified low-stage cases;
- False negative rate: Missed high-stage predictions;
- False positive rate: Misclassified early-stage patients.
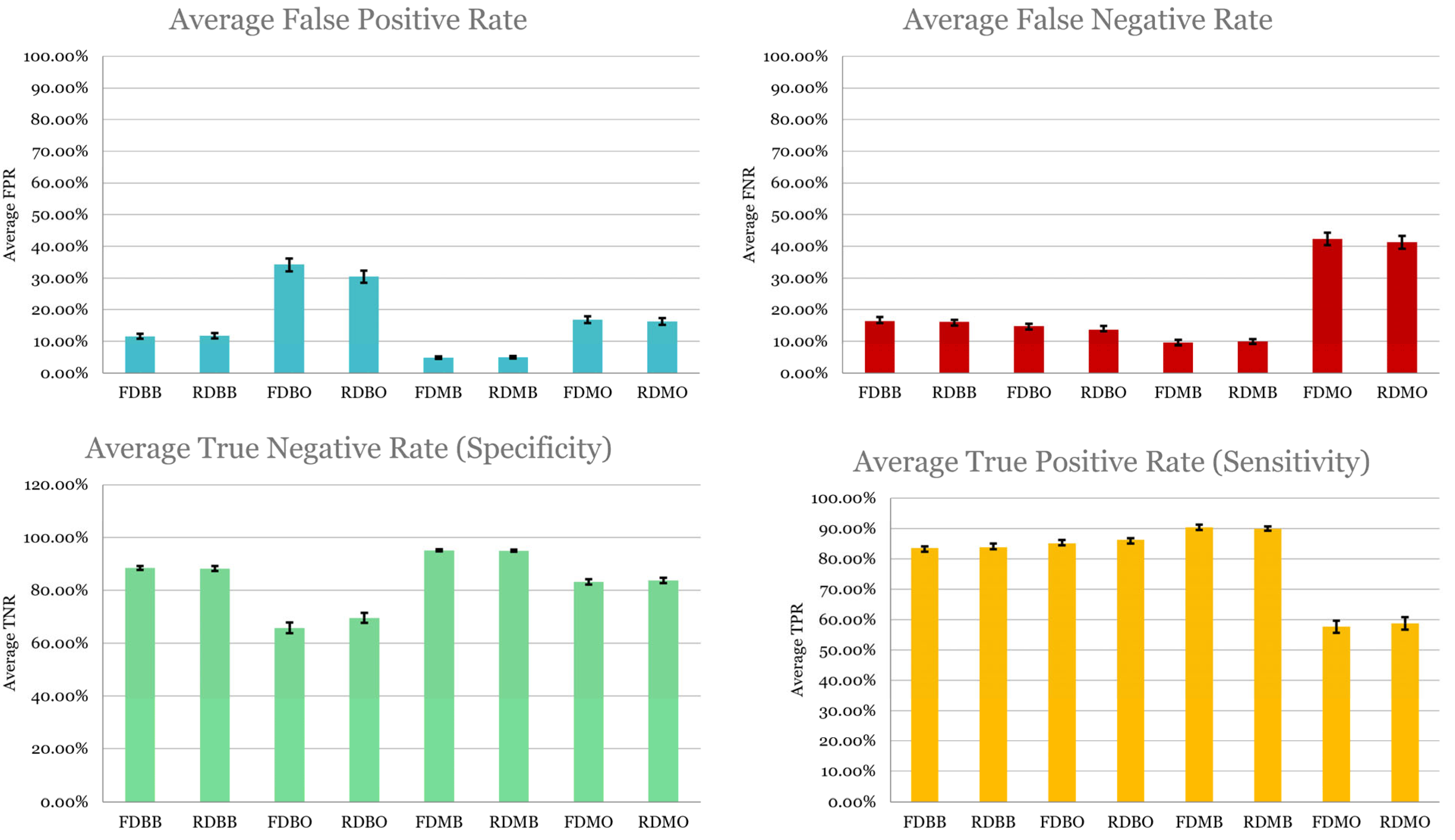
- Sensitivity was highest in FDMB and RDMB, confirming their strength in detecting advanced disease, clinically critical since undertreating high-stage cancer can be fatal;
- Specificity was higher in binary models (RDBB, FDBB), possibly due to a clearer separation of low vs. high stages;
- False negative rates were highest in post-SMOTE pipelines, underscoring the risk of training on imbalanced data;
- Models using a reduced set of radiomics retained comparable sensitivity to that of the full set.
