TIMP1 Overexpression in Ovarian Cancer Spheroids: Implications for Prognosis, Resistance, and Metastatic Potential
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Ovarian Cancer Spheroids
2.3. Cell Treatments and Reagents
2.4. Cell Viability Assays
2.5. Ovarian Cancer Chemoresistant Cells
2.6. Ovarian Anoikis-Resistant Cells
2.7. Production of TIMP1 Vector
2.8. Ovarian Cells Overexpressing TIMP1
2.9. RNA Extraction and Real-Time PCR
2.10. Bulk RNA Sequencing
2.11. Western Blot
2.12. Enzyme-Linked Immunosorbent Assay (ELISA)
2.13. Wound Healing Assay
2.14. Soft Agar
2.15. Zebrafish Model
2.16. Bioinformatic Analysis
2.17. Statistical Analysis
3. Results
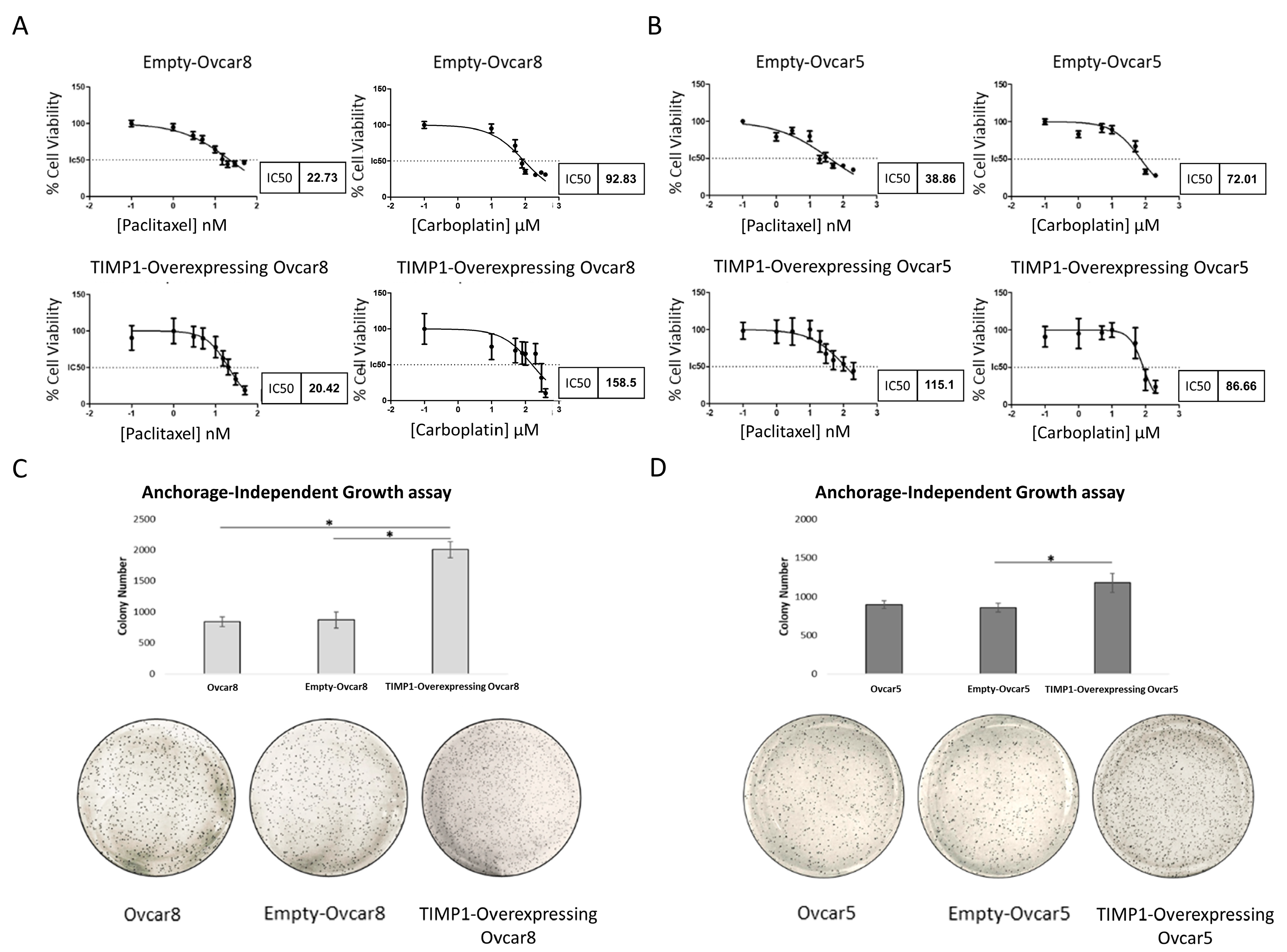
3.1. Ovarian Cancer Spheroids Are Characterized by Low Sensitivity to Standard Therapy and Resistance to Apoptosis
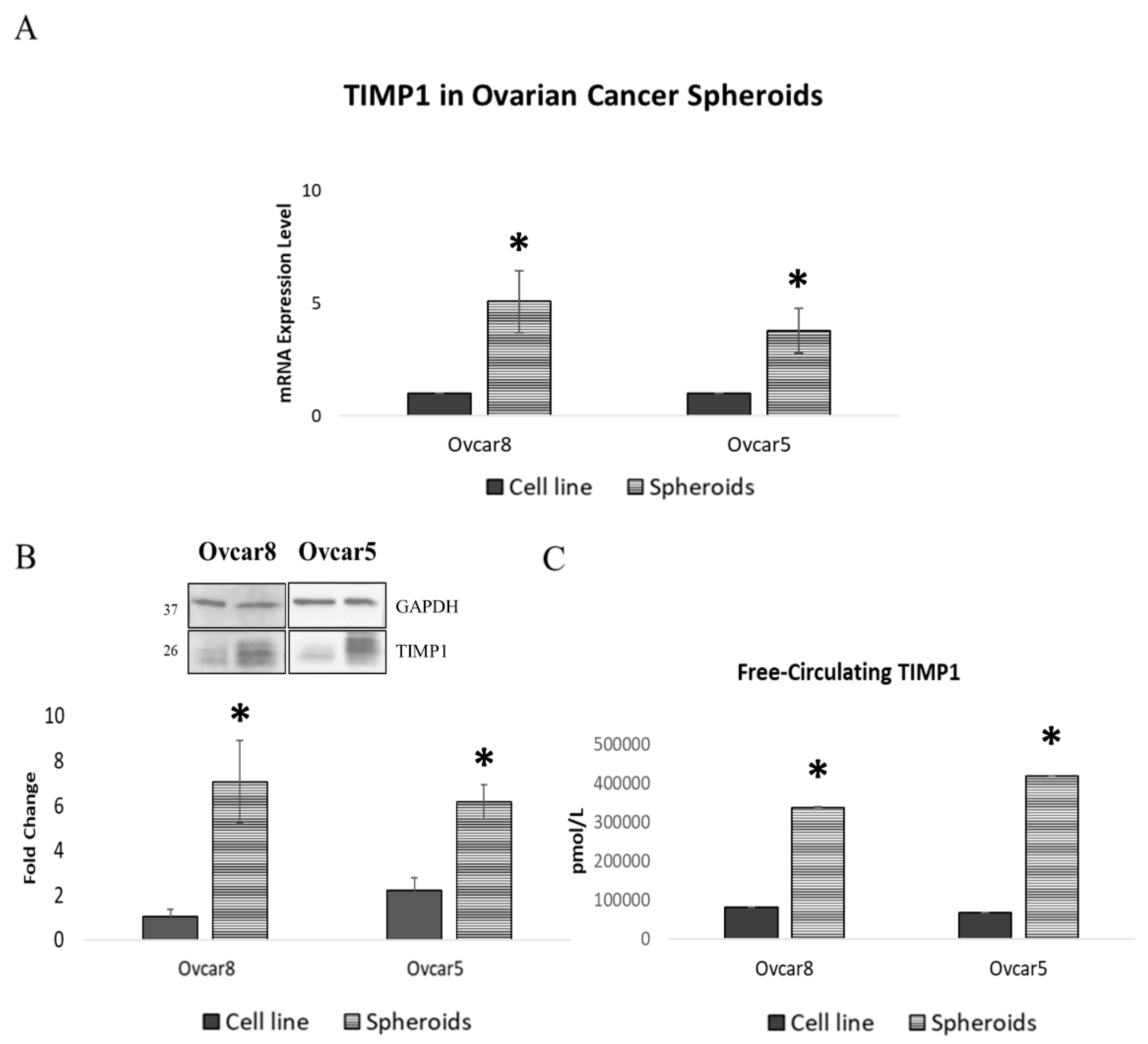
3.2. Ovarian Cancer Spheroids Show Increased TIMP1 Expression Levels
3.3. High TIMP1 Expression Correlates with Worse Overall Survival in Patients
3.4. Chemoresistant Ovarian Cancer Cell Lines Show Increased TIMP1 Expression
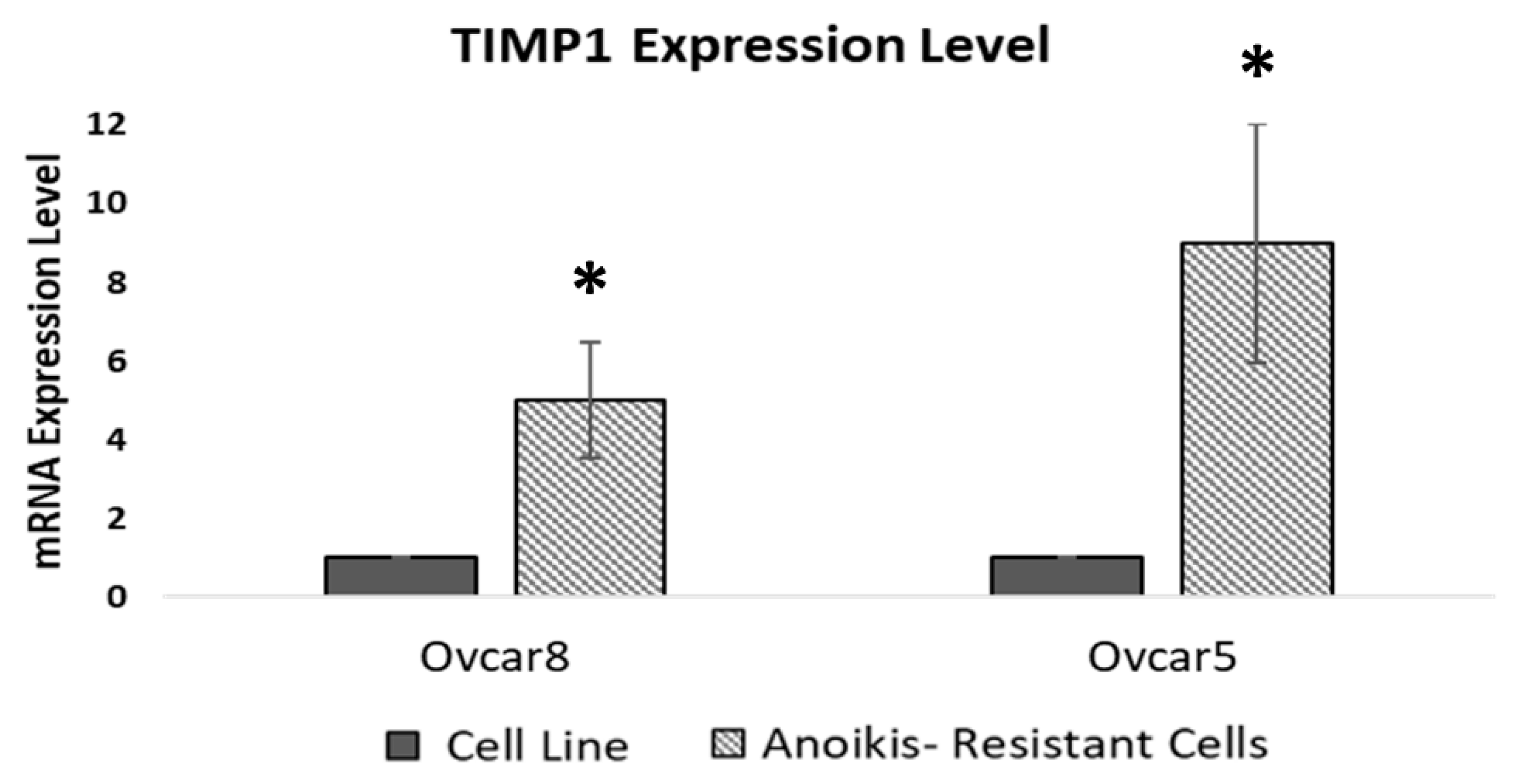
3.5. Anoikis-Resistant Cell Lines Display Increased TIMP1 Expression
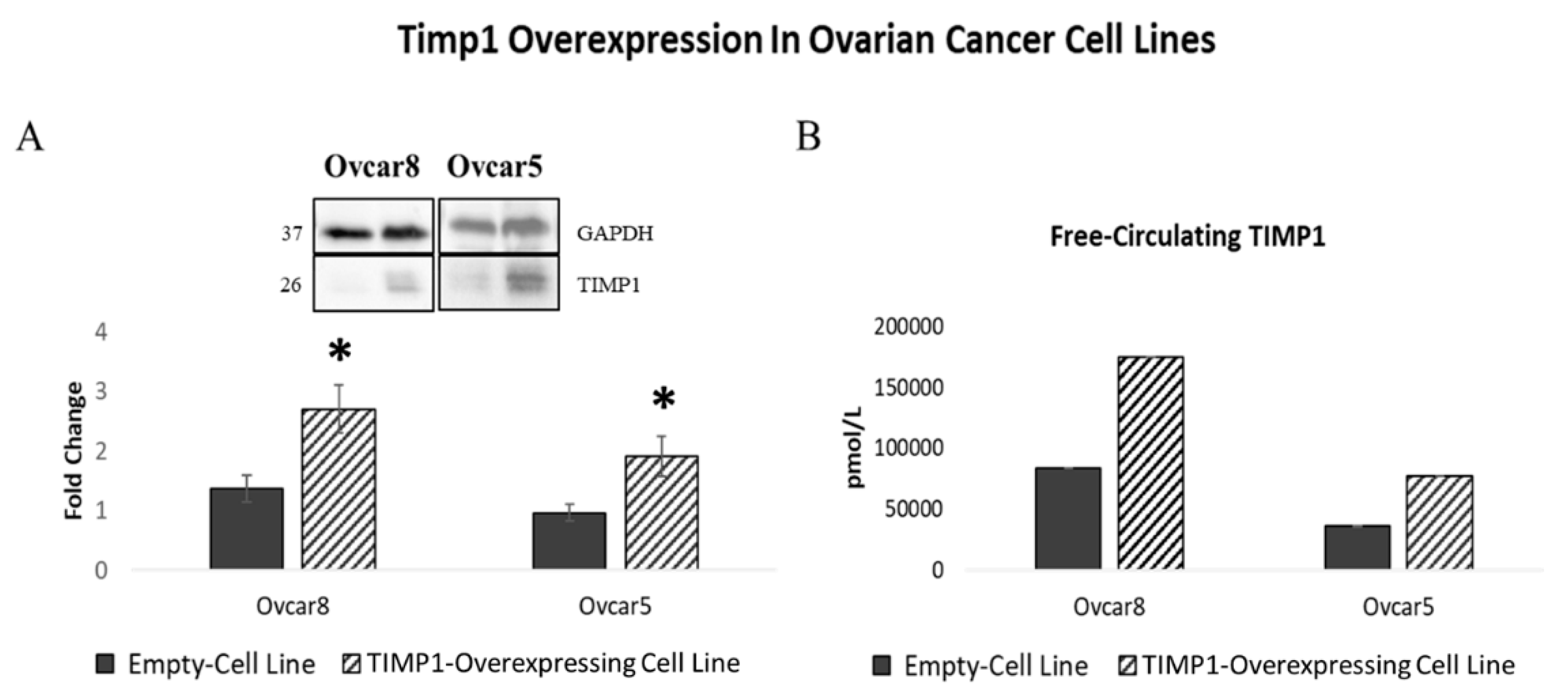
3.6. TIMP1 Overexpression Is Linked to Therapy and Anoikis Resistance
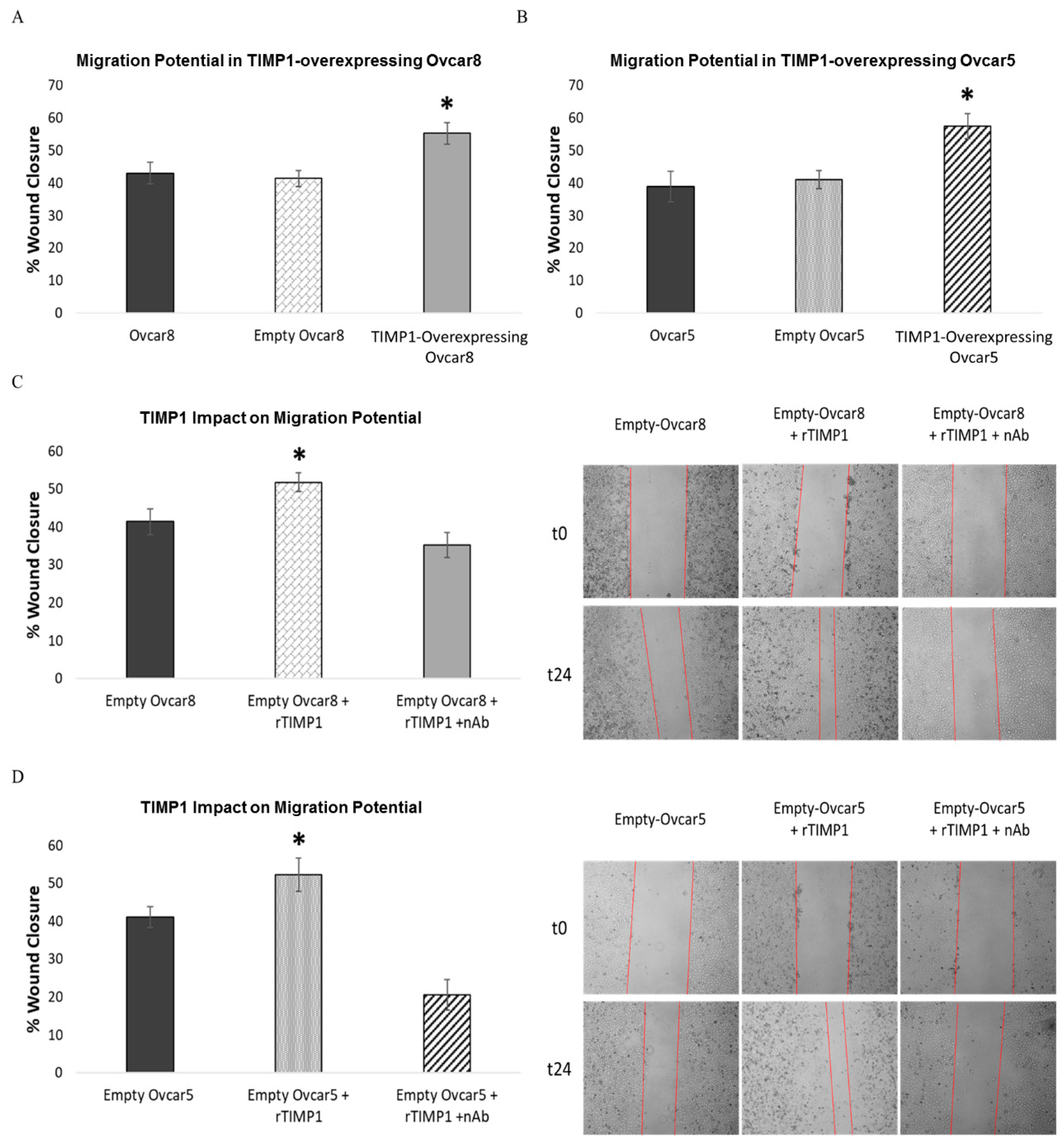
3.7. TIMP1 Overexpression Increases Ovarian Cancer Cell Migration
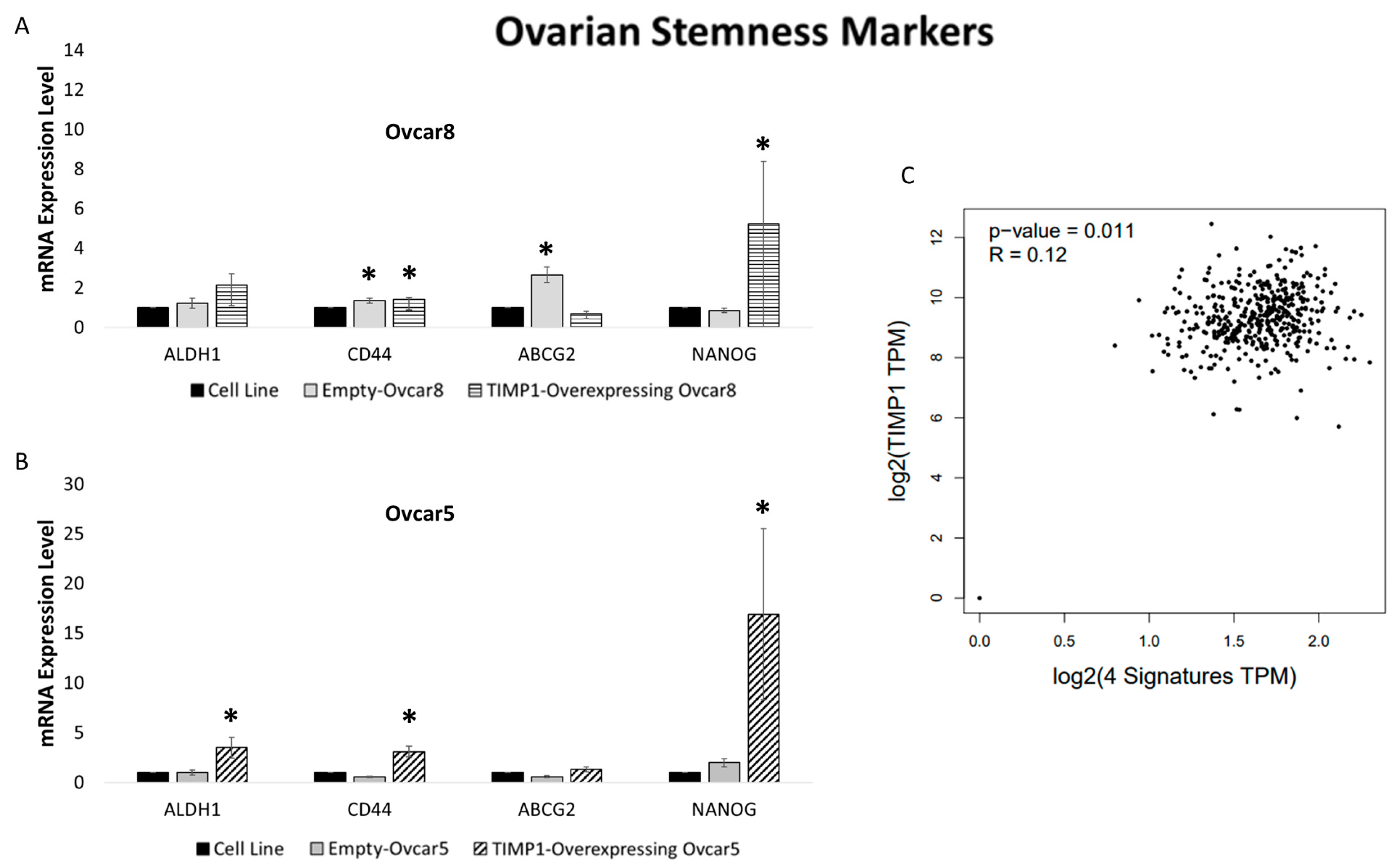
3.8. TIMP1 Overexpression in Ovarian Cancer Cell Lines Is Related to Stem Cell Marker Expression
3.9. TIMP1-Overexpressing Ovcar5 Cells Exhibit Powered Metastatic Potential In Vivo
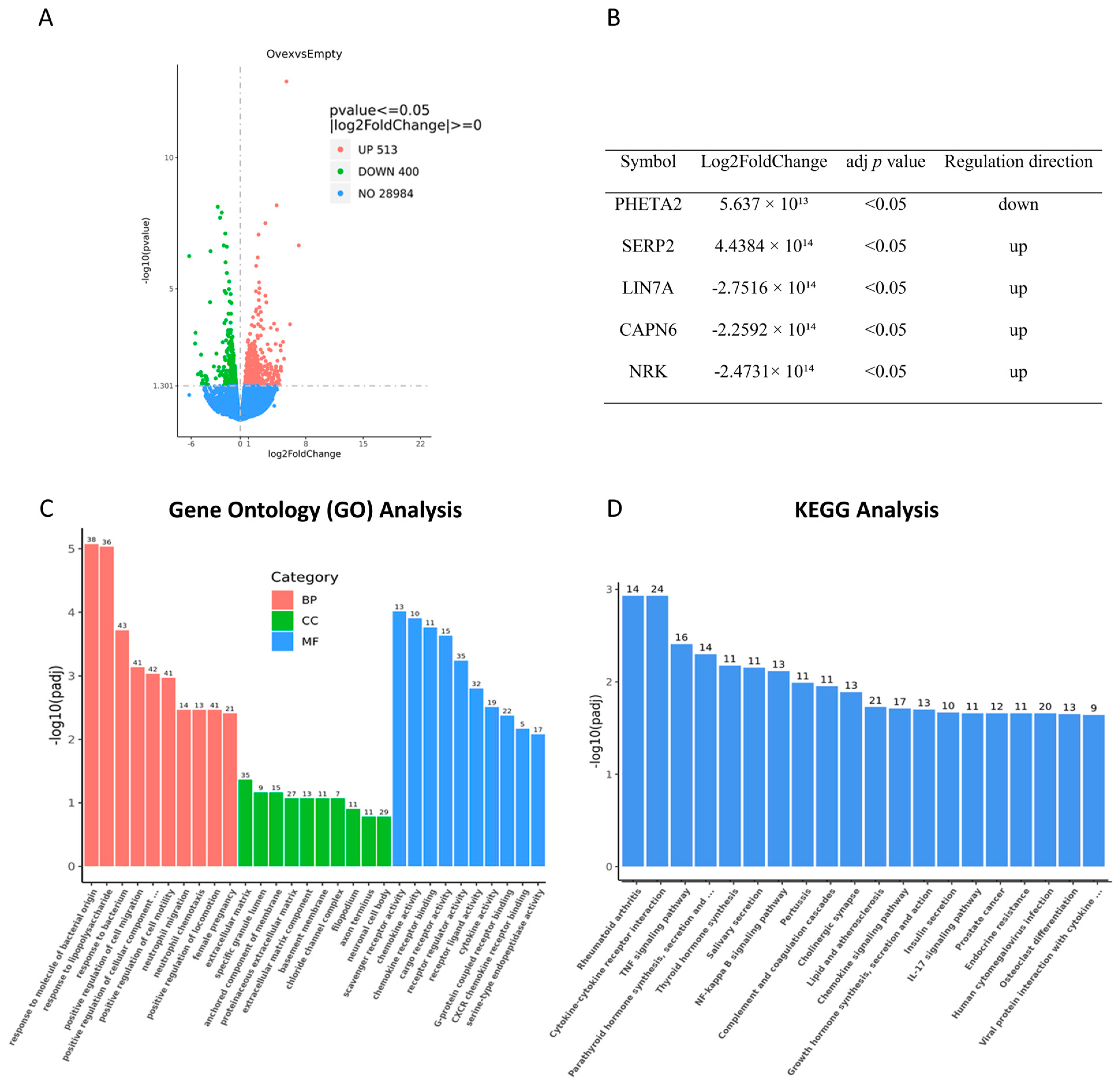
3.10. TIMP1 Overexpression Significantly Alters the Transcriptome of Ovarian Cancer Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gaona-Luviano, P.; Medina-Gaona, L.A.; Magaña-Pérez, K. Epidemiology of ovarian cancer. Chin. Clin. Oncol. 2020, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Kossaï, M.; Leary, A.; Scoazec, J.Y.; Genestie, C. Ovarian Cancer: A Heterogeneous Disease. Pathobiology 2018, 85, 41–49. [Google Scholar] [CrossRef]
