Functional Classification of Fusion Proteins in Sarcoma
Abstract
Simple Summary
Abstract
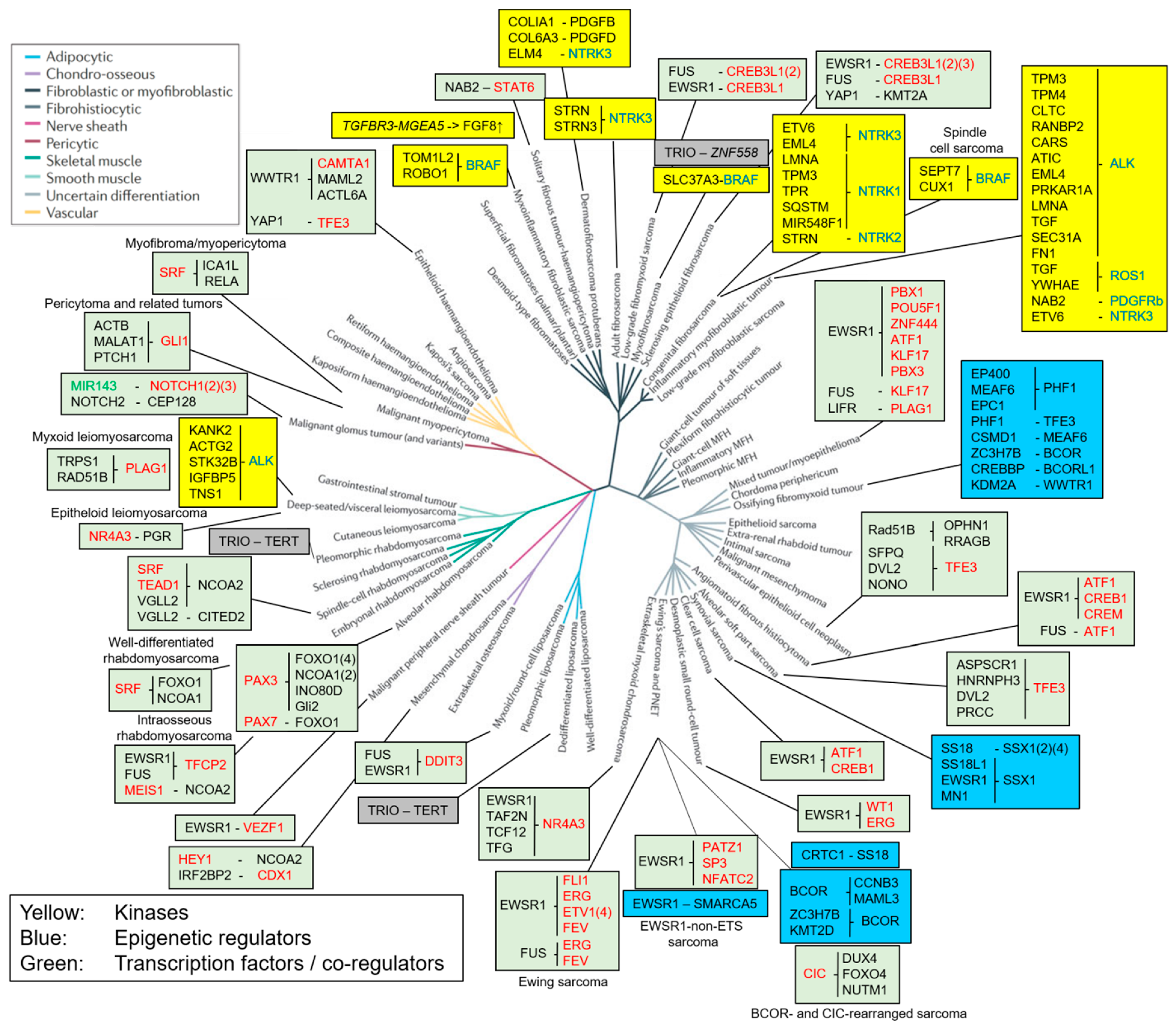
1. Introduction
2. Fusions of Kinases
2.1. ALK/ROS1/PDGFRβ Translocations in Inflammatory Myofibroblastic Tumors and Leiomyosarcoma
2.2. NTRK Translocations in Congenital and Adult Fibrosarcoma
2.3. BRAF Fusions in Myxoinflammatory Fibroblastic Sarcoma, Myxofibrosarcoma and Infantile Fibrosarcoma
2.4. FGFR1 Translocations in Phosphaturic Mesenchymal Tumor
3. Fusions of Growth Factors
PDGFB/D Translocations in Dermatofibrosarcoma Protuberans
4. Fusion Proteins Affecting Transcription
4.1. Fusions of Components of Epigenetic Complexes
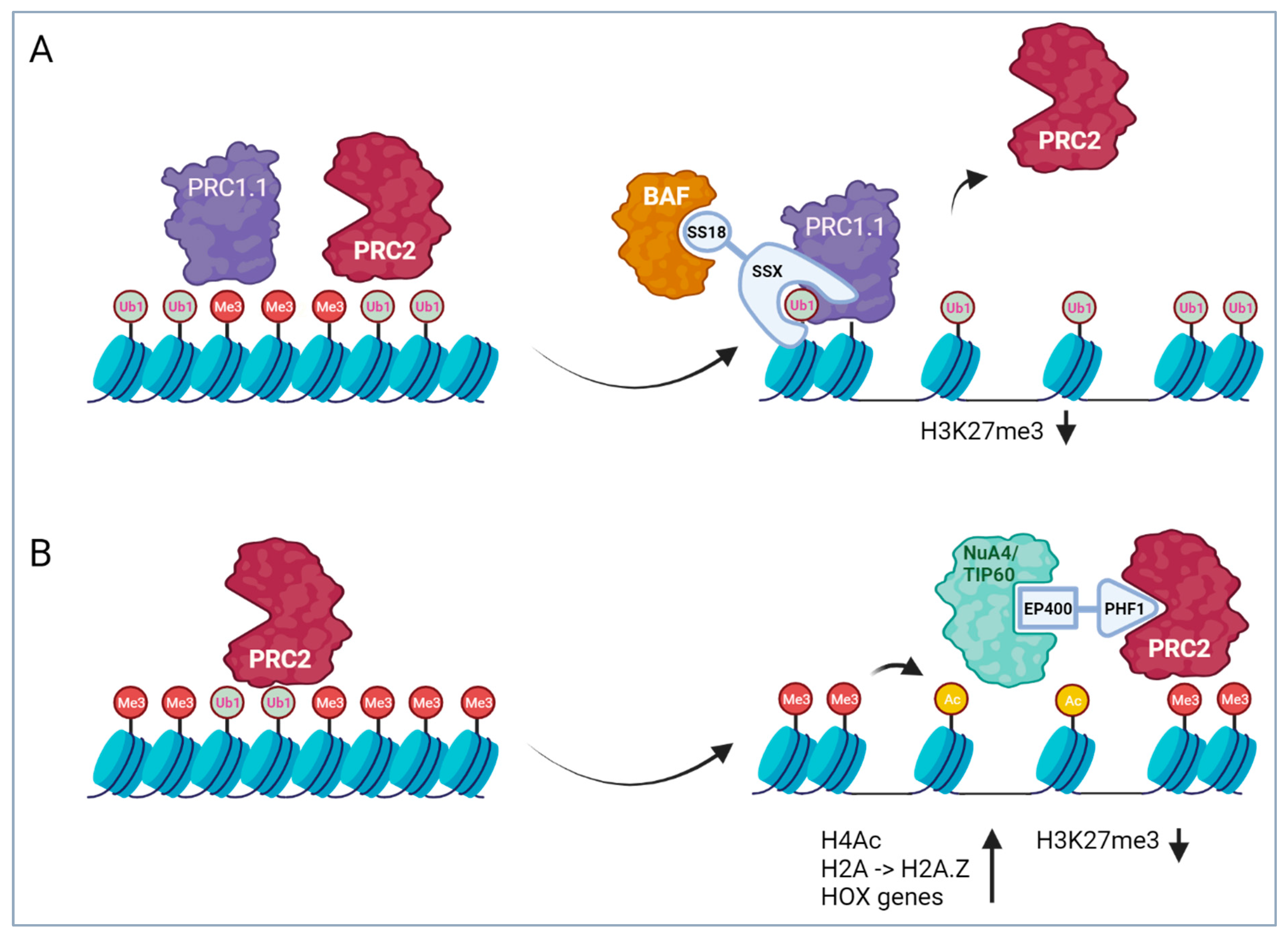
4.1.1. SS18 Translocations in Synovial Sarcoma
4.1.2. PHF1 Translocations in OFMT and Endometrial Stromal Sarcoma (ESS)
4.1.3. BCOR Translocations in Undifferentiated Small Round Cell Sarcomas of Bone and Soft Tissue and OFMT
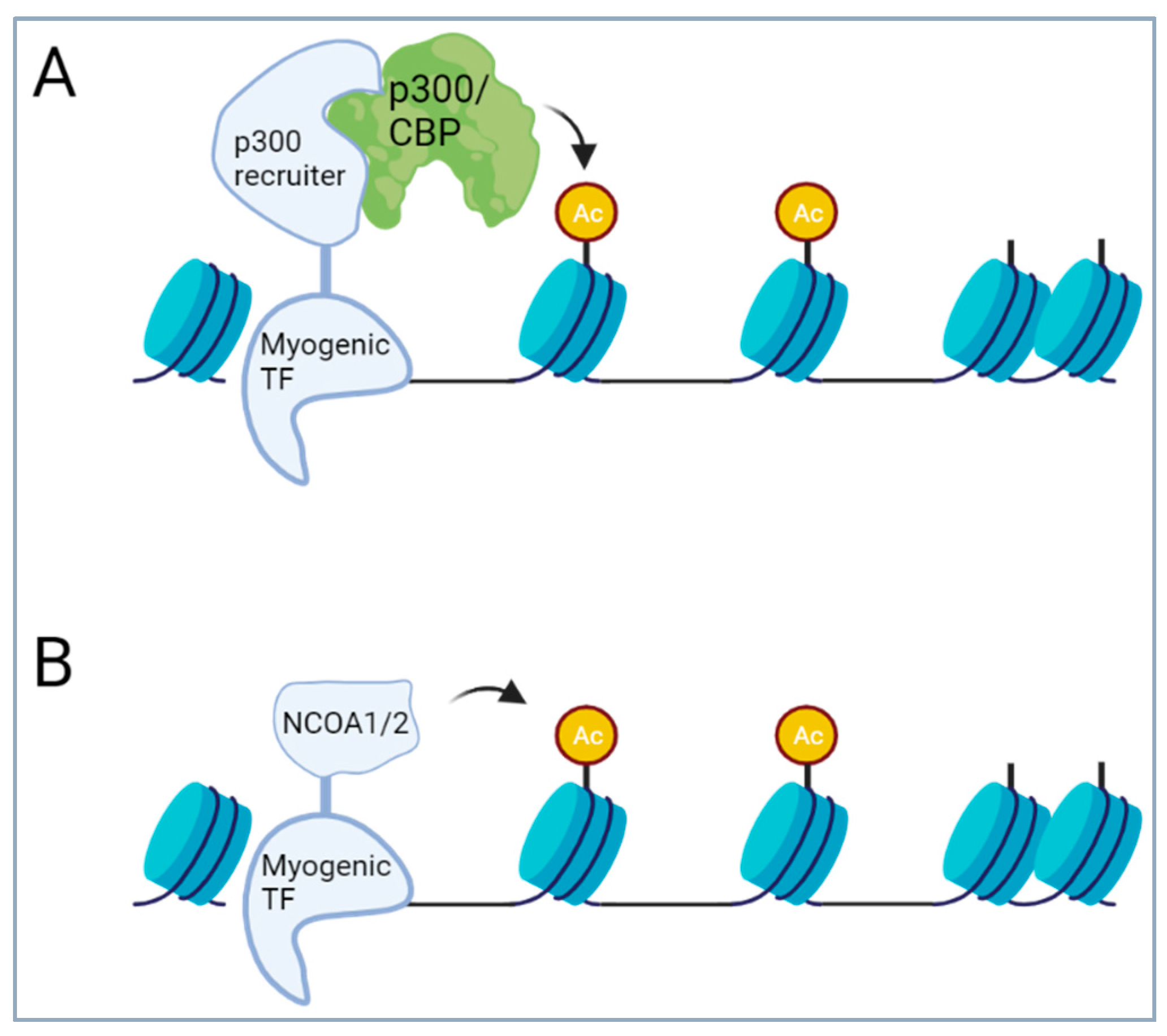
4.2. Fusions of Transcription Factors
4.2.1. EWSR1 Translocations in 11 Different Sarcoma Entities
4.2.2. CIC Translocations in Undifferentiated Small Round Cell Sarcomas of Soft Tissue and Bone
4.2.3. Translocations of Transcription Factors of the Skeletal Muscle Lineages
4.2.4. Translocations Activating the Notch Pathway in Mesenchymal Chondrosarcoma and Malignant Glomus Tumors
4.2.5. NAB2::STAT6 in Solitary Fibrous Tumor/Hemangiopericytoma
5. Fusions of Transcription Factors with Non-Transcriptional Regulators
5.1. TFE3 Translocations in Three Different Sarcoma Types
5.2. Translocations Activating Hedgehog Signaling
6. Fusion Proteins with an Unclear Mechanism of Action
Trio Translocations Found in Different Sarcomas
7. Therapeutic Perspectives
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Weinberg, R.A. The Biology of Cancer, 2nd ed.; Garland Science: New York, NY, USA, 2014. [Google Scholar]
- Teicher, B.A. Searching for molecular targets in sarcoma. Biochem. Pharmacol. 2012, 84, 1–10. [Google Scholar] [CrossRef]
- Taylor, B.S.; Barretina, J.; Maki, R.G.; Antonescu, C.R.; Singer, S.; Ladanyi, M. Advances in sarcoma genomics and new therapeutic targets. Nat. Rev. Cancer 2011, 11, 541–557. [Google Scholar] [CrossRef]
- Burningham, Z.; Hashibe, M.; Spector, L.; Schiffman, J.D. The epidemiology of sarcoma. Clin. Sarcoma Res. 2012, 2, 14. [Google Scholar] [CrossRef]
- Fletcher, C.B.M.; Hogendoorn, P.C.W.; Mertens, F. WHO Classification of Tumours of Soft Tissue and Bone. 2013. Available online: https://publications.iarc.fr/Book-And-Report-Series/Who-Classification-Of-Tumours/WHO-Classification-Of-Tumours-Of-Soft-Tissue-And-Bone-2013 (accessed on 28 February 2024).
- Helman, L.J.; Meltzer, P. Mechanisms of sarcoma development. Nat. Rev. Cancer 2003, 3, 685–694. [Google Scholar] [CrossRef]
- Mertens, F.; Antonescu, C.R.; Mitelman, F. Gene fusions in soft tissue tumors: Recurrent and overlapping pathogenetic themes. Genes Chromosomes Cancer 2016, 55, 291–310. [Google Scholar] [CrossRef]
- Azorsa, D.O.; Bode, P.K.; Wachtel, M.; Cheuk, A.T.C.; Meltzer, P.S.; Vokuhl, C.; Camenisch, U.; Khov, H.L.; Bode, B.; Schäfer, B.W.; et al. Immunohistochemical detection of PAX-FOXO1 fusion proteins in alveolar rhabdomyosarcoma using breakpoint specific monoclonal antibodies. Mod. Pathol. 2021, 34, 748–757. [Google Scholar] [CrossRef]
- Baranov, E.; McBride, M.J.; Bellizzi, A.M.; Ligon, A.H.; Fletcher, C.D.; Kadoch, C.; Hornick, J.L. A Novel SS18-SSX Fusion-specific Antibody for the Diagnosis of Synovial Sarcoma. Am. J. Surg. Pathol. 2020, 44, 922–933. [Google Scholar] [CrossRef]
- Zaborowski, M.; Vargas, A.C.; Pulvers, J.; Clarkson, A.; de Guzman, D.; Sioson, L.; Maclean, F.; Chou, A.; Gill, A.J. When used together SS18-SSX fusion-specific and SSX C-terminus immunohistochemistry are highly specific and sensitive for the diagnosis of synovial sarcoma and can replace FISH or molecular testing in most cases. Histopathology 2020, 77, 588–600. [Google Scholar] [CrossRef]
- Miura, K.; Shimizu, K.; Eguchi, T.; Koike, S.; Matsuoka, S.; Takeda, T.; Hamanaka, K.; Uehara, T. Usefulness of SS18-SSX antibody as a diagnostic marker for pulmonary metastatic synovial sarcoma. Diagn. Pathol. 2021, 16, 54. [Google Scholar] [CrossRef]
- Lasota, J.; Chłopek, M.; Kaczorowski, M.; Natálie, K.; Ryś, J.; Kopczyński, J.; Sulaieva, O.; Michal, M.; Kruczak, A.; Harazin-Lechowska, A.; et al. Utility of Immunohistochemistry with Antibodies to SS18-SSX Chimeric Proteins and C-Terminus of SSX Protein for Synovial Sarcoma Differential Diagnosis. Am. J. Surg. Pathol. 2024, 48, 97–105. [Google Scholar] [CrossRef]
- Doyle, L.A.; Vivero, M.; Fletcher, C.D.; Mertens, F.; Hornick, J.L. Nuclear expression of STAT6 distinguishes solitary fibrous tumor from histologic mimics. Mod. Pathol. 2014, 27, 390–395. [Google Scholar] [CrossRef]
- Demicco, E.G.; Harms, P.W.; Patel, R.M.; Smith, S.C.; Ingram, D.; Torres, K.; Carskadon, S.L.; Camelo-Piragua, S.; McHugh, J.B.; Siddiqui, J.; et al. Extensive survey of STAT6 expression in a large series of mesenchymal tumors. Am. J. Clin. Pathol. 2015, 143, 672–682. [Google Scholar] [CrossRef]
- Yoshida, A.; Tsuta, K.; Ohno, M.; Yoshida, M.; Narita, Y.; Kawai, A.; Asamura, H.; Kushima, R. STAT6 immunohistochemistry is helpful in the diagnosis of solitary fibrous tumors. Am. J. Surg. Pathol. 2014, 38, 552–559. [Google Scholar] [CrossRef]
- Doyle, L.A.; Fletcher, C.D.; Hornick, J.L. Nuclear Expression of CAMTA1 Distinguishes Epithelioid Hemangioendothelioma from Histologic Mimics. Am. J. Surg. Pathol. 2016, 40, 94–102. [Google Scholar] [CrossRef]
- Shibuya, R.; Matsuyama, A.; Shiba, E.; Harada, H.; Yabuki, K.; Hisaoka, M. CAMTA1 is a useful immunohistochemical marker for diagnosing epithelioid haemangioendothelioma. Histopathology 2015, 67, 827–835. [Google Scholar] [CrossRef]
- Baranov, E.; Black, M.A.; Fletcher, C.D.M.; Charville, G.W.; Hornick, J.L. Nuclear expression of DDIT3 distinguishes high-grade myxoid liposarcoma from other round cell sarcomas. Mod. Pathol. 2021, 34, 1367–1372. [Google Scholar] [CrossRef]
