Unlocking Overexpressed Membrane Proteins to Guide Breast Cancer Precision Medicine
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
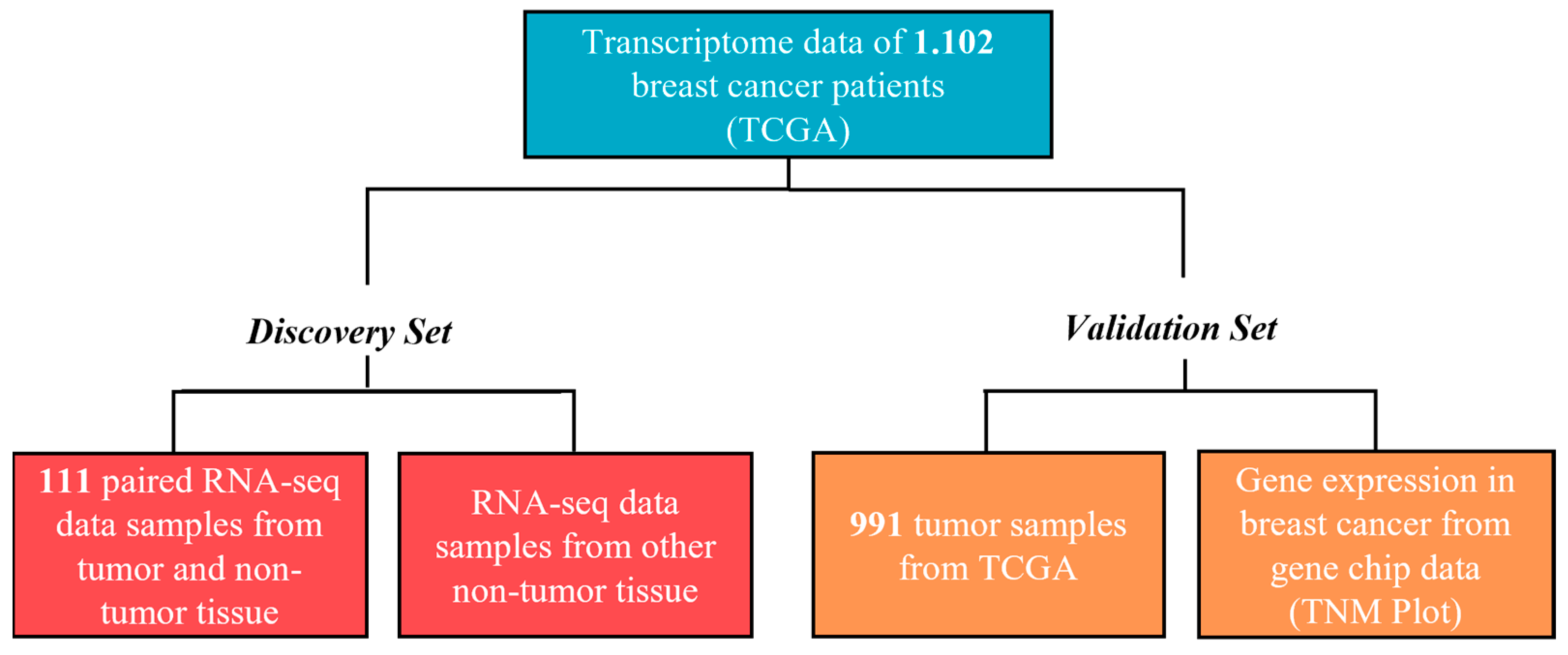
2.1. Data Processing, Samples, and Databases
