Unclassified Chromosomal Abnormalities as an Indicator of Genomic Damage in Survivors of Hodgkin’s Lymphoma
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Lymphocyte Culture
2.3. Cell Analysis
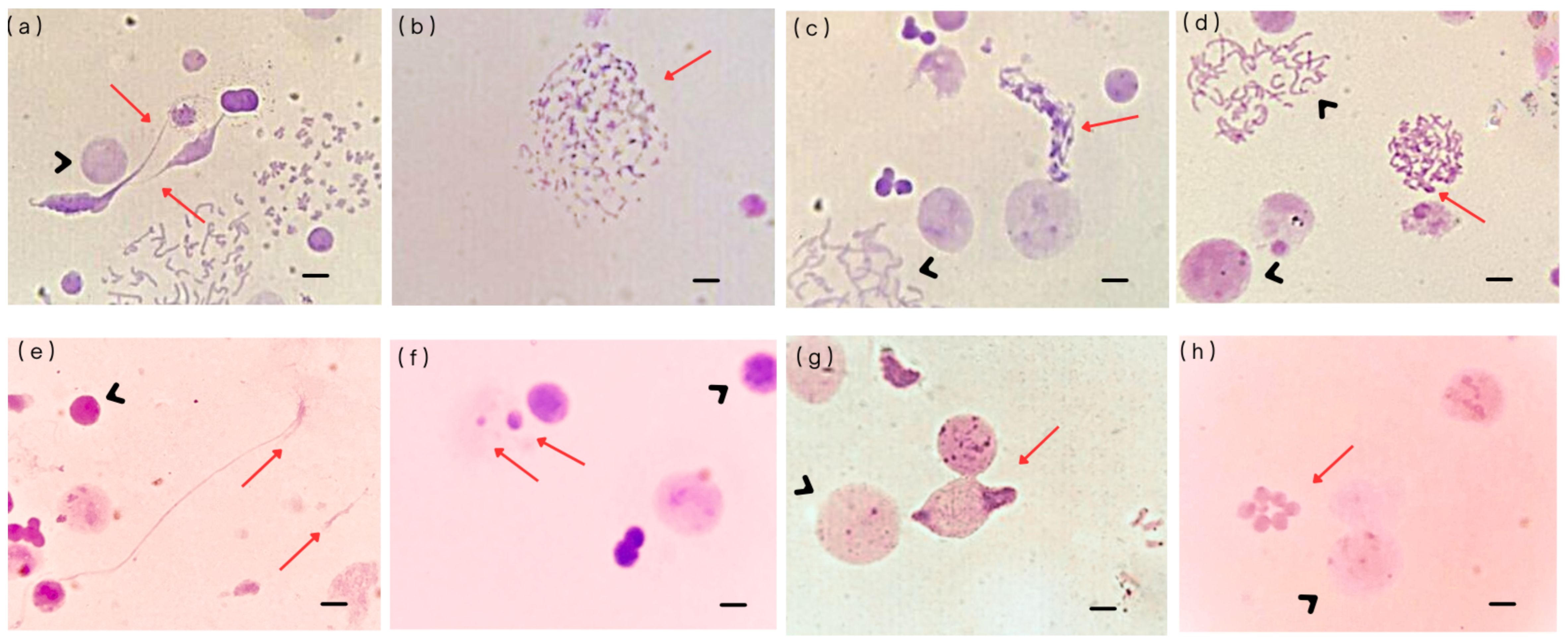
- (a)
- Free chromatin: These structures consist of interphase chromatin that is not enclosed by a nuclear envelope. They have been observed in various cell lines exposed to chemotherapeutic agents, showing a clear dose–response relationship [12]. Their formation is likely related to nuclear envelope instability, primarily because of underlying genomic instability. Another possible origin of free chromatin is the disruption of micronuclei (MN), which has been identified as one of their possible fates [20].
- (b)
- Defective mitotic figures (DMFs): These structures are characterized by abnormal mitotic figures in which chromosomes and chromatin fibers coexist, indicating incomplete or faulty chromatin condensation. The primary mechanism underlying DMF formation involves defects in chromatin condensation combined with G2–M checkpoint failure. DMFs have been frequently observed in oncological cell lines, supporting the hypothesis that aberrant chromosomal condensation contributes to cancer development.
- (c)
- Sticky chromosomes: These aberrations involve chromosomes that adhere to one another, entangled by chromatin fibers, often appearing as fuzzy or clumped structures. Their formation is linked to abnormal methylation patterns that disrupt proper chromatin condensation. There appears to be a strong association between sticky chromosomes and defective mitotic figure formation. Sticky chromosomes are commonly observed in cells treated with ethidium bromide for high-resolution chromosome preparations, or in cells exposed to agents that interfere with DNA replication, methylation, or chromosomal compaction.
- (d)
- Unit fibers: These are substructures of metaphase chromosomes with a diameter approximately five times smaller than that of a fully condensed chromatid, around 0.4 µm. Unit fibers were first observed by Back [21], who interpreted them as intermediates in chromosome condensation, likely resulting from the coiling of the 250–300 Å solenoid fiber. Unit fibers represent a disruption in normal chromosomal condensation. They have been observed in short-term lymphocyte cultures treated with topoisomerase II inhibitors [15], and their appearance likely reflects incomplete chromatid condensation during metaphase, as result of genotoxic treatment.