- De Leo, A.; Santini, D.; Ceccarelli, C.; Santandrea, G.; Palicelli, A.; Acquaviva, G.; Chiarucci, F.; Rosini, F.; Ravegnini, G.; Pession, A.; et al. What Is New on Ovarian Carcinoma: Integrated Morphologic and Molecular Analysis Following the New 2020 World Health Organization Classification of Female Genital Tumors. Diagnostics 2021, 11, 697. [Google Scholar] [CrossRef] [PubMed]
- Salomon-Perzyński, A.; Salomon-Perzyńska, M.; Michalski, B.; Skrzypulec-Plinta, V. High-grade serous ovarian cancer: The clone wars. Arch. Gynecol. Obstet. 2017, 295, 569–576. [Google Scholar] [CrossRef]
- Li, S.S.; Ma, J.; Wong, A.S.T. Chemoresistance in ovarian cancer: Exploiting cancer stem cell metabolism. J. Gynecol. Oncol. 2018, 29, e32. [Google Scholar] [CrossRef]
- Ardizzoia, A.; Jemma, A.; Redaelli, S.; Silva, M.; Bentivegna, A.; Lavitrano, M.; Conconi, D. AhRR and PPP1R3C: Potential Prognostic Biomarkers for Serous Ovarian Cancer. Int. J. Mol. Sci. 2023, 24, 11455. [Google Scholar] [CrossRef]
- Shapira, I.; Oswald, M.; Lovecchio, J.; Khalili, H.; Menzin, A.; Whyte, J.; Dos Santos, L.; Liang, S.; Bhuiya, T.; Keogh, M.; et al. Circulating biomarkers for detection of ovarian cancer and predicting cancer outcomes. Br. J. Cancer 2014, 110, 976–983. [Google Scholar] [CrossRef]
- Punzón-Jiménez, P.; Lago, V.; Domingo, S.; Simón, C.; Mas, A. Molecular Management of High-Grade Serous Ovarian Carcinoma. Int. J. Mol. Sci. 2022, 23, 13777. [Google Scholar] [CrossRef]
- Al Habyan, S.; Kalos, C.; Szymborski, J.; McCaffrey, L. Multicellular detachment generates metastatic spheroids during intra-abdominal dissemination in epithelial ovarian cancer. Oncogene 2018, 37, 5127–5135. [Google Scholar] [CrossRef]
- Saed, G.M.; Diamond, M.P.; Fletcher, N.M. Updates of the role of oxidative stress in the pathogenesis of ovarian cancer. Gynecol. Oncol. 2017, 145, 595–602. [Google Scholar] [CrossRef]
