CT Texture Patterns Reflect HPV Status but Not Histological Differentiation in Oropharyngeal Squamous Cell Carcinoma
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Patient Selection
2.3. HPV Detection
2.4. Lymph Node Staging
2.5. CT Image Acquisition and Processing
2.6. Texture Feature Extraction
2.7. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chaturvedi, A.K.; Engels, E.A.; Pfeiffer, R.M.; Hernandez, B.Y.; Xiao, W.; Kim, E.; Jiang, B.; Goodman, M.T.; Sibug-Saber, M.; Cozen, W.; et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J. Clin. Oncol. 2011, 29, 4294–4301. [Google Scholar] [CrossRef] [PubMed]
- Gillison, M.L.; Koch, W.M.; Capone, R.B.; Spafford, M.; Westra, W.H.; Wu, L.; Zahurak, M.L.; Daniel, R.W.; Viglione, M.; Symer, D.E.; et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 2000, 92, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.I.; Nguyen-Tân, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.S., Jr.; Beadle, B.; Bishop, J.A.; Chernock, R.D.; Colasacco, C.; Lacchetti, C.; Moncur, J.T.; Rocco, J.W.; Schwartz, M.R.; Seethala, R.R.; et al. Human Papillomavirus Testing in Head and Neck Carcinomas: Guideline From the College of American Pathologists. Arch. Pathol. Lab. Med. 2018, 142, 559–597. [Google Scholar] [CrossRef]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Buch, K.; Fujita, A.; Li, B.; Kawashima, Y.; Qureshi, M.M.; Sakai, O. Using Texture Analysis to Determine Human Papillomavirus Status of Oropharyngeal Squamous Cell Carcinomas on CT. AJNR Am. J. Neuroradiol. 2015, 36, 1343–1348. [Google Scholar] [CrossRef]
- Leijenaar, R.T.; Bogowicz, M.; Jochems, A.; Hoebers, F.J.; Wesseling, F.W.; Huang, S.H.; Chan, B.; Waldron, J.N.; O’Sullivan, B.; Rietveld, D.; et al. Development and validation of a radiomic signature to predict HPV (p16) status from standard CT imaging: A multicenter study. Br. J. Radiol. 2018, 91, 20170498. [Google Scholar] [CrossRef]
- Mungai, F.; Verrone, G.B.; Pietragalla, M.; Berti, V.; Addeo, G.; Desideri, I.; Bonasera, L.; Miele, V. CT assessment of tumor heterogeneity and the potential for the prediction of human papillomavirus status in oropharyngeal squamous cell carcinoma. Radiol. Med. 2019, 124, 804–811. [Google Scholar] [CrossRef]
- Bicci, E.; Nardi, C.; Calamandrei, L.; Pietragalla, M.; Cavigli, E.; Mungai, F.; Bonasera, L.; Miele, V. Role of Texture Analysis in Oropharyngeal Carcinoma: A Systematic Review of the Literature. Cancers 2022, 14, 2445. [Google Scholar] [CrossRef]