- (e)
- Chromosome fragmentation (C-Frag): C-Frag is a distinct form of cell death during mitosis, characterized by the progressive degradation of chromosomes. It was previously misidentified as chromatin pulverization or premature chromosome condensation. However, unlike apoptosis or mitotic catastrophe, C-Frag arises in response to various cellular stressors, such as gene mutations, infections, drug treatments, and centrosomal dysfunction, and is now recognized as a general stress response mechanism. Some studies suggest that C-Frag may lead to aneuploidy; when it affects a single chromosome, it can trigger genomic chaos, including chromothripsis, ultimately contributing to karyotypic abnormalities [22].
- (f)
- Micronuclei (MN): These are small nuclei containing one or a few whole chromosomes, chromosomal fragments, or both. They form through various mechanisms, including chromosome displacement during metaphase, slow chromosome separation during transitions from anaphase to telophase, both bipolar and multipolar, fragments from broken chromosome bridges, inheritance of MN from mother cells, separated nuclear fragments during anaphase, extrusion of chromosomes into a mini cell that fuses with a daughter cell, and formation of nuclear buds during the interphase. MN formation is linked to dysfunction in gene networks responsible for DNA damage response, commonly observed in cancers.
- (g)
- Micronuclei cluster: This is a group of micronuclei of varying sizes that typically form within a single cell, which can be either diploid or polyploid. These clusters can evolve over time through processes of cell fusion and fission, acting as transitional structures during cancer macroevolution (large-scale genomic changes driving cancer progression) [30]. Their formation depends on both the intensity and duration of stress or damaging exposure on cells, which triggers diverse cellular responses, including activation of apoptotic pathways. The formation and evolution of micronuclei clusters may be connected to genomic chaos, reflecting massive genomic rearrangements and instability [23,30]. It has been proposed that MN clusters arise as a consequence of frustrated cell death. These clusters of MN undergo repeated cycles of fusion and fission, reflecting a trial-and-error process in the search for viable genomic configurations. Through these dynamic events, the genomic content is rearranged, leading to the generation of novel, unstable genomes a phenomenon known as genomic chaos. While most MN clusters do not persist, a small subset may survive and initiate macroevolutionary changes that contribute to cancer development [22,23].
- (h)
- Abnormal nuclear morphology: It is characterized by markedly altered chromatin condensation patterns within the nucleus and it exhibits a dose-dependent response to chemotherapeutic agents such as doxorubicin. These nuclear changes reflect underlying damage to DNA and disruptions in nuclear organization, often linked to the cytotoxicity of chemotherapy [15].