- Gunay, G.; Kirit, H.A.; Kamatar, A.; Baghdasaryan, O.; Hamsici, S.; Acar, H. The effects of size and shape of the ovarian cancer spheroids on the drug resistance and migration. Gynecol. Oncol. 2020, 159, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Iżycka, N.; Zaborowski, M.P.; Ciecierski, Ł.; Jaz, K.; Szubert, S.; Miedziarek, C.; Rezler, M.; Piątek-Bajan, K.; Synakiewicz, A.; Jankowska, A.; et al. Cancer Stem Cell Markers-Clinical Relevance and Prognostic Value in High-Grade Serous Ovarian Cancer (HGSOC) Based on The Cancer Genome Atlas Analysis. Int. J. Mol. Sci. 2023, 24, 12746. [Google Scholar] [CrossRef] [PubMed]
- Justo, B.L.; Jasiulionis, M.G. Characteristics of TIMP1, CD63, and β1-Integrin and the Functional Impact of Their Interaction in Cancer. Int. J. Mol. Sci. 2021, 22, 9319. [Google Scholar] [CrossRef]
- Ishiguro, T.; Ohata, H.; Sato, A.; Yamawaki, K.; Enomoto, T.; Okamoto, K. Tumor-derived spheroids: Relevance to cancer stem cells and clinical applications. Cancer Sci. 2017, 108, 283–289. [Google Scholar] [CrossRef]
- Halim, H.; Luanpitpong, S.; Chanvorachote, P. Acquisition of anoikis resistance up-regulates caveolin-1 expression in human non-small cell lung cancer cells. Anticancer Res. 2012, 32, 1649–1658. [Google Scholar]
- Ryan, M.D.; Drew, J. Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J. 1994, 13, 928–937. [Google Scholar] [CrossRef]
- Villa, C.; Colombo, G.; Meneghini, S.; Gotti, C.; Moretti, M.; Ferini-Strambi, L.; Chisci, E.; Giovannoni, R.; Becchetti, A.; Combi, R.; et al. CHRNA2 and nocturnal frontal lobe epilepsy: Identification and characterization of a novel loss of function mutation. Front. Mol. Neurosci. 2019, 12, 17. [Google Scholar] [CrossRef]
- Khin, M.; Davis, L.J.; Lantvit, D.D.; Orjala, J.; Burdette, J.E. Aulosirazole stimulates FOXO3a nuclear translocation to regulate apoptosis and cell-cycle progression in high-grade serous ovarian cancer (HGSOC) cells. Mol. Pharmacol. 2024, 106, 145–154. [Google Scholar] [CrossRef]
