Simple Summary
Prostate cancer is a leading cause of death among men worldwide. Some researchers have speculated that the prostatic microbiome is involved in prostatic inflammation and the pathogenesis of prostate cancer; however, there has not been consensus regarding specific organisms or their overall impact on this process. In order to synthesize the data that exists on the prostate microbiome, we performed a systematic review of the literature. In this review, we concluded that the methods that have been used to identify microbes within the prostate are highly variable and therefore do not constitute robust evidence. Further research is necessary to refine the methods used and to better understand which organisms may play a role in the development of prostate cancer.
Abstract
Some researchers have speculated that the prostatic microbiome is involved in the development of prostate cancer (PCa) but there is no consensus on certain microbiota in the prostatic tissue of PCa vs. healthy controls. This systematic review aims to investigate and compare the microbiome of PCa and healthy tissue to determine the microbial association with the pathogenesis of PCa. We searched MEDLINE, Embase, and Scopus databases. Articles were screened by two independent and blinded reviewers. Literature that compared the prostatic tissue microbiome of patients with PCa with benign controls was included. We found that PCa may be associated with increased Propionibacterium acnes, the herpesviridae and papillomaviridae families, and Mycoplasma genitalium, but definitive conclusions cannot be drawn from the existing data. Challenges include the difficulty of obtaining uncontaminated tissue samples and securing tissue from healthy controls. As a result, methods are varied with many studies using cancerous and “healthy” tissue from the same prostate. The organisms chosen for each study were also highly variable, making it difficult to compare studies. These issues have led to lower confidence in our results. Overall, further work is warranted to better understand the implications of the prostatic microbiome in the pathogenesis of PCa.
1. Introduction
Prostate cancer continues to be the second leading cause of cancer-related deaths among men in the United States, with current statistics reporting that 1 in every 44 men will die of prostate cancer and 1 in every 8 men will develop prostate cancer in their lifetime [1]. Our understanding of prostate cancer has evolved over the years, showing the connection between many risk factors not limited to age, ethnicity, family history, and diet [2]. Current treatment options, according to the latest guidelines, include radical prostatectomy, radiotherapy, and hormone therapy as standard treatment options for patients presenting with local or regional disease, resulting in 5-year relative survival rates of greater than 99% in local and regional disease [3]. Despite the high rates of disease control with current treatment options, recurrence and disease progression to metastatic prostate cancer continue to pose challenges to patients as well as physicians [4]. To shed light on the etiology, multiple factors are being actively investigated for potential drivers of cancer progression. Prostate cancer carcinogenesis and disease progression are complex processes with numerous intrinsic and environmental factors substantially affecting these processes, including diet, obesity, inherited genetic risk alleles, smoking, social determinants of health, inflammation, and possibly infectious agents, among others [2,5,6].
More recently, the human microbiome has become an area of increasing interest owing to its potential role in various disease processes, specifically in cancers [7,8,9,10,11]. The microbiome consists of bacterial, viral, and fungal taxa that are often associated with health and wellness. However, with the advances in technology, negative impacts on the body have also been discovered in relation to the human microbiome. The major underlying mechanism for such impacts is believed to be caused by inflammation and alterations to host immunity [12]. One of the most studied areas is the gut microbiome, which has been shown to play a role in digestive tract pathologies such as inflammatory bowel disease and colorectal cancer as well as many other diseases like Alzheimer’s disease and rheumatoid arthritis [13,14,15]. The gut microbiota has also been linked to prostate cancer progression and treatment response. by metabolizing chemotherapeutic drugs and influencing resistance development [16,17,18]. This could be due to various reasons, including the gut microbiome’s ability to metabolize chemotherapeutic drugs through biotransformation, like gemcitabine, which has showed diminished effectivity when metabolized by Mycoplasma hyorhinis [19]. The gut microbiome also plays a role in modifying the immune function, which can lead to variation of the response and efficacy of popular immunological therapies like checkpoint inhibitors [13,20,21]. It is speculated to be caused by alterations in the intestinal microbiome causing an increased abundance of bacteria that have androgenic functions. Recent studies have found specific microbial species and described their suspected mechanism for contributing to the aforementioned effects. Ruminococcus spp. has been found to be enriched in patients with castration resistant prostate cancer [20,22]. This is thought to be due to the ability of this bacterium to synthesize dehydroepiandrosterone (DHEA) from pregnenolone. Thus aiding tumor growth even in an androgen-deprived environment [22]. Similarly, several studies have described the increased abundance of specific bacterial strains, such as Akkermansia muciniphila in the gut of patients with prostate cancer that receive treatment with androgen deprivation therapy (ADT), hence it is theorized that this bacterium might promote the antitumor effects of ADT [22,23,24]. This data proves that the microbiome can have significant effects on disease progression aside from providing new therapeutic targets. However, it is important to explore the effects that the microbiota of other areas like the urinary tract and prostatic tissue might have on this disease. Especially because these areas are more closely related to the affected organ, the prostate. Because of this, a similar link has been investigated between the genitourinary microbiome in men with prostatitis and prostate cancer. The most commonly found organisms in the urinary microbiome of patients with prostate cancer are proinflammatory organisms, including Cutibacterium, Streptococcus anginosus and Escherichia coli [19]. The pathogenesis of prostate cancer progression, in this case, is theorized to be the result of microorganisms producing reactive oxygen species, resulting in local inflammation that ultimately drives the carcinogenic process [25]. Previous studies have found ties between the microbiome of prostatic tissue, seminal fluid, and urine and various pathological processes such as prostatitis [26], and some studies have found a possible link between prostatitis and an increased risk of the development of cancer [27]. Furthermore, to our knowledge, there is no consensus on significant microbiota differences found in prostate cancer compared to healthy prostatic tissue controls that may be directly associated with carcinogenesis or disease progression despite tantalizing individual studies.