2.2. Identification of Overexpressed Transcripts Encoding Membrane Proteins
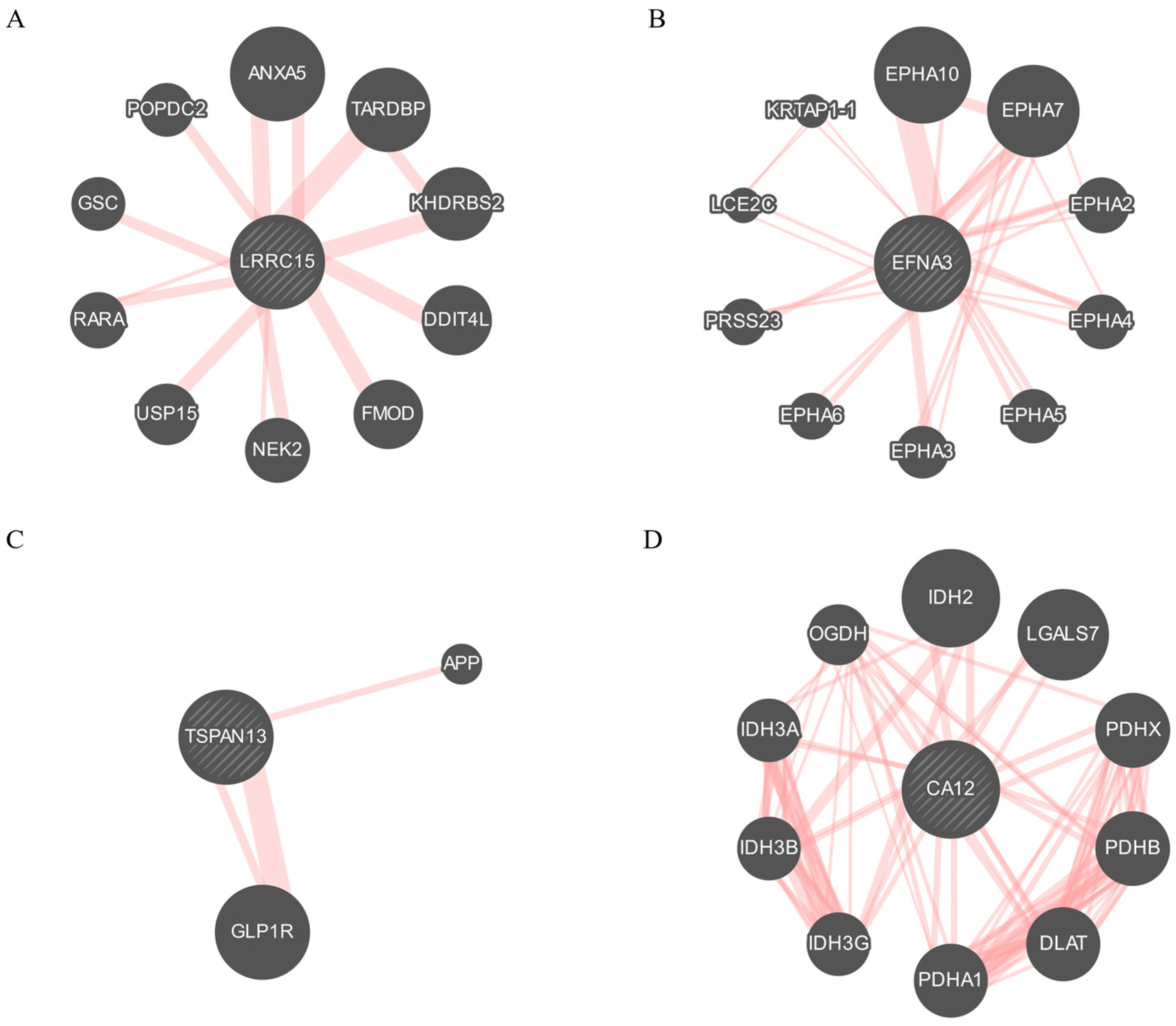
2.3. Protein–Protein Interaction (PPI) Network
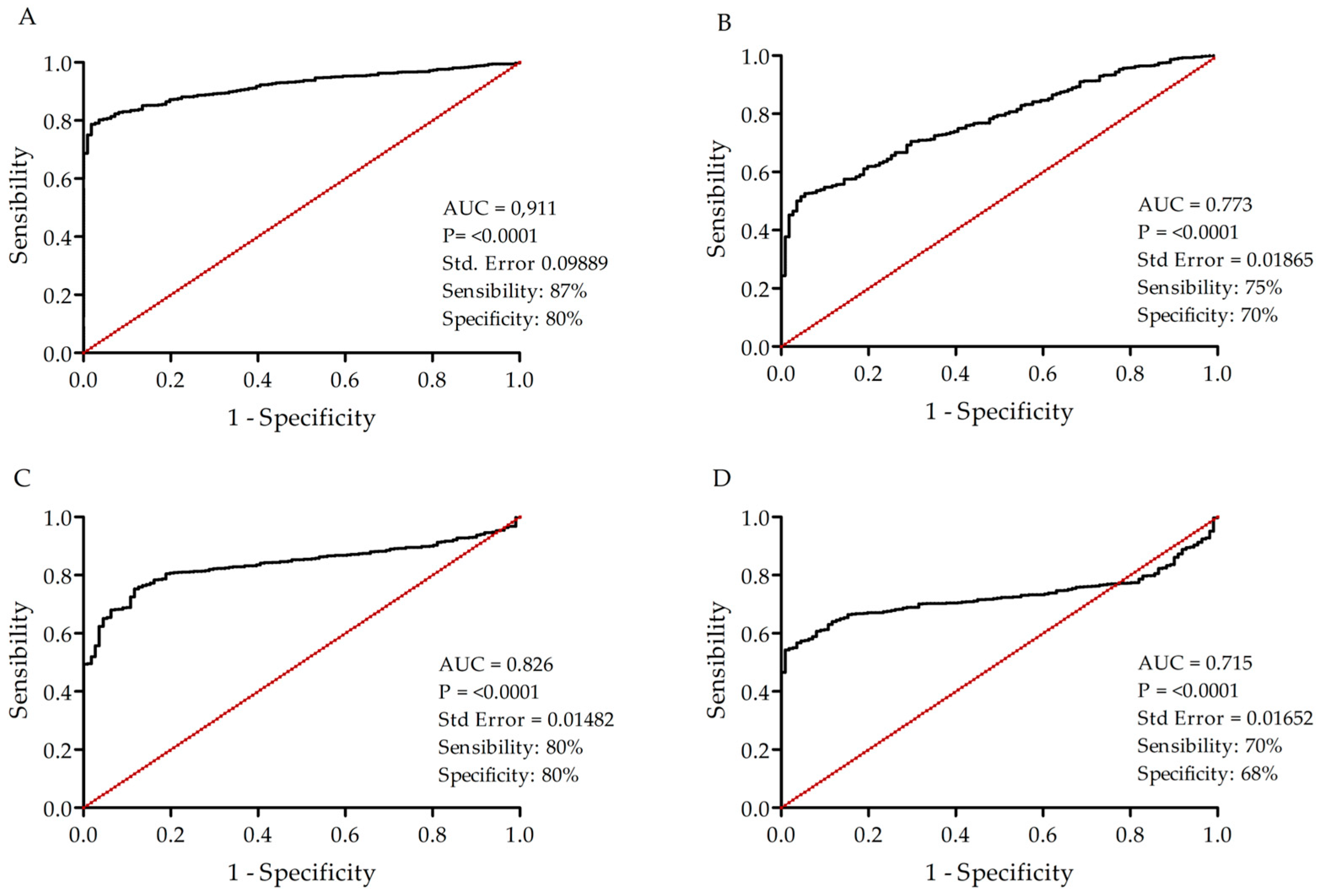
2.4. Analysis of the Diagnostic Value of the Selected Targets