- Ebott, J.; McAdams, J.; Kim, C.; Jansen, C.; Woodman, M.; De La Cruz, P.; Schrol, C.; Ribeiro, J.; James, N. Enhanced amphiregulin exposure promotes modulation of the high grade serous ovarian cancer tumor immune microenvironment. Front. Pharmacol. 2024, 15, 1375421. [Google Scholar] [CrossRef]
- Valdivia, A.; Cowan, M.; Cardenas, H.; Isac, A.; Zhao, G.; Huang, H. E2F1 mediates competition, proliferation and response to cisplatin in cohabitating resistant and sensitive ovarian cancer cells. Front. Oncol. 2024, 14, 1304691. [Google Scholar] [CrossRef]
- Mitra, A.K.; Davis, D.A.; Tomar, S.; Roy, L.; Gurler, H.; Xie, J.; Lantvit, D.D.; Cardenas, H.; Fang, F.; Liu, Y.; et al. In vivo tumor growth of high-grade serous ovarian cancer cell lines. Gynecol. Oncol. 2015, 138, 372–377. [Google Scholar] [CrossRef]
- Fogg, K.C.; Miller, A.E.; Li, Y.; Flanigan, W.; Walker, A.; O’Shea, A.; Kendziorski, C.; Kreeger, P.K. Ovarian cancer cells direct monocyte differentiation through a non-canonical pathway. BMC Cancer 2020, 20, 1008. [Google Scholar] [CrossRef]
- Kim, Y.N.; Koo, K.H.; Sung, J.Y.; Yun, U.J.; Kim, H. Anoikis resistance: An essential prerequisite for tumor metastasis. Int. J. Cell Biol. 2012, 2012, 306879. [Google Scholar] [CrossRef]
- Pieterse, Z.; Amaya-Padilla, M.A.; Singomat, T.; Binju, M.; Madjid, B.D.; Yu, Y.; Kaur, P. Ovarian cancer stem cells and their role in drug resistance. Int. J. Biochem. Cell Biol. 2019, 106, 117–126. [Google Scholar] [CrossRef]
- Kenda Suster, N.; Virant-Klun, I. Presence and role of stem cells in ovarian cancer. World J. Stem Cells. 2019, 11, 383–397. [Google Scholar] [CrossRef]
- Liao, J.; Qian, F.; Tchabo, N.; Mhawech-Fauceglia, P.; Beck, A.; Qian, Z.; Wang, X.; Huss, W.J.; Lele, S.B.; Morrison, C.D.; et al. Ovarian cancer spheroid cells with stem cell-like properties contribute to tumor generation, metastasis and chemotherapy resistance through hypoxia-resistant metabolism. PLoS ONE 2014, 9, e84941. [Google Scholar] [CrossRef]
- Frąszczak, K.; Barczyński, B. The role of cancer stem cell markers in ovarian cancer. Cancers 2023, 16, 40. [Google Scholar] [CrossRef]