Understanding the mechanisms by which the microbiome influences prostate cancer is crucial in developing targeted therapeutic strategies. The microbiome has been shown to affect the local immune response within the prostate gland, with dysbiosis facilitating local chronic inflammation and potentially promoting tumor growth [7,25,28]. Exploiting these interactions may offer novel therapeutic approaches, including the exploration of interventions like probiotics, prebiotics, postbiotics and microbiota transplantation, with promising potential to prevent disease progression, alleviate chemotherapy side effects, and personalize treatment plans [29,30]. In addition to these new therapeutic options, leveraging the relationship between the microbiome and local immune processes might increase the efficacy of immunotherapies for prostate cancer such as immune checkpoint inhibitors (like anti PD-1 agents), Sipuleucel-T, and CAR-T therapy [31].
Deciphering the unique “fingerprint” of the prostatic microbiome and its interplay with prostate cancer is crucial for this personalized approach. This systematic review delves into a vital comparison, the microbial composition of prostate cancer versus noncancerous prostatic tissue, to shed light on specific microorganisms that may be involved in prostate carcinogenesis or impact disease progression or treatment response as potential areas of future medical intervention.
2. Methods
This review was conducted according to a PROSPERO-published protocol (CRD42023438939). In the reporting of our findings, some deviations from the protocol occurred, as the review team found no meta-analyzable data. For this reason, we instead qualitatively assessed the data and reported the species that were correlated and inversely correlated with prostate cancer development in multiple studies.
MEDLINE, Embase, and Scopus were searched in April 2023 to capture studies relevant to the prostatic microbiome. In consultation with an expert librarian, search terms were devised and utilized to search the databases as follows: “prostate cancer” AND (microbiota OR microbiome OR metagenome OR microbial). This study was conducted by following the preferred reporting items for systematic reviews and meta-analyses (PRISMA). A tiered approach was used to review articles according to PRISMA guidelines using the Covidence platform [32]. The study screening and selection process is demonstrated in a PRISMA flow diagram (Figure 1). All authors screened articles by title and abstract to identify articles for full-text review. Each article title and abstract were reviewed by two blinded reviewers. Conflicts were then resolved by arbitration by a third reviewer. Select articles were then reviewed in full, again with two reviewers screening each text.
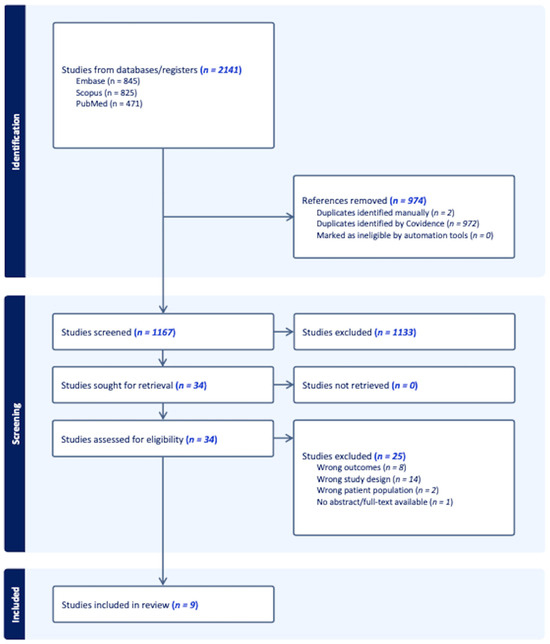
Figure 1.
A PRISMA flow diagram.
Studies were included if they described a comparison between the microbiome of prostatic tissue in patients with prostate cancer and that of benign control samples. We excluded studies that were case reports, case series, systematic reviews, or meta-analyses. Studies that did not specifically look at tissue samples, those that were ex-vivo studies, and those that were not human tissue were also excluded. Data from the included studies was extracted by two authors and discussed to reach a consensus. Any disagreements were discussed with the entire review team. Data included participants/samples, species found in the control group, species found in the malignant group, and whether there were subgroup analyses to stratify tumors by Gleason grade. Select studies were also discussed with an expert in the field of metagenomics to better understand study results and applicability.
3. Results
3.1. Article Retrieval
In this review, our particular interest was that of assessing the relationship that exists between the prostatic tissue microbiome and prostate cancer. In that sense, we primarily focused on defining the differences, if any, between the microbiome of benign prostatic tissue and prostate cancer. The systematic search that was conducted resulted in 2141 records, which were uploaded to the Covidence web-based platform for screening. A total of 974 duplicate records were identified and removed. During the process of title and abstract screening, 1133 articles were deemed to be irrelevant and were therefore excluded. The 34 remaining records were included for full-text review. In the end and after careful consideration, nine articles published between 2015 and 2023 were included. The remaining 24 articles were excluded due to a variety of reasons, including but not limited to: inappropriate outcome measures, inappropriate study population, or lack of full-text availability. A summary of relevant data from the included studies is presented in Table 1.

Table 1.
Summary of microbiome detection.