- Trotta, B.M.; Pease, C.S.; Rasamny, J.J.; Raghavan, P.; Mukherjee, S. Oral cavity and oropharyngeal squamous cell cancer: Key imaging findings for staging and treatment planning. Radiographics 2011, 31, 339–354. [Google Scholar] [CrossRef]
- Subramaniam, N.; Poptani, H.; Schache, A.; Bhat, V.; Iyer, S.; Sunil, H.V.; Chandrasekhar Naveen, H.; Pillai, V.; Chaturvedi, P.; Krishna Shri, H.; et al. Imaging advances in oral cavity cancer and perspectives from a population in need: Consensus from the UK-India Oral Cancer Imaging Group. J. Head. Neck Physicians Surg. 2021, 9, 4–12. [Google Scholar] [CrossRef]
- M.D. Anderson Cancer Center Head and Neck Quantitative Imaging Working Group. Investigation of radiomic signatures for local recurrence using primary tumor texture analysis in oropharyngeal head and neck cancer patients. Sci. Rep. 2018, 8, 1524. [Google Scholar] [CrossRef]
- Elhalawani, H.; Mohamed, A.S.; White, A.L.; Zafereo, J.; Wong, A.J.; Berends, J.E.; AboHashem, S.; Williams, B.; Aymard, J.M.; Kanwar, A.; et al. Matched computed tomography segmentation and demographic data for oropharyngeal cancer radiomics challenges. Sci. Data 2017, 4, 170077. [Google Scholar] [CrossRef]
- Singhi, A.D.; Westra, W.H. Comparison of in situ HPV hybridization and p16 immunohistochemistry for detection of HPV-associated head and neck cancer based on a prospective clinical experience. Cancer 2010, 116, 2166–2173. [Google Scholar] [CrossRef]
- Ferreira, A.M.C.; Altemani, J.M.C.; Macedo, L.T.; Lourenço, G.J.; Lima, C.S.P. Genetic variability in cisplatin metabolic pathways and outcome of locally advanced head and neck squamous cell carcinoma patients. Sci. Rep. 2023, 13, 16762. [Google Scholar] [CrossRef]
- Brierley, J.D.; Gospodarowicz, M.K.; Wittekind, C. TNM Classification of Malignant Tumours, 8th ed.; Wiley-Blackwell: Oxford, UK, 2017. [Google Scholar]
- Nussi, A.D.; de Castro Lopes, S.L.P.; de Rosa, C.S.; Gomes, J.P.P.; Ogawa, C.M.; Braz-Silva, P.H.; Costa, A.L.F. In vivo study of cone beam computed tomography texture analysis of mandibular condyle and its correlation with gender and age. Oral. Radiol. 2023, 39, 191–197. [Google Scholar] [CrossRef]
- Costa, A.L.F.; Fardim, K.A.C.; Ribeiro, I.T.; Jardini, M.A.N.; Braz-Silva, P.H.; Orhan, K.; de Castro Lopes, S.L.P. Cone-beam computed tomography texture analysis can help differentiate odontogenic and non-odontogenic maxillary sinusitis. Imaging Sci. Dent. 2023, 53, 43–51. [Google Scholar] [CrossRef]
- De Rosa, C.S.; Bergamini, M.L.; Palmieri, M.; Sarmento, D.J.S.; de Carvalho, M.O.; Ricardo, A.L.F.; Hasseus, B.; Jonasson, P.; Braz-Silva, P.H.; Ferreira Costa, A.L. Differentiation of periapical granuloma from radicular cyst using cone beam computed tomography images texture analysis. Heliyon 2020, 6, e05194. [Google Scholar] [CrossRef]