- Scapa, J.V.; Cloutier, J.M.; Raghavan, S.S.; Peters-Schulze, G.; Varma, S.; Charville, G.W. DDIT3 Immunohistochemistry Is a Useful Tool for the Diagnosis of Myxoid Liposarcoma. Am. J. Surg. Pathol. 2021, 45, 230–239. [Google Scholar] [CrossRef]
- Cessna, M.H.; Zhou, H.; Sanger, W.G.; Perkins, S.L.; Tripp, S.; Pickering, D.; Daines, C.; Coffin, C.M. Expression of ALK1 and p80 in inflammatory myofibroblastic tumor and its mesenchymal mimics: A study of 135 cases. Mod. Pathol. 2002, 15, 931–938. [Google Scholar] [CrossRef]
- Cook, J.R.; Dehner, L.P.; Collins, M.H.; Ma, Z.; Morris, S.W.; Coffin, C.M.; Hill, D.A. Anaplastic lymphoma kinase (ALK) expression in the inflammatory myofibroblastic tumor: A comparative immunohistochemical study. Am. J. Surg. Pathol. 2001, 25, 1364–1371. [Google Scholar] [CrossRef]
- Dancey, J.E.; Chen, H.X. Strategies for optimizing combinations of molecularly targeted anticancer agents. Nat. Rev. Drug Discov. 2006, 5, 649–659. [Google Scholar] [CrossRef]
- Lovly, C.M.; Gupta, A.; Lipson, D.; Otto, G.; Brennan, T.; Chung, C.T.; Borinstein, S.C.; Ross, J.S.; Stephens, P.J.; Miller, V.A.; et al. Inflammatory myofibroblastic tumors harbor multiple potentially actionable kinase fusions. Cancer Discov. 2014, 4, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.C.; Zhang, L.; Drilon, A.E.; Chi, P.; Alaggio, R.; Borsu, L.; Benayed, R.; Travis, W.D.; Ladanyi, M.; Antonescu, C.R. Expanding the Molecular Characterization of Thoracic Inflammatory Myofibroblastic Tumors beyond ALK Gene Rearrangements. J. Thorac. Oncol. 2019, 14, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Coffin, C.M.; Patel, A.; Perkins, S.; Elenitoba-Johnson, K.S.; Perlman, E.; Griffin, C.A. ALK1 and p80 expression and chromosomal rearrangements involving 2p23 in inflammatory myofibroblastic tumor. Mod. Pathol. 2001, 14, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Antonescu, C.R.; Suurmeijer, A.J.; Zhang, L.; Sung, Y.-S.; Jungbluth, A.A.; Travis, W.D.; Al-Ahmadie, H.; Fletcher, C.D.; Alaggio, R. Molecular characterization of inflammatory myofibroblastic tumors with frequent ALK and ROS1 gene fusions and rare novel RET rearrangement. Am. J. Surg. Pathol. 2015, 39, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Davis, L.E.; Nusser, K.D.; Przybyl, J.; Pittsenbarger, J.; Hofmann, N.E.; Varma, S.; Vennam, S.; Debiec-Rychter, M.; van de Rijn, M.; Davare, M.A. Discovery and Characterization of Recurrent, Targetable ALK Fusions in Leiomyosarcoma. Mol. Cancer Res. 2019, 17, 676–685. [Google Scholar] [CrossRef] [PubMed]
- Mano, H. ALKoma: A cancer subtype with a shared target. Cancer Discov. 2012, 2, 495–502. [Google Scholar] [CrossRef]
- Ono, A.; Murakami, H.; Serizawa, M.; Wakuda, K.; Kenmotsu, H.; Naito, T.; Taira, T.; Koh, Y.; Ohde, Y.; Nakajima, T.; et al. Drastic initial response and subsequent response to two ALK inhibitors in a patient with a highly aggressive ALK-rearranged inflammatory myofibroblastic tumor arising in the pleural cavity. Lung Cancer 2016, 99, 151–154. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Butrynski, J.E.; D’Adamo, D.R.; Hornick, J.L.; Dal Cin, P.; Antonescu, C.R.; Jhanwar, S.C.; Ladanyi, M.; Capelletti, M.; Rodig, S.J.; Ramaiya, N.; et al. Crizotinib in ALK-rearranged inflammatory myofibroblastic tumor. N. Engl. J. Med. 2010, 363, 1727–1733. [Google Scholar] [CrossRef] [PubMed]
- Theilen, T.; Soerensen, J.; Bochennek, K.; Becker, M.; Schwabe, D.; Rolle, U.; Klingebiel, T.; Lehrnbecher, T. Crizotinib in ALK(+) inflammatory myofibroblastic tumors-Current experience and future perspectives. Pediatr. Blood Cancer 2018, 65, e26920. [Google Scholar] [CrossRef]
- Bourgeois, J.M.; Knezevich, S.R.; Mathers, J.A.; Sorensen, P.H. Molecular detection of the ETV6-NTRK3 gene fusion differentiates congenital fibrosarcoma from other childhood spindle cell tumors. Am. J. Surg. Pathol. 2000, 24, 937–946. [Google Scholar] [CrossRef]
- Knezevich, S.R.; McFadden, D.E.; Tao, W.; Lim, J.F.; Sorensen, P.H. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat. Genet. 1998, 18, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Tannenbaum-Dvir, S.; Glade Bender, J.L.; Church, A.J.; Janeway, K.A.; Harris, M.H.; Mansukhani, M.M.; Nagy, P.L.; Andrews, S.J.; Murty, V.V.; Kadenhe-Chiweshe, A.; et al. Characterization of a novel fusion gene EML4-NTRK3 in a case of recurrent congenital fibrosarcoma. Cold Spring Harb. Mol. Case Stud. 2015, 1, a000471. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.; Pavlick, D.; Brennan, T.; Yelensky, R.; Crawford, J.; Ross, J.S.; Miller, V.A.; Malicki, D.; Stephens, P.J.; Ali, S.M.; et al. Evaluation of a Congenital Infantile Fibrosarcoma by Comprehensive Genomic Profiling Reveals an LMNA-NTRK1 Gene Fusion Responsive to Crizotinib. J. Natl. Cancer Inst. 2016, 108, djv307. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.C.; Fletcher, C.D.; Alaggio, R.; Wexler, L.; Zhang, L.; Sung, Y.S.; Orhan, D.; Chang, W.C.; Swanson, D.; Dickson, B.C.; et al. Recurrent BRAF Gene Fusions in a Subset of Pediatric Spindle Cell Sarcomas: Expanding the Genetic Spectrum of Tumors With Overlapping Features With Infantile Fibrosarcoma. Am. J. Surg. Pathol. 2018, 42, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, F.; Nakatani, F.; Asano, N.; Wakai, S.; Sekimizu, M.; Mitani, S.; Kubo, T.; Kawai, A.; Ichikawa, H.; Yoshida, A. Novel NTRK3 Fusions in Fibrosarcomas of Adults. Am. J. Surg. Pathol. 2019, 43, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Coppola, V.; Barrick, C.A.; Southon, E.A.; Celeste, A.; Wang, K.; Chen, B.; Haddad, E.-B.; Yin, J.; Nussenzweig, A.; Subramaniam, A.; et al. Ablation of TrkA function in the immune system causes B cell abnormalities. Development 2004, 131, 5185–5195. [Google Scholar] [CrossRef] [PubMed]