3.2. Diversity Assessment
Microbial diversity and the approach taken to determine it varied from article to article. Alpha diversity is a measure of the diversity or total number of organisms and their relative proportions within a defined community or space [40,42]. Beta diversity is a measurement that can differ greatly between studies as it compares the diversity of species across different samples or communities [42]. Only five out of the nine included articles, mention performing a diversity analysis. Out of those five articles, Sarkar et al., Salachan et al., and Gonçalves et al. reported that their alpha diversity assessment indicated a lower species richness in samples from the malignant group than those in the benign (control) groups [38,40,41]. However, Feng et al. and Cavarretta et al. were not able to distinguish between benign and malignant groups with alpha or beta diversity [35,37]. Four out of the five included articles did not include or mention diversity analysis. Refer to Table 1 for a summary of the diversity assessment findings of the included studies.
3.3. Study Outcomes
Across the nine included studies in this analysis, a total of 22 genera were found to be significantly increased in patients with prostate cancer groups compared to benign controls. Four out of the nine included studies obtained both their prostate cancer tissue samples and their non-prostate cancer controls (benign tissue) from radical prostatectomies of adult males with a clinically localized prostate cancer diagnosis [33,35,37,40]. Two studies obtained their prostate cancer tissue samples from patients who underwent radical prostatectomy for prostate cancer. In contrast, they obtained the benign tissue controls from patients with benign prostatic hyperplasia (BPH) who underwent transurethral resection of the prostate [34,39]. Two studies used transrectal ultrasound-guided prostate biopsies to obtain tissue samples for both their benign control and experimental groups [38,41]. Only one study used mRNA-seq data downloaded from EBI ENA [36].
At the genus level, Rickettsia [34], Mycobacterium [34], Bordetella [34], Mycoplasma [34,39], Sphingomonas [34], Bartonella [34], Helicobacter [34], Bacillus [34], Porphyromonas [34], Salmonella [34], Aeromonas [34], Brevundimonas [34], Shigella [34], Staphylococcus [35], Streptococcus [35], Cutibacterium [36,38,41], Lawsonella [38], Shewanella [40], Prevotella [41], Cupriavidus [41], and Methylobacterium [41] were found to be significantly increased in the prostate cancer cohort in several of the studies that were analyzed. However, in two studies, Prevotella and Staphylococcus were reported to be significantly more abundant in the benign groups versus the prostate cancer groups [38,40]. The other genera found to be particularly increased in the benign cohorts were Cellvibrio [41], Kocuria [41], Vibrio [40], Bacteroides [40], Chlamydia [34], Pseudomonas [34], Burkholderia [34], Campylobacter [34]. Of note, Pseudomonas was identified in four of the nine included studies [34,37,38,41]. In two of these studies, Pseudomonas was found to be present in both the benign and malignant cohorts [37,38]. In the remaining two studies, Pseudomonas was reported to be present exclusively in the benign cohorts [34,41]. It was significantly increased in the benign group in the Banerjee study [34]. Gonçalves et al. found that Pseudomonas was the most prevalent genus in the benign group, although this finding was not reported as statistically significant [38]. Aside from bacteria, four out of the nine articles also investigated the presence of viral organisms in their samples. At the family level, Banerjee et al. found Poxviridae, Reoviridae, Papillomaviridae, and Herpesviridae to have a high hybridization signal in the cancer cohort versus in their control population [34]. EBV, HBV, HPV16, and HPV18 were also found to be significantly increased in the malignant cohort in the study by Sarkar et al. [41]. Finally, Saimiriine betaherpesvirus was exclusively found in the benign cohort by Salachan et al. [40]. Only Banerjee et al. explored other types of organisms, including viruses, parasites and fungi [34]. Of note, this study defined results as significant if they had a log2 fold change > 1 and an adjusted p value < 0.05. They further classified these significant results into High, Mid, and Low depending on the size of their hybridization signal. For the purpose of our study, we only considered those with a high hybridization signal (Log2 ≥ 10) as significant for our analysis.
3.4. Quality Assessment
Authors working in pairs assessed the risk of bias independently. Discrepancies were resolved by discussion or arbitration with a third author. Although the authors’ judgments were favorable in most of the risk of bias domains, our confidence in the evidence in the literature remains low due to the nature of the studies included in this review supported by the fact that they were all non-randomized and retrospective studies. These articles varied greatly in their methodology in terms of what was used for sample and control cohorts, as well as which types of organisms were of interest. We followed guidance from the Cochrane collaboration on how to assess individual study level risk of bias and utilized the “risk of bias in non-randomized studies of exposures” [43]. Each study was evaluated for seven domains of potential risk of bias and each one was categorized as low, some concerns, high risk, and very high risk of bias. The highest concern of risk of bias in any domain constituted the overall risk of bias for each study. A summary of the judgment for these domains and the overall risk of bias is depicted in Figure 2.
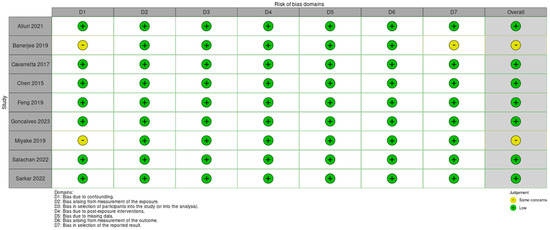
Figure 2.
Risk of bias domains [33,34,35,36,37,38,39,40,41].
Utilizing the Cochrane guidance for risk of bias, we deemed most of the included studies low risk of bias for their good account for major known confounders, their well-defined patient cohorts, their consistently classified exposures across study populations, their handling of missing data, their measurement methods to eliminate assessor effects, and their systematic reporting of their results even in cases when there were no significant findings.