- De Oliveira, L.A.P.; Lopes, D.L.G.; Gomes, J.P.P.; da Silveira, R.V.; Nozaki, D.V.A.; Santos, L.F.; Castellano, G.; de Castro Lopes, S.L.P.; Costa, A.L.F. Enhanced Diagnostic Precision: Assessing Tumor Differentiation in Head and Neck Squamous Cell Carcinoma Using Multi-Slice Spiral CT Texture Analysis. J. Clin. Med. 2024, 13, 4038. [Google Scholar] [CrossRef]
- Castellano, G.; Bonilha, L.; Li, L.M.; Cendes, F. Texture analysis of medical images. Clin. Radiol. 2004, 59, 1061–1069. [Google Scholar] [CrossRef]
- Lubner, M.G.; Smith, A.D.; Sandrasegaran, K.; Sahani, D.V.; Pickhardt, P.J. CT texture analysis: Definitions, Applications, Biologic Correlates, and Challenges. Radiographics 2017, 37, 1483–1503. [Google Scholar] [CrossRef] [PubMed]
- Parmar, C.; Grossmann, P.; Rietveld, D.; Michelle, M.R.; Lambin, P.; Aerts, H.J.W.L. Radiomic Machine-Learning Classifiers for Prognostic Biomarkers of Head and Neck Cancer. Front. Oncol. 2015, 5, 272. [Google Scholar] [CrossRef] [PubMed]
- Bagher-Ebadian, H.; Siddiqui, F.; Ghanem, A.I.; Zhu, S.; Lu, M.; Movsas, B.; Chetty, I.J. Radiomics outperforms clinical factors in characterizing human papilloma virus (HPV) for patients with oropharyngeal squamous cell carcinomas. Biomed. Phys. Eng. Express 2022, 8, 044001. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Han, M.; Kim, K.S.; Shin, S.J.; Choi, J.W.; Ha, E.J. Discrimination of HPV status using CT texture analysis: Tumour heterogeneity in oropharyngeal squamous cell carcinomas. Neuroradiology 2019, 61, 1415–1424. [Google Scholar] [CrossRef]
- Queiroz, P.M.; Fardim, K.C.; Costa, A.L.F.; Matheus, R.A.; Lopes, S.L.P.C. Texture analysis in cone-beam computed tomographic images of medication-related osteonecrosis of the jaw. Imaging Sci. Dent. 2023, 53, 109–115. [Google Scholar] [CrossRef]
- Jung, Y.J.; Han, M.; Ha, E.J.; Choi, J.W. Differentiation of salivary gland tumors through tumor heterogeneity: A comparison between pleomorphic adenoma and Warthin tumor using CT texture analysis. Neuroradiology 2020, 62, 1451–1458. [Google Scholar] [CrossRef]
- Succaria, F.; Kvistborg, P.; Stein, J.E.; Engle, E.L.; McMiller, T.L.; Rooper, L.M.; Thompson, E.; Berger, A.E.; van den Brekel, M.; Zuur, C.L.; et al. Characterization of the tumor immune microenvironment in human papillomavirus-positive and -negative head and neck squamous cell carcinomas. Cancer Immunol. Immunother. 2021, 70, 1227–1237. [Google Scholar] [CrossRef]