- Jemma, A.; Ardizzoia, A.; Redaelli, S.; Bentivegna, A.; Lavitrano, M.; Conconi, D. Prognostic relevance of copy number losses in ovarian cancer. Genes 2024, 15, 1487. [Google Scholar] [CrossRef]
- Wilczyński, J.R.; Wilczyński, M.; Paradowska, E. Cancer stem cells in ovarian cancer—A source of tumor success and a challenging target for novel therapies. Int. J. Mol. Sci. 2022, 23, 2496. [Google Scholar] [CrossRef] [PubMed]
- Keyvani, V.; Farshchian, M.; Esmaeili, S.-A.; Yari, H.; Moghbeli, M.; Kazemi Nezhad, S.-R.; Abbaszadegan, M.R. Ovarian cancer stem cells and targeted therapy. J. Ovarian Res. 2019, 12, 120. [Google Scholar] [CrossRef] [PubMed]
- Abreu, M.; Cabezas-Sainz, P.; Alonso-Alconada, L.; Ferreirós, A.; Mondelo-Macía, P.; Lago-Lestón, R.M.; Abalo, A.; Díaz, E.; Palacios-Zambrano, S.; Rojo-Sebastian, A.; et al. Circulating tumor cells characterization revealed TIMP1 as a potential therapeutic target in ovarian cancer. Cells 2020, 9, 1218. [Google Scholar] [CrossRef]
- Song, G.; Xu, S.; Zhang, H.; Wang, Y.; Xiao, C.; Jiang, T.; Wu, L.; Zhang, T.; Sun, X.; Zhong, L.; et al. TIMP1 is a prognostic marker for the progression and metastasis of colon cancer through FAK-PI3K/AKT and MAPK pathway. J. Exp. Clin. Cancer Res. 2016, 35, 148. [Google Scholar] [CrossRef]
- Würtz, S.Ø.; Schrohl, A.-S.; Sørensen, N.M.; Lademann, U.; Christensen, I.J.; Mouridsen, H.; Brünner, N. Tissue inhibitor of metalloproteinases-1 in breast cancer. Endocr. Relat. Cancer 2005, 12, 215–227. [Google Scholar] [CrossRef]
- Escalona, R.M.; Kannourakis, G.; Findlay, J.K.; Ahmed, N. Expression of TIMPs and MMPs in ovarian tumors, ascites, ascites-derived cells, and cancer cell lines: Characteristic modulatory response before and after chemotherapy treatment. Front. Oncol. 2022, 11, 796588. [Google Scholar] [CrossRef]
- Dorna, D.; Paluszczak, J. Targeting cancer stem cells as a strategy for reducing chemotherapy resistance in head and neck cancers. J. Cancer Res. Clin. Oncol. 2023, 149, 13417–13435. [Google Scholar] [CrossRef]