- Lannon, C.L.; Sorensen, P.H. ETV6-NTRK3: A chimeric protein tyrosine kinase with transformation activity in multiple cell lineages. Semin. Cancer Biol. 2005, 15, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Tognon, C.E.; Mackereth, C.D.; Somasiri, A.M.; McIntosh, L.P.; Sorensen, P.H. Mutations in the SAM domain of the ETV6-NTRK3 chimeric tyrosine kinase block polymerization and transformation activity. Mol. Cell. Biol. 2004, 24, 4636–4650. [Google Scholar] [CrossRef]
- Morrison, K.B.; Tognon, C.E.; Garnett, M.J.; Deal, C.; Sorensen, P.H. ETV6-NTRK3 transformation requires insulin-like growth factor 1 receptor signaling and is associated with constitutive IRS-1 tyrosine phosphorylation. Oncogene 2002, 21, 5684–5695. [Google Scholar] [CrossRef]
- Vaishnavi, A.; Le, A.T.; Doebele, R.C. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov. 2015, 5, 25–34. [Google Scholar] [CrossRef]
- Drilon, A.; Laetsch, T.W.; Kummar, S.; DuBois, S.G.; Lassen, U.N.; Demetri, G.D.; Nathenson, M.; Doebele, R.C.; Farago, A.F.; Pappo, A.S.; et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N. Engl. J. Med. 2018, 378, 731–739. [Google Scholar] [CrossRef]
- Laetsch, T.W.; DuBois, S.G.; Mascarenhas, L.; Turpin, B.; Federman, N.; Albert, C.M.; Nagasubramanian, R.; Davis, J.L.; Rudzinski, E.; Feraco, A.M.; et al. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: Phase 1 results from a multicentre, open-label, phase 1/2 study. Lancet Oncol. 2018, 19, 705–714. [Google Scholar] [CrossRef]
- Kao, Y.C.; Ranucci, V.; Zhang, L.; Sung, Y.S.; Athanasian, E.A.; Swanson, D.; Dickson, B.C.; Antonescu, C.R. Recurrent BRAF Gene Rearrangements in Myxoinflammatory Fibroblastic Sarcomas, but Not Hemosiderotic Fibrolipomatous Tumors. Am. J. Surg. Pathol. 2017, 41, 1456–1465. [Google Scholar] [CrossRef]
- Ogura, K.; Hosoda, F.; Arai, Y.; Nakamura, H.; Hama, N.; Totoki, Y.; Yoshida, A.; Nagai, M.; Kato, M.; Arakawa, E.; et al. Integrated genetic and epigenetic analysis of myxofibrosarcoma. Nat. Commun. 2018, 9, 2765. [Google Scholar] [CrossRef]
- Arbajian, E.; Hofvander, J.; Magnusson, L.; Mertens, F. Deep sequencing of myxoinflammatory fibroblastic sarcoma. Genes Chromosomes Cancer 2020, 59, 309–317. [Google Scholar] [CrossRef]
- Hallor, K.H.; Sciot, R.; Staaf, J.; Heidenblad, M.; Rydholm, A.; Bauer, H.C.; Åström, K.; Domanski, H.A.; Meis, J.M.; Kindblom, L.G.; et al. Two genetic pathways, t(1;10) and amplification of 3p11-12, in myxoinflammatory fibroblastic sarcoma, haemosiderotic fibrolipomatous tumour, and morphologically similar lesions. J. Pathol. 2009, 217, 716–727. [Google Scholar] [CrossRef]
- Karoulia, Z.; Gavathiotis, E.; Poulikakos, P.I. New perspectives for targeting RAF kinase in human cancer. Nat. Rev. Cancer 2017, 17, 676–691. [Google Scholar] [CrossRef]
- Morimoto, T.; Takenaka, S.; Hashimoto, N.; Araki, N.; Myoui, A.; Yoshikawa, H. Malignant phosphaturic mesenchymal tumor of the pelvis: A report of two cases. Oncol. Lett. 2014, 8, 67–71. [Google Scholar] [CrossRef]
- Lee, J.-C.; Jeng, Y.-M.; Su, S.-Y.; Wu, C.-T.; Tsai, K.-S.; Lee, C.-H.; Lin, C.-Y.; Carter, J.M.; Huang, J.-W.; Chen, S.-H.; et al. Identification of a novel FN1-FGFR1 genetic fusion as a frequent event in phosphaturic mesenchymal tumour. J. Pathol. 2015, 235, 539–545. [Google Scholar] [CrossRef]
- Lee, J.-C.; Su, S.-Y.; Changou, C.A.; Yang, R.-S.; Tsai, K.-S.; Collins, M.T.; Orwoll, E.S.; Lin, C.-Y.; Chen, S.-H.; Shih, S.-R.; et al. Characterization of FN1-FGFR1 and novel FN1-FGF1 fusion genes in a large series of phosphaturic mesenchymal tumors. Mod. Pathol. 2016, 29, 1335–1346. [Google Scholar] [CrossRef]
- Ren, H.; Tan, Z.P.; Zhu, X.; Crosby, K.; Haack, H.; Ren, J.M.; Beausoleil, S.; Moritz, A.; Innocenti, G.; Rush, J.; et al. Identification of anaplastic lymphoma kinase as a potential therapeutic target in ovarian cancer. Cancer Res. 2012, 72, 3312–3323. [Google Scholar] [CrossRef]
- Chong, W.H.; Molinolo, A.A.; Chen, C.C.; Collins, M.T. Tumor-induced osteomalacia. Endocr. Relat. Cancer 2011, 18, R53–R77. [Google Scholar] [CrossRef]
- Dickson, B.C.; Hornick, J.L.; Fletcher, C.D.M.; Demicco, E.G.; Howarth, D.J.; Swanson, D.; Zhang, L.; Sung, Y.; Antonescu, C.R. Dermatofibrosarcoma protuberans with a novel COL6A3-PDGFD fusion gene and apparent predilection for breast. Genes Chromosomes Cancer 2018, 57, 437–445. [Google Scholar] [CrossRef]
- Shimuzu, A.; O’Brien, K.; Sjoblum, T.; Pietras, K. The dermatofibrosarcoma protuberans-associated collagen type Ialpha1/platelet-derived growth factor (PDGF) B-chain fusion gene generates a transforming protein that is processed to functional PDGF-BB. Cancer Res. 1999, 59, 3719–3723. [Google Scholar]
- Heinrich, M.C.; Joensuu, H.; Demetri, G.D.; Corless, C.L.; Apperley, J.; Fletcher, J.A.; Soulieres, D.; Dirnhofer, S.; Harlow, A.; Town, A.; et al. Phase II, open-label study evaluating the activity of imatinib in treating life-threatening malignancies known to be associated with imatinib-sensitive tyrosine kinases. Clin. Cancer Res. 2008, 14, 2717–2725. [Google Scholar] [CrossRef]
- Rutkowski, P.; Debiec-Rychter, M. Current treatment options for dermatofibrosarcoma protuberans. Expert Rev. Anticancer. Ther. 2015, 15, 901–909. [Google Scholar] [CrossRef]
- McArthur, G.A.; Demetri, G.D.; van Oosterom, A.; Heinrich, M.C.; Debiec-Rychter, M.; Corless, C.L.; Nikolova, Z.; Dimitrijevic, S.; Fletcher, J.A. Molecular and clinical analysis of locally advanced dermatofibrosarcoma protuberans treated with imatinib: Imatinib Target Exploration Consortium Study B2225. J. Clin. Oncol. 2005, 23, 866–873. [Google Scholar] [CrossRef]
- Rutkowski, P.; Van Glabbeke, M.; Rankin, C.J.; Ruka, W.; Rubin, B.P.; Debiec-Rychter, M.; Lazar, A.; Gelderblom, H.; Sciot, R.; Lopez-Terrada, D.; et al. Imatinib mesylate in advanced dermatofibrosarcoma protuberans: Pooled analysis of two phase II clinical trials. J. Clin. Oncol. 2010, 28, 1772–1779. [Google Scholar] [CrossRef]