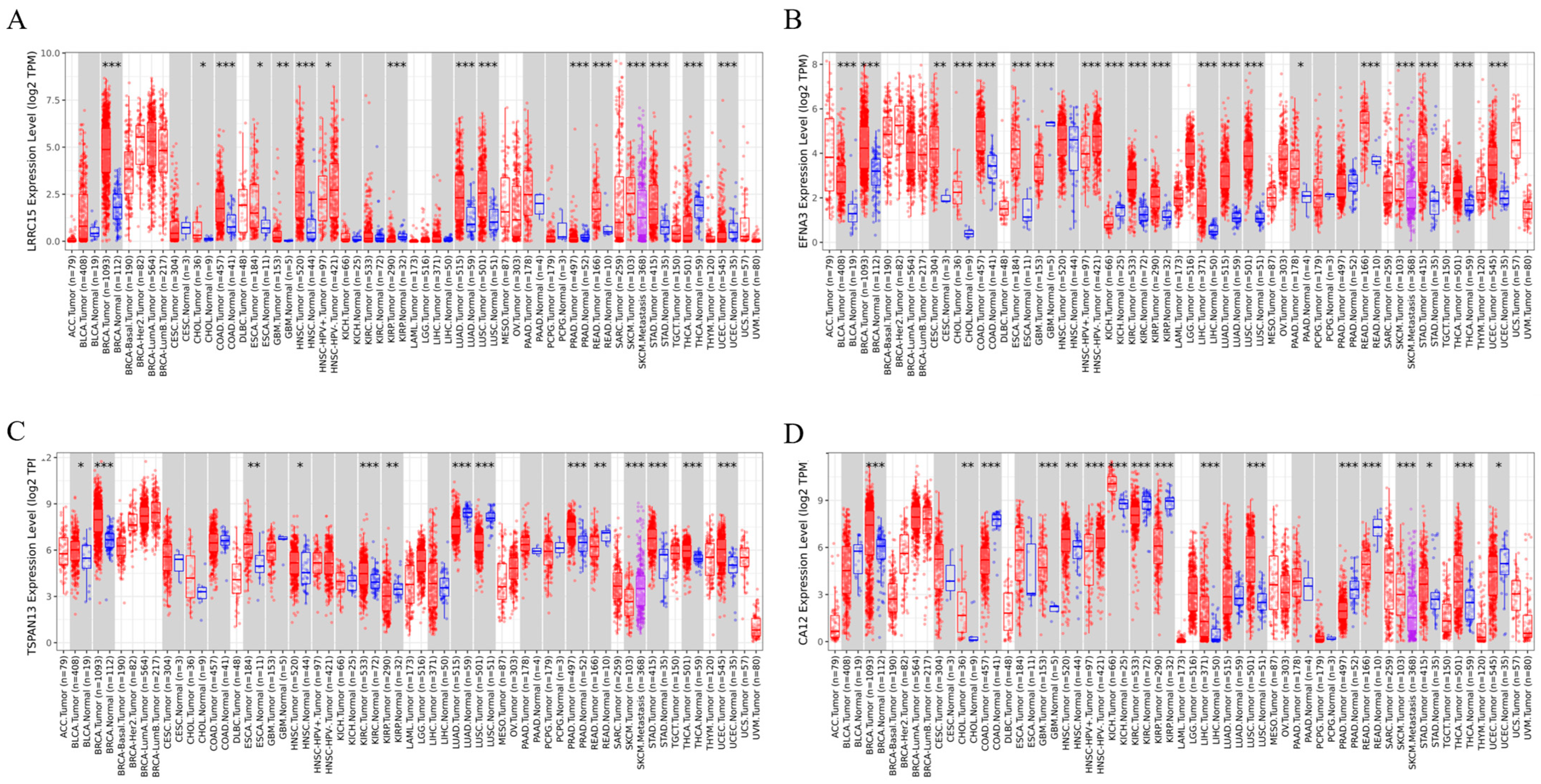
2.5. Pan-Cancer Analysis of the Selected Genes in Tumor and Adjacent Non-Tumor Tissues
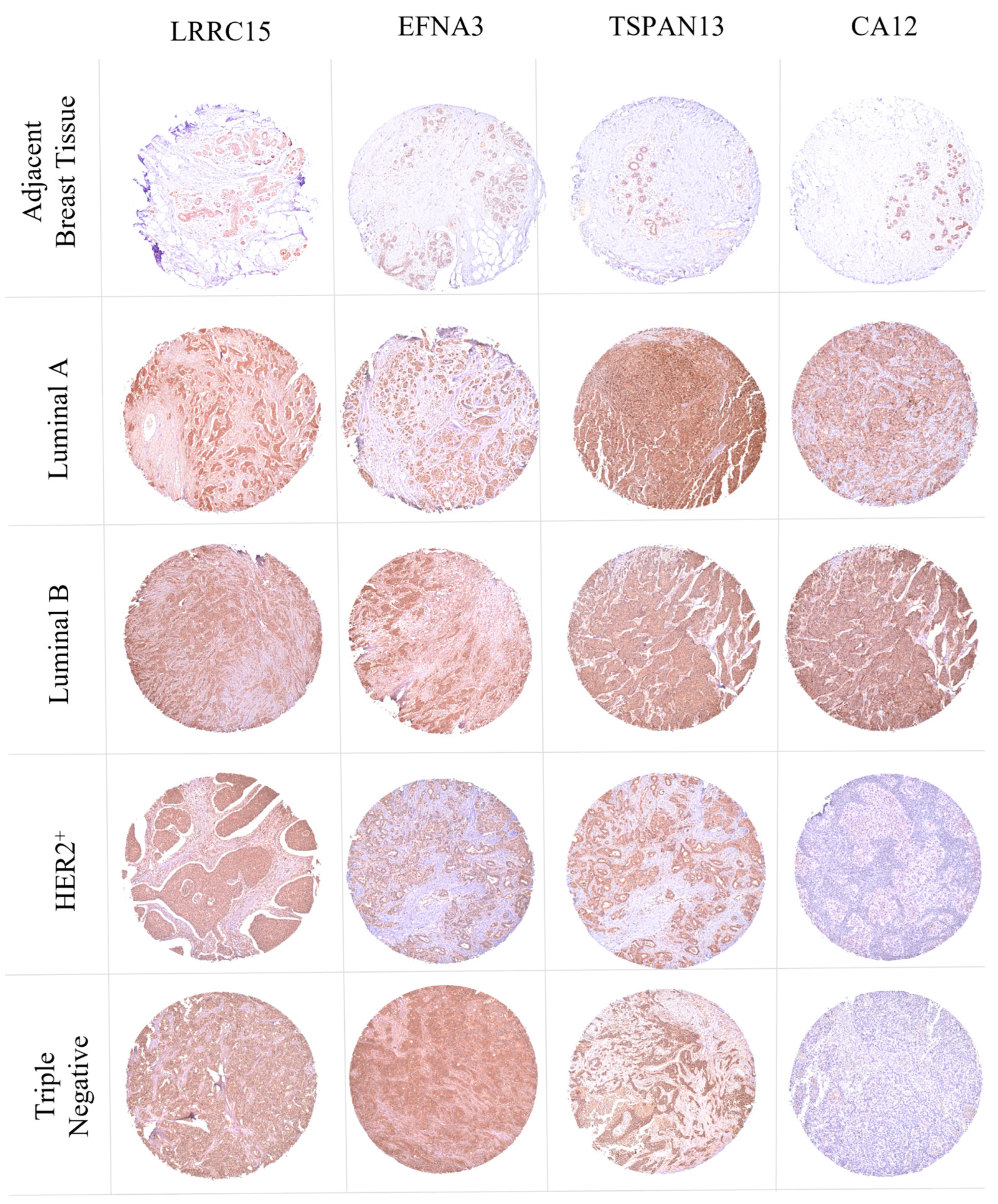
2.6. Tissue Microarray
- 0: incomplete, weak, and scanty staining in the membranes of <10% of tumor cells.