- Sonego, M.; Poletto, E.; Pivetta, E.; Nicoloso, M.S.; Pellicani, R.; Rampioni Vinciguerra, G.L.; Citron, F.; Sorio, R.; Mongiat, M.; Baldassarre, G. TIMP-1 is overexpressed and secreted by platinum-resistant epithelial ovarian cancer cells. Cells 2019, 9, 6. [Google Scholar] [CrossRef]
- Zhu, D.; Zha, X.; Hu, M.; Tao, A.; Zhou, H.; Zhou, X.; Sun, Y. High expression of TIMP-1 in human breast cancer tissues is a predictive factor for resistance to paclitaxel-based chemotherapy. Med. Oncol. 2012, 29, 3207–3215. [Google Scholar] [CrossRef]
- Wang, T.; Lv, J.-H.; Zhang, X.-F.; Li, C.-J.; Han, X.; Sun, Y.-J. Tissue inhibitor of metalloproteinase-1 protects MCF-7 breast cancer cells from paclitaxel-induced apoptosis by decreasing the stability of cyclin B1. Int. J. Cancer. 2010, 126, 362–370. [Google Scholar] [CrossRef]
- Hermawan, A.; Putri, H. Bioinformatics analysis of TIMP1, HK2, and IGFBP7 as potential biomarkers and therapeutic targets of paclitaxel resistance in breast cancer. Middle East J. Cancer 2021, 12, 198–207. [Google Scholar] [CrossRef]
- Paoli, P.; Giannoni, E.; Chiarugi, P. Anoikis molecular pathways and its role in cancer progression. Biochim. Biophys. Acta. 2013, 1833, 3481–3498. [Google Scholar] [CrossRef] [PubMed]
- Sattari Fard, F.; Jalilzadeh, N.; Mehdizadeh, A.; Sajjadian, F.; Velaei, K. Understanding and targeting anoikis in metastasis for cancer therapies. Cell Biol. Int. 2023, 47, 683–698. [Google Scholar] [CrossRef]
- Ricca, T.I.; Liang, G.; Suenaga, A.P.M.; Han, S.W.; Jones, P.A.; Jasiulionis, M.G. Tissue inhibitor of metalloproteinase 1 expression associated with gene demethylation confers anoikis resistance in early phases of melanocyte malignant transformation. Transl. Oncol. 2009, 2, 329–340. [Google Scholar] [CrossRef]
- Stuelten, C.H.; Parent, C.A.; Montell, D.J. Cell motility in cancer invasion and metastasis: Insights from simple model organisms. Nat. Rev. Cancer. 2018, 18, 296–312. [Google Scholar] [CrossRef]