4.4.3. Overall Accuracy
4.5. Summary
- A small set of “shape” and “first-order” radiomics can yield high diagnostic performance;
- SMOTE-before-split is superior for balanced learning but may risk data leakage if not carefully managed;
- Multiclass classification adds complexity but improves sensitivity;
- XGBoost offers transparent, high-performing modeling well suited for radiomics-based clinical tools.
5. Discussion
5.1. Efficacy of Feature Selection and Reduction
- Reduced computational complexity: Faster training and inference, especially beneficial in clinical settings with constrained resources;
- Improved interpretability: Clinicians can more easily trace model decisions to specific, biologically relevant features;
- Lower overfitting risk: A compact model generalizes better, especially in modest sample sizes typical of medical datasets.
5.2. Impact of Class Structure and Oversampling Strategy
5.2.1. Multiclass Classification Enhances Sensitivity
5.2.2. Order of Balancing Data Procedure (SMOTE) Affects Generalization
- Pre-split SMOTE may be preferred in exploratory settings focused on identifying informative features and best-case performance;
- Post-split SMOTE may be more appropriate for validating generalization and real-world applicability.
5.3. Advantages of XGBoost in Clinical Modeling
- Interpretable decision trees: As shown in Figure 4, XGBoost trees allowed transparent inspection of classification logic;
- Native handling of missing data and heterogeneity: The model’s robustness is well suited for radiomics, which often contains irregular feature patterns;
- Integrated feature selection: Feature importance scores were easily extracted and validated, facilitating the reduced model.
5.4. Limitations and Future Directions
- Sample size: While relatively large with 398 records, the dataset remains modest in the machine learning domains considering the large number of predictors (including 107 radiomic features). Larger, multi-center datasets would help validate the generalizability of the identified features and the model architecture;
- Ground truth labeling: Staging labels are derived from clinical records and subject to inter-observer variability. Automated image-based staging is only as accurate as its reference labels;
- Feature standardization: Radiomic feature extraction depends on imaging parameters (e.g., voxel size, reconstruction kernel). Though PyRadiomics ensures some reproducibility [25], cross-platform consistency remains a challenge;
- Limited scope: The scope of this study was confined to a preliminary examination of eight data pipelines in conjunction with the standard XGBoost model.
- Incorporating multi-modal data, including PET scans and genomic markers;
- Exploring longitudinal features to predict stage progression over time;
- Evaluating generalization across external validation sets. The future work is focused on further generalization of the proposed method using other lung cancer datasets. Collaboration with cancer centers will be established to externally validate the proposed model utilizing their lung cancer data for further validation and potential customization;
- Hyperparameter tuning of the proposed MCGBT model to improve performance.
6. Conclusions
- Classification granularity (binary vs. multiclass);
- Feature dimensionality (full- vs. reduced-feature sets);
- Oversampling strategy (SMOTE before vs. after train–test split).
- Identifying shape-based radiomic features (sphericity, flatness, elongation) as predictive factors for lung cancer stage;
- Extracting a reduced set of 12 radiomics with comparable cancer staging accuracy as that of the full set of radiomics, enabling lean and deployable classifiers;
- Establishing that the order of a data-balancing procedure must be carefully contemplated. Pre-split oversampling improves the learning process, but it requires caution to avoid data leakage;
- Introducing Monte Carlo XGBoost as an efficient, interpretable, and scalable classifier with applications to medical data.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Implementation of XGBoost and Hyperparameters Setup
- objective: “binary:logistic” for binary classification; “multi:softmax” for multiclass;
- eval_metric: “logloss” for probability calibration and “mlogloss” for multiclass;
- max_depth: 6 (to balance expressiveness and overfitting);
- eta (learning rate): 0.1 (to control step size during updates);
- subsample: 0.8 (randomly sample 80% of rows per tree);
- colsample_bytree: 0.8 (randomly sample 80% of features per tree);
- lambda: 1 (L2 regularization);
- alpha: 0 (no L1 regularization);
- n_estimators: 100 (maximum number of boosting iterations).