- 1+: incomplete, weak, and scanty staining in the membranes of >10% of tumor cells.
- 2+: circumferential and incomplete staining and/or weak/moderate membrane staining in more than 10% of tumor cells, or complete and/or intense membrane staining in more than 10% of tumor cells.
- 3+: uniform and intense membrane staining of tumor cells.
2.7. Statistical Analysis
3. Results
3.1. Clinical, Pathological, and Molecular Characteristics of the Cohort from TCGA Database
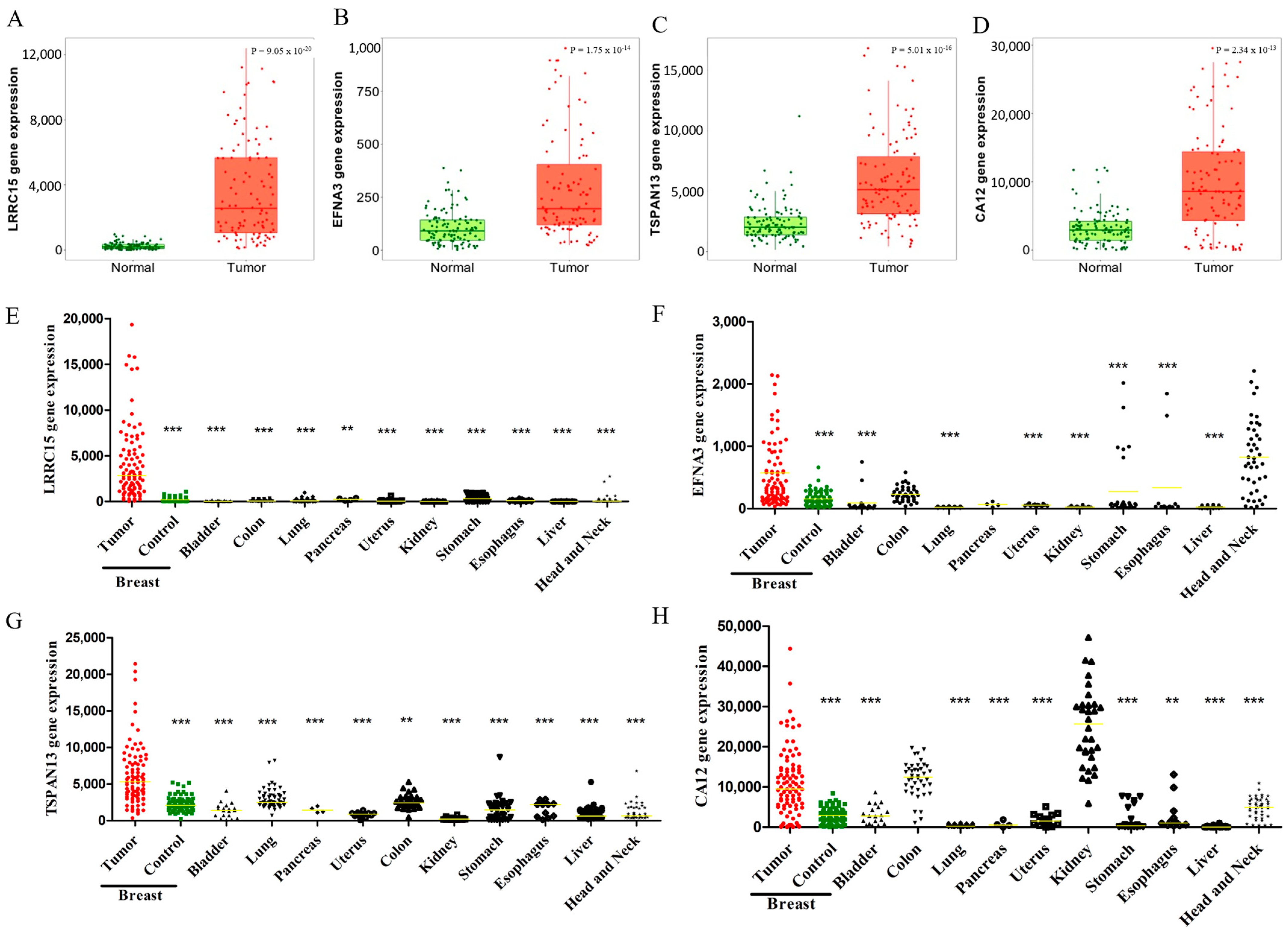
3.2. LRRC15, EFNA3, TSPAN13, and CA12 Are Highly Expressed in BC Patients
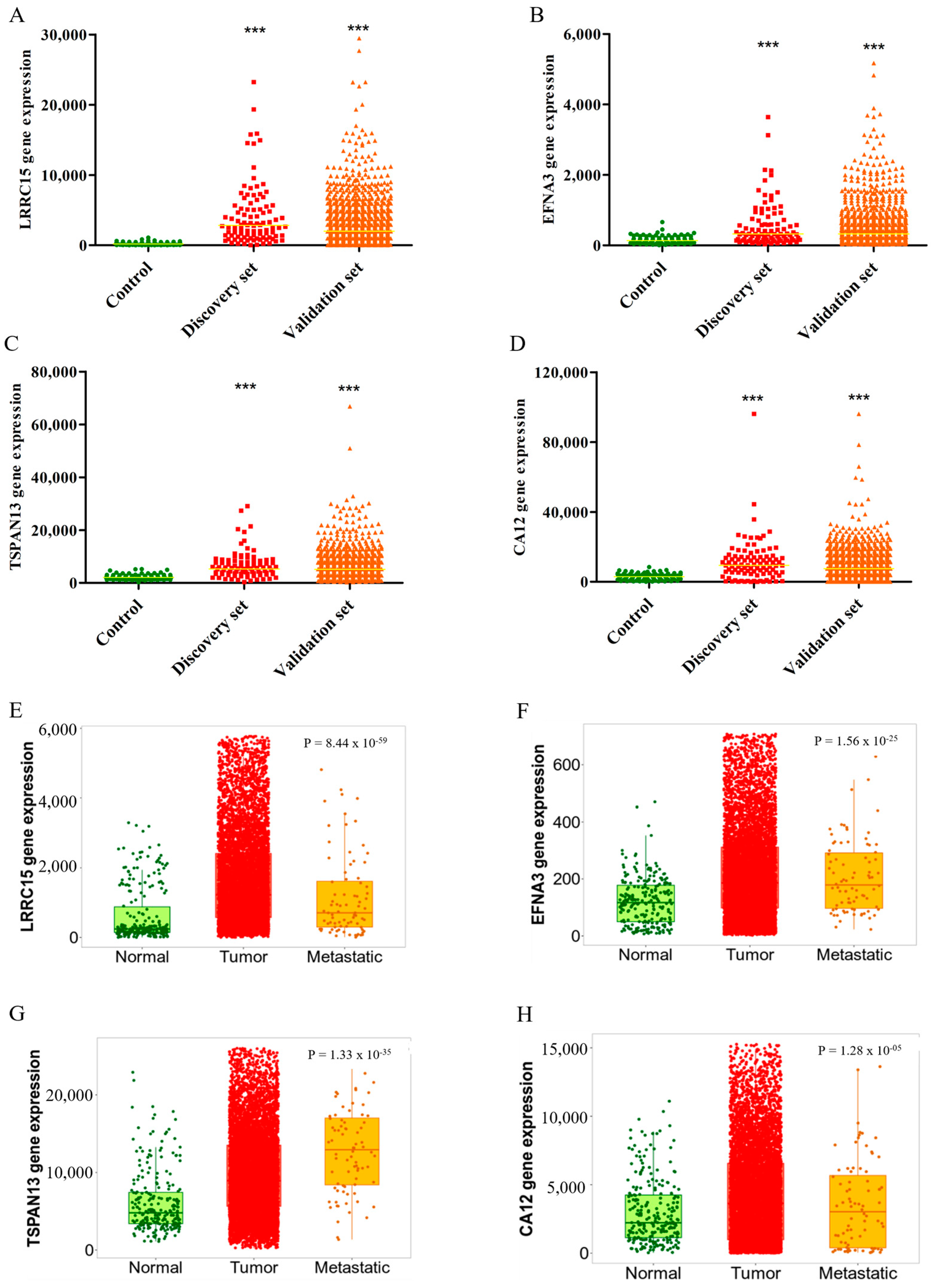
3.3. Validation of LRRC15, EFNA3, TSPAN13, and CA12 Overexpressions in BC Tissues
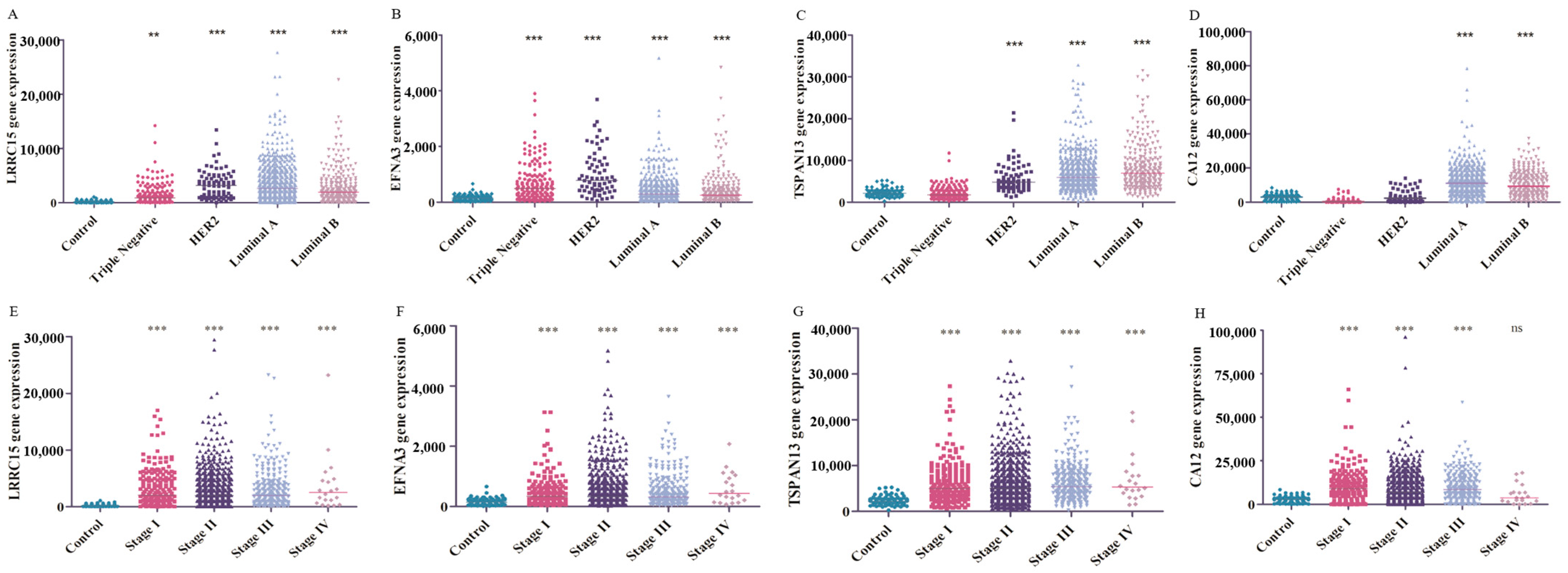
3.4. Potential Use of the Four Targets for BC Molecular Classification and Precision Diagnosis
3.5. LRRC15, EFNA3, TSPAN13, and CA12 as Potential Novel Biomarkers for BC
3.6. Cross-Cancer Overexpression beyond Breast Tumors
3.7. Exploring the Signaling Pathways and Protein Interactions of Identified Membrane Proteins in Tumorigenesis
3.8. LRRC15, EFNA3, TSPAN13, and CA12 Are Overexpressed in BC Clinical Samples
3.9. Validation of LRRC15, TSPAN13, and CA12 Protein Expression Profiles in Breast Cancer Patients: Insights from the CPTAC Dataset
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Zubair, M.; Wang, S.; Ali, N. Advanced Approaches to Breast Cancer Classification and Diagnosis. Front. Pharmacol. 2021, 11, 2487. [Google Scholar] [CrossRef]
- Prat, A.; Pineda, E.; Adamo, B.; Galván, P.; Fernández, A.; Gaba, L.; Díez, M.; Viladot, M.; Arance, A.; Muñoz, M. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 2015, 24 (Suppl. S2), S26–S35. [Google Scholar] [CrossRef]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef]
- Tang, Y.; Wang, Y.; Kiani, M.F.; Wang, B. Classification, Treatment Strategy, and Associated Drug Resistance in Breast Cancer. Clin. Breast Cancer 2016, 16, 335–343. [Google Scholar] [CrossRef]
- Lovelace, D.L.; McDaniel, L.R.; Golden, D. Long-Term Effects of Breast Cancer Surgery, Treatment, and Survivor Care. J. Midwifery Womens Health 2019, 64, 713–724. [Google Scholar] [CrossRef]
- Wang, R.; Zhu, Y.; Liu, X.; Liao, X.; He, J.; Niu, L. The Clinicopathological features and survival outcomes of patients with different metastatic sites in stage IV breast cancer. BMC Cancer 2019, 19, 1091. [Google Scholar] [CrossRef]
- Nakhjavani, M.; Samarasinghe, R.M.; Shigdar, S. Triple-negative breast cancer brain metastasis: An update on druggable targets, current clinical trials, and future treatment options. Drug Discov. Today 2022, 27, 1298–1314. [Google Scholar] [CrossRef]
- Quezada, H.; Guzmán-Ortiz, A.L.; Díaz-Sánchez, H.; Valle-Rios, R.; Aguirre-Hernández, J. Omics-based biomarkers: Current status and potential use in the clinic. Bol. Med. Hosp. Infant. Mex. 2017, 74, 219–226. [Google Scholar] [CrossRef]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef]
- Dunn, B.K.; Meerzaman, D. Editorial: Using Cancer ‘Omics’ to Understand Cancer. Front. Oncol. 2020, 10, 1201. [Google Scholar] [CrossRef]
- Neagu, A.N.; Whitham, D.; Seymour, L.; Haaker, N.; Pelkey, I.; Darie, C.C. Proteomics-Based Identification of Dysregulated Proteins and Biomarker Discovery in Invasive Ductal Carcinoma, the Most Common Breast Cancer Subtype. Proteomes 2023, 11, 13. [Google Scholar] [CrossRef]