4. Discussion
In recent years, the effect of the microbiome on cancer pathobiology has led to more focused work regarding the relationship between the genitourinary microbiome and prostate cancer. While our review highlights several relevant studies regarding the prostate microbiome specifically, it also emphasizes the need for additional prostate tissue microbiome studies to increase the power and quality of these findings. Studies to date focus on exploring bacterial and viral signatures in prostate cancer tissue in a broad manner.
It is known that the male genitourinary tract is commonly colonized by Corynebacterium, Streptococcus, Staphylococcus, Finegoldia, Peptoniphilus, Anaerococcus, and Lactobicillus [38]. The bacterial taxa that were found to be significantly increased in prostate cancer tissue amongst the nine studies had varied results with minimal overlap. Propionibacterium acnes (P. acnes) was the only bacterium found to be commonly increased in prostatic tissue in three of the nine studies. Several other studies have demonstrated significant expression of P. acnes in prostate cancer and prostatitis in human and rodent cell lines [44,45]. The pathophysiology of prostate cancer progression from P. acnes inoculation is theorized to be due to a chronic inflammatory state as a result of increased production of proinflammatory cytokines and neutrophil recruitment [46,47]. Furthermore, P. acnes has been proposed to induce cancer progression through an immunosuppressive environment as a result of macrophage gene alterations causing increased recruitment of T regulatory cells [48,49]. A study conducted by Yow et al. found that cutibacterium (previously known as propionibacterium) was also significantly increased in aggressive prostate cancer tissue compared to the healthy control [50]. Conversely, previous literature including some of the cited studies reported P. acnes to be prevalent in both benign and malignant prostate tissue [45,51]. Therefore, further explorations of the bacterial taxa are warranted to determine their significance in the progression of prostate cancer.
In our review, the viral taxa were only investigated in four out of the nine studies. Most commonly found were the Herpesviridae and Papillomaviridae (HPV) strains. HPV is a well-studied virus that has been shown to be associated with numerous malignant processes including cervical, penile, anal, vaginal, and various head and neck cancers [52]. A commonly proposed mechanism in cancer development via HPV is related to the virus’s ability to inactivate tumor suppressor genes p53 and pRb via the E6 and E7 oncoproteins [53]. There have been several studies that explored an association between oncogenic viruses and prostate cancer, with only some being able to report a clear association with HPV and others showing mixed results as the virus was found in both prostatic cancer and benign tissue [54,55,56,57,58]. It has been speculated that these varied results are the result of differing methodologies, specifically in the collection, primers, and assays used for viral detection. Nonetheless, this emphasizes the need for a standard methodology when examining microbiomes in relation to human cancer cells, to have a clearer understanding of viral associations in the progression of prostate cancer.
Herpesviridae was also found to be increased in two of the four studies that looked at viruses in our review with Epstein–Barr virus (EBV) being one of the commonly found strains [40,41]. EBV has been extensively studied and has been shown, in addition to HPV, to be one of the larger contributors to virus-associated cancers [59]. Generally, these viruses are commonly found in benign tissue as a result of sexual transmission but are reported to be the cause of several carcinomas [60,61,62]. EBV was found to have carcinogenic potential by inhibiting apoptosis and stimulating cell survival via the EBV nuclear antigen 1 and latent membrane protein 1 [63,64]. While the direct association of EBV to PCa has not been elucidated, there has been literature that showed co-inoculation of prostate tissue with HPV to be associated with PCa progression [54,65]. A study performed by de Lima et al. examined HPV and EBV co-inoculation of cervical carcinoma, and proposed that EBV infection accelerated the integration of HPV genomes into the normal cell genomes [66]. This has further been shown in a study by Nahand et al. that reported maximum integration of the HPV genome when coinfected by EBV compared to the control [65]. Therefore, the assumption is that the viruses act simultaneously to increase cell proliferation and inhibit cell apoptosis via different mechanisms.
Future studies may consider focusing on these specific bacterial and viral families to validate their association with prostate cancer in addition to exploring other novel microorganisms. It is also necessary to point out that only when the literature reaches a consensus on which microorganisms form the benign prostate microbiome, will we be able to elucidate the full extent and consequences of the modifications that it suffers during the development of cancer.
Similarly, the reviewed works call attention to the need for standardized outcome reporting when assessing biodiversity and microorganism abundance in the prostate cancer tissue microbiome. Of the studies we reviewed, each one utilized a different outcome measure, including but not limited to bacterial reads per human genome reads, percent RNA reads matched to the human genome, percent abundance, CT values for bacterial load/abundance, percent of samples with specific organismal DNA, and hybridization signal intensity. The lack of standardization and consensus in methodology for reporting these outcomes further complicates our ability to compare results and draw conclusions from the existing literature. Based on the microbiome work available to date, future studies should focus on reporting outcomes that are standardized and reproducible to best characterize the prostate cancer microbiome in an easily understandable, comparable, and potentially clinically relevant manner. We also need to consider the particular limitations of the detection techniques when analyzing microbiome research. Amplicon sequencing is currently one of the most used techniques of next generation sequencing, it relies on PCR amplification of the bacterial 16s ribosomal RNA, which contains nine hypervariable regions. However, only a subset of these hypervariable regions is targeted in most studies for cost reduction. This action can result in bias introduction and inaccurate representation of the abundance of the studied bacteria. In a similar manner, DNA/RNA extraction methods, the sequencing platform used, and even storage conditions may add variability to the results. Because of this, the same technique needs to be used for all samples in the study so as to avoid unnecessary confounders during analysis. Each one of these aspects needs to be clearly stated in the manuscripts for transparency and clarity [67].
