A Metastatic Cancer Expression Generator (MetGen): A Generative Contrastive Learning Framework for Metastatic Cancer Generation
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Challenges in the Current State
1.2. Our Contributions
1.2.1. MetGen Avoids Mode Collapse
1.2.2. MetGen Ensures Stable Training
1.2.3. MetGen Has an Interpretable Latent Space
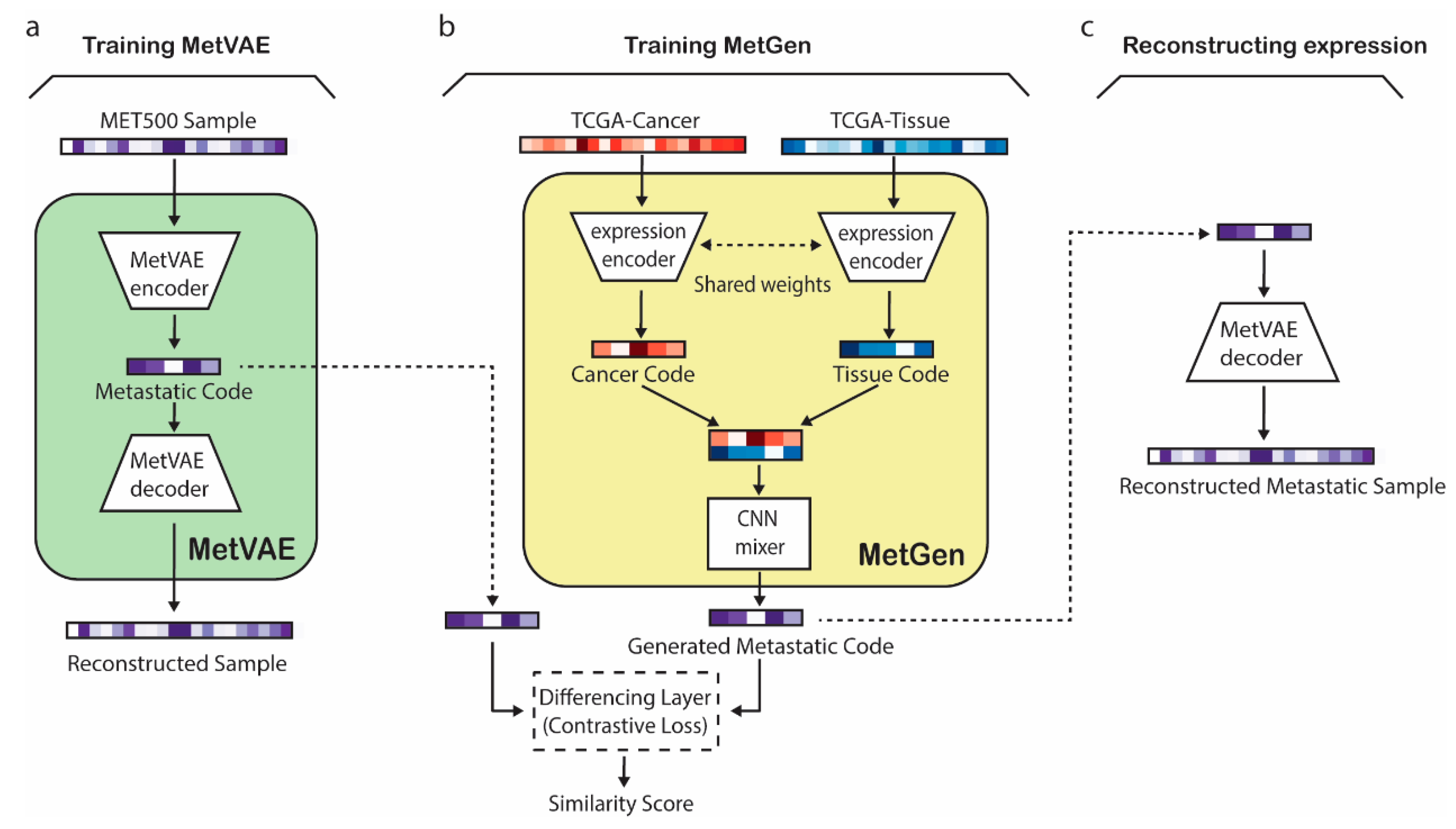
2. Materials and Methods
2.1. Data Preprocessing
Training and Testing Data Preparation
2.2. MetVAE Model
2.3. Contrastive Learning for MetGen
2.3.1. TCGA Encoder
2.3.2. CNN Mixer
2.3.3. Contrastive Learner
2.4. Overview of MetGen Framework
2.5. Statistical Tools
2.5.1. Standard DNN Classifiers
2.5.2. GSVA
2.5.3. Differential Analysis
3. Results and Discussion
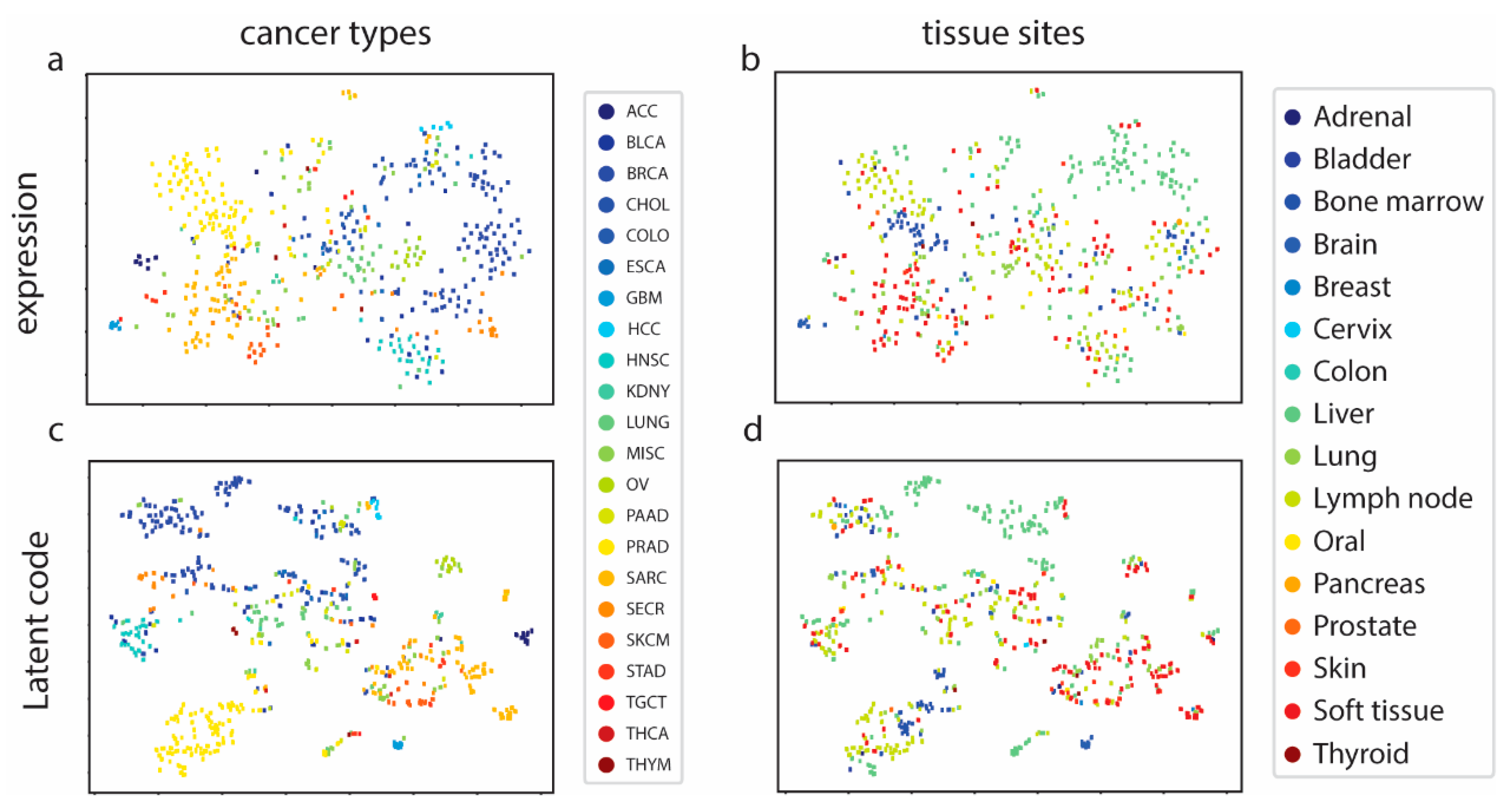
3.1. MetVAE Codes Capture Cancer and Tissue Features of Metastatic Samples
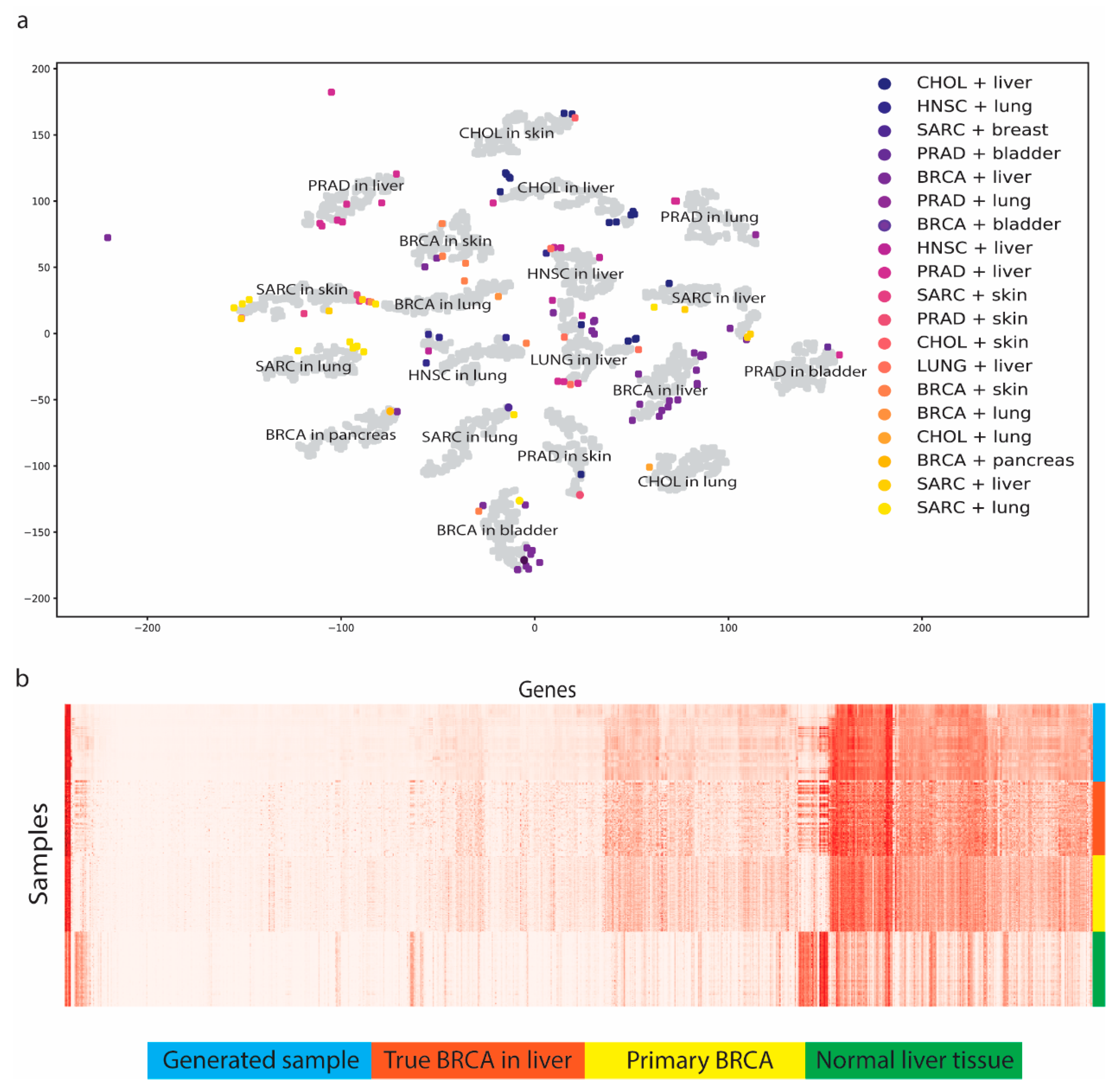
3.2. MetGen Generates High-Quality Metastatic Cancer Samples
3.3. Benchmark Using Stand-Alone MetVAE
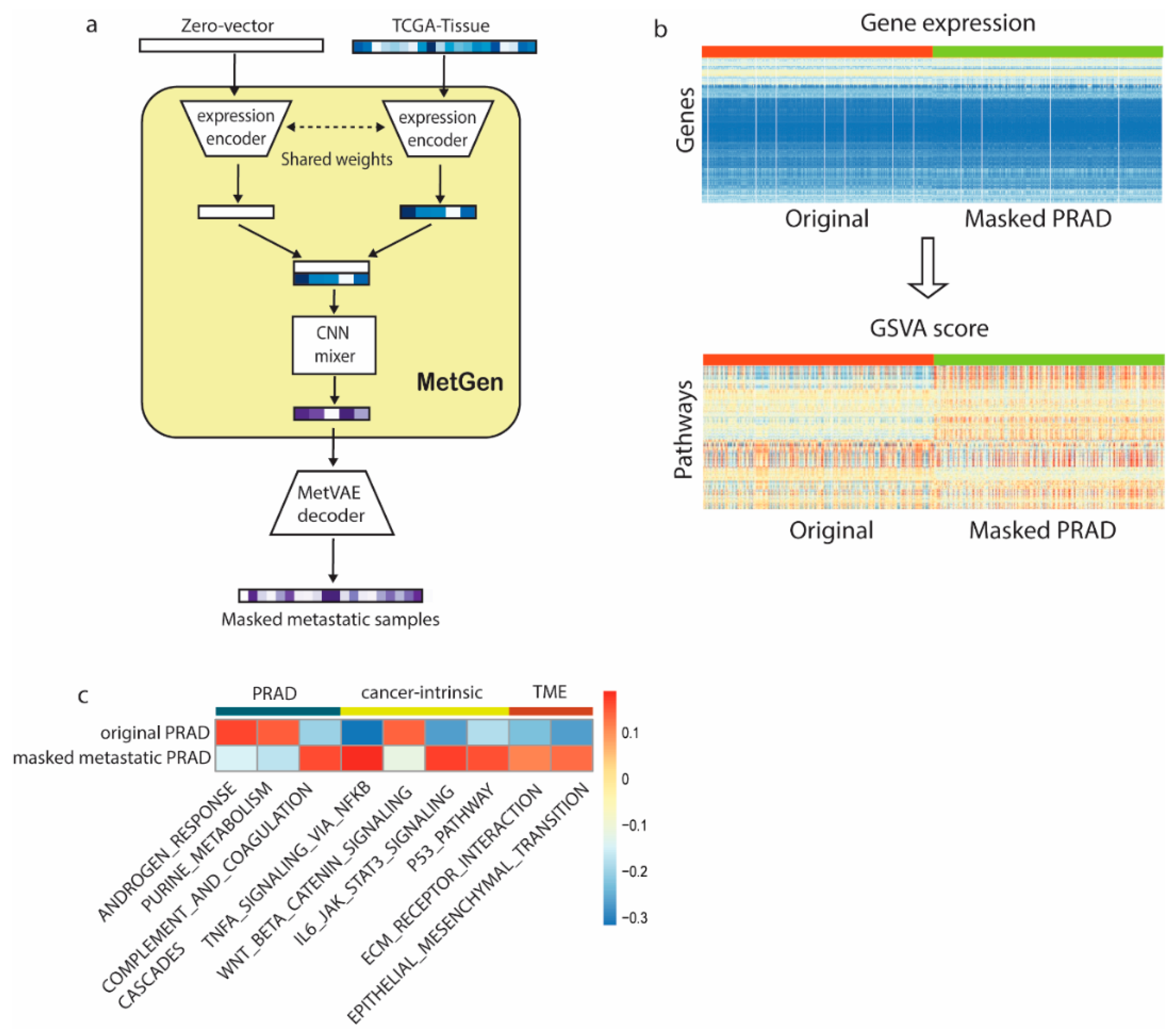