2.4. Statistical Analysis
3. Results
3.1. Patients
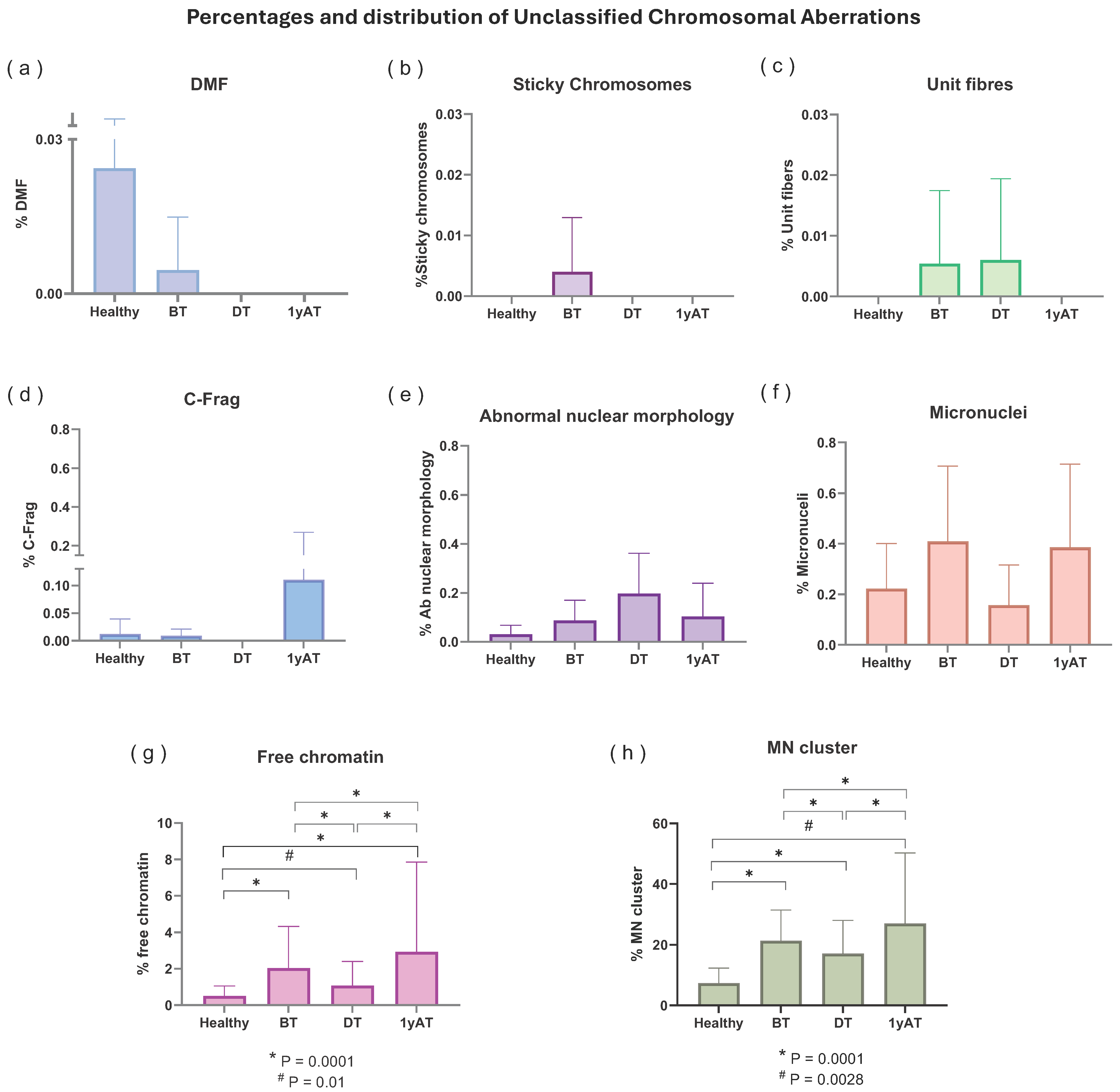
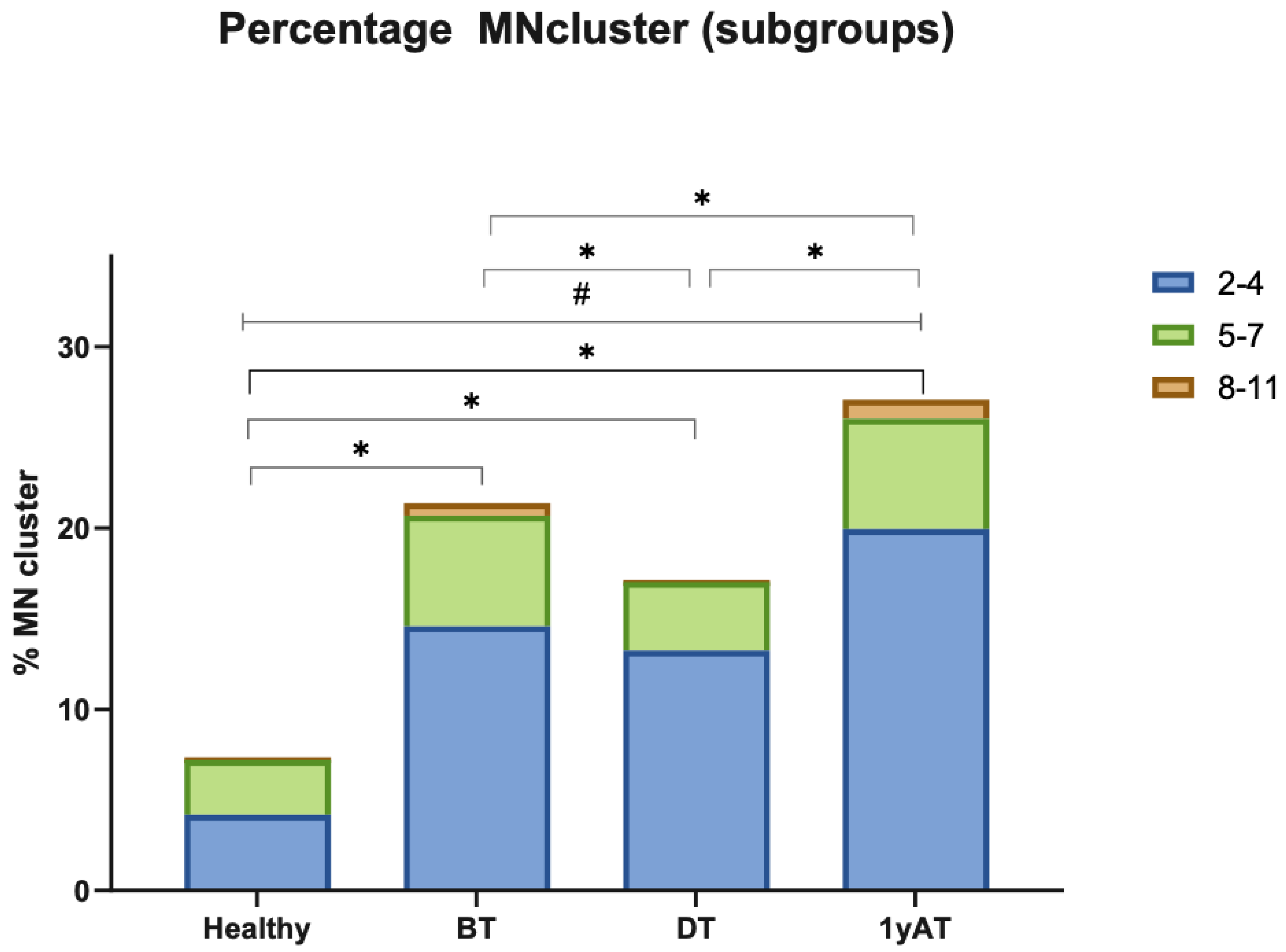
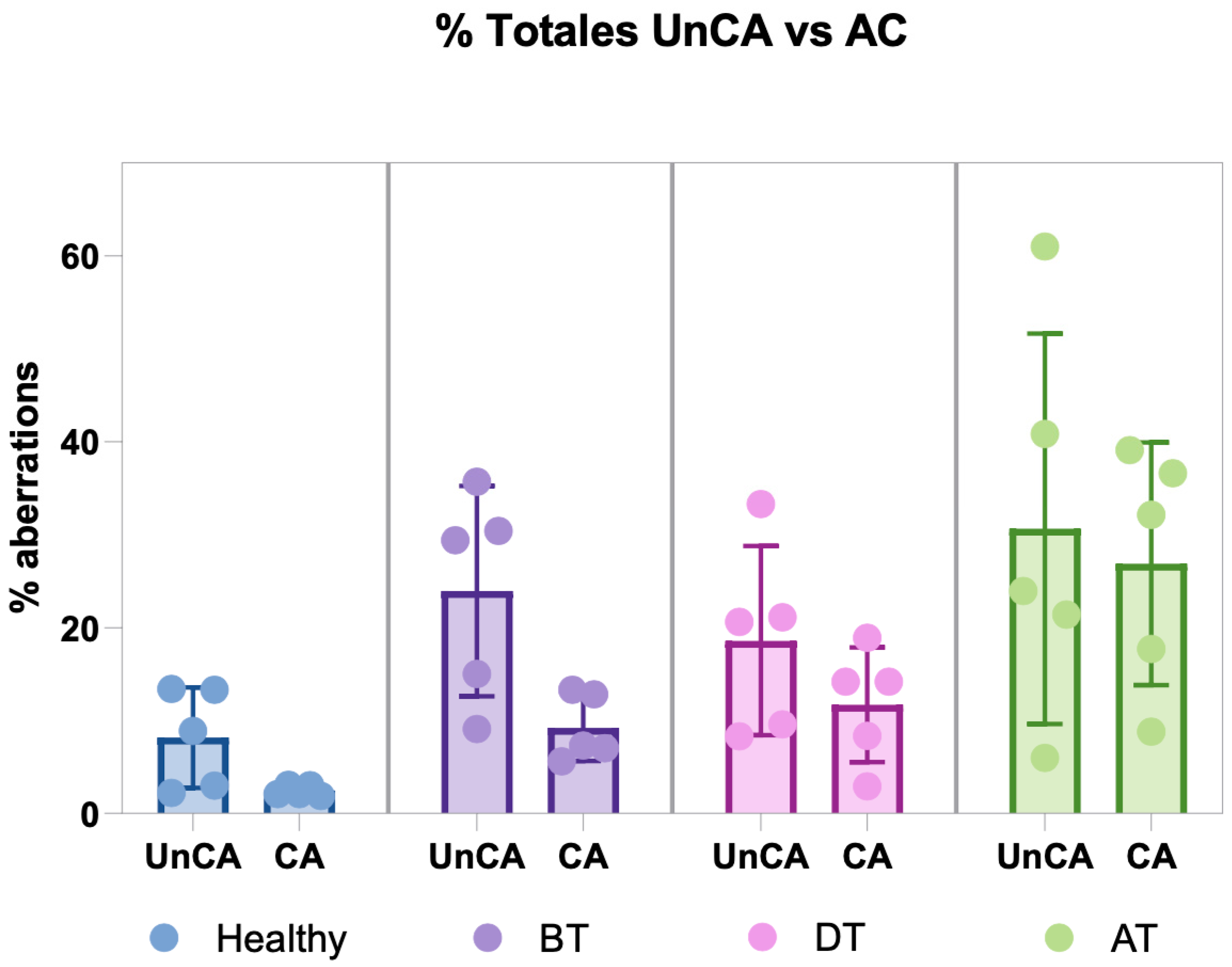
3.2. Unclassified Chromosomal Aberrations (UnCAs)
3.2.1. UnCAs in Hodgkin’s Lymphoma Patients Versus Healthy Individuals
3.2.2. UnCA vs. Chromosomal Aberrations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HL | Hodgkin’s lymphoma |
| BT | Before treatment |
| DT | During treatment |
| AT | After treatment |
| 1yAT | One year after treatment |
| ABVD | Adriamycin, Bleomycin, Vinblastine, Dacarbazine |
| UnCA | Unclassified chromosomal aberrations |
| NCCA | Non-clonal chromosomal aberrations |
| SMN | Second malignant neoplasm |
| DMFs | Defective mitotic figures |
| C-Frag | Chromosome fragmentation |
| MN | Micronuclei |
| MN cluster | Micronuclei cluster |
| Gy | Gray |
| M-FISH | Multiplex fluorescence in situ hybridization |
| SSC | Saline sodium citrate |
| SD | Standard deviation |
| CA | Chromosomal aberrations |
| MOPP | Mechlorethamine, Oncovine, Procarbazine, Prednisone |
References
- Kaatsch, P.; Byrne, J.; Grabow, D. Managing a Pan-European Consortium on Late Effects among Long-Term Survivors of Childhood and Adolescent Cancer—The Pancarelife Project. Int. J. Environ. Res. Public. Health 2021, 18, 3918. [Google Scholar] [CrossRef] [PubMed]
- Connors, J.M.; Cozen, W.; Steidl, C.; Carbone, A.; Hoppe, R.T.; Flechtner, H.H.; Bartlett, N.L. Hodgkin Lymphoma. Nat. Rev. Dis. Primers 2020, 6, 61. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Zahnreich, S.; Schmidberger, H. Childhood Cancer: Occurrence, Treatment and Risk of Second Primary Malignancies. Cancers 2021, 13, 2607. [Google Scholar] [CrossRef] [PubMed]
- Farnetani, G.; Vannucci, M.; Fino, M.G.; Cioppi, F.; Rosta, V.; Palma, M.; Tamburrino, L.; Vinci, S.; Casamonti, E.; Degl’Innocenti, S.; et al. Severe Sperm DNA Fragmentation May Persist for up to 3 Years after Cytotoxic Therapy in Patients Affected by Hodgkin Lymphoma and Non-Hodgkin Lymphoma. Hum. Reprod. 2024, 39, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Engert, A.; Younes, A. Hodgkin Lymphoma: A Comprehensive Update on Diagnostics and Clinics, 2nd ed.; Hodgkin Lymphoma: A Comprehensive Overview; Springer: New York, NY, USA, 2015. [Google Scholar] [CrossRef]
- Ramos, S.; Navarrete-Meneses, P.; Molina, B.; Cervantes-Barragán, D.E.; Lozano, V.; Gallardo, E.; Marchetti, F.; Frias, S. Genomic Chaos in Peripheral Blood Lymphocytes of Hodgkin’s Lymphoma Patients One Year After ABVD Chemotherapy/Radiotherapy. Environ. Mol. Mutagen. 2018, 59, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Frias, S.; Ramos, S.; Salas, C.; Molina, B.; Sánchez, S.; Rivera-Luna, R. Nonclonal Chromosome Aberrations and Genome Chaos in Somatic and Germ Cells from Patients and Survivors of Hodgkin Lymphoma. Genes 2019, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.J.; Sharpe, Z.; Heng, H.H. Origins and Consequences of Chromosomal Instability: From Cellular Adaptation to Genome Chaos-mediated System Survival. Genes 2020, 11, 1162. [Google Scholar] [CrossRef] [PubMed]