- Liao, K.Y.; Chiu, C.C.; Chiang, W.C.; Yu-Rou, C.; Geoffrey, Z.; Shih-Neng, Y.; Tzung-Chi, H. Radiomics Features Analysis of PET Images in Oropharyngeal and Hypopharyngeal Cancer. Medicine 2019, 98, e15446. [Google Scholar] [CrossRef]
- Meyer, H.J.; Hamerla, G.; Höhn, A.K.; Surov, A. CT Texture Analysis-Correlations With Histopathology Parameters in Head and Neck SCC. Front. Oncology. 2019, 9, 444. [Google Scholar] [CrossRef]
- Kim, T.Y.; Lee, J.Y.; Lee, Y.J.; Dong Woo, P.; Kyung, T.; Yun Young, C. CT Texture Analysis of Tonsil Cancer: Discrimination from Normal Palatine Tonsils. PLoS ONE 2021, 16, e0255835. [Google Scholar] [CrossRef]

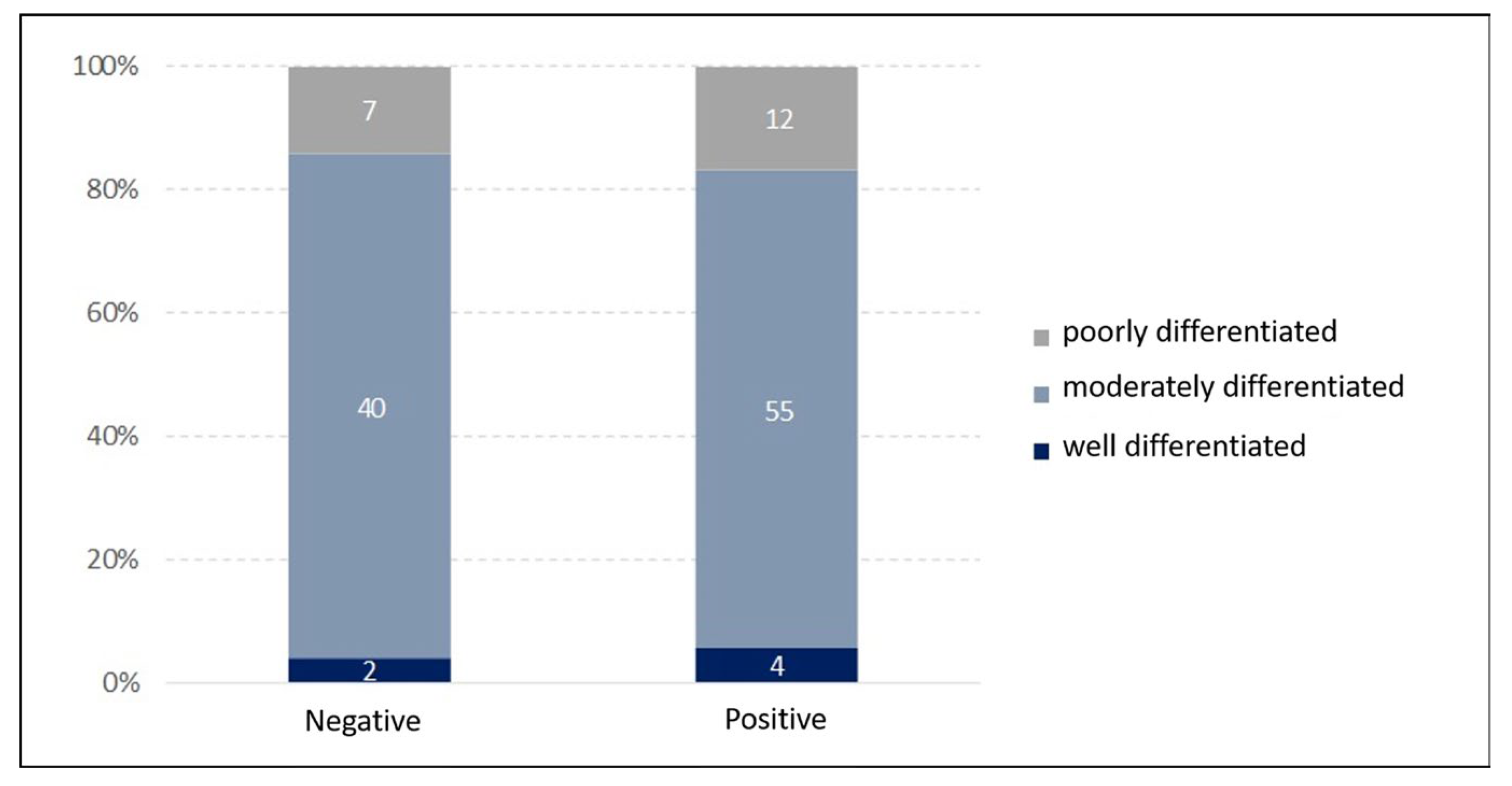
| Variable | Negative HPV Status | Positive HPV Status | ||||
|---|---|---|---|---|---|---|
| Good Differentiation (n = 2) | Moderated Differentiation (n = 40) | Poor Differentiation (n = 7) | Good Differentiation (n = 4) | Moderated Differentiation (n = 55) | Poor Differentiation (n = 12) | |
| Smoking: | ||||||
| No | 0 | 2 | 1 | 0 | 6 | 3 |
| Yes | 2 | 38 | 6 | 4 | 49 | 9 |
| Alcohol use: | ||||||
| No | 0 | 6 | 0 | 1 | 11 | 2 |
| Yes | 2 | 34 | 7 | 3 | 44 | 10 |
| Gender: | ||||||
| Female | 0 | 8 | 0 | 0 | 5 | 1 |
| Male | 2 | 32 | 7 | 4 | 50 | 11 |
| Clinical stage: | ||||||
| I | 1 | 1 | 0 | 0 | 1 | 1 |
| II | 0 | 0 | 0 | 0 | 2 | 0 |
| III | 0 | 7 | 0 | 1 | 9 | 2 |
| IVA | 1 | 20 | 5 | 1 | 24 | 6 |
| IVB | 0 | 8 | 1 | 1 | 12 | 1 |
| IVC | 0 | 4 | 1 | 1 | 6 | 2 |
| X | 0 | 0 | 0 | 0 | 1 | 0 |
| Variable | Negative | Positive | |||
|---|---|---|---|---|---|
| Moderate Differentiation (n = 40) | Poor Differentiation (n = 7) | Good Differentiation (n = 4) | Moderate Differentiation (n = 55) | Poor Differentiation (n = 12) | |
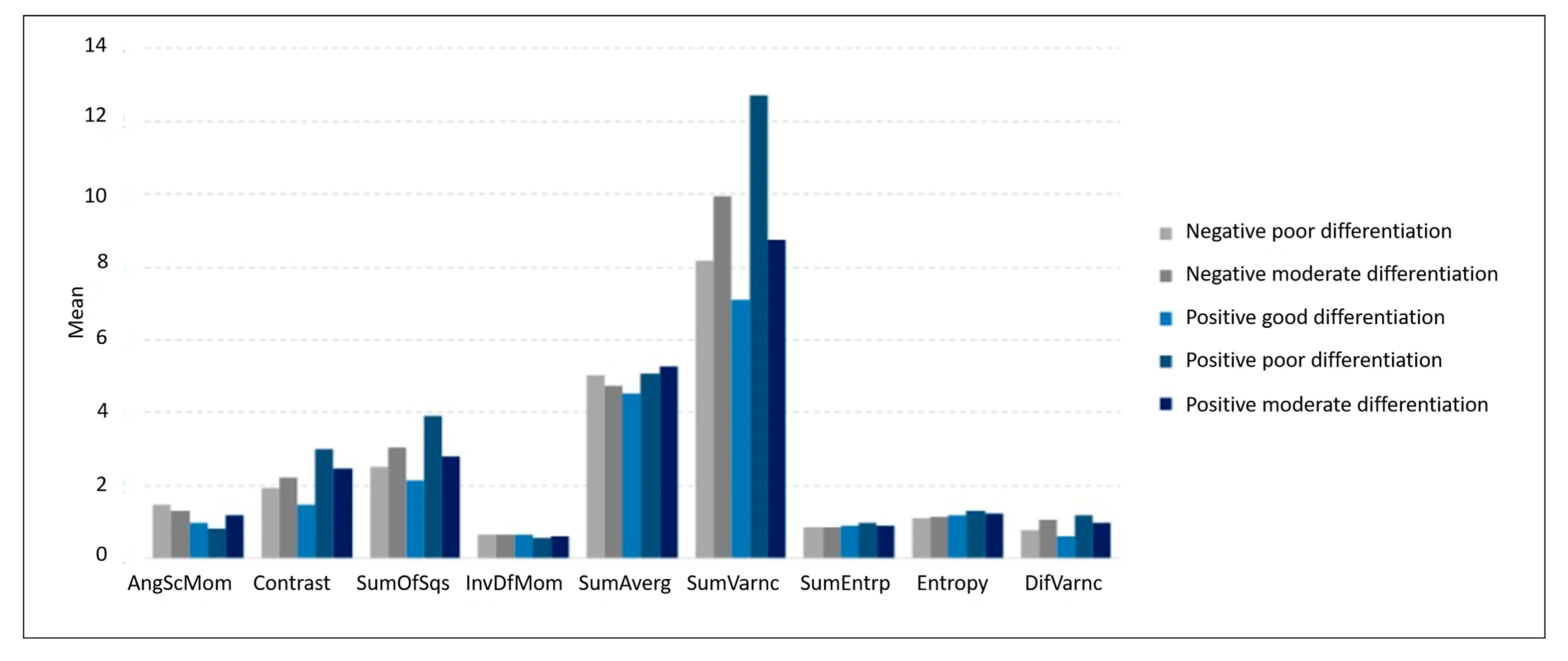
| AngScMom | 0.15 (0.14) | 0.13 (0.07) | 0.10 (0.05) | 0.08 (0.07) | 0.12 (0.11) |
| Contrast | 1.93 (2.12) | 2.22 (2.15) | 1.47 (0.73) | 3.00 (3.18) | 2.49 (2.88) |
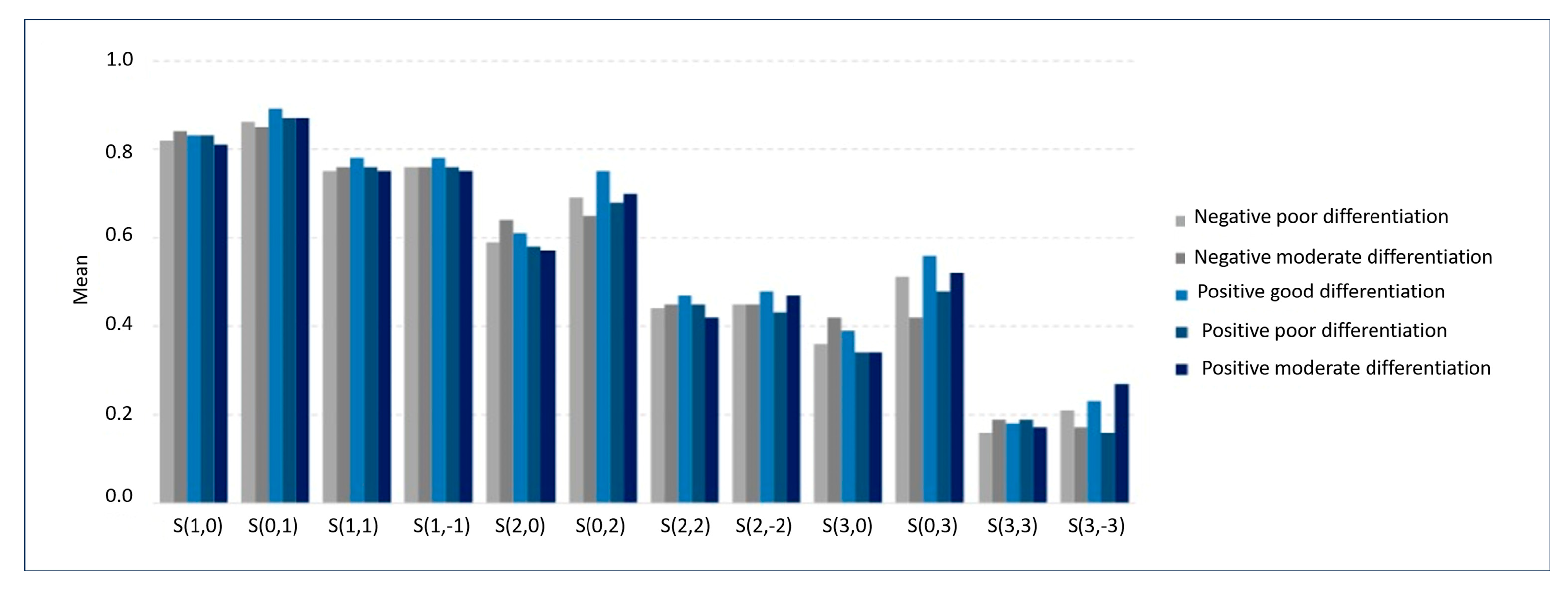
| S (1, 0) Correlat | 0.82 (0.08) | 0.84 (0.03) | 0.83 (0.12) | 0.83 (0.10) | 0.81 (0.10) |
| S (0, 1) Correlat | 0.86 (0.06) | 0.85 (0.05) | 0.89 (0.03) | 0.87 (0.08) | 0.87 (0.06) |
| S (1, 1) Correlat | 0.75 (0.10) | 0.76 (0.05) | 0.78 (0.13) | 0.76 (0.12) | 0.75 (0.12) |
| S (1, −1) Correlat | 0.76 (0.10) | 0.76 (0.06) | 0.78 (0.12) | 0.76 (0.13) | 0.75 (0.15) |
| S (2, 0) Correlat | 0.59 (0.18) | 0.64 (0.09) | 0.61 (0.29) | 0.58 (0.21) | 0.57 (0.23) |
| S (0, 2) Correlat | 0.69 (0.12) | 0.65 (0.07) | 0.75 (0.07) | 0.68 (0.14) | 0.70 (0.14) |
| S (2, 2) Correlat | 0.44 (0.19) | 0.45 (0.10) | 0.47 (0.30) | 0.45 (0.22) | 0.42 (0.20) |