3.4. MetGen Model Learns Metastatic Prostate Cancer Characteristics
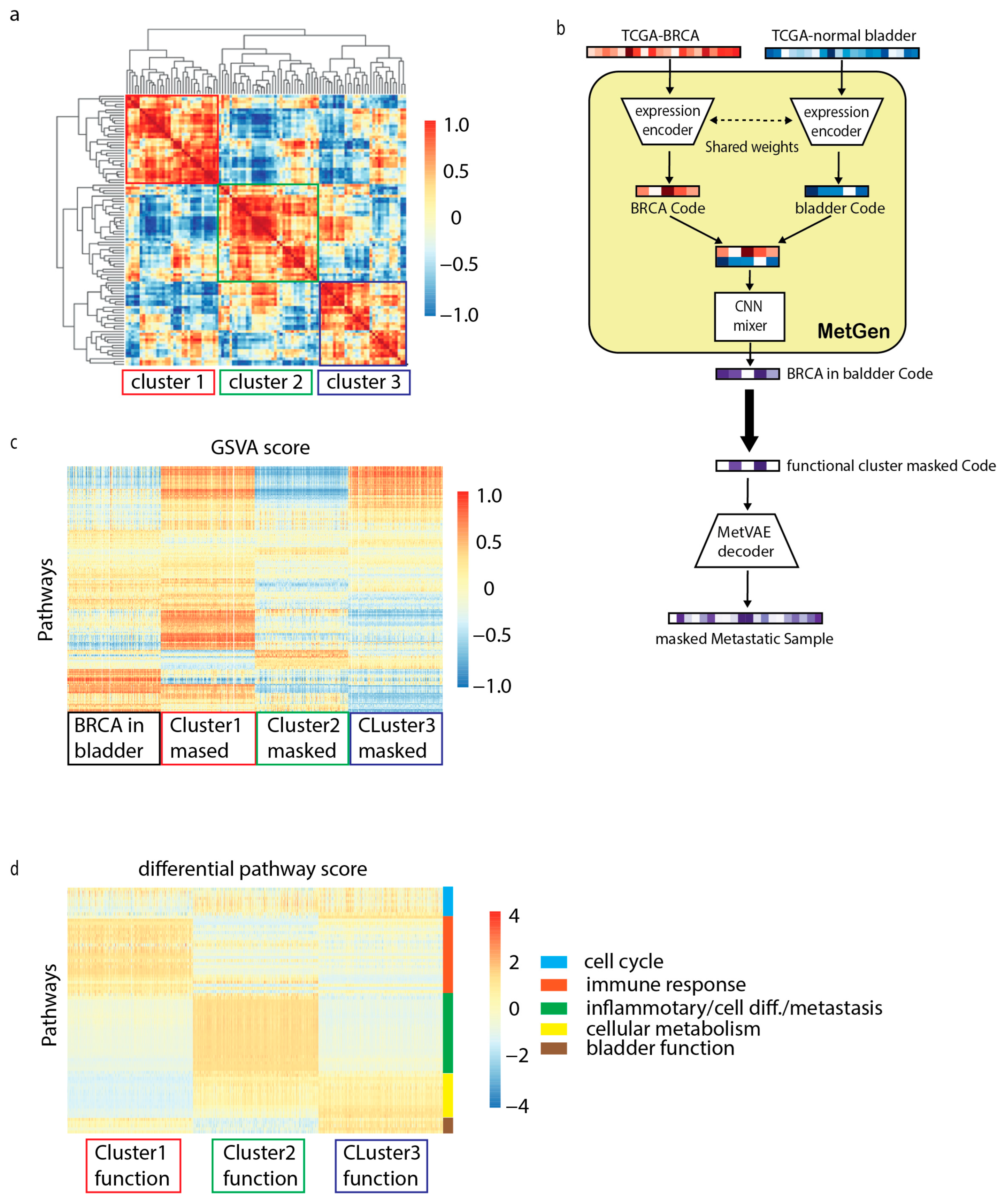
3.5. MetGen Latent Components Learn Functional Clusters
3.5.1. Cell Cycle
3.5.2. Immune Response
3.5.3. Inflammatory/Cell Differentiation/Metastasis
3.5.4. Cellular Metabolism
3.5.5. Bladder Function
4. Conclusions
- MetVAE can encode metastatic cancer and tissue site information faithfully into latent code. We investigated MetVAE for cancer and tissue type classification using MET500 data, and our model had good performance in both tasks.
- MetGen can generate metastatic cancer expressions from primary tumor tissues and normal tissues. We generated 19 metastatic cancer types using TCGA data. Our generated samples stored essential metastatic cancer information and achieved good performance in multiple classification tests.
- We demonstrated the interpretability of our models using samples of metastatic prostate cancer and metastatic breast cancer in the bladder. Highly relevant functions were learned from primary cancer and tissue sites, which further affirmed the power of our model.