Appendix A.2. Complete List of Radiomics by Category
| First-order statistics | Energy |
| Total energy | |
| Entropy | |
| Minimum | |
| 10th percentile | |
| 90th percentile | |
| Maximum | |
| Mean | |
| Median | |
| Interquartile range | |
| Range | |
| Mean absolute deviation | |
| Robust mean absolute deviation | |
| Root mean squared | |
| Skewness | |
| Kurtosis | |
| Variance | |
| Uniformity | |
| Shape features | Mesh volume |
| Voxel volume | |
| Surface area | |
| Surface-area-to-volume ratio | |
| Sphericity | |
| Maximum 3D diameter | |
| Maximum 2D diameter slice | |
| Maximum 2D diameter column | |
| Maximum 2D diameter row | |
| Major axis length | |
| Minor axis length | |
| Least axis length | |
| Elongation | |
| Flatness | |
| Gray-level co-occurrence matrix (GLCM) features | Autocorrelation |
| Joint average | |
| Cluster prominence | |
| Cluster shade | |
| Cluster tendency | |
| Contrast | |
| Correlation | |
| Difference average | |
| Difference entropy | |
| Difference variance | |
| Joint energy | |
| Joint entropy | |
| Informational measure of correlation (IMC) 1 | |
| Informational measure of correlation (IMC) 2 | |
| Inverse difference moment (IDM) | |
| Maximal correlation coefficient (MCC) | |
| Inverse difference moment normalized (IDMN) | |
| Inverse difference (ID) | |
| Inverse difference normalized (IDN) | |
| Inverse variance | |
| Maximum probability | |
| Sum average | |
| Sum entropy | |
| Sum of squares | |
| Gray-level size zone matrix (GLSZM) features | Small area emphasis (SAE) |
| Large area emphasis (LAE) | |
| Gray-level non-uniformity (GLN) | |
| Gray-level non-uniformity normalized (GLNN) | |
| Size zone non-uniformity (SZN) | |
| Size zone non-uniformity normalized (SZNN) | |
| Zone percentage | |
| Gray-level variance (GLV) | |
| Zone variance (ZV) | |
| Zone entropy (ZE) | |
| Low gray-level zone emphasis (LGLZE) | |
| High gray-level zone emphasis (HGLZE) | |
| Small area low gray-level emphasis (SALGLE) | |
| Small area high gray-level emphasis (SAHGLE) | |
| Large area low gray-level emphasis (LALGLE) | |
| Large area high gray-level emphasis (LAHGLE) | |
| Gray-level run length matrix (GLRLM) features | Short-run emphasis (SRE) |
| Long-run emphasis (LRE) | |
| Gray-level non-uniformity 1 (GLN1) | |
| Gray-level non-uniformity normalized 1 (GLNN1) | |
| Run length non-uniformity (RLN) | |
| Run length non-uniformity normalized (RLNN) | |
| Run percentage (RP) | |
| Gray-level variance 1 (GLV1) | |
| Run variance (RV) | |
| Run entropy (RE) | |
| Low gray-level run emphasis (LGLRE) | |
| High gray-level run emphasis (HGLRE) | |
| Short-run low gray-level emphasis (SRLGLE) | |
| Short-run high gray-level emphasis (SRHGLE) | |
| Neighboring gray tone difference matrix (NGTDM) features | Long-run low gray-level emphasis (LRLGLE) |
| Long-run high gray-level emphasis (LRHGLE) | |
| Coarseness | |
| Contrast 1 | |
| Busyness | |
| Gray-level dependence matrix (GLDM) features | Complexity |
| Strength | |
| Small-dependence emphasis (SDE) | |
| Large-dependence emphasis (LDE) | |
| Gray-level non-uniformity 2 (GLN2) | |
| Dependence non-uniformity (DN) | |
| Dependence non-uniformity normalized (DNN) | |
| Gray-level variance 2 (GLV2) | |
| Dependence variance (DV) | |
| Dependence entropy (DE) | |
| Low gray-level emphasis (LGLE) | |
| High gray-level emphasis (HGLE) | |
| Small dependence low gray-level emphasis (SDLGLE) | |
| Small-dependence high gray-level emphasis (SDHGLE) | |
| Large-dependence low gray-level emphasis (LDLGLE) | |
| Large-dependence high gray-level emphasis (LDHGLE) |
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA A Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- World Health Organization. Lung Cancer. 2023. Available online: https://www.who.int/news-room/fact-sheets/detail/lung-cancer (accessed on 6 June 2024).