The current standard for “healthy or non-cancerous” prostate tissue and prostate cancer tissue should be examined as well. Most “healthy” prostate tissue samples that were utilized as benign controls were taken from a region of the prostate without cancerous cells from the prostate glands of patients diagnosed with prostate cancer. This method not only risks missing key microbiota changes that may occur in precancerous tissue but may also confound the data with an individual’s unique microbiome. Although difficult to obtain, prostate tissue from healthy controls without prostate cancer, rather than cancerous prostate tissue, would be ideal to identify the differences in the microbiome. The best example of this is the techniques used by some of the included studies, which utilized benign tissue samples from patients with benign prostatic hyperplasia who underwent transrectal resection of the prostate.
Although the limitations outlined above led to lower quality data in this review, the studies we reviewed and included have provided a more focused microbiome to be explored in future studies. Additionally, as this area of research is fairly new, the literature reviewed yields important insight into the best way to structure novel studies and report findings in a significant and uniform way.
5. Conclusions
With the current evidence available, we are able to elucidate a likely relationship between microorganisms, like Cutibacterium and Herpesviridae, and PCa, as they seem to be increased in this population. However, there are major limitations to the current research, which makes it difficult to generalize or draw extensive conclusions about these relationships. Further research is needed to test these associations by focusing on implementing a standard approach to outcomes reporting and obtaining benign prostatic tissue from healthy patients to serve as an optimal control group.
Author Contributions
Conceptualization, D.F.W.G. and O.E.; methodology, C.D.M. and O.E.; validation, N.A.F., P.G. and O.S.; formal analysis, D.F.W.G. and O.E.; investigation, D.F.W.G.; resources, C.D.M.; data curation, P.G.; writing—original draft preparation, D.F.W.G., O.E., C.D.M., P.G. and N.A.F.; writing—review and editing, C.A.W. and D.W-G.; visualization, O.E.; supervision, C.A.W.; project administration, D.F.W.G. and O.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- National Cancer Institute. SEER Cancer Stat Facts: Prostate Cancer. Available online: https://seer.cancer.gov/statfacts/html/prost.html (accessed on 3 April 2024).
- Barry, M.J.; Simmons, L.H. Prevention of Prostate Cancer Morbidity and Mortality: Primary Prevention and Early Detection. Med. Clin. N. Am. 2017, 101, 787–806. [Google Scholar] [CrossRef] [PubMed]
- Survival Rates for Prostate Cancer. Available online: https://www.cancer.org/cancer/types/prostate-cancer/detection-diagnosis-staging/survival-rates.html (accessed on 12 March 2024).
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhao, D.; Spring, D.J.; DePinho, R.A. Genetics and Biology of Prostate Cancer. Genes Dev. 2018, 32, 1105–1140. [Google Scholar] [CrossRef] [PubMed]
- Vesely, M.D.; Kershaw, M.H.; Schreiber, R.D.; Smyth, M.J. Natural Innate and Adaptive Immunity to Cancer. Annu. Rev. Immunol. 2011, 29, 235–271. [Google Scholar] [CrossRef] [PubMed]
- Porter, C.M.; Shrestha, E.; Peiffer, L.B.; Sfanos, K.S. The Microbiome in Prostate Inflammation and Prostate Cancer. Prostate Cancer Prostatic Dis. 2018, 21, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Cimadamore, A.; Santoni, M.; Massari, F.; Gasparrini, S.; Cheng, L.; Lopez-Beltran, A.; Montironi, R.; Scarpelli, M. Microbiome and Cancers, with Focus on Genitourinary Tumors. Front. Oncol. 2019, 9, 178. [Google Scholar] [CrossRef] [PubMed]
- Pleguezuelos-Manzano, C.; Puschhof, J.; Rosendahl Huber, A.; van Hoeck, A.; Wood, H.M.; Nomburg, J.; Gurjao, C.; Manders, F.; Dalmasso, G.; Stege, P.B.; et al. Mutational Signature in Colorectal Cancer Caused by Genotoxic Pks+ E. coli. Nature 2020, 580, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Barrett, M.; Hand, C.K.; Shanahan, F.; Murphy, T.; O’Toole, P.W. Mutagenesis by Microbe: The Role of the Microbiota in Shaping the Cancer Genome. Trends Cancer Res. 2020, 6, 277–287. [Google Scholar] [CrossRef] [PubMed]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans, Biological Agents. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans; IARC: Lyon, France, 2012; Volume 100B, pp. 1–441. [Google Scholar]