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seyfried, T.N.; Huysentruyt, L.C. On the origin of cancer metastasis. Crit. Rev. Oncog. 2013, 18, 43–73. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef] [PubMed]
- Crea, F.; Clermont, P.L.; Parolia, A.; Wang, Y.; Helgason, C.D. The non-coding transcriptome as a dynamic regulator of cancer metastasis. Cancer Metastasis Rev. 2014, 33, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Crnic, I.; Christofori, G. Novel technologies and recent advances in metastasis research. Int. J. Dev. Biol. 2004, 48, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Welch, D.R. Microarrays bring new insights into understanding of breast cancer metastasis to bone. Breast Cancer Res. 2004, 6, 61. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Caswell-Jin, J.L.; Plevritis, S.K.; Tian, L.; Cadham, C.J.; Xu, C.; Stout, N.K.; Sledge, G.W.; Mandelblatt, J.S.; Kurian, A.W. Change in Survival in Metastatic Breast Cancer with Treatment Advances: Meta-Analysis and Systematic Review. JNCI Cancer Spectr. 2018, 2, pky062. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Robinson, D.R.; Wu, Y.-M.; Lonigro, R.J.; Vats, P.; Cobain, E.; Everett, J.; Cao, X.; Rabban, E.; Kumar-Sinha, C.; Raymond, V. Integrative clinical genomics of metastatic cancer. Nature 2017, 548, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Durgadevi, M. Generative Adversarial Network (GAN): A general review on different variants of GAN and applications. In Proceedings of the 2021 6th International Conference on Communication and Electronics Systems (ICCES), Coimbatre, India, 8–10 July 2021. [Google Scholar]
- Chicco, D. Siamese neural networks: An overview. In Artificial Neural Networks; Humana: New York, NY, USA, 2021; pp. 73–94. [Google Scholar]
- Kushwaha, V.; Nandi, G. Study of prevention of mode collapse in generative adversarial network (gan). In Proceedings of the 2020 IEEE 4th Conference on Information & Communication Technology (CICT), Chennai, India, 3–5 December 2020. [Google Scholar]
- Higgins, I.; Matthey, L.; Pal, A.; Burgess, C.; Glorot, X.; Botvinick, M.; Mohamed, S.; Lerchner, A. beta-vae: Learning basic visual concepts with a constrained variational framework. In Proceedings of the International Conference on Learning Representations, Toulon, France, 24–26 April 2017. [Google Scholar]
- Kingma, D.P.; Welling, M. Auto-encoding variational bayes. arXiv 2013, arXiv:1312.6114. [Google Scholar]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A Simple Framework for Contrastive Learning of Visual Representations. In Proceedings of the 37th International Conference on Machine Learning, Virtual, 13–18 July 2020; pp. 1597–1607, PMLR: Proceedings of Machine Learning Research. [Google Scholar]
- Mostavi, M.; Chiu, Y.-C.; Huang, Y.; Chen, Y. Convolutional neural network models for cancer type prediction based on gene expression. BMC Med. Genom. 2020, 13, 44. [Google Scholar] [CrossRef]
- Chollet, F. Keras: Deep Learning Library. 2015. Available online: https://github.com/fchollet/keras (accessed on 9 April 2024).