- Rutkowski, P.; Dębiec-Rychter, M.; Nowecki, Z.I.; Michej, W.; Symonides, M.; Ptaszynski, K.; Ruka, W. Treatment of advanced dermatofibrosarcoma protuberans with imatinib mesylate with or without surgical resection. J. Eur. Acad. Dermatol. Venereol. 2011, 25, 264–270. [Google Scholar] [CrossRef]
- Bradner, J.E.; Hnisz, D.; Young, R.A. Transcriptional Addiction in Cancer. Cell 2017, 168, 629–643. [Google Scholar] [CrossRef]
- Gonda, T.J.; Ramsay, R.G. Directly targeting transcriptional dysregulation in cancer. Nat. Rev. Cancer 2015, 15, 686–694. [Google Scholar] [CrossRef]
- Gröbner, S.N.; Worst, B.C.; Weischenfeldt, J.; Buchhalter, I.; Kleinheinz, K.; Rudneva, V.A.; Johann, P.D.; Balasubramanian, G.P.; Segura-Wang, M.; Brabetz, S.; et al. The landscape of genomic alterations across childhood cancers. Nature 2018, 555, 321–327. [Google Scholar] [CrossRef]
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018, 174, 1034–1035. [Google Scholar] [CrossRef]
- Pfister, S.X.; Ashworth, A. Marked for death: Targeting epigenetic changes in cancer. Nat. Rev. Drug Discov. 2017, 16, 241–263. [Google Scholar] [CrossRef]
- Kennison, J.A.; Tamkun, J.W. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc. Natl. Acad. Sci. USA 1988, 85, 8136–8140. [Google Scholar] [CrossRef]
- Tamkun, J.W.; Deuring, R.; Scott, M.P.; Kissinger, M.; Pattatucci, A.M.; Kaufman, T.C.; Kennison, J.A. Brahma: A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 1992, 68, 561–572. [Google Scholar] [CrossRef]
- Clark, J.; Rocques, P.J.; Crew, A.J.; Gill, S.; Shipley, J.; Chan, A.M.-L.; Gusterson, B.A.; Cooper, C.S. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat. Genet. 1994, 7, 502–508. [Google Scholar] [CrossRef]
- Crew, A.; Clark, J.; Fisher, C.; Gill, S.; Grimer, R.; Chand, A.; Shipley, J.; Gusterson, B.; Cooper, C. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. EMBO J. 1995, 14, 2333–2340. [Google Scholar] [CrossRef]
- Kao, Y.; Sung, Y.; Zhang, L.; Kenan, S.; Singer, S.; Tap, W.D.; Swanson, D.; Dickson, B.C.; Antonescu, C.R. BCOR upregulation in a poorly differentiated synovial sarcoma with SS18L1-SSX1 fusion-A pathologic and molecular pitfall. Genes Chromosomes Cancer 2016, 56, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Storlazzi, C.T.; Mertens, F.; Mandahl, N.; Gisselsson, D.; Isaksson, M.; Gustafson, P.; Domanski, H.A.; Panagopoulos, I. A novel fusion gene, SS18L1/SSX1, in synovial sarcoma. Genes Chromosomes Cancer 2003, 37, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Kadoch, C.; Hargreaves, D.C.; Hodges, C.; Elias, L.; Ho, L.; Ranish, J.; Crabtree, G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013, 45, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.A.; McNeel, D.G. The SSX family of cancer-testis antigens as target proteins for tumor therapy. Clin. Dev. Immunol. 2010, 2010, 150591. [Google Scholar] [CrossRef] [PubMed]
- Barco, R.; Garcia, C.B.; Eid, J.E. The synovial sarcoma-associated SYT-SSX2 oncogene antagonizes the polycomb complex protein Bmi1. PLoS ONE 2009, 4, e5060. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Banito, A.; Li, X.; Laporte, A.N.; Roe, J.-S.; Sanchez-Vega, F.; Huang, C.-H.; Dancsok, A.R.; Hatzi, K.; Chen, C.-C.; Tschaharganeh, D.F.; et al. The SS18-SSX Oncoprotein Hijacks KDM2B-PRC1.1 to Drive Synovial Sarcoma. Cancer Cell 2018, 34, 346–348. [Google Scholar] [CrossRef] [PubMed]
- Kadoch, C.; Williams, R.T.; Calarco, J.P.; Miller, E.L.; Weber, C.M.; Braun, S.M.G.; Pulice, J.L.; Chory, E.J.; Crabtree, G.R. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat. Genet. 2017, 49, 213–222. [Google Scholar] [CrossRef]
- Kadoch, C.; Crabtree, G.R. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 2013, 153, 71–85. [Google Scholar] [CrossRef] [PubMed]
- McBride, M.J.; Pulice, J.L.; Beird, H.C.; Ingram, D.R.; D’Avino, A.R.; Shern, J.F.; Charville, G.W.; Hornick, J.L.; Nakayama, R.T.; Garcia-Rivera, E.M.; et al. The SS18-SSX Fusion Oncoprotein Hijacks BAF Complex Targeting and Function to Drive Synovial Sarcoma. Cancer Cell 2018, 33, 1128–1141.e7. [Google Scholar] [CrossRef]
- Benabdallah, N.S.; Dalal, V.; Scott, R.W.; Marcous, F.; Sotiriou, A.; Kommoss, F.K.F.; Pejkovska, A.; Gaspar, L.; Wagner, L.; Sánchez-Rivera, F.J.; et al. Aberrant gene activation in synovial sarcoma relies on SSX specificity and increased PRC1.1 stability. Nat. Struct. Mol Biol. 2023, 30, 1640–1652. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, A.; Arai, Y.; Satomi, K.; Kubo, T.; Ryo, E.; Matsushita, Y.; Hama, N.; Sudo, K.; Komiyama, M.; Yatabe, Y.; et al. Identification of novel SSX1 fusions in synovial sarcoma. Mod. Pathol. 2022, 35, 228–239. [Google Scholar] [CrossRef]
- Sullivan, L.M.; Folpe, A.L.; Pawel, B.R.; Judkins, A.R.; Biegel, J.A. Epithelioid sarcoma is associated with a high percentage of SMARCB1 deletions. Mod. Pathol. 2013, 26, 385–392. [Google Scholar] [CrossRef]
- Versteege, I.; Sévenet, N.; Lange, J.; Rousseau-Merck, M.F.; Ambros, P.; Handgretinger, R.; Aurias, A.; Delattre, O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 1998, 394, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Grünewald, T.G.; Postel-Vinay, S.; Nakayama, R.T.; Berlow, N.E.; Bolzicco, A.; Cerullo, V.; Dermawan, J.K.; Frezza, A.M.; Italiano, A.; Jin, J.X.; et al. Translational Aspects of Epithelioid Sarcoma-Current Consensus. Clin. Cancer Res. 2023, 30, 1079–1092. [Google Scholar] [CrossRef] [PubMed]
- Chai, J.; Charboneau, A.L.; Betz, B.L.; Weissman, B.E. Loss of the hSNF5 gene concomitantly inactivates p21CIP/WAF1 and p16INK4a activity associated with replicative senescence in A204 rhabdoid tumor cells. Cancer Res. 2005, 65, 10192–10198. [Google Scholar] [CrossRef] [PubMed]
- Betz, B.L.; Strobeck, M.W.; Reisman, D.N.; Knudsen, E.S.; Weissman, B.E. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene 2002, 21, 5193–5203. [Google Scholar] [CrossRef]
- Knutson, S.K.; Warholic, N.M.; Wigle, T.J.; Klaus, C.R.; Allain, C.J.; Raimondi, A.; Scott, M.P.; Chesworth, R.; Moyer, M.P.; Copeland, R.A.; et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc. Natl. Acad. Sci. USA 2013, 110, 7922–7927. [Google Scholar] [CrossRef]