- De Vries, S.; Schaapveld, M.; Janus, C.P.M.; Daniëls, L.A.; Petersen, E.J.; Van Der Maazen, R.W.M.; Zijlstra, J.M.; Beijert, M.; Nijziel, M.R.; Verschueren, K.M.S.; et al. Long-Term Cause-Specific Mortality in Hodgkin Lymphoma Patients. J. Natl. Cancer Inst. 2021, 113, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Heng, E.; Thanedar, S.; Henrg, H.H. The Importance of Monitoring Non-Clonal Chromosome Aberrations (NCCAs) in Cancer Research. Methods Mol. Biol. 2024, 2825, 79–111. [Google Scholar] [PubMed]
- Liu, G.; Stevens, J.B.; Horne, S.D.; Abdallah, B.Y.; Ye, K.J.; Bremer, S.W.; Ye, C.J.; Chen, D.J.; Heng, H.H. Genome Chaos: Survival Strategy during Crisis. Cell Cycle 2014, 13, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H.Q.; Bremer, S.W.; Stevens, J.; Ye, K.J.; Miller, F.; Liu, G.; Ye, C.J. Cancer Progression by Non-Clonal Chromosome Aberrations. J. Cell. Biochem. 2006, 98, 1424–1435. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H.Q.; Stevens, J.B.; Liu, G.; Bremer, S.W.; Ye, K.J.; Reddy, P.V.; Wu, G.S.; Wang, Y.A.; Tainsky, M.A.; Ye, C.J. Stochastic Cancer Progression Driven by Non-Clonal Chromosome Aberrations. J. Cell Physiol. 2006, 208, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H.; Liu, G.; Stevens, J.B.; Abdallah, B.Y.; Horne, S.D.; Ye, K.J.; Bremer, S.W.; Chowdhury, S.K.; Ye, C.J. Karyotype Heterogeneity and Unclassified Chromosomal Abnormalities. Cytogenet. Genome Res. 2013, 139, 144–157. [Google Scholar] [CrossRef] [PubMed]
- Rangel, N.; Forero-Castro, M.; Rondon-Lagos, M. New Insights in the Cytogenetic Practice: Karyotypic Chaos, Non-Clonal Chromosomal Alterations and Chromosomal Instability in Human Cancer and Therapy Response. Genes 2017, 8, 155. [Google Scholar] [CrossRef] [PubMed]
- Heng, J.; Heng, H.H. Genome Chaos: Creating New Genomic Information Essential for Cancer Macroevolution. Semin. Cancer Biol. 2022, 81, 160–175. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M.; Kirsch-Volders, M.; Natarajan, A.T.; Surralles, J.; Crott, J.W.; Parry, J.; Norppa, H.; Eastmond, D.A.; Tucker, J.D.; Thomas, P. Molecular Mechanisms of Micronucleus, Nucleoplasmic Bridge and Nuclear Bud Formation in Mammalian and Human Cells. Mutagenesis 2011, 26, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M.; Knasmueller, S.; Bolognesi, C.; Bonassi, S.; Holland, N.; Migliore, L.; Palitti, F.; Natarajan, A.T.; Kirsch-Volders, M. Molecular Mechanisms by Which in Vivo Exposure to Exogenous Chemical Genotoxic Agents Can Lead to Micronucleus Formation in Lymphocytes in Vivo and Ex Vivo in