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdottir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Zhang, Z.; Hernandez, K.; Savage, J.; Li, S.; Miller, D.; Agrawal, S.; Ortuno, F.; Staudt, L.M.; Heath, A.; Grossman, R.L. Uniform genomic data analysis in the NCI Genomic Data Commons. Nat. Commun. 2021, 12, 1226. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The molecular signatures database hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Alumkal, J.J.; Sun, D.; Lu, E.; Beer, T.M.; Thomas, G.V.; Latour, E.; Aggarwal, R.; Cetnar, J.; Ryan, C.J.; Tabatabaei, S. Transcriptional profiling identifies an androgen receptor activity-low, stemness program associated with enzalutamide resistance. Proc. Natl. Acad. Sci. USA 2020, 117, 12315–12323. [Google Scholar] [CrossRef]
- Formaggio, N.; Rubin, M.A.; Theurillat, J.-P. Loss and revival of androgen receptor signaling in advanced prostate cancer. Oncogene 2021, 40, 1205–1216. [Google Scholar] [CrossRef]
- Davies, A.; Nouruzi, S.; Ganguli, D.; Namekawa, T.; Thaper, D.; Linder, S.; Karaoğlanoğlu, F.; Omur, M.E.; Kim, S.; Kobelev, M. An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat. Cell Biol. 2021, 23, 1023–1034. [Google Scholar] [CrossRef]
- Yin, J.; Ren, W.; Huang, X.; Deng, J.; Li, T.; Yin, Y. Potential mechanisms connecting purine metabolism and cancer therapy. Front. Immunol. 2018, 9, 1697. [Google Scholar] [CrossRef]
- De Vitto, H.; Arachchige, D.B.; Richardson, B.C.; French, J.B. The intersection of purine and mitochondrial metabolism in cancer. Cells 2021, 10, 2603. [Google Scholar] [CrossRef]
- Ahmad, F.; Cherukuri, M.K.; Choyke, P.L. Metabolic reprogramming in prostate cancer. Br. J. Cancer 2021, 125, 1185–1196. [Google Scholar] [CrossRef]
- Revel, M.; Daugan, M.V.; Sautés-Fridman, C.; Fridman, W.H.; Roumenina, L.T. Complement system: Promoter or suppressor of cancer progression? Antibodies 2020, 9, 57. [Google Scholar] [CrossRef]
- Stallone, G.; Netti, G.S.; Cormio, L.; Castellano, G.; Infante, B.; Pontrelli, P.; Divella, C.; Selvaggio, O.; Spadaccino, F.; Ranieri, E. Modulation of complement activation by pentraxin-3 in prostate cancer. Sci. Rep. 2020, 10, 18400. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X. Targeting the Wnt/β-catenin signaling pathway in cancer. J. Hematol. Oncol. 2020, 13, 165. [Google Scholar] [CrossRef]
- Wen, X.; Wu, Y.; Awadasseid, A.; Tanaka, Y.; Zhang, W. New advances in canonical Wnt/β-catenin signaling in cancer. Cancer Manag. Res. 2020, 12, 6987. [Google Scholar] [CrossRef]
- Brassart-Pasco, S.; Brézillon, S.; Brassart, B.; Ramont, L.; Oudart, J.-B.; Monboisse, J.C. Tumor microenvironment: Extracellular matrix alterations influence tumor progression. Front. Oncol. 2020, 10, 397. [Google Scholar] [CrossRef]
- Bao, Y.; Wang, L.; Shi, L.; Yun, F.; Liu, X.; Chen, Y.; Chen, C.; Ren, Y.; Jia, Y. Transcriptome profiling revealed multiple genes and ECM-receptor interaction pathways that may be associated with breast cancer. Cell. Mol. Biol. Lett. 2019, 24, 38. [Google Scholar] [CrossRef]
- Jing, Y.; Han, Z.; Zhang, S.; Liu, Y.; Wei, L. Epithelial-Mesenchymal Transition in tumor microenvironment. Cell Biosci. 2011, 1, 29. [Google Scholar] [CrossRef]
- Gao, D.; Mittal, V. Tumor microenvironment regulates epithelial–mesenchymal transitions in metastasis. Expert Rev. Anticancer Ther. 2012, 12, 857–859. [Google Scholar] [CrossRef]