- Nicolaro, M.; Portal, D.E.; Shinder, B.; Patel, H.V.; Singer, E.A. The Human Microbiome and Genitourinary Malignancies. Ann. Transl. Med. 2020, 8, 1245. [Google Scholar] [CrossRef]
- Fujita, K.; Matsushita, M.; Banno, E.; De Velasco, M.A.; Hatano, K.; Nonomura, N.; Uemura, H. Gut Microbiome and Prostate Cancer. Int. J. Urol. 2022, 29, 793–798. [Google Scholar] [CrossRef]
- Nishida, A.; Inoue, R.; Inatomi, O.; Bamba, S.; Naito, Y.; Andoh, A. Gut Microbiota in the Pathogenesis of Inflammatory Bowel Disease. Clin. J. Gastroenterol. 2018, 11, 1–10. [Google Scholar] [CrossRef]
- Lin, C.; Cai, X.; Zhang, J.; Wang, W.; Sheng, Q.; Hua, H.; Zhou, X. Role of Gut Microbiota in the Development and Treatment of Colorectal Cancer. Digestion 2019, 100, 72–78. [Google Scholar] [CrossRef]
- Cogdill, A.P.; Gaudreau, P.O.; Arora, R.; Gopalakrishnan, V.; Wargo, J.A. The Impact of Intratumoral and Gastrointestinal Microbiota on Systemic Cancer Therapy. Trends Immunol. 2018, 39, 900–920. [Google Scholar] [CrossRef]
- Roy, S.; Trinchieri, G. Microbiota: A Key Orchestrator of Cancer Therapy. Nat. Rev. Cancer 2017, 17, 271–285. [Google Scholar] [CrossRef]
- García-González, A.P.; Ritter, A.D.; Shrestha, S.; Andersen, E.C.; Yilmaz, L.S.; Walhout, A.J.M. Bacterial Metabolism Affects the C. elegans Response to Cancer Chemotherapeutics. Cell 2017, 169, 431–441.e8. [Google Scholar] [CrossRef]
- Rizzo, A.; Santoni, M.; Mollica, V.; Fiorentino, M.; Brandi, G.; Massari, F. Microbiota and prostate cancer. Semin. Cancer Biol. 2022, 86, 1058–1065. [Google Scholar] [CrossRef]
- El Tekle, G.; Garrett, W.S. Bacteria in Cancer Initiation, Promotion and Progression. Nat. Rev. Cancer 2023, 23, 600–618. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut Microbiome Influences Efficacy of PD-1-Based Immunotherapy against Epithelial Tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Pernigoni, N.; Zagato, E.; Calcinotto, A.; Troiani, M.; Mestre, R.P.; Calì, B.; Attanasio, G.; Troisi, J.; Minini, M.; Mosole, S.; et al. Commensal Bacteria Promote Endocrine Resistance in Prostate Cancer through Androgen Biosynthesis. Science 2021, 374, 216–224. [Google Scholar] [CrossRef]
- Terrisse, S.; Goubet, A.-G.; Ueda, K.; Thomas, A.M.; Quiniou, V.; Thelemaque, C.; Dunsmore, G.; Clave, E.; Gamat-Huber, M.; Yonekura, S.; et al. Immune System and Intestinal Microbiota Determine Efficacy of Androgen Deprivation Therapy against Prostate Cancer. J. Immunother. Cancer 2022, 10, e004191. [Google Scholar] [CrossRef]
- Daisley, B.A.; Chanyi, R.M.; Abdur-Rashid, K.; Al, K.F.; Gibbons, S.; Chmiel, J.A.; Wilcox, H.; Reid, G.; Anderson, A.; Dewar, M.; et al. Abiraterone Acetate Preferentially Enriches for the Gut Commensal Akkermansia Muciniphila in Castrate-Resistant Prostate Cancer Patients. Nat. Commun. 2020, 11, 4822. [Google Scholar] [CrossRef] [PubMed]
- Sfanos, K.S.; Yegnasubramanian, S.; Nelson, W.G.; De Marzo, A.M. The Inflammatory Microenvironment and Microbiome in Prostate Cancer Development. Nat. Rev. Urol. 2018, 15, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Puerta Suárez, J.; Cardona Maya, W.D. Microbiota, Prostatitis, and Fertility: Bacterial Diversity as a Possible Health Ally. Adv. Urol. 2021, 2021, 1007366. [Google Scholar] [CrossRef] [PubMed]
- Jung, G.; Kim, J.K.; Kim, H.; Lee, J.; Hong, S.K. The Association between Prostatitis and Risk of Prostate Cancer: A National Health Insurance Database Study. World J. Urol. 2022, 40, 2781–2787. [Google Scholar] [CrossRef]
- Platz, E.A.; Kulac, I.; Barber, J.R.; Drake, C.G.; Joshu, C.E.; Nelson, W.G.; Lucia, M.S.; Klein, E.A.; Lippman, S.M.; Parnes, H.L.; et al. A Prospective Study of Chronic Inflammation in Benign Prostate Tissue and Risk of Prostate Cancer: Linked PCPT and SELECT Cohorts. Cancer Epidemiol. Biomark. Prev. 2017, 26, 1549–1557. [Google Scholar] [CrossRef]
- Ranjbar, M.; Salehi, R.; Haghjooy Javanmard, S.; Rafiee, L.; Faraji, H.; Jafarpor, S.; Ferns, G.A.; Ghayour-Mobarhan, M.; Manian, M.; Nedaeinia, R. The Dysbiosis Signature of Fusobacterium Nucleatum in Colorectal Cancer-Cause or Consequences? A Systematic Review. Cancer Cell Int. 2021, 21, 194. [Google Scholar] [CrossRef]
- McQuade, J.L.; Daniel, C.R.; Helmink, B.A.; Wargo, J.A. Modulating the Microbiome to Improve Therapeutic Response in Cancer. Lancet Oncol. 2019, 20, e77–e91. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.-L.; et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Covidence—Better Systematic Review Management. Available online: http://www.covidence.org (accessed on 12 March 2024).