| S (2, −2) Correlat | 0.45 (0.20) | 0.45 (0.09) | 0.48 (0.24) | 0.43 (0.19) | 0.47 (0.25) |
| S (3, 0) Correlat | 0.36 (0.24) | 0.42 (0.12) | 0.39 (0.36) | 0.34 (0.25) | 0.34 (0.26) |
| S (0, 3) Correlat | 0.51 (0.15) | 0.42 (0.11) | 0.56 (0.12) | 0.48 (0.18) | 0.52 (0.19) |
| S (3, 3) Correlat | 0.16 (0.25) | 0.19 (0.11) | 0.18 (0.33) | 0.19 (0.25) | 0.17 (0.19) |
| S (3, −3) Correlat | 0.21 (0.23) | 0.17 (0.08) | 0.23 (0.18) | 0.16 (0.19) | 0.27 (0.27) |
| SumOfSqs | 2.52 (2.94) | 3.04 (3.41) | 2.14 (1.56) | 3.91 (5.43) | 2.81 (2.17) |
| InvDfMom | 0.67 (0.14) | 0.67 (0.12) | 0.65 (0.09) | 0.59 (0.13) | 0.63 (0.15) |
| SumAverg | 50.3 (12.7) | 47.5 (11.6) | 45.3 (12.0) | 50.8 (10.8) | 52.8 (11.8) |
| SumVarnc | 8.16 (10.1) | 9.93 (11.5) | 7.10 (5.71) | 12.7 (18.8) | 8.76 (6.31) |
| SumEntrp | 0.85 (0.24) | 0.87 (0.19) | 0.92 (0.14) | 0.97 (0.19) | 0.92 (0.21) |
| Entropy | 1.12 (0.38) | 1.14 (0.30) | 1.20 (0.21) | 1.33 (0.32) | 1.24 (0.36) |
| DifVarnc | 0.78 (0.73) | 1.05 (1.07) | 0.62 (0.27) | 1.18 (1.17) | 0.99 (1.02) |
| Variable | Negative | Positive | p-Value | |||
|---|---|---|---|---|---|---|
| Moderate Differentiation (n = 40) | Poor Differentiation (n = 7) | Good Differentiation (n = 4) | Moderate Differentiation (n = 55) | Poor Differentiation (n = 12) | ||
| AngScMom | 0.09 [0.06; 0.18] | 0.13 [0.08; 0.17] | 0.09 [0.08; 0.11] | 0.07 [0.04; 0.12] | 0.08 [0.06; 0.15] | 0.038 |
| Contrast | 1.26 [0.63; 2.31] | 0.96 [0.73; 3.57] | 1.57 [1.13; 1.91] | 1.89 [1.05; 3.87] | 1.50 [0.97; 2.34] | 0.206 |
| S (1, 0) Correlat | 0.83 [0.79; 0.87] | 0.82 [0.81; 0.86] | 0.87 [0.79; 0.91] | 0.84 [0.80; 0.89] | 0.85 [0.76; 0.88] | 0.797 |
| S (0, 1) Correlat | 0.87 [0.82; 0.90] | 0.84 [0.82; 0.87] | 0.89 [0.87; 0.92] | 0.89 [0.86; 0.91] | 0.87 [0.84; 0.91] | 0.283 |
| S (1, 1) Correlat | 0.77 [0.68; 0.81] | 0.75 [0.72; 0.79] | 0.82 [0.73; 0.87] | 0.79 [0.72; 0.84] | 0.76 [0.65; 0.85] | 0.685 |
| S (1, −1) Correlat | 0.76 [0.70; 0.81] | 0.75 [0.72; 0.79] | 0.83 [0.77; 0.84] | 0.77 [0.74; 0.83] | 0.80 [0.71; 0.84] | 0.692 |
| S (2, 0) Correlat | 0.61 [0.49; 0.68] | 0.62 [0.58; 0.71] | 0.72 [0.56; 0.77] | 0.63 [0.50; 0.72] | 0.66 [0.51; 0.71] | 0.888 |
| S (0, 2) Correlat | 0.69 [0.64; 0.77] | 0.67 [0.59; 0.70] | 0.77 [0.73; 0.79] | 0.72 [0.61; 0.77] | 0.67 [0.62; 0.80] | 0.577 |
| S (2, 2) Correlat | 0.43 [0.34; 0.56] | 0.50 [0.43; 0.52] | 0.60 [0.43; 0.64] | 0.44 [0.34; 0.59] | 0.47 [0.28; 0.57] | 0.921 |
| S (2, −2) Correlat | 0.44 [0.33; 0.60] | 0.44 [0.42; 0.48] | 0.57 [0.44; 0.61] | 0.44 [0.35; 0.54] | 0.56 [0.38; 0.61] | 0.698 |
| S (3, 0) Correlat | 0.38 [0.22; 0.53] | 0.39 [0.34; 0.51] | 0.55 [0.35; 0.59] | 0.37 [0.19; 0.51] | 0.44 [0.24; 0.50] | 0.819 |