- Stark, G.R.; Taylor, W.R. Analyzing the G2/M checkpoint. In Checkpoint Controls and Cancer: Volume 1: Reviews and Model Systems; Humana Press: Totowa, NJ, USA, 2004; pp. 51–82. [Google Scholar]
- Eastman, A. Cell cycle checkpoints and their impact on anticancer therapeutic strategies. J. Cell. Biochem. 2004, 91, 223–231. [Google Scholar] [CrossRef]
- Ren, B.; Cam, H.; Takahashi, Y.; Volkert, T.; Terragni, J.; Young, R.A.; Dynlacht, B.D. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 2002, 16, 245–256. [Google Scholar] [CrossRef]
- Dang, C.V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 1999, 19, 1–11. [Google Scholar] [CrossRef]
- Xie, S.; Chen, M.; Yan, B.; He, X.; Chen, X.; Li, D. Identification of a role for the PI3K/AKT/mTOR signaling pathway in innate immune cells. PLoS ONE 2014, 9, e94496. [Google Scholar] [CrossRef]
- Belardelli, F.; Ferrantini, M.; Proietti, E.; Kirkwood, J.M. Interferon-alpha in tumor immunity and immunotherapy. Cytokine Growth Factor Rev. 2002, 13, 119–134. [Google Scholar] [CrossRef]
- Cutolo, M.; Sulli, A.; Seriolo, B.; Accardo, S.; Masi, A. Estrogens, the immune response and autoimmunity. Clin. Exp. Rheumatol. 1995, 13, 217–226. [Google Scholar]
- Kovats, S. Estrogen receptors regulate innate immune cells and signaling pathways. Cell. Immunol. 2015, 294, 63–69. [Google Scholar] [CrossRef]
- Lin, G.L.; Hankenson, K.D. Integration of BMP, Wnt, and notch signaling pathways in osteoblast differentiation. J. Cell. Biochem. 2011, 112, 3491–3501. [Google Scholar] [CrossRef]
- Jia, Y.; Wang, Y.; Xie, J. The Hedgehog pathway: Role in cell differentiation, polarity and proliferation. Arch. Toxicol. 2015, 89, 179–191. [Google Scholar] [CrossRef]
- Lawson, N.D.; Vogel, A.M.; Weinstein, B.M. sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev. Cell 2002, 3, 127–136. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, Z.; Zhang, A.; Gupte, A.A.; Hamilton, D.J. The role of calcium signaling in melanoma. Int. J. Mol. Sci. 2022, 23, 1010. [Google Scholar] [CrossRef]
- Jurewicz, M.M.; Stern, L.J. Class II MHC antigen processing in immune tolerance and inflammation. Immunogenetics 2019, 71, 171–187. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. T cells and MHC proteins. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Barber, G.N. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr. Opin. Immunol. 2011, 23, 10–20. [Google Scholar] [CrossRef]
- McDonnell, A.M.; Dang, C.H. Basic review of the cytochrome p450 system. J. Adv. Pract. Oncol. 2013, 4, 263. [Google Scholar]
- Liu, K.; Czaja, M. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013, 20, 3–11. [Google Scholar] [CrossRef]
- Nelson, D.R. The cytochrome p450 homepage. Hum. Genom. 2009, 4, 59. [Google Scholar] [CrossRef]
- Carattino, M.D.; Prakasam, H.S.; Ruiz, W.G.; Clayton, D.R.; McGuire, M.; Gallo, L.I.; Apodaca, G. Bladder filling and voiding affect umbrella cell tight junction organization and function. Am. J. Physiol.-Ren. Physiol. 2013, 305, F1158–F1168. [Google Scholar] [CrossRef]
- Kuo, I.Y.; Ehrlich, B.E. Signaling in muscle contraction. Cold Spring Harb. Perspect. Biol. 2015, 7, a006023. [Google Scholar] [CrossRef]
| TCGA Cancer Type | Number of Samples | TCGA Normal Tissue Sites | Number of Samples |
|---|---|---|---|
| BRCA | 159 | bladder | 8 |
| CHOL | 45 | breast | 5 |
| HNSC | 45 | liver | 243 |