- Forte, D.; Salvestrini, V.; Corradi, G.; Rossi, L.; Catani, L.; Lemoli, R.M.; Cavo, M.; Curti, A. The tissue inhibitor of metalloproteinases-1 (TIMP-1) promotes survival and migration of acute myeloid leukemia cells through CD63/PI3K/Akt/p21 signaling. Oncotarget 2017, 8, 2261–2274. [Google Scholar] [CrossRef] [PubMed]
- Roth, S.M.; Berens, E.B.; Sharif, G.M.; Glasgow, E.; Wellstein, A. Cancer Cell Invasion and Metastasis in Zebrafish Models (Danio rerio). Methods Mol. Biol. 2021, 2294, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Raldúa, D.; Piña, B. In vivo zebrafish assays for analyzing drug toxicity. Expert Opin. Drug Metab. Toxicol. 2014, 10, 685–697. [Google Scholar] [CrossRef]
- Lindahl, G.; Fjellander, S.; Selvaraj, K.; Vildeval, M.; Ali, Z.; Almter, R.; Erkstam, A.; Rodriguez, G.V.; Abrahamsson, A.; Kersley, A.R.; et al. Zebrafish tumour xenograft models: A prognostic approach to epithelial ovarian cancer. NPJ Precis Oncol. 2024, 8, 53. [Google Scholar] [CrossRef]
- Abreu, M.; Cabezas-Sainz, P.; Pereira-Veiga, T.; Falo, C.; Abalo, A.; Morilla, I.; Curiel, T.; Cueva, J.; Rodríguez, C.; Varela-Pose, V.; et al. Looking for a better characterization of triple-negative breast cancer by means of circulating tumor cells. J. Clin. Med. 2020, 9, 353. [Google Scholar] [CrossRef]
- Cheng, G.; Fan, X.; Hao, M.; Wang, J.; Zhou, X.; Sun, X. Higher levels of TIMP-1 expression are associated with a poor prognosis in triple-negative breast cancer. Mol. Cancer 2016, 15, 30. [Google Scholar] [CrossRef]
- Chang, Y.-H.; Chiu, Y.-J.; Cheng, H.-C.; Liu, F.-J.; Lai, W.-W.; Chang, H.-J.; Liao, P.-C. Down-regulation of TIMP-1 inhibits cell migration, invasion, and metastatic colonization in lung adenocarcinoma. Tumour Biol. 2015, 36, 3957–3967. [Google Scholar] [CrossRef]