- Italiano, A.; Soria, J.C.; Toulmonde, M.; Michot, J.M.; Lucchesi, C.; Varga, A.; Coindre, J.M.; Blakemore, S.J.; Clawson, A.; Suttle, B.; et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: A first-in-human, open-label, phase 1 study. Lancet Oncol. 2018, 19, 649–659. [Google Scholar] [CrossRef]
- Stacchiotti, S.; Schoffski, P.; Jones, R.; Agulnik, M.; Villalobos, V.M.; Jahan, T.M.; Chen, T.W.-W.; Italiano, A.; Demetri, G.D.; Cote, G.M.; et al. Safety and efficacy of tazemetostat, a first-in-class EZH2 inhibitor, in patients (pts) with epithelioid sarcoma (ES) (NCT02601950). J. Clin. Oncol. 2019, 37 (Suppl. S15), 11003. [Google Scholar] [CrossRef]
- Helming, K.C.; Wang, X.; Roberts, C.W. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell 2014, 26, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Antonescu, C.R.; Sung, Y.S.; Chen, C.L.; Zhang, L.; Chen, H.W.; Singer, S.; Agaram, N.P.; Sboner, A.; Fletcher, C.D. Novel ZC3H7B-BCOR, MEAF6-PHF1, and EPC1-PHF1 fusions in ossifying fibromyxoid tumors--molecular characterization shows genetic overlap with endometrial stromal sarcoma. Genes Chromosomes Cancer 2014, 53, 183–193. [Google Scholar] [CrossRef]
- Cai, L.; Rothbart, S.B.; Lu, R.; Xu, B.; Chen, W.-Y.; Tripathy, A.; Rockowitz, S.; Zheng, D.; Patel, D.J.; Allis, C.D.; et al. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol. Cell 2013, 49, 571–582. [Google Scholar] [CrossRef]
- Musselman, C.A.; Avvakumov, N.; Watanabe, R.; Abraham, C.G.; Lalonde, M.-E.; Hong, Z.; Allen, C.; Roy, S.; Nuñez, J.K.; Nickoloff, J.; et al. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat. Struct. Mol. Biol. 2012, 19, 1266–1272. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Bachmann, A.L.; Tauscher, K.; Benda, C.; Fierz, B.; Muller, J. DNA binding by PHF1 prolongs PRC2 residence time on chromatin and thereby promotes H3K27 methylation. Nat. Struct. Mol. Biol. 2017, 24, 1039–1047. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Xu, M.; Huang, C.; Liu, N.; Chen, S.; Zhu, B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 2011, 286, 7983–7989. [Google Scholar] [CrossRef] [PubMed]
- Brien, G.L.; Gambero, G.; O’Connell, D.J.; Jerman, E.; Turner, S.A.; Egan, C.M.; Dunne, E.J.; Jurgens, M.C.; Wynne, K.; Piao, L.; et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat. Struct. Mol. Biol. 2012, 19, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Ballaré, C.; Lange, M.; Lapinaite, A.; Martin, G.M.; Morey, L.; Pascual, G.; Liefke, R.; Simon, B.; Shi, Y.; Gozani, O.; et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat. Struct. Mol. Biol. 2012, 19, 1257–1265. [Google Scholar] [CrossRef] [PubMed]
- Abed, J.A.; Jones, R.S. H3K36me3 key to Polycomb-mediated gene silencing in lineage specification. Nat. Struct. Mol. Biol. 2012, 19, 1214–1215. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Kohashi, K.; Yamamoto, H.; Ishii, T.; Yoshida, T.; Matsunobu, T.; Iwamoto, Y.; Oda, Y. Ossifying fibromyxoid tumor presenting EP400-PHF1 fusion gene. Hum. Pathol. 2013, 44, 2603–2608. [Google Scholar] [CrossRef] [PubMed]
- Gebre-Medhin, S.; Nord, K.H.; Möller, E.; Mandahl, N.; Magnusson, L.; Nilsson, J.; Jo, V.Y.; von Steyern, F.V.; Brosjö, O.; Larsson, O.; et al. Recurrent rearrangement of the PHF1 gene in ossifying fibromyxoid tumors. Am. J. Pathol. 2012, 181, 1069–1077. [Google Scholar] [CrossRef]
- Kao, Y.C.; Sung, Y.S.; Zhang, L.; Chen, C.L.; Huang, S.C.; Antonescu, C.R. Expanding the molecular signature of ossifying fibromyxoid tumors with two novel gene fusions: CREBBP-BCORL1 and KDM2A-WWTR1. Genes Chromosomes Cancer 2017, 56, 42–50. [Google Scholar] [CrossRef]
- Workman, J.L.; Abmayr, S.M. (Eds.) Fundamentals of Chromatin; Springer: New York, NY, USA, 2014. [Google Scholar]
- Auger, A.; Galarneau, L.; Altaf, M.; Nourani, A.; Doyon, Y.; Utley, R.T.; Cronier, D.; Allard, S.; Côté, J. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol. Cell. Biol. 2008, 28, 2257–2270. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Su, T.; Yen, L.; Jacquet, K.; Huang, C.; Côté, J.; Kurdistani, S.K.; Carey, M.F. EP400 Deposits H3.3 into Promoters and Enhancers during Gene Activation. Mol. Cell 2016, 61, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Suurmeijer, A.J.H.; Song, W.; Sung, Y.; Zhang, L.; Swanson, D.; Fletcher, C.D.M.; Dickson, B.C.; Antonescu, C.R. Novel recurrent PHF1-TFE3 fusions in ossifying fibromyxoid tumors. Genes Chromosomes Cancer 2019, 58, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Hofvander, J.; Jo, V.Y.; Fletcher, C.D.M.; Puls, F.; Flucke, U.; Nilsson, J.; Magnusson, L.; Mertens, F. PHF1 fusions cause distinct gene expression and chromatin accessibility profiles in ossifying fibromyxoid tumors and mesenchymal cells. Mod. Pathol. 2020, 33, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Micci, F.; Panagopoulos, I.; Bjerkehagen, B.; Heim, S. Consistent rearrangement of chromosomal band 6p21 with generation of fusion genes JAZF1/PHF1 and EPC1/PHF1 in endometrial stromal sarcoma. Cancer Res. 2006, 66, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Panagopoulos, I.; Micci, F.; Thorsen, J.; Gorunova, L.; Eibak, A.M.; Bjerkehagen, B.; Davidson, B.; Heim, S. Novel fusion of MYST/Esa1-associated factor 6 and PHF1 in endometrial stromal sarcoma. PLoS ONE 2012, 7, e39354. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, M.; Gorunova, L.; Davidson, B.; Heim, S.; Panagopoulos, I.; Micci, F. Identification of an EPC2-PHF1 fusion transcript in low-grade endometrial stromal sarcoma. Oncotarget 2018, 9, 19203–19208. [Google Scholar] [CrossRef] [PubMed]
- Koontz, J.I.; Soreng, A.L.; Nucci, M.; Kuo, F.C.; Pauwels, P.; Berghe, H.v.D.; Cin, P.D.; Fletcher, J.A.; Sklar, J. Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proc. Natl. Acad. Sci. USA 2001, 98, 6348–6353. [Google Scholar] [CrossRef] [PubMed]