| S (0, 3) Correlat | 0.51 [0.42; 0.63] | 0.38 [0.34; 0.51] | 0.62 [0.55; 0.63] | 0.50 [0.34; 0.60] | 0.47 [0.40; 0.67] | 0.565 |
| S (3, 3) Correlat | 0.15 [0.03; 0.31] | 0.20 [0.17; 0.25] | 0.29 [0.13; 0.34] | 0.18 [0.02; 0.34] | 0.17 [0.05; 0.28] | 0.989 |
| S (3, −3) Correlat | 0.23 [0.07; 0.37] | 0.18 [0.14; 0.21] | 0.25 [0.18; 0.30] | 0.17 [0.03; 0.28] | 0.34 [0.10; 0.46] | 0.418 |
| SumOfSqs | 1.57 [0.86; 3.28] | 1.27 [0.75; 4.91] | 1.75 [1.16; 2.73] | 2.33 [1.33; 4.38] | 2.12 [1.35; 4.37] | 0.330 |
| InvDfMom | 0.67 [0.59; 0.77] | 0.69 [0.62; 0.73] | 0.63 [0.60; 0.68] | 0.59 [0.49; 0.69] | 0.64 [0.59; 0.70] | 0.095 |
| SumAverg | 51.1 [44.6; 57.6] | 51.0 [42.9; 54.2] | 45.7 [37.4; 53.6] | 50.6 [44.7; 56.5] | 56.0 [48.4; 61.1] | 0.729 |
| SumVarnc | 5.11 [2.74; 10.4] | 4.11 [2.27; 15.8] | 5.42 [3.15; 9.37] | 7.63 [4.18; 12.7] | 7.33 [3.96; 15.0] | 0.374 |
| SumEntrp | 0.88 [0.74; 1.01] | 0.85 [0.75; 0.92] | 0.92 [0.82; 1.01] | 0.97 [0.85; 1.10] | 0.94 [0.80; 1.08] | 0.111 |
| Entropy | 1.19 [0.92; 1.36] | 1.05 [0.96; 1.26] | 1.24 [1.13; 1.31] | 1.34 [1.08; 1.57] | 1.27 [1.04; 1.41] | 0.085 |
| DifVarnc | 0.50 [0.32; 0.96] | 0.42 [0.37; 1.50] | 0.68 [0.49; 0.81] | 0.77 [0.49; 1.47] | 0.66 [0.46; 0.98] | 0.275 |
| Variable | Good (n = 6) | Differentiation Grade Moderate (n = 95) | Poor (n = 19) | p-Value | |||
|---|---|---|---|---|---|---|---|
| Mean (S.D.) | Median [Q1; Q3] | Mean (S.D.) | Median [Q1; Q3] | Mean (S.D.) | Median [Q1; Q3] | ||
| AngScMom | 0.12 (0.06) | 0.09 [0.08; 0.15] | 0.11 (0.11) | 0.08 [0.05; 0.14] | 0.12 (0.09) | 0.09 [0.05; 0.17] | 0.453 |
| Contrast | 1.21 (0.70) | 1.12 [0.61; 1.69] | 2.55 (2.82) | 1.64 [0.81; 3.47] | 2.39 (2.58) | 1.35 [0.86; 2.44] | 0.538 |
| S (1, 0) Correlat | 0.84 (0.10) | 0.87 [0.82; 0.92] | 0.83 (0.09) | 0.84 [0.80; 0.89] | 0.82 (0.08) | 0.84 [0.80; 0.88] | 0.729 |
| S (0, 1) Correlat | 0.90 (0.04) | 0.89 [0.87; 0.92] | 0.87 (0.07) | 0.88 [0.84; 0.90] | 0.86 (0.06) | 0.86 [0.83; 0.90] | 0.334 |
| S (1, 1) Correlat | 0.80 (0.11) | 0.83 [0.78; 0.88] | 0.76 (0.12) | 0.78 [0.71; 0.83] | 0.75 (0.10) | 0.75 [0.69; 0.82] | 0.407 |
| S (1, −1) Correlat | 0.79 (0.11) | 0.83 [0.76; 0.86] | 0.76 (0.12) | 0.76 [0.72; 0.82] | 0.76 (0.12) | 0.79 [0.72; 0.84] | 0.571 |
| S (2, 0) Correlat | 0.64 (0.24) | 0.72 [0.65; 0.79] | 0.58 (0.20) | 0.62 [0.49; 0.71] | 0.60 (0.19) | 0.66 [0.55; 0.71] | 0.376 |
| S (0, 2) Correlat | 0.77 (0.08) | 0.77 [0.74; 0.80] | 0.69 (0.13) | 0.70 [0.62; 0.77] | 0.68 (0.12) | 0.67 [0.61; 0.74] | 0.170 |