- Hu, X.; Li, Y.; Kong, D.; Hu, L.; Liu, D.; Wu, J. Long noncoding RNA CASC9 promotes LIN7A expression via miR-758-3p to facilitate the malignancy of ovarian cancer. J. Cell Physiol. 2019, 234, 10800–10808. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xiao, D.; Tang, F.; Gao, H.; Li, X. CAPN6 in disease: An emerging therapeutic target. Int. J. Mol. Med. 2020, 46, 1644–1652. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Gu, L.J.; Zhu, N.; Wang, L.; Cai, M.C.; Jia, J.S.; Rong, S.; Yuan, W.J. Calpain 6 inhibits autophagy in inflammatory environments: A preliminary study on myoblasts and a chronic kidney disease rat model. Int. J. Mol. Med. 2021, 48, 194. [Google Scholar] [CrossRef] [PubMed]
- Dantas, E.; Murthy, A.; Ahmed, T.; Ahmed, M.; Ramsamooj, S.; Hurd, M.A.; Lam, T.; Malbari, M.; Agrusa, C.; Elemento, O.; et al. TIMP1 is an early biomarker for detection and prognosis of lung cancer. Clin. Transl. Med. 2023, 13, e1391. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jemma, A.; Ardizzoia, A.; Villa, C.; Bonomo, S.; Mauri, M.; Reale, C.; Ambrosino, C.; Cadamuro, M.; Lavitrano, M.; Conconi, D. TIMP1 Overexpression in Ovarian Cancer Spheroids: Implications for Prognosis, Resistance, and Metastatic Potential. Cancers 2025, 17, 1605. https://doi.org/10.3390/cancers17101605
Jemma A, Ardizzoia A, Villa C, Bonomo S, Mauri M, Reale C, Ambrosino C, Cadamuro M, Lavitrano M, Conconi D. TIMP1 Overexpression in Ovarian Cancer Spheroids: Implications for Prognosis, Resistance, and Metastatic Potential. Cancers. 2025; 17(10):1605. https://doi.org/10.3390/cancers17101605
Chicago/Turabian StyleJemma, Andrea, Alessandra Ardizzoia, Chiara Villa, Sara Bonomo, Mario Mauri, Carla Reale, Concetta Ambrosino, Massimiliano Cadamuro, Marialuisa Lavitrano, and Donatella Conconi. 2025. "TIMP1 Overexpression in Ovarian Cancer Spheroids: Implications for Prognosis, Resistance, and Metastatic Potential" Cancers 17, no. 10: 1605. https://doi.org/10.3390/cancers17101605
APA StyleJemma, A., Ardizzoia, A., Villa, C., Bonomo, S., Mauri, M., Reale, C., Ambrosino, C., Cadamuro, M., Lavitrano, M., & Conconi, D. (2025). TIMP1 Overexpression in Ovarian Cancer Spheroids: Implications for Prognosis, Resistance, and Metastatic Potential. Cancers, 17(10), 1605. https://doi.org/10.3390/cancers17101605