- Micci, F.; Brunetti, M.; Cin, P.D.; Nucci, M.R.; Gorunova, L.; Heim, S.; Panagopoulos, I. Fusion of the genes BRD8 and PHF1 in endometrial stromal sarcoma. Genes Chromosomes Cancer 2017, 56, 841–845. [Google Scholar] [CrossRef]
- Piunti, A.; Smith, E.R.; Morgan, M.A.J.; Ugarenko, M.; Khaltyan, N.; Helmin, K.A.; Ryan, C.A.; Murray, D.C.; Rickels, R.A.; Yilmaz, B.D.; et al. CATACOMB: An endogenous inducible gene that antagonizes H3K27 methylation activity of Polycomb repressive complex 2 via an H3K27M-like mechanism. Sci. Adv. 2019, 5, eaax2887. [Google Scholar] [CrossRef]
- Sudarshan, D.; Avvakumov, N.; Lalonde, M.E.; Alerasool, N.; Joly-Beauparlant, C.; Jacquet, K.; Mameri, A.; Lambert, J.P.; Rousseau, J.; Lachance, C.; et al. Recurrent chromosomal translocations in sarcomas create a megacomplex that mislocalizes NuA4/TIP60 to Polycomb target loci. Genes Dev. 2022, 36, 664–683. [Google Scholar] [CrossRef]
- Pierron, G.; Tirode, F.; Lucchesi, C.; Reynaud, S.; Ballet, S.; Cohen-Gogo, S.; Perrin, V.; Coindre, J.-M.; Delattre, O. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat. Genet. 2012, 44, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Specht, K.; Zhang, L.; Sung, Y.-S.M.; Nucci, M.; Dry, S.; Vaiyapuri, S.; Richter, G.H.; Fletcher, C.D.; Antonescu, C.R. Novel BCOR-MAML3 and ZC3H7B-BCOR Gene Fusions in Undifferentiated Small Blue Round Cell Sarcomas. Am. J. Surg. Pathol. 2016, 40, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Cidre-Aranaz, F.; Watson, S.; Amatruda, J.F.; Nakamura, T.; Delattre, O.; de Alava, E.; Dirksen, U.; Grünewald, T.G.P. Small round cell sarcomas. Nat. Rev. Dis. Prim. 2022, 8, 66. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.C.; Sung, Y.S.; Zhang, L.; Huang, S.C.; Argani, P.; Chung, C.T.; Graf, N.S.; Wright, D.C.; Kellie, S.J.; Agaram, N.P.; et al. Recurrent BCOR Internal Tandem Duplication and YWHAE-NUTM2B Fusions in Soft Tissue Undifferentiated Round Cell Sarcoma of Infancy: Overlapping Genetic Features with Clear Cell Sarcoma of Kidney. Am. J. Surg. Pathol. 2016, 40, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Ueno-Yokohata, H.; Okita, H.; Nakasato, K.; Akimoto, S.; Hata, J.-I.; Koshinaga, T.; Fukuzawa, M.; Kiyokawa, N. Consistent in-frame internal tandem duplications of BCOR characterize clear cell sarcoma of the kidney. Nat. Genet. 2015, 47, 861–863. [Google Scholar] [CrossRef]
- Watson, S.; Perrin, V.; Guillemot, D.; Reynaud, S.; Coindre, J.; Karanian, M.; Guinebretière, J.; Freneaux, P.; Le Loarer, F.; Bouvet, M.; et al. Transcriptomic definition of molecular subgroups of small round cell sarcomas. J. Pathol. 2018, 245, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Bourbon, H.M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef]
- Farcas, A.M.; Blackledge, N.P.; Sudbery, I.; Long, H.K.; McGouran, J.F.; Rose, N.R.; Lee, S.; Sims, D.; Cerase, A.; Sheahan, T.W.; et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife 2012, 1, e00205. [Google Scholar] [CrossRef]
- Lo, W.-C.; Chou, C.-S.; Gokoffski, K.K.; Wan, F.Y.-M.; Lander, A.D.; Calof, A.L.; Nie, Q. Feedback regulation in multistage cell lineages. Math. Biosci. Eng. 2009, 6, 59–82. [Google Scholar]
- Xu, J.; Du, Y.; Deng, H. Direct lineage reprogramming: Strategies, mechanisms, and applications. Cell Stem. Cell 2015, 16, 119–134. [Google Scholar] [CrossRef]
- Knott, M.M.L.; Holting, T.L.B.; Ohmura, S.; Kirchner, T.; Cidre-Aranaz, F.; Grunewald, T.G.P. Targeting the undruggable: Exploiting neomorphic features of fusion oncoproteins in childhood sarcomas for innovative therapies. Cancer Metastasis Rev. 2019, 38, 625–642. [Google Scholar] [CrossRef] [PubMed]
- Ohno, T.; Ouchida, M.; Lee, L.; Gatalica, Z.; Rao, V.N.; Reddy, E.S. The EWS gene, involved in Ewing family of tumors, malignant melanoma of soft parts and desmoplastic small round cell tumors, codes for an RNA binding protein with novel regulatory domains. Oncogene 1994, 9, 3087–3097. [Google Scholar] [PubMed]
- Andersson, M.K.; Ståhlberg, A.; Arvidsson, Y.; Olofsson, A.; Semb, H.; Stenman, G.; Nilsson, O.; Åman, P. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 2008, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Prieur, A.; Tirode, F.; Cohen, P.; Delattre, O. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol. Cell. Biol. 2004, 24, 7275–7283. [Google Scholar] [CrossRef] [PubMed]
- Sankar, S.; Bell, R.; Stephens, B.; Zhuo, R.; Sharma, S.; Bearss, D.J.; Lessnick, S.L. Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene 2013, 32, 5089–5100. [Google Scholar] [CrossRef] [PubMed]
- Riggi, N.; Knoechel, B.; Gillespie, S.M.; Rheinbay, E.; Boulay, G.; Suvà, M.L.; Rossetti, N.E.; Boonseng, W.E.; Oksuz, O.; Cook, E.B.; et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 2014, 26, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Vibert, J.; Saulnier, O.; Collin, C.; Petit, F.; Borgman, K.J.; Vigneau, J.; Gautier, M.; Zaidi, S.; Pierron, G.; Watson, S.; et al. Oncogenic chimeric transcription factors drive tumor-specific transcription, processing, and translation of silent genomic regions. Mol Cell 2022, 82, 2458–2471.e9. [Google Scholar] [CrossRef]
- Boulay, G.; Sandoval, G.J.; Riggi, N.; Iyer, S.; Buisson, R.; Naigles, B.; Awad, M.E.; Rengarajan, S.; Volorio, A.; McBride, M.J.; et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell 2017, 171, 163–178.e19. [Google Scholar] [CrossRef]
- Iglesias, V.; Paladin, L.; Juan-Blanco, T.; Pallares, I.; Aloy, P.; Tosatto, S.C.E.; Ventura, S. In silico Characterization of Human Prion-Like Proteins: Beyond Neurological Diseases. Front. Physiol. 2019, 10, 314. [Google Scholar] [CrossRef]
- Chong, S.; Dugast-Darzacq, C.; Liu, Z.; Dong, P.; Dailey, G.M.; Cattoglio, C.; Heckert, A.; Banala, S.; Lavis, L.; Darzacq, X.; et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 2018, 361, eaar2555. [Google Scholar] [CrossRef]
- Behbahanipour, M.; Garcia-Pardo, J.; Ventura, S. Decoding the role of coiled-coil motifs in human prion-like proteins. Prion 2021, 15, 143–154. [Google Scholar] [CrossRef]