- Boonstra, M.C.; De Geus, S.W.; Prevoo, H.A.; Hawinkels, L.J.; Van De Velde, C.J.; Kuppen, P.J.; Vahrmeijer, A.L.; Sier, C.F. Selecting Targets for Tumor Imaging: An Overview of Cancer-Associated Membrane Proteins. Biomark Cancer 2016, 8, BIC.S38542. [Google Scholar] [CrossRef]
- Yin, H.; Flynn, A.D. Drugging Membrane Protein Interactions. Annu. Rev. Biomed. Eng. 2016, 18, 51–76. [Google Scholar] [CrossRef]
- Kampen, K.R. Membrane proteins: The key players of a cancer cell. J. Membr. Biol. 2011, 242, 69–74. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Lee, C.-H.; Chuang, Y.-H.; Lee, J.-Y.; Chiu, Y.-Y.; Wu Lee, Y.-H.; Jong, Y.-J.; Hwang, J.-K.; Huang, S.-H.; Chen, L.-C.; et al. Membrane protein-regulated networks across human cancers. Nat. Commun. 2019, 10, 3131. [Google Scholar] [CrossRef]
- Poudineh, M.; Sargent, E.H.; Pantel, K.; Kelley, S.O. Profiling circulating tumour cells and other biomarkers of invasive cancers. Nat. Biomed. Eng. 2018, 2, 72–84. [Google Scholar] [CrossRef]
- Bartha, Á.; Győrffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Chandrashekar, D.S.; Varambally, S.; Creighton, C.J. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat. Commun. 2019, 10, 5679. [Google Scholar] [CrossRef]
- Pontén, F.; Jirström, K.; Uhlen, M. The Human Protein Atlas—A tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Al-Mahmood, S.; Sapiezynski, J.; Garbuzenko, O.B.; Minko, T. Metastatic and triple-negative breast cancer: Challenges and treatment options. Drug Deliv. Transl. Res. 2018, 8, 1483–1507. [Google Scholar] [CrossRef]
- Dong, P.; Yu, B.; Pan, L.; Tian, X.; Liu, F. Identification of Key Genes and Pathways in Triple-Negative Breast Cancer by Integrated Bioinformatics Analysis. Biomed. Res. Int. 2018, 2018, 2760918. [Google Scholar] [CrossRef]
- Wang, Z.; Deisboeck, T.S. Dynamic targeting in cancer treatment. Front. Physiol. 2019, 10, 96. [Google Scholar] [CrossRef]
- Pasto, A.; Giordano, F.; Evangelopoulos, M.; Amadori, A.; Tasciotti, E. Cell membrane protein functionalization of nanoparticles as a new tumor-targeting strategy. Clin. Transl. Med. 2019, 8, 8. [Google Scholar] [CrossRef]
- Jones, M.L.; Alfaleh, M.A.; Kumble, S.; Zhang, S.; Osborne, G.W.; Yeh, M.; Arora, N.; Hou, J.J.C.; Howard, C.B.; Chin, D.Y.; et al. Targeting membrane proteins for antibody discovery using phage display. Sci. Rep. 2016, 6, 26240. [Google Scholar] [CrossRef]
- Mercier, M.C.; Dontenwill, M.; Choulier, L. Selection of nucleic acid aptamers targeting tumor cell-surface protein biomarkers. Cancers 2017, 9, 69. [Google Scholar] [CrossRef]
- Ray, U.; Jung, D.-B.; Jin, L.; Xiao, Y.; Dasari, S.; Bhattacharya, S.S.; Thirusangu, P.; Staub, J.K.; Roy, D.; Roy, B.; et al. Targeting LRRC15 Inhibits Metastatic Dissemination of Ovarian Cancer. Cancer Res. 2022, 82, 1038–1054. [Google Scholar] [CrossRef]
- Satoh, K.; Hata, M.; Shimizu, T.; Yokota, H.; Akatsu, H.; Yamamoto, T.; Kosaka, K.; Yamada, T. Lib, transcriptionally induced in senile plaque-associated astrocytes, promotes glial migration through extracellular matrix. Biochem. Biophys. Res. Commun. 2005, 335, 631–636. [Google Scholar] [CrossRef]
- O’Prey, J.; Wilkinson, S.; Ryan, K.M. Tumor antigen LRRC15 impedes adenoviral infection: Implications for virus-based cancer therapy. J. Virol. 2008, 82, 5933–5939. [Google Scholar] [CrossRef] [PubMed]