Humans. Mutat. Res. Rev. Mutat. Res. 2016, 770, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Ni, J.; Liang, Z.; Xue, J.; Fenech, M.F.; Wang, X. The Molecular Origins and Pathophysiological Consequences of Micronuclei: New Insights into an Age-Old Problem. Mutat. Res. Rev. Mutat. Res. 2019, 779, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Bak, A.L.; Bak, P.; Zeuthen, J. Higher Levels of Organization in Chromosomes. J. Theor. Biol. 1979, 76, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.B.; Horne, S.D.; Abdallah, B.Y.; Ye, C.J.; Heng, H.H. Chromosomal Instability and Transcriptome Dynamics in Cancer. Cancer Metastasis Rev. 2013, 32, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.J.; Sharpe, Z.; Alemara, S.; Mackenzie, S.; Liu, G.; Abdallah, B.; Horne, S.; Regan, S.; Heng, H.H. Micronuclei and Genome Chaos: Changing the System Inheritance. Genes 2019, 10, 366. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Dai, X.; Zhou, T.; Wang, H.; Ni, J.; Xue, J.; Wang, X. Mosaic Loss of Human Y Chromosome: What, How and Why. Hum. Genet. 2020, 139, 421–446. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Dai, X.; Wu, X.; Zhou, T.; Ni, J.; Xue, J.; Wang, X. Understanding the Birth of Rupture-Prone and Irreparable Micronuclei. Chromosoma 2020, 129, 181–200. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Rodger, E.J.; Eccles, M.R. Epigenetic Drivers of Tumourigenesis and Cancer Metastasis. Semin. Cancer Biol. 2018, 51, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Luzhna, L.; Kathiria, P.; Kovalchuk, O. Micronuclei in Genotoxicity Assessment: From Genetics to Epigenetics and Beyond. Front. Genet. 2013, 4, 131. [Google Scholar] [CrossRef] [PubMed]
- Horne, S.D.; Chowdhury, S.K.; Heng, H.H.Q. Stress, Genomic Adaptation, and the Evolutionary Trade-Off. Front. Genet. 2014, 5, 92. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M. Cytokinesis-Block Micronucleus Cytome Assay Evolution into a More Comprehensive Method to Measure Chromosomal Instability. Genes 2020, 11, 1203. [Google Scholar] [CrossRef] [PubMed]
- Heng, E.; Moy, A.; Liu, G.; Heng, H.H.; Zhang, K. ER Stress and Micronuclei Cluster: Stress Response Contributes to Genome Chaos in Cancer. Front. Cell Dev. Biol. 2021, 9, 673188. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.A.; Teixeira, A.C.; Mayorano, M.B.; Carrara, H.H.A.; Andrade, J.M.; Takahashi, C.S. Basal Levels of DNA Damage Detected by Micronuclei and Comet Assays in Untreated Breast Cancer Patients and Healthy Women. Clin. Exp. Med. 2010, 10, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Pelevina, I.I.; Aleshchenko, A.V.; Antoshchina, M.M.; Vorob’eva, N.I.; Kudriashova, O.V.; Lashkova, O.E.; Lizunova, E.I.; Osipov, A.N.; Riabchenko, N.I.; Serebrianyĭ, A.M.; et al. Molecular-biological properties of blood lymphocytes of Hodgkin’s lymphoma patients. Plausible possibility of treatment effect prognosis. Radiats Biol. Radioecol. 2012, 52, 142–148. [Google Scholar] [PubMed]