- Cheng, Y.; Shen, Z.; Gao, Y.; Chen, F.; Xu, H.; Mo, Q.; Chu, X.; Peng, C.-L.; McKenzie, T.T.; Palacios, B.E.; et al. Phase transition and remodeling complex assembly are important for SS18-SSX oncogenic activity in synovial sarcomas. Nat. Commun. 2022, 13, 2724. [Google Scholar] [CrossRef]
- Soto, L.F.; Li, Z.; Santoso, C.S.; Berenson, A.; Ho, I.; Shen, V.X.; Yuan, S.; Bass, J.I.F. Compendium of human transcription factor effector domains. Mol. Cell 2022, 82, 514–526. [Google Scholar] [CrossRef]
- Boija, A.; Klein, I.A.; Young, R.A. Biomolecular Condensates and Cancer. Cancer Cell 2021, 39, 174–192. [Google Scholar] [CrossRef]
- Harlow, M.L.; Maloney, N.; Roland, J.; Guillen Navarro, M.J.; Easton, M.K.; Kitchen-Goosen, S.M.; Boguslawski, E.A.; Madaj, Z.B.; Johnson, B.K.; Bowman, M.J.; et al. Lurbinectedin Inactivates the Ewing Sarcoma Oncoprotein EWS-FLI1 by Redistributing It within the Nucleus. Cancer Res. 2016, 76, 6657–6668. [Google Scholar] [CrossRef]
- Kawamura-Saito, M.; Yamazaki, Y.; Kaneko, K.; Kawaguchi, N.; Kanda, H.; Mukai, H.; Gotoh, T.; Motoi, T.; Fukayama, M.; Aburatani, H.; et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol. Genet. 2006, 15, 2125–2137. [Google Scholar] [CrossRef]
- Italiano, A.; Sung, Y.S.; Zhang, L.; Singer, S.; Maki, R.G.; Coindre, J.; Antonescu, C.R. High prevalence of CIC fusion with double-homeobox (DUX4) transcription factors in EWSR1-negative undifferentiated small blue round cell sarcomas. Genes Chromosomes Cancer 2012, 51, 207–218. [Google Scholar] [CrossRef]
- Sugita, S.; Arai, Y.; Tonooka, A.; Hama, N.; Totoki, Y.; Fujii, T.; Aoyama, T.; Asanuma, H.; Tsukahara, T.; Kaya, M.; et al. A novel CIC-FOXO4 gene fusion in undifferentiated small round cell sarcoma: A genetically distinct variant of Ewing-like sarcoma. Am. J. Surg. Pathol. 2014, 38, 1571–1576. [Google Scholar] [CrossRef]
- Le Loarer, F.; Pissaloux, D.; Watson, S.; Godfraind, C.; Galmiche-Rolland, L.; Silva, K.; Mayeur, L.; Italiano, A.; Michot, A.; Pierron, G.; et al. Clinicopathologic Features of CIC-NUTM1 Sarcomas, a New Molecular Variant of the Family of CIC-Fused Sarcomas. Am. J. Surg. Pathol. 2019, 43, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Bosnakovski, D.; Ener, E.T.; Cooper, M.S.; Gearhart, M.D.; Knights, K.A.; Xu, N.C.; Palumbo, C.A.; Toso, E.A.; Marsh, G.P.; Maple, H.J.; et al. Inactivation of the CIC-DUX4 oncogene through P300/CBP inhibition, a therapeutic approach for CIC-DUX4 sarcoma. Oncogenesis 2021, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Okimoto, R.A.; Wu, W.; Nanjo, S.; Olivas, V.; Lin, Y.K.; Ponce, R.K.; Oyama, R.; Kondo, T.; Bivona, T.G. CIC-DUX4 oncoprotein drives sarcoma metastasis and tumorigenesis via distinct regulatory programs. J. Clin. Investig. 2019, 129, 3401–3406. [Google Scholar] [CrossRef]
- Shiota, H.; Barral, S.; Buchou, T.; Tan, M.; Coute, Y.; Charbonnier, G.; Reynoird, N.; Boussouar, F.; Gerard, M.; Zhu, M.; et al. Nut Directs p300-Dependent, Genome-Wide H4 Hyperacetylation in Male Germ Cells. Cell Rep. 2018, 24, 3477–3487.e6. [Google Scholar] [CrossRef]
- Sugita, S.; Arai, Y.; Aoyama, T.; Asanuma, H.; Mukai, W.; Hama, N.; Emori, M.; Shibata, T.; Hasegawa, T. NUTM2A-CIC fusion small round cell sarcoma: A genetically distinct variant of CIC-rearranged sarcoma. Hum. Pathol. 2017, 65, 225–230. [Google Scholar] [CrossRef]
- Sorensen, P.H.; Lynch, J.C.; Qualman, S.J.; Tirabosco, R.; Lim, J.F.; Maurer, H.M.; Bridge, J.A.; Crist, W.M.; Triche, T.J.; Barr, F.G. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: A report from the children’s oncology group. J. Clin. Oncol. 2002, 20, 2672–2679. [Google Scholar] [CrossRef]
- Barr, F.G.; Qualman, S.J.; Macris, M.H.; Melnyk, N.; Lawlor, E.R.; Strzelecki, D.M.; Triche, T.J.; Bridge, J.A.; Sorensen, P.H. Genetic heterogeneity in the alveolar rhabdomyosarcoma subset without typical gene fusions. Cancer Res. 2002, 62, 4704–4710. [Google Scholar]
- Wachtel, M.; Dettling, M.; Koscielniak, E.; Stegmaier, S.; Treuner, J.; Simon-Klingenstein, K.; Bühlmann, P.; Niggli, F.K.; Schäfer, B.W. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res. 2004, 64, 5539–5545. [Google Scholar] [CrossRef]
- Sumegi, J.; Streblow, R.; Frayer, R.W.; Cin, P.D.; Rosenberg, A.; Meloni-Ehrig, A.; Bridge, J.A. Recurrent t(2;2) and t(2;8) translocations in rhabdomyosarcoma without the canonical PAX-FOXO1 fuse PAX3 to members of the nuclear receptor transcriptional coactivator family. Genes Chromosomes Cancer 2010, 49, 224–236. [Google Scholar] [CrossRef]
- Shern, J.F.; Chen, L.; Chmielecki, J.; Wei, J.S.; Patidar, R.; Rosenberg, M.; Ambrogio, L.; Auclair, D.; Wang, J.; Song, Y.K.; et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014, 4, 216–231. [Google Scholar] [CrossRef]
- Chmielecki, J.; Bailey, M.; He, J.; Elvin, J.; Vergilio, J.A.; Ramkissoon, S.; Suh, J.; Frampton, G.M.; Sun, J.X.; Morley, S.; et al. Genomic Profiling of a Large Set of Diverse Pediatric Cancers Identifies Known and Novel Mutations across Tumor Spectra. Cancer Res. 2017, 77, 509–519. [Google Scholar] [CrossRef]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef]
- Fritchie, K.J.; Jin, L.; Wang, X.; Graham, R.P.; Torbenson, M.S.; Lewis, J.E.; Rivera, M.; Garcia, J.J.; Schembri-Wismayer, D.J.; Westendorf, J.J.; et al. Fusion gene profile of biphenotypic sinonasal sarcoma: An analysis of 44 cases. Histopathology 2016, 69, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bledsoe, K.L.; Graham, R.P.; Asmann, Y.W.; Viswanatha, D.S.; Lewis, J.E.; Lewis, J.T.; Chou, M.M.; Yaszemski, M.J.; Jen, J.; et al. Recurrent PAX3-MAML3 fusion in biphenotypic sinonasal sarcoma. Nat. Genet. 2014, 46, 666–668. [Google Scholar] [CrossRef] [PubMed]
- Miano, J.M. Serum response factor: Toggling between disparate programs of gene expression. J. Mol. Cell Cardiol. 2003, 35, 577–593. [Google Scholar] [CrossRef] [PubMed]