- Satoh, K.; Hata, M.; Yokota, H. A novel member of the leucine-rich repeat superfamily induced in rat astrocytes by β-amyloid. Biochem. Biophys. Res. Commun. 2002, 290, 756–762. [Google Scholar] [CrossRef] [PubMed]
- Satoh, K.; Hata, M.; Yokota, H. High Lib mRNA Expression in Breast Carcinomas. DNA Res. 2004, 11, 199–203. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Purcell, J.W.; Tanlimco, S.G.; Hickson, J.A.; Fox, M.; Sho, M.; Durkin, L.; Uziel, T.; Powers, R.; Foster, K.; McGonigal, T.; et al. LRRC15 is a novel mesenchymal protein and stromal target for antibody–drug conjugates. Cancer Res. 2018, 78, 4059–4072. [Google Scholar] [CrossRef] [PubMed]
- Hingorani, P.; Roth, M.E.; Wang, Y.; Zhang, W.; Gill, J.B.; Harrison, D.J.; Teicher, B.; Erickson, S.; Gatto, G.; Smith, M.A.; et al. ABBV-085, antibody-drug conjugate targeting LRRC15, is effective in osteosarcoma: A report by the pediatric preclinical testing consortium. Mol. Cancer Ther. 2021, 20, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Li, Y.; Wang, H.; Qin, T.; Yin, X.; Ma, X. Therapeutic progress and challenges for triple negative breast cancer: Targeted therapy and immunotherapy. Mol. Biomed. 2022, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Ray, U.; Pathoulas, C.L.; Thirusangu, P.; Purcell, J.W.; Kannan, N.; Shridhar, V. Exploiting LRRC15 as a Novel Therapeutic Target in Cancer. Cancer Res. 2022, 82, 1675–1681. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Dean, D.; Wei, R.; Hornicek, F.J.; Ulmert, D.; Duan, Z. Expression and clinical implications of leucine-rich repeat containing 15 (LRRC15) in osteosarcoma. J. Orthop. Res. 2020, 38, 2362–2372. [Google Scholar] [CrossRef]
- Demetri, G.D.; Luke, J.J.; Hollebecque, A.; Powderly, J.D.; Spira, A.I.; Subbiah, V.; Naumovski, L.; Chen, C.; Fang, H.; Lai, D.W.; et al. First-in-human phase I study of ABBV-085, an antibody–drug conjugate targeting LRRC15, in sarcomas and other advanced solid tumors. Clin. Cancer Res. 2021, 27, 3556–3566. [Google Scholar] [CrossRef]
- Nievergall, E.; Lackmann, M.; Janes, P.W. Eph-dependent cell-cell adhesion and segregation in development and cancer. Cell. Mol. Life Sci. 2012, 69, 1813–1842. [Google Scholar] [CrossRef]
- Gómez-Maldonado, L.; Tiana, M.; Roche, O.; Prado-Cabrero, A.; Jensen, L.; Fernandez-Barral, A.; Guijarro-Muñoz, I.; Favaro, E.; Moreno-Bueno, G.; Sanz, L.; et al. EFNA3 long noncoding RNAs induced by hypoxia promote metastatic dissemination. Oncogene 2015, 34, 2609–2620. [Google Scholar] [CrossRef]
- Liang, Z.; Wang, X.; Dong, K.; Li, X.; Qin, C.; Zhou, H. Expression Pattern and Prognostic Value of EPHA/EFNA in Breast Cancer by Bioinformatics Analysis: Revealing Its Importance in Chemotherapy. Biomed. Res. Int. 2021, 2021, 5575704. [Google Scholar] [CrossRef]
- Cherukuri, A.; Shoham, T.; Sohn, H.W.; Levy, S.; Brooks, S.; Carter, R.; Pierce, S.K. The tetraspanin CD81 is necessary for partitioning of coligated CD19/CD21-B cell antigen receptor complexes into signaling-active lipid rafts. J. Immunol. 2004, 172, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, N.; Wang, Z.; Ashman, L.K.; Brady-Kalnay, S.M.; Kreidberg, J.A. α3β1 integrin-CD151, a component of the cadherin-catenin complex, regulates PTPμ expression and cell-cell adhesion. J. Cell Biol. 2003, 163, 1351–1362. [Google Scholar] [CrossRef] [PubMed]