| S (2, 2) Correlat | 0.54 (0.26) | 0.62 [0.57; 0.66] | 0.45 (0.21) | 0.44 [0.34; 0.58] | 0.43 (0.17) | 0.50 [0.32; 0.56] | 0.227 |
| S (2, −2) Correlat | 0.51 (0.22) | 0.57 [0.48; 0.63] | 0.44 (0.19) | 0.44 [0.34; 0.57] | 0.46 (0.21) | 0.51 [0.42; 0.60] | 0.325 |
| S (3, 0) Correlat | 0.42 (0.29) | 0.55 [0.41; 0.59] | 0.35 (0.24) | 0.37 [0.20; 0.52] | 0.37 (0.22) | 0.41 [0.28; 0.50] | 0.414 |
| S (0, 3) Correlat | 0.61 (0.14) | 0.62 [0.60; 0.64] | 0.49 (0.17) | 0.50 [0.38; 0.62] | 0.48 (0.17) | 0.46 [0.38; 0.55] | 0.200 |
| S (3, 3) Correlat | 0.26 (0.30) | 0.30 [0.28; 0.40] | 0.18 (0.25) | 0.17 [0.02; 0.34] | 0.17 (0.16) | 0.19 [0.10; 0.28] | 0.476 |
| S (3, −3) Correlat | 0.27 (0.18) | 0.25 [0.20; 0.38] | 0.18 (0.21) | 0.19 [0.04; 0.29] | 0.23 (0.22) | 0.20 [0.13; 0.41] | 0.435 |
| SumOfSqs | 1.90 (1.35) | 1.74 [0.89; 2.20] | 3.33 (4.58) | 1.94 [1.13; 3.88] | 2.90 (2.60) | 1.92 [0.88; 4.56] | 0.800 |
| InvDfMom | 0.68 (0.09) | 0.67 [0.62; 0.76] | 0.62 (0.14) | 0.61 [0.53; 0.73] | 0.64 (0.14) | 0.65 [0.58; 0.73] | 0.502 |
| SumAverg | 46.0 (14.0) | 45.7 [33.7; 56.7] | 50.6 (11.6) | 50.6 [44.6; 57.2] | 50.9 (11.7) | 52.5 [42.9; 58.6] | 0.719 |
| SumVarnc | 6.40 (4.88) | 5.42 [2.73; 7.73] | 10.8 (15.8) | 5.96 [3.60; 12.2] | 9.19 (8.30) | 6.05 [2.65; 15.4] | 0.830 |
| SumEntrp | 0.89 (0.15) | 0.91 [0.78; 0.99] | 0.92 (0.22) | 0.94 [0.81; 1.06] | 0.90 (0.20) | 0.88 [0.76; 1.04] | 0.784 |
| Entropy | 1.14 (0.22) | 1.20 [0.98; 1.26] | 1.24 (0.36) | 1.27 [1.00; 1.48] | 1.20 (0.34) | 1.19 [0.96; 1.39] | 0.595 |
| DifVarnc | 0.51 (0.26) | 0.46 [0.29; 0.74] | 1.01 (1.02) | 0.65 [0.38; 1.28] | 1.01 (1.01) | 0.53 [0.40; 1.02] | 0.522 |
| Variable | Negative (n = 49) | Positive (n = 71) | p-Value | ||
|---|---|---|---|---|---|
| Mean (S.D.) | Median [Q1; Q3] | Median (S.D.) | Median [Q1; Q3] | ||
| AngScMom | 0.15 (0.13) | 0.10 [0.06; 0.17] | 0.09 (0.07) | 0.07 [0.04; 0.12] | 0.003 |
| Contrast | 1.93 (2.07) | 1.03 [0.68; 2.20] | 2.83 (3.05) | 1.81 [0.99; 3.60] | 0.016 |
| S (1, 0) Correlat | 0.83 (0.07) | 0.83 [0.80; 0.87] | 0.83 (0.10) | 0.84 [0.80; 0.89] | 0.273 |
| S (0, 1) Correlat | 0.86 (0.06) | 0.86 [0.82; 0.90] | 0.87 (0.07) | 0.89 [0.85; 0.91] | 0.079 |
| S (1, 1) Correlat | 0.76 (0.10) | 0.77 [0.69; 0.81] | 0.76 (0.12) | 0.79 [0.71; 0.84] | 0.287 |
| S (1, −1) Correlat | 0.76 (0.10) | 0.75 [0.70; 0.81] | 0.76 (0.13) | 0.79 [0.74; 0.83] | 0.276 |
| S (2, 0) Correlat | 0.60 (0.17) | 0.62 [0.52; 0.70] | 0.58 (0.22) | 0.64 [0.50; 0.72] | 0.981 |
| S (0, 2) Correlat | 0.69 (0.11) | 0.69 [0.63; 0.74] | 0.69 (0.14) | 0.72 [0.62; 0.77] | 0.652 |
| S (2, 2) Correlat | 0.45 (0.18) | 0.45 [0.36; 0.56] | 0.45 (0.22) | 0.45 [0.32; 0.59] | 0.887 |