- Alluri, L.S.C.; Paes Batista da Silva, A.; Verma, S.; Fu, P.; Shen, D.L.; MacLennan, G.; Gupta, S.; Bissada, N.F. Presence of Specific Periodontal Pathogens in Prostate Gland Diagnosed with Chronic Inflammation and Adenocarcinoma. Cureus 2021, 13, e17742. [Google Scholar] [CrossRef]
- Banerjee, S.; Alwine, J.C.; Wei, Z.; Tian, T.; Shih, N.; Sperling, C.; Guzzo, T.; Feldman, M.D.; Robertson, E.S. Microbiome Signatures in Prostate Cancer. Carcinogenesis 2019, 40, 749–764. [Google Scholar] [CrossRef]
- Cavarretta, I.; Ferrarese, R.; Cazzaniga, W.; Saita, D.; Lucianò, R.; Ceresola, E.R.; Locatelli, I.; Visconti, L.; Lavorgna, G.; Briganti, A.; et al. The Microbiome of the Prostate Tumor Microenvironment. Eur. Urol. 2017, 72, 625–631. [Google Scholar] [CrossRef]
- Chen, Y.; Wei, J. Identification of Pathogen Signatures in Prostate Cancer Using RNA-Seq. PLoS ONE 2015, 10, e0128955. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Ramnarine, V.R.; Bell, R.; Volik, S.; Davicioni, E.; Hayes, V.M.; Ren, S.; Collins, C.C. Metagenomic and Metatranscriptomic Analysis of Human Prostate Microbiota from Patients with Prostate Cancer. BMC Genom. 2019, 20, 146. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, M.F.M.; Pina-Vaz, T.; Fernandes, Â.R.; Miranda, I.M.; Silva, C.M.; Rodrigues, A.G.; Lisboa, C. Microbiota of Urine, Glans and Prostate Biopsies in Patients with Prostate Cancer Reveals a Dysbiosis in the Genitourinary System. Cancers 2023, 15, 1423. [Google Scholar] [CrossRef] [PubMed]
- Miyake, M.; Ohnishi, K.; Hori, S.; Nakano, A.; Nakano, R.; Yano, H.; Ohnishi, S.; Owari, T.; Morizawa, Y.; Itami, Y.; et al. Mycoplasma Genitalium Infection and Chronic Inflammation in Human Prostate Cancer: Detection Using Prostatectomy and Needle Biopsy Specimens. Cells 2019, 8, 212. [Google Scholar] [CrossRef] [PubMed]
- Salachan, P.V.; Rasmussen, M.; Fredsøe, J.; Ulhøi, B.; Borre, M.; Sørensen, K.D. Microbiota of the Prostate Tumor Environment Investigated by Whole-Transcriptome Profiling. Genome Med. 2022, 14, 9. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, P.; Malik, S.; Banerjee, A.; Datta, C.; Pal, D.K.; Ghosh, A.; Saha, A. Differential Microbial Signature Associated with Benign Prostatic Hyperplasia and Prostate Cancer. Front. Cell. Infect. Microbiol. 2022, 12, 894777. [Google Scholar] [CrossRef] [PubMed]
- Andermann, T.; Antonelli, A.; Barrett, R.L.; Silvestro, D. Estimating Alpha, Beta, and Gamma Diversity Through Deep Learning. Front. Plant Sci. 2022, 13, 839407. [Google Scholar] [CrossRef]
- ROBINS-E Tool. Available online: https://www.riskofbias.info/welcome/robins-e-tool (accessed on 12 March 2024).
- Shinohara, D.B.; Vaghasia, A.M.; Yu, S.-H.; Mak, T.N.; Brüggemann, H.; Nelson, W.G.; De Marzo, A.M.; Yegnasubramanian, S.; Sfanos, K.S. A Mouse Model of Chronic Prostatic Inflammation Using a Human Prostate Cancer-Derived Isolate of Propionibacterium Acnes. Prostate 2013, 73, 1007–1015. [Google Scholar] [CrossRef]
- Fassi Fehri, L.; Mak, T.N.; Laube, B.; Brinkmann, V.; Ogilvie, L.A.; Mollenkopf, H.; Lein, M.; Schmidt, T.; Meyer, T.F.; Brüggemann, H. Prevalence of Propionibacterium Acnes in Diseased Prostates and Its Inflammatory and Transforming Activity on Prostate Epithelial Cells. Int. J. Med. Microbiol. 2011, 301, 69–78. [Google Scholar] [CrossRef]
- Kim, J.; Ochoa, M.-T.; Krutzik, S.R.; Takeuchi, O.; Uematsu, S.; Legaspi, A.J.; Brightbill, H.D.; Holland, D.; Cunliffe, W.J.; Akira, S.; et al. Activation of Toll-like Receptor 2 in Acne Triggers Inflammatory Cytokine Responses. J. Immunol. 2002, 169, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Webster, G.F.; Leyden, J.J. Characterization of Serum-Independent Polymorphonuclear Leukocyte Chemotactic Factors Produced by Propionibacterium Acnes. Inflammation 1980, 4, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Davidsson, S.; Carlsson, J.; Greenberg, L.; Wijkander, J.; Söderquist, B.; Erlandsson, A. Cutibacterium Acnes Induces the Expression of Immunosuppressive Genes in Macrophages and Is Associated with an Increase of Regulatory T-Cells in Prostate Cancer. Microbiol. Spectr. 2021, 9, e0149721. [Google Scholar] [CrossRef] [PubMed]