- Carloni, V.; Mazzocca, A.; Ravichandran, K.S. Tetraspanin CD81 is linked to ERK/MAPKinase signaling by Shc in liver tumor cells. Oncogene 2004, 23, 1566–1574. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, S.; Yuan, X.; Liu, Y.; Zhang, K.; Wang, J.; Zhu, J.; Ma, R. miR-4732-5p promotes breast cancer progression by targeting TSPAN13. J. Cell. Mol. Med. 2019, 23, 2549–2557. [Google Scholar] [CrossRef] [PubMed]
- Berditchevski, F.; Odintsova, E.; Sawada, S.; Gilbert, E. Expression of the palmitoylation-deficient CD151 weakens the association of α3β1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling. J. Biol. Chem. 2002, 277, 36991–37000. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Sossey-Alaoui, K.; Beachy, S.H.; Geradts, J. The tetraspanin superfamily member NET-6 is a new tumor suppressor gene. J. Cancer Res. Clin. Oncol. 2007, 133, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zhang, X.; Geradts, J.; Wei, Q.; Hochwald, S.; Xu, H.; Huang, H. Expression of tetraspanins NET-6 and CD151 in breast cancer as a potential tumor biomarker. Clin. Exp. Med. 2019, 19, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Waheed, A.; Sly, W.S. Carbonic anhydrase XII functions in health and disease. Gene 2017, 623, 33–40. [Google Scholar] [CrossRef]
- Li, Y.; Lei, B.; Zou, J.; Wang, W.; Chen, A.; Zhang, J.; Fu, Y.; Li, Z. High expression of CA12 and good prognosis in breast cancer High expression of carbonic anhydrase 12 (CA12) is associated with good prognosis in breast cancer. Neoplasma 2019, 66, 420–426. [Google Scholar] [CrossRef]
- Barnett, D.H.; Sheng, S.; Charn, T.H.; Waheed, A.; Sly, W.S.; Lin, C.-Y.; Liu, E.T.; Katzenellenbogen, B.S. Estrogen receptor regulation of carbonic anhydrase XII through a distal enhancer in breast cancer. Cancer Res. 2008, 68, 3505–3515. [Google Scholar] [CrossRef] [PubMed]
- Franke, C.M.; Gu, V.W.; Grimm, B.G.; Cassady, V.C.; White, J.R.; Weigel, R.J.; Kulak, M.V. TFAP2C regulates carbonic anhydrase XII in human breast cancer. Oncogene 2019, 39, 1290–1301. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Tang, L.; Wang, H.; Lin, L.; Fu, J. Carbonic anhydrase 12 gene silencing reverses the sensitivity of paclitaxel in drug-resistant breast cancer cells. Bioengineered 2021, 12, 9806–9818. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, Y.S.; Moresco, J.J.; Tu, P.G.; Yates, J.R.; Nardulli, A.M. Proteomic analysis identifies highly expressed plasma membrane proteins for detection and therapeutic targeting of specific breast cancer subtypes. Clin. Proteom. 2018, 15, 30. [Google Scholar] [CrossRef] [PubMed]
- Sever, R.; Brugge, J.S. Signal transduction in cancer. Cold Spring Harb. Perspect. Med. 2015, 5, a006098. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ivanov, A.A.; Su, R.; Gonzalez-Pecchi, V.; Qi, Q.; Liu, S.; Webber, P.; McMillan, E.; Rusnak, L.; Pham, C.; et al. The OncoPPi network of cancer-focused protein-protein interactions to inform biological insights and therapeutic strategies. Nat. Commun. 2017, 8, 14356. [Google Scholar] [CrossRef]
| Characteristics | n | % |
|---|---|---|
| No. of patients | 1102 | 100 |
| Age (years) | 1096 | 100 |
| 30–50 | 332 | 30 |
| 51–70 | 554 | 50 |
| 71–90 | 210 | 20 |
| Menopause | 1096 | 100 |