- American Cancer Society. Key Statistics for Lung Cancer. 2023. Available online: https://www.cancer.org/cancer/types/lung-cancer/about/key-statistics.html (accessed on 6 June 2024).
- Rosell, R.; Cecere, F.; Santarpia, M.; Reguart, N.; Taron, M. Predicting the outcome of chemotherapy for lung cancer. Curr. Opin. Pharmacol. 2006, 6, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Tsim, S.; O’dOwd, C.A.; Milroy, R.; Davidson, S. Staging of non-small cell lung cancer (NSCLC): A review. Respir. Med. 2010, 104, 1767–1774. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Cancer Staging. 2024. Available online: https://www.cancer.org/content/dam/CRC/PDF/Public/6682.00.pdf (accessed on 15 December 2024).
- Chen, B.; Zhang, R.; Gan, Y.; Yang, L.; Li, W. Development and clinical application of radiomics in lung cancer. Radiat. Oncol. 2017, 12, 154. [Google Scholar] [CrossRef] [PubMed]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef]
- Coroller, T.P.; Grossmann, P.; Hou, Y.; Rios Velazquez, E.; Leijenaar, R.T.H.; Hermann, G.; Lambin, P.; Haibe-Kains, B.; Mak, R.H.; Aerts, H.J.W.L. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother. Oncol. 2015, 114, 345–350. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, Y.; Canes, A.; Steinberg, D.; Lyashevska, O. Predictive analytics with gradient boosting in clinical medicine. Ann. Transl. Med. 2019, 7, 152. [Google Scholar] [CrossRef]
- Natekin, A.; Knoll, A. Gradient boosting machines, a tutorial. Front. Neurorobot. 2013, 7, 21. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Ogunleye, A.A.; Wang, Q.-G. XGBoost model for chronic kidney disease diagnosis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 17, 2131–2140. [Google Scholar] [CrossRef]
- Li, S.; Zhang, X. Research on orthopedic auxiliary classification and prediction model based on XGBoost algorithm. Neural Comput. Appl. 2020, 32, 1971–1979. [Google Scholar] [CrossRef]
- Liew, X.Y.; Hameed, N.; Clos, J. An investigation of XGBoost-based algorithm for breast cancer classification. Mach. Learn. Appl. 2021, 6, 100154. [Google Scholar] [CrossRef]
- Nagpal, K.; Foote, D.; Liu, Y.; Chen, P.-H.C.; Wulczyn, E.; Tan, F.; Olson, N.; Smith, J.L.; Mohtashamian, A.; Wren, J.H.; et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit. Med. 2019, 2, 48. [Google Scholar] [CrossRef]
- Kachouie, N.N.; Deebani, W.; Shutaywi, M.; Christiani, D.C. Lung cancer clustering by identification of similarities and discrepancies of DNA copy numbers using maximal information coefficient. PLoS ONE 2024, 19, e0301131. [Google Scholar] [CrossRef]
- Kachouie, N.N.; Deebani, W.; Christiani, D.C. Christiani. Identifying similarities and disparities between DNA copy number changes in cancer and matched blood samples. Cancer Investig. 2019, 37, 535–545. [Google Scholar] [CrossRef]
- Kachouie, N.N.; Shutaywi, M.; Christiani, D.C. Christiani. Discriminant analysis of lung cancer using nonlinear clustering of copy numbers. Cancer Investig. 2020, 38, 102–112. [Google Scholar] [CrossRef]
- Kachouie, N.N.; Lin, X.; Christiani, D.C.; Schwartzman, A. Detection of local DNA copy number changes in lung cancer population analyses using a multi-scale approach. Commun. Stat. Case Stud. Data Anal. Appl. 2015, 1, 206–216. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Wee, L.; Rios Velazquez, E.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B. Data from NSCLC-Radiomics, The Cancer Imaging Archive, 2019.
- PyRadiomics. Radiomic Features. 2016. Available online: https://pyradiomics.readthedocs.io/en/latest/features.html (accessed on 15 January 2025).