| LUNG | 52 | lung | 75 |
| PRAD | 155 | pancreas | 1 |
| SARC | 100 | skin | 40 |
| Layer (Type) | Output Shape | Number of Param | Connected to |
|---|---|---|---|
| encoder_input | (None, 7312, 1) | 0 | |
| Flatten_1 | (None, 7312) | 0 | encoder_input |
| dense | (None, 1000) | 7,313,000 | Flatten_1 |
| batch_normalization | (None, 1000) | 4000 | Dense |
| Flatten_2 | (None, 1000) | 0 | batch_normalization |
| z_mean | (None, 100) | 100,100 | Flatten_2 |
| z_log_var | (None, 100) | 100,100 | Flatten_2 |
| z_sample | (None, 100) | 0 | z_mean, z_log_var |
| Layer (Type) | Output Shape | Number of Param |
|---|---|---|
| z_sampleing(input) | (None, 100) | 0 |
| batch_normalization | (None, 100) | 400 |
| dense | (None, 1000) | 101,000 |
| batch_normalization | (None, 1000) | 4000 |
| dense | (None, 7312) | 7,319,312 |
| reshape | (None, 7312, 1) | 0 |
| Layer (Type) | Output Shape | Number of Param |
|---|---|---|
| TCGA_input | (None, 7312, 1) | 0 |
| conv1d | (None, 228, 64) | 2112 |
| batch_normalization | (None, 228, 64) | 256 |
| activation | (None, 228, 64) | 0 |
| dense | (None, 57, 16) | 4112 |
| batch_normalization | (None, 57, 16) | 64 |
| activation | (None, 57, 16) | 0 |
| flatten | (None, 912) | 0 |
| dropout | (None, 912) | 0 |
| dense | (None, 912) | 467,456 |
| Layer (Type) | Output Shape | Number of Param | Connected to |
|---|---|---|---|
| Cancer_input | (None, 512) | 0 | |
| Tissue_input | (None, 512) | 0 | |
| stack | (None, 2, 512, 1) | 0 | Cancer_input, Tissue_input |
| Conv2d_1 | (None, 1, 512, 64) | 192 | stack |
| batch_normalization_1 | (None, 1, 512, 64) | 256 | Conv2d_1 |
| Conv2d_2 | (None, 1, 16, 32) | 65,568 | batch_normalization_1 |
| batch_normalization_2 | (None, 1, 16, 32) | 128 | Conv2d_2 |
| flatten | (None, 512) | 0 | batch_normalization_2 |
| Dropout_1 | (None, 512) | 0 | flatten |
| dense | (None, 200) | 102,600 | Dropout_1 |
| Dropout_2 | (None, 200) | 0 | dense |
| MET500_code(input) | (None, 100, 1) | 0 | |
| Learned_code | (None, 100) | 20,100 | Dropout_2 |
| Layer (Type) | Output Shape | Number of Param |
|---|---|---|
| embeddings (input) | (None, 100) | 0 |
| dense | (None, 60) | 6060 |
| dropout | (None, 60) | 0 |
| dense | (None, 40) | 2440 |
| dropout | (None, 40) | 0 |
| dense | (None, number of classes) | |
| classifier | (None, number of classes) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Chiu, Y.-C.; Chen, Y.; Huang, Y. A Metastatic Cancer Expression Generator (MetGen): A Generative Contrastive Learning Framework for Metastatic Cancer Generation. Cancers 2024, 16, 1653. https://doi.org/10.3390/cancers16091653
Liu Z, Chiu Y-C, Chen Y, Huang Y. A Metastatic Cancer Expression Generator (MetGen): A Generative Contrastive Learning Framework for Metastatic Cancer Generation. Cancers. 2024; 16(9):1653. https://doi.org/10.3390/cancers16091653
Chicago/Turabian StyleLiu, Zhentao, Yu-Chiao Chiu, Yidong Chen, and Yufei Huang. 2024. "A Metastatic Cancer Expression Generator (MetGen): A Generative Contrastive Learning Framework for Metastatic Cancer Generation" Cancers 16, no. 9: 1653. https://doi.org/10.3390/cancers16091653
APA StyleLiu, Z., Chiu, Y.-C., Chen, Y., & Huang, Y. (2024). A Metastatic Cancer Expression Generator (MetGen): A Generative Contrastive Learning Framework for Metastatic Cancer Generation. Cancers, 16(9), 1653. https://doi.org/10.3390/cancers16091653