- Falzetti, D.; Crescenzi, B.; Matteucci, C.; Falini, B.; Martelli, M.F.; den Berghe, M.; Mecucci, C. Genomic Instability and Recurrent Breakpoints Are Main Cytogenetic Findings in Hodgkin’s Disease. Haematologica 1999, 84, 298–305. [Google Scholar] [PubMed]
- Montgomery, N.D.; Coward, W.B.; Johnson, S.; Yuan, J.; Gulley, M.L.; Mathews, S.P.; Kaiser-Rogers, K.; Rao, K.W.; Sanger, W.G.; Sanmann, J.N.; et al. Karyotypic Abnormalities Associated with Epstein-Barr Virus Status in Classical Hodgkin Lymphoma. Cancer Genet. 2016, 209, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Tijhuis, A.E.; Foijer, F. Characterizing Chromosomal Instability-Driven Cancer Evolution and Cell Fitness at a Glance. J. Cell Sci. 2024, 137, jcs260199. [Google Scholar] [CrossRef] [PubMed]
- Pentzold, C.; Kokal, M.; Pentzold, S.; Weise, A. Sites of Chromosomal Instability in the Context of Nuclear Architecture and Function. Cell. Mol. Life Sci. 2021, 78, 2095–2103. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Garg, M.; Chandra, A.B.; Mayorga, V.S.; Ahmed, S.; Ailawadhi, S. Trends in the Risks of Secondary Cancers in Patients With Hodgkin Lymphoma. Clin. Lymphoma Myeloma Leuk. 2018, 18, 576–589.e1. [Google Scholar] [CrossRef] [PubMed]
- Salas, C.; Perez-Vera, P.; Frias, S. Genetic Abnormalities in Leukemia Secondary to Treatment in Patients with Hodgkin’s Disease. Rev. De Investig. Clin.-Clin. Transl. Investig. 2011, 63, 53–63. [Google Scholar]
- Bilban-Jakopin, C.; Bilban, M. Genotoxic Effects of Radiotherapy and Chemotherapy on Circulating Lymphocytes in Patients with Hodgkin’s Disease. Mutat. Res.-Genet. Toxicol. Environ. Mutagen. 2001, 497, 81–88. [Google Scholar] [CrossRef] [PubMed]

| ID Patient | 66 | 111 | 118 | 122 | 109 | |
|---|---|---|---|---|---|---|
| Gender | Male | Male | Female | Male | Male | |
| Diagnostic age | 18 | 22 | 25 | 28 | 33 | |
| Subtype HD | Mixed cellularity | Mixed cellularity | Nodular sclerosis | Mixed cellularity | Mixed cellularity | |
| Stage | IIB | IB | IIB | IIB | IIIA | |
| Radiotherapy | Field | Neck | Neck | Neck and mediastinum | Mediastinum | No data available |
| Dose | 30 Gy | 30 Gy | 36 Gy | 45 Gy | ||
| Sessions | 20 | 10 | 20 | 25 | ||
| Months after chemotherapy | 5 | 4 | 6 | 6 | ||
| ABVD cycles | 8 | 8 | 6 | 6 | ||
| Before treatment | Medical assessment at blood sampling | Leukopenia due to lymphopenia. | Microcytosis. High level of B-globulin. | Macrocytosis. Elevated monocyte levels | Iron deficiency anemia. Lymphopenia possibly due to invasion of cancer cells, and high levels of eosinophils, neutrophils and monocytes. | |