| S (2, −2) Correlat | 0.46 (0.19) | 0.44 [0.36; 0.60] | 0.44 (0.20) | 0.46 [0.35; 0.57] | 0.731 |
| S (3, 0) Correlat | 0.37 (0.22) | 0.39 [0.26; 0.53] | 0.34 (0.25) | 0.38 [0.20; 0.51] | 0.522 |
| S (0, 3) Correlat | 0.50 (0.15) | 0.51 [0.39; 0.60] | 0.49 (0.17) | 0.50 [0.38; 0.63] | 0.875 |
| S (3, 3) Correlat | 0.18 (0.23) | 0.18 [0.04; 0.29] | 0.19 (0.24) | 0.18 [0.02; 0.33] | 0.985 |
| S (3, −3) Correlat | 0.21 (0.22) | 0.21 [0.10; 0.35] | 0.18 (0.21) | 0.20 [0.03; 0.29] | 0.401 |
| SumOfSqs | 2.55 (2.93) | 1.53 [0.79; 3.16] | 3.63 (4.89) | 2.32 [1.29; 4.27] | 0.034 |
| InvDfMom | 0.67 (0.14) | 0.69 [0.59; 0.76] | 0.60 (0.13) | 0.60 [0.49; 0.69] | 0.006 |
| SumAverg | 49.8 (12.7) | 51.0 [43.9; 57.5] | 50.8 (11.0) | 51.2 [43.9; 56.9] | 0.763 |
| SumVarnc | 8.28 (10.0) | 5.08 [2.37; 10.2] | 11.7 (16.9) | 7.49 [4.15; 13.6] | 0.044 |
| SumEntrp | 0.85 (0.23) | 0.87 [0.73; 0.98] | 0.96 (0.19) | 0.97 [0.83; 1.09] | 0.008 |
| Entropy | 1.12 (0.36) | 1.13 [0.93; 1.36] | 1.31 (0.32) | 1.29 [1.08; 1.52] | 0.005 |
| DifVarnc | 0.80 (0.77) | 0.46 [0.33; 0.94] | 1.11 (1.12) | 0.77 [0.48; 1.33] | 0.024 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Oliveira, L.A.P.; Peresi, C.E.I.L.; Nozaki, D.V.A.; Costa, E.F.D.; Santos, L.F.; Lima, C.S.P.; Lopes, S.L.P.d.C.; Costa, A.L.F. CT Texture Patterns Reflect HPV Status but Not Histological Differentiation in Oropharyngeal Squamous Cell Carcinoma. Cancers 2025, 17, 2317. https://doi.org/10.3390/cancers17142317
de Oliveira LAP, Peresi CEIL, Nozaki DVA, Costa EFD, Santos LF, Lima CSP, Lopes SLPdC, Costa ALF. CT Texture Patterns Reflect HPV Status but Not Histological Differentiation in Oropharyngeal Squamous Cell Carcinoma. Cancers. 2025; 17(14):2317. https://doi.org/10.3390/cancers17142317
Chicago/Turabian Stylede Oliveira, Lays Assolini Pinheiro, Caio Elias Irajaya Lobo Peresi, Daniel Vitor Aguiar Nozaki, Ericka Francislaine Dias Costa, Lana Ferreira Santos, Carmen Silvia Passos Lima, Sérgio Lúcio Pereira de Castro Lopes, and Andre Luiz Ferreira Costa. 2025. "CT Texture Patterns Reflect HPV Status but Not Histological Differentiation in Oropharyngeal Squamous Cell Carcinoma" Cancers 17, no. 14: 2317. https://doi.org/10.3390/cancers17142317
APA Stylede Oliveira, L. A. P., Peresi, C. E. I. L., Nozaki, D. V. A., Costa, E. F. D., Santos, L. F., Lima, C. S. P., Lopes, S. L. P. d. C., & Costa, A. L. F. (2025). CT Texture Patterns Reflect HPV Status but Not Histological Differentiation in Oropharyngeal Squamous Cell Carcinoma. Cancers, 17(14), 2317. https://doi.org/10.3390/cancers17142317