| Pre | 230 | 21 |
| Post | 742 | 68 |
| Unknown | 125 | 11 |
| Pathologic T stage | 1102 | 100 |
| T1 | 284 | 25 |
| T2 | 640 | 58 |
| T3 | 138 | 12 |
| T4 | 40 | 4 |
| Pathologic N stage | 1102 | 100 |
| N0 | 516 | 47 |
| N1 | 367 | 33 |
| N2 | 120 | 11 |
| N3 | 79 | 7 |
| NX | 20 | 2 |
| Pathologic M stage | 1102 | 100 |
| M0 | 917 | 83 |
| M1 | 22 | 2 |
| MX | 163 | 15 |
| Grade | 1102 | 100 |
| I | 186 | 17 |
| II | 628 | 57 |
| III | 254 | 23 |
| IV | 20 | 2 |
| Molecular subtype | 1042 | 100 |
| Luminal A | 535 | 51 |
| Luminal B | 250 | 24 |
| HER2 | 77 | 7 |
| TNBC | 180 | 17 |
| Sensibility | Specificity | Accuracy | |
|---|---|---|---|
| LRRC15 | 87% | 80% | 86% |
| LRRC15 + EFNA3 | 96% | 99% | 96% |
| LRRC15 + TSPAN13 | 96% | 96% | 96% |
| LRRC15 + CA12 | 94% | 95% | 94% |
| Staining of BC Specimens | |||||
|---|---|---|---|---|---|
| LRRC15 | EFNA3 | TSPAN13 | CA12 | ||
| Positive Samples (%) | 100 | 90 | 99 | 68 | |
| Strong | Moderate | Weak | Negative | ||
| +3 | +2 | +1 | 0 | ||
| LRRC15 | 99 | 1 | - | - | |
| EFNA3 | 51 | 22 | 17 | 10 | |
| TSPAN13 | 81 | 9 | 9 | 1 | |
| CA12 | 41 | 14 | 13 | 32 | |
| Molecular Subtypes (%) | |||||
| Luminal A | Luminal B | HER2+ | TNBC | ||
| Number of Samples | 39 | 27 | 17 | 17 | |
| LRRC15 | +3 | 100 | 100 | 100 | 96 |
| +2 | - | - | - | 4 | |
| +1 | - | - | - | - | |
| 0 | - | - | - | - | |
| EFNA3 | +3 | 41 | 63 | 47 | 59 |
| +2 | 28 | 15 | 24 | 18 | |
| +1 | 25 | 19 | 12 | - | |
| 0 | 5 | 3 | 18 | 24 | |
| TSPAN13 | +3 | 85 | 85 | 71 | 76 |
| +2 | 8 | 7 | 18 | 6 | |
| +1 | 8 | 7 | 6 | 18 | |
| 0 | - | - | 6 | - | |
| CA12 | +3 | 72 | 37 | 19 | - |
| +2 | 13 | 33 | - | - | |
| +1 | 13 | 7 | 6 | 29 | |
| 0 | 3 | 22 | 76 | 71 | |
| BC Staging (%) | |||||
| I | II | III | |||
| Number of Samples | 6 | 72 | 22 | ||
| LRRC15 | +3 | 100 | 99 | 100 | |
| +2 | - | 1 | - | ||
| +1 | - | - | - | ||
| 0 | - | - | - | ||
| EFNA3 | +3 | 33 | 50 | 9 | |
| +2 | 33 | 19 | 5 | ||
| +1 | 33 | 19 | 27 | ||
| 0 | - | 12 | 59 | ||
| TSPAN13 | +3 | 80 | 81 | 86 | |
| +2 | 20 | 7 | 9 | ||
| +1 | - | 11 | 5 | ||
| 0 | - | 1 | - | ||
| CA12 | +3 | 80 | 37 | 45 | |
| +2 | 20 | 10 | 23 | ||
| +1 | - | 16 | 9 | ||
| 0 | - | 37 | 23 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendonça, J.B.; Fernandes, P.V.; Fernandes, D.C.; Rodrigues, F.R.; Waghabi, M.C.; Tilli, T.M. Unlocking Overexpressed Membrane Proteins to Guide Breast Cancer Precision Medicine. Cancers 2024, 16, 1402. https://doi.org/10.3390/cancers16071402
Mendonça JB, Fernandes PV, Fernandes DC, Rodrigues FR, Waghabi MC, Tilli TM. Unlocking Overexpressed Membrane Proteins to Guide Breast Cancer Precision Medicine. Cancers. 2024; 16(7):1402. https://doi.org/10.3390/cancers16071402
Chicago/Turabian StyleMendonça, Júlia Badaró, Priscila Valverde Fernandes, Danielle C. Fernandes, Fabiana Resende Rodrigues, Mariana Caldas Waghabi, and Tatiana Martins Tilli. 2024. "Unlocking Overexpressed Membrane Proteins to Guide Breast Cancer Precision Medicine" Cancers 16, no. 7: 1402. https://doi.org/10.3390/cancers16071402
APA StyleMendonça, J. B., Fernandes, P. V., Fernandes, D. C., Rodrigues, F. R., Waghabi, M. C., & Tilli, T. M. (2024). Unlocking Overexpressed Membrane Proteins to Guide Breast Cancer Precision Medicine. Cancers, 16(7), 1402. https://doi.org/10.3390/cancers16071402