| During treatment (between 3rd–4th cycle) | Leukopenia due to lymphopenia, this sample showed the lowest levels of cellularity in this patient. | Normal lymphocyte levels. Elevated monocyte levels. | Macrocytosis. Leukopenia due to lymphopenia. Elevated monocytes levels. | Anaphylactic reaction to bleomycin with skin toxicity (treated with anti-steroids). Elevated eosinophil counts (commonly associated with allergic clinical features). | ||
| After treatment | Leukopenia due to lymphopenia. | Normal leukocyte levels with a slight decrease in mean platelet volume. | Mild lymphopenia. Mild macrocytosis. | Values of hematocrit and monocytes slightly above normal, while eosinophils raised very close to the value of the sample before treatment. | ||
| Months after radiotherapy/chemotherapy (sample AT) | 12/17 | 12/16 | 12/18 | 9/15 | ||
| Remission | After finished anticancer treatment, FDG-PET was negative for tumor activity. Without relapse and asymptomatic | |||||
| Sampling Time | Patient ID | # Studied Cells | DMF | Sticky Chromosomes | Free Chromatin | Unit Fiber | C-Frag | AbNM | MN | MN Cluster | TOTAL UnCA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Before Treatment | 66-I | 4295 | 0.02 | 0.00 | 0.95 | 0.00 | 0.02 | 0.00 | 0.35 | 28.03 | 29.40 |
| 109-I | 3688 | 0.00 | 0.00 | 2.03 | 0.03 | 0.00 | 0.00 | 0.70 | 12.30 | 15.00 | |
| 111-I | 3355 | 0.00 | 0.00 | 0.14 | 0.00 | 0.00 | 0.18 | 0.12 | 8.61 | 9.10 | |
| 118-I | 4762 | 0.00 | 0.02 | 5.96 | 0.00 | 0.02 | 0.14 | 0.74 | 28.85 | 35.70 | |
| 122-I | 4315 | 0.00 | 0.00 | 1.06 | 0.00 | 0.00 | 0.12 | 0.14 | 29.04 | 30.40 | |
| Mean | 4083 | 0.004 | 0.004 | 2.03 | 0.01 | 0.01 | 0.09 | 0.41 | 21.37 | 23.92 | |
| SD | 558.08 | 0.01 | 0.01 | 2.30 | 0.01 | 0.01 | 0.08 | 0.30 | 10.05 | 11.29 | |
| During Treatment | 66-II | 3903 | 0.00 | 0.00 | 0.10 | 0.00 | 0.00 | 0.13 | 0.23 | 20.60 | 21.10 |
| 109-II | 3339 | 0.00 | 0.00 | 0.09 | 0.03 | 0.00 | 0.48 | 0.00 | 8.99 | 9.60 | |
| 111-II | 4501 | 0.00 | 0.00 | 0.20 | 0.00 | 0.00 | 0.07 | 0.00 | 33.04 | 33.30 | |
| 118-II | 3778 | 0.00 | 0.00 | 2.09 | 0.00 | 0.00 | 0.11 | 0.37 | 18.03 | 20.60 | |
| 122-II | 3278 | 0.00 | 0.00 | 2.90 | 0.00 | 0.00 | 0.21 | 0.18 | 5.00 | 8.30 | |
| Mean | 3760 | 0.00 | 0.00 | 1.08 | 0.01 | 0.00 | 0.20 | 0.16 | 17.13 | 18.58 | |
| SD | 494.83 | 0.00 | 0.00 | 1.33 | 0.01 | 0.00 | 0.17 | 0.16 | 10.95 | 10.17 | |
| 1yAfter Treatment | 66-III | 3822 | 0.00 | 0.00 | 11.72 | 0.00 | 0.21 | 0.31 | 0.98 | 8.16 | 21.38 |
| 109-III | 3203 | 0.00 | 0.00 | 0.94 | 0.00 | 0.34 | 0.00 | 0.31 | 4.40 | 5.99 | |
| 111-III | 5092 | 0.00 | 0.00 | 0.10 | 0.00 | 0.00 | 0.04 | 0.20 | 40.46 | 40.79 | |
| 118-III | 7760 | 0.00 | 0.00 | 0.90 | 0.00 | 0.00 | 0.00 | 0.26 | 59.86 | 61.02 | |
| 122-III | 4067 | 0.00 | 0.00 | 0.98 | 0.00 | 0.00 | 0.17 | 0.20 | 22.57 | 23.92 | |
| Mean | 4789 | 0.00 | 0.00 | 2.93 | 0.00 | 0.11 | 0.10 | 0.39 | 27.09 | 30.62 | |
| SD | 1795.16 | 0.00 | 0.00 | 4.93 | 0.00 | 0.16 | 0.14 | 0.33 | 23.16 | 21.00 | |
| Healthy individuals | NL-31 | 3468 | 0.06 | 0.00 | 0.35 | 0.00 | 0.00 | 0.00 | 0.38 | 12.66 | 13.44 |