- Radej, S.; Płaza, P.; Olender, A.; Szewc, M.; Bar, K.; Maciejewski, R. Infiltrating Treg and Th17 Cells of the Prostate Hypertrophy Gland Associated with Propionibacterium Acnes Infection. Res. Rep. Urol. 2020, 12, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Yow, M.A.; Tabrizi, S.N.; Severi, G.; Bolton, D.M.; Pedersen, J.; Australian Prostate Cancer BioResource; Giles, G.G.; Southey, M.C. Characterisation of Microbial Communities within Aggressive Prostate Cancer Tissues. Infect. Agent. Cancer 2017, 12, 4. [Google Scholar] [CrossRef]
- Alexeyev, O.A.; Marklund, I.; Shannon, B.; Golovleva, I.; Olsson, J.; Andersson, C.; Eriksson, I.; Cohen, R.; Elgh, F. Direct Visualization of Propionibacterium Acnes in Prostate Tissue by Multicolor Fluorescent in Situ Hybridization Assay. J. Clin. Microbiol. 2007, 45, 3721–3728. [Google Scholar] [CrossRef]
- Graham, S.V. The Human Papillomavirus Replication Cycle, and Its Links to Cancer Progression: A Comprehensive Review. Clin. Sci. 2017, 131, 2201–2221. [Google Scholar] [CrossRef]
- McLaughlin-Drubin, M.E.; Münger, K. Oncogenic Activities of Human Papillomaviruses. Virus Res. 2009, 143, 195–208. [Google Scholar] [CrossRef]
- Whitaker, N.J.; Glenn, W.K.; Sahrudin, A.; Orde, M.M.; Delprado, W.; Lawson, J.S. Human Papillomavirus and Epstein Barr Virus in Prostate Cancer: Koilocytes Indicate Potential Oncogenic Influences of Human Papillomavirus in Prostate Cancer. Prostate 2013, 73, 236–241. [Google Scholar] [CrossRef]
- McNicol, P.J.; Dodd, J.G. High Prevalence of Human Papillomavirus in Prostate Tissues. J. Urol. 1991, 145, 850–853. [Google Scholar] [CrossRef]
- McNicol, P.J.; Dodd, J.G. Detection of Human Papillomavirus DNA in Prostate Gland Tissue by Using the Polymerase Chain Reaction Amplification Assay. J. Clin. Microbiol. 1990, 28, 409–412. [Google Scholar] [CrossRef]
- Wideroff, L.; Schottenfeld, D.; Carey, T.E.; Beals, T.; Fu, G.; Sakr, W.; Sarkar, F.; Schork, A.; Grossman, H.B.; Shaw, M.W. Human Papillomavirus DNA in Malignant and Hyperplastic Prostate Tissue of Black and White Males. Prostate 1996, 28, 117–123. [Google Scholar] [CrossRef]
- Tu, H.; Jacobs, S.C.; Mergner, W.J.; Kyprianou, N. Rare Incidence of Human Papillomavirus Types 16 and 18 in Primary and Metastatic Human Prostate Cancer. Urology 1994, 44, 726–731. [Google Scholar] [CrossRef]
- Shi, Y.; Peng, S.-L.; Yang, L.-F.; Chen, X.; Tao, Y.-G.; Cao, Y. Co-Infection of Epstein-Barr Virus and Human Papillomavirus in Human Tumorigenesis. Chin. J. Cancer 2016, 35, 16. [Google Scholar] [CrossRef]
- Khenchouche, A.; Sadouki, N.; Boudriche, A.; Houali, K.; Graba, A.; Ooka, T.; Bouguermouh, A. Human Papillomavirus and Epstein-Barr Virus Co-Infection in Cervical Carcinoma in Algerian Women. Virol. J. 2013, 10, 340. [Google Scholar] [CrossRef]
- Polz-Gruszka, D.; Morshed, K.; Stec, A.; Polz-Dacewicz, M. Prevalence of Human Papillomavirus (HPV) and Epstein-Barr Virus (EBV) in Oral and Oropharyngeal Squamous Cell Carcinoma in South-Eastern Poland. Infect. Agents Cancer 2015, 10, 37. [Google Scholar] [CrossRef]
- Al Moustafa, A.-E.; Chen, D.; Ghabreau, L.; Akil, N. Association between Human Papillomavirus and Epstein-Barr Virus Infections in Human Oral Carcinogenesis. Med. Hypotheses 2009, 73, 184–186. [Google Scholar] [CrossRef]
- Boudreault, S.; Armero, V.E.S.; Scott, M.S.; Perreault, J.-P.; Bisaillon, M. The Epstein-Barr Virus EBNA1 Protein Modulates the Alternative Splicing of Cellular Genes. Virol. J. 2019, 16, 29. [Google Scholar] [CrossRef]
- Moghoofei, M.; Mostafaei, S.; Nesaei, A.; Etemadi, A.; Sadri Nahand, J.; Mirzaei, H.; Rashidi, B.; Babaei, F.; Khodabandehlou, N. Epstein-Barr Virus and Thyroid Cancer: The Role of Viral Expressed Proteins. J. Cell. Physiol. 2019, 234, 3790–3799. [Google Scholar] [CrossRef]
- Nahand, J.S.; Khanaliha, K.; Mirzaei, H.; Moghoofei, M.; Baghi, H.B.; Esghaei, M.; Khatami, A.R.; Fatemipour, M.; Bokharaei-Salim, F. Possible Role of HPV/EBV Coinfection in Anoikis Resistance and Development in Prostate Cancer. BMC Cancer 2021, 21, 926. [Google Scholar] [CrossRef]
- de Lima, M.A.P.; Neto, P.J.N.; Lima, L.P.M.; Gonçalves Júnior, J.; Teixeira Junior, A.G.; Teodoro, I.P.P.; Facundo, H.T.; da Silva, C.G.L.; Lima, M.V.A. Association between Epstein-Barr Virus (EBV) and Cervical Carcinoma: A Meta-Analysis. Gynecol. Oncol. 2018, 148, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Wensel, C.R.; Pluznick, J.L.; Salzberg, S.L.; Sears, C.L. Next-generation sequencing: Insights to advance clinical investigations of the microbiome. J. Clin. Investig. 2022, 132, e154944. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).