- National Cancer Institute. Definition of Stage IIIA Non-Small Cell Lung Cancer. 2011. Available online: https://www.cancer.gov/publications/dictionaries/cancer-terms/def/stage-iiia-non-small-cell-lung-cancer (accessed on 29 June 2024).
- National Cancer Institute. Definition of Stage IIIB Non-Small Cell Lung Cancer. 2011. Available online: https://www.cancer.gov/publications/dictionaries/cancer-terms/def/stage-iiib-non-small-cell-lung-cancer (accessed on 29 June 2024).
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Huang, Z.; Hu, C.; Chi, C.; Jiang, Z.; Tong, Y.; Zhao, C. An artificial intelligence model for predicting 1-year survival of bone metastases in non-small-cell lung cancer patients based on XGBoost algorithm. BioMed Res. Int. 2020, 2020, 3462363. [Google Scholar] [CrossRef]
- Altuhaifa, F.A.; Win, K.T.; Su, G. Predicting lung cancer survival based on clinical data using machine learning: A review. Comput. Biol. Med. 2023, 165, 107338. [Google Scholar] [CrossRef]
- Li, Q.; Yang, H.; Wang, P.; Liu, X.; Lv, K.; Ye, M. XGBoost-based and tumor-immune characterized gene signature for the prediction of metastatic status in breast cancer. J. Transl. Med. 2022, 20, 177. [Google Scholar] [CrossRef]




| Pipeline | Feature Set | Class Type | SMOTE Technique |
|---|---|---|---|
| FDBB | Full | Binary | Before split |
| FDBO | Full | Binary | After split |
| RDBB | Reduced | Binary | Before split |
| RDBO | Reduced | Binary | After split |
| FDMB | Full | Multiclass | Before split |
| FDMO | Full | Multiclass | After split |
| RDMB | Reduced | Multiclass | Before split |
| RDMO | Reduced | Multiclass | After split |
| Radiomic Type | Identified Features |
|---|---|
| Shape | Sphericity, elongation, flatness, maximum 3D diameter, maximum 2D diameter column, maximum 2D diameter slice |
| First-order stats | Maximum, minimum, median, 90th percentile |
| GLDM/GLCM | Dependence variance, zone entropy, energy, maximal correlation coefficient (MCC), gray-level variance 2, large-dependence low gray-level emphasis (LDLGLE) |
| GLRLM | Long-run low gray-level emphasis (LRLGLE) |
| NGTDM | Strength |
| Pipeline | Feature Set | Class Type | SMOTE Strategy | Accuracy (%) |
|---|---|---|---|---|
| FDMB | Full | Multiclass | Before Split | 90.3 |
| RDMB | Reduced | Multiclass | Before Split | 89.98 |
| RDBB | Reduced | Binary | Before Split | 86.17 |
| FDBB | Full | Binary | Before Split | 85.85 |
| RDBO | Reduced | Binary | After Split | 80.81 |
| FDBO | Full | Binary | After Split | 79.26 |
| RDMO | Reduced | Multiclass | After Split | 76.1 |
| FDMO | Full | Multiclass | After Split | 75.96 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eley, A.; Hlaing, T.T.; Breininger, D.; Helforoush, Z.; Kachouie, N.N. Monte Carlo Gradient Boosted Trees for Cancer Staging: A Machine Learning Approach. Cancers 2025, 17, 2452. https://doi.org/10.3390/cancers17152452
Eley A, Hlaing TT, Breininger D, Helforoush Z, Kachouie NN. Monte Carlo Gradient Boosted Trees for Cancer Staging: A Machine Learning Approach. Cancers. 2025; 17(15):2452. https://doi.org/10.3390/cancers17152452
Chicago/Turabian StyleEley, Audrey, Thu Thu Hlaing, Daniel Breininger, Zarindokht Helforoush, and Nezamoddin N. Kachouie. 2025. "Monte Carlo Gradient Boosted Trees for Cancer Staging: A Machine Learning Approach" Cancers 17, no. 15: 2452. https://doi.org/10.3390/cancers17152452
APA StyleEley, A., Hlaing, T. T., Breininger, D., Helforoush, Z., & Kachouie, N. N. (2025). Monte Carlo Gradient Boosted Trees for Cancer Staging: A Machine Learning Approach. Cancers, 17(15), 2452. https://doi.org/10.3390/cancers17152452