| NL-32 | 3105 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 | 0.03 | 0.10 | 2.71 | 2.96 | |
| NL-34 | 3464 | 0.00 | 0.00 | 1.18 | 0.00 | 0.00 | 0.03 | 0.09 | 12.04 | 13.34 | |
| NL-35 | 3114 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.10 | 2.06 | 2.15 | |
| NL-36 | 3293 | 0.00 | 0.00 | 0.97 | 0.00 | 0.061 | 0.09 | 0.46 | 7.32 | 8.90 | |
| Mean | 3289 | 0.02 | 0.00 | 0.51 | 0.00 | 0.012 | 0.03 | 0.22 | 7.36 | 8.16 | |
| SD | 178.30 | 0.03 | 0.00 | 0.54 | 0.00 | 0.027 | 0.04 | 0.18 | 4.99 | 5.44 |
| Healthy | Hodgkin’s Patients | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NL ID | Healthy | ID | BT | DT | AT | ||||||||||||
| Abnormal Metaphases | Abnormal Nuclei | UnCA | CA | Abnormal Metaphases | Abnormal Nuclei | UnCA | CA | Abnormal Metaphases | Abnormal Nuclei | UnCA | CA | Abnormal Metaphases | Abnormal Nuclei | UnCA | CA | ||
| 31 | 0.06 | 13.3 | 13.4 | 2.1 | 66 | 0.05 | 29.3 | 29.4 | 5.6 | 0.00 | 21.1 | 21.1 | 2.9 | 0.20 | 21.2 | 21.4 | 17.7 |
| 32 | 0.06 | 2.9 | 3 | 3.03 | 109 | 0.03 | 15.0 | 15 | 12.8 | 0.03 | 9.5 | 9.6 | 18.9 | 0.3 | 5.6 | 6 | 36.58 |
| 34 | 0.00 | 13.3 | 13.3 | 1.9 | 111 | 0.00 | 9.1 | 9.1 | 7.31 | 0.00 | 33.3 | 33.3 | 14.2 | 0.00 | 40.8 | 40.8 | 8.82 |
| 35 | 0.00 | 2.15 | 2.2 | 2.1 | 118 | 0.04 | 35.7 | 35.7 | 13.3 | 0.00 | 20.6 | 20.6 | 14.2 | 0.00 | 61.0 | 61 | 32.14 |
| 36 | 0.06 | 8.84 | 8.9 | 3.1 | 122 | 0.00 | 30.4 | 30.4 | 7.04 | 0.00 | 8.3 | 8.3 | 8.33 | 0.00 | 23.9 | 23.9 | 39.1 |
| Mean ± SD | 0.04 ± 0.03 | 8.12 ± 5.44 | 8.2 ± 5.4 | 2.4 ± 0.57 | Mean ± SD | 0.02 ± 0.02 | 23.8 ± 11.2 | 23.92 ± 11.29 | 9.21 ± 3.57 | 0.005 ± 0.01 | 18.5 ± 10.1 | 18.58 ± 10.17 | 11.7 ± 6.19 | 0.11 ± 0.15 | 30.5 ± 21.1 | 30.62 ± 20.99 | 26.87 ± 13.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos, S.; Molina, B.; Navarrete-Meneses, M.d.P.; Cervantes-Barragan, D.E.; Lozano, V.; Frias, S. Unclassified Chromosomal Abnormalities as an Indicator of Genomic Damage in Survivors of Hodgkin’s Lymphoma. Cancers 2025, 17, 2437. https://doi.org/10.3390/cancers17152437
Ramos S, Molina B, Navarrete-Meneses MdP, Cervantes-Barragan DE, Lozano V, Frias S. Unclassified Chromosomal Abnormalities as an Indicator of Genomic Damage in Survivors of Hodgkin’s Lymphoma. Cancers. 2025; 17(15):2437. https://doi.org/10.3390/cancers17152437
Chicago/Turabian StyleRamos, Sandra, Bertha Molina, María del Pilar Navarrete-Meneses, David E. Cervantes-Barragan, Valentín Lozano, and Sara Frias. 2025. "Unclassified Chromosomal Abnormalities as an Indicator of Genomic Damage in Survivors of Hodgkin’s Lymphoma" Cancers 17, no. 15: 2437. https://doi.org/10.3390/cancers17152437
APA StyleRamos, S., Molina, B., Navarrete-Meneses, M. d. P., Cervantes-Barragan, D. E., Lozano, V., & Frias, S. (2025). Unclassified Chromosomal Abnormalities as an Indicator of Genomic Damage in Survivors of Hodgkin’s Lymphoma. Cancers, 17(15), 2437. https://doi.org/10.3390/cancers17152437