- Coletti, D.; Daou, N.; Hassani, M.; Li, Z.; Parlakian, A. Serum Response Factor in Muscle Tissues: From Development to Ageing. Eur. J. Transl. Myol. 2016, 26, 6008. [Google Scholar] [CrossRef] [PubMed]
- Gualdrini, F.; Esnault, C.; Horswell, S.; Stewart, A.; Matthews, N.; Treisman, R. SRF Co-factors Control the Balance between Cell Proliferation and Contractility. Mol Cell 2016, 64, 1048–1061. [Google Scholar] [CrossRef] [PubMed]
- Miano, J.M. Myocardin in biology and disease. J. Biomed. Res. 2015, 29, 3–19. [Google Scholar] [PubMed]
- Karanian, M.; Pissaloux, D.; Gomez-Brouchet, A.; Chevenet, C.; Le Loarer, F.; Fernandez, C.; Minard, V.; Corradini, N.; Castex, M.-P.; Duc-Gallet, A.M.; et al. SRF-FOXO1 and SRF-NCOA1 Fusion Genes Delineate a Distinctive Subset of Well-differentiated Rhabdomyosarcoma. Am. J. Surg. Pathol. 2020, 44, 607–616. [Google Scholar] [CrossRef]
- Alaggio, R.; Zhang, L.; Sung, Y.S.; Huang, S.C.; Chen, C.L.; Bisogno, G.; Zin, A.; Agaram, N.P.; LaQuaglia, M.P.; Wexler, L.H.; et al. A Molecular Study of Pediatric Spindle and Sclerosing Rhabdomyosarcoma: Identification of Novel and Recurrent VGLL2-related Fusions in Infantile Cases. Am. J. Surg. Pathol. 2016, 40, 224–235. [Google Scholar] [CrossRef]
- Mosquera, J.M.; Sboner, A.; Zhang, L.; Kitabayashi, N.; Chen, C.L.; Sung, Y.S.; Wexler, L.H.; LaQuaglia, M.P.; Edelman, M.; Sreekantaiah, C.; et al. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes Chromosomes Cancer 2013, 52, 538–550. [Google Scholar] [CrossRef]
- Antonescu, C.R.; Sung, Y.S.; Zhang, L.; Agaram, N.P.; Fletcher, C.D. Recurrent SRF-RELA Fusions Define a Novel Subset of Cellular Myofibroma/Myopericytoma: A Potential Diagnostic Pitfall with Sarcomas with Myogenic Differentiation. Am. J. Surg. Pathol. 2017, 41, 677–684. [Google Scholar] [CrossRef]
- Suurmeijer, A.J.; Dickson, B.C.; Swanson, D.; Sung, Y.S.; Zhang, L.; Antonescu, C.R. Novel SRF-ICA1L Fusions in Cellular Myoid Neoplasms with Potential for Malignant Behavior. Am. J. Surg. Pathol. 2020, 44, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.H.; Maeda, T.; Mullett, S.J.; Stewart, A.F. Transcription cofactor Vgl-2 is required for skeletal muscle differentiation. Genesis 2004, 39, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Chapman, D.L.; Stewart, A.F. Mammalian vestigial-like 2, a cofactor of TEF-1 and MEF2 transcription factors that promotes skeletal muscle differentiation. J. Biol. Chem. 2002, 277, 48889–48898. [Google Scholar] [CrossRef] [PubMed]
- Pobbati, A.V.; Chan, S.W.; Lee, I.; Song, H.; Hong, W. Structural and functional similarity between the Vgll1-TEAD and the YAP-TEAD complexes. Structure 2012, 20, 1135–1140. [Google Scholar] [CrossRef] [PubMed]
- Figeac, N.; Mohamed, A.D.; Sun, C.; Schönfelder, M.; Matallanas, D.; Garcia-Munoz, A.; Missiaglia, E.; Collie-Duguid, E.; De Mello, V.; Pobbati, A.V.; et al. VGLL3 operates via TEAD1, TEAD3 and TEAD4 to influence myogenesis in skeletal muscle. J. Cell Sci. 2019, 132, jcs225946. [Google Scholar] [CrossRef] [PubMed]
- Spencer, T.E.; Jenster, G.; Burcin, M.M.; Allis, C.D.; Zhou, J.; Mizzen, C.A.; McKenna, N.J.; Onate, S.A.; Tsai, S.Y.; Tsai, M.-J.; et al. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 1997, 389, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Asante, Y.; Benischke, K.; Osman, I.; Ngo, Q.A.; Wurth, J.; Laubscher, D.; Kim, H.; Udhayakumar, B.; Khan, I.H.; Chin, D.H.; et al. PAX3-FOXO1 uses its activation domain to recruit CBP/P300 and shape RNA Pol2 cluster distribution. Nat. Commun. 2023, 14, 8361. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Tindall, D.J. Dynamic FoxO transcription factors. J. Cell Sci. 2007, 120 Pt 15, 2479–2487. [Google Scholar] [CrossRef]
- Braganca, J.; Eloranta, J.J.; Bamforth, S.D.; Ibbitt, J.C.; Hurst, H.C.; Bhattacharya, S. Physical and functional interactions among AP-2 transcription factors, p300/CREB-binding protein, and CITED2. J. Biol. Chem. 2003, 278, 16021–16029. [Google Scholar] [CrossRef]
- van Tetering, G.; Vooijs, M. Proteolytic cleavage of Notch: “HIT and RUN”. Curr. Mol. Med. 2011, 11, 255–269. [Google Scholar] [CrossRef]
- Mosquera, J.; Sboner, A.; Zhang, L.; Chen, C.; Sung, Y.; Chen, H.; Agaram, N.P.; Briskin, D.; Basha, B.M.; Singer, S.; et al. Novel MIR143-NOTCH fusions in benign and malignant glomus tumors. Genes Chromosomes Cancer 2013, 52, 1075–1087. [Google Scholar] [CrossRef]
- Oldershaw, R.A.; Hardingham, T.E. Notch signaling during chondrogenesis of human bone marrow stem cells. Bone 2010, 46, 286–293. [Google Scholar] [CrossRef]
- Oldershaw, R.A.; Tew, S.R.; Russell, A.M.; Meade, K.; Hawkins, R.; McKay, T.R.; Brennan, K.R.; Hardingham, T.E. Notch signaling through Jagged-1 is necessary to initiate chondrogenesis in human bone marrow stromal cells but must be switched off to complete chondrogenesis. Stem Cells 2008, 26, 666–674. [Google Scholar] [CrossRef]
- Robinson, D.R.; Wu, Y.-M.; Kalyana-Sundaram, S.; Cao, X.; Lonigro, R.J.; Sung, Y.-S.; Chen, C.-L.; Zhang, L.; Wang, R.; Su, F.; et al. Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat. Genet. 2013, 45, 180–185. [Google Scholar] [CrossRef]
- Chmielecki, J.; Crago, A.M.; Rosenberg, M.; O’Connor, R.; Walker, S.R.; Ambrogio, L.; Auclair, D.; McKenna, A.; Heinrich, M.C.; Frank, D.A.; et al. Whole-exome sequencing identifies a recurrent NAB2-STAT6 fusion in solitary fibrous tumors. Nat. Genet. 2013, 45, 131–132. [Google Scholar] [CrossRef]
- Tai, H.C.; Chuang, I.C.; Chen, T.C.; Li, C.F.; Huang, S.C.; Kao, Y.C.; Lin, P.C.; Tsai, J.W.; Lan, J.; Yu, S.C.; et al. NAB2-STAT6 fusion types account for clinicopathological variations in solitary fibrous tumors. Mod. Pathol. 2015, 28, 1324–1335. [Google Scholar] [CrossRef] [PubMed]
- Georgiesh, T.; Namløs, H.M.; Sharma, N.; Lorenz, S.; Myklebost, O.; Bjerkehagen, B.; Meza-Zepeda, L.A.; Boye, K. Clinical and molecular implications of NAB2-STAT6 fusion variants in solitary fibrous tumour. Pathology 2021, 53, 713–719. [Google Scholar] [CrossRef]
- Huang, S.C.; Li, C.F.; Kao, Y.C.; Chuang, I.C.; Tai, H.C.; Tsai, J.W.; Yu, S.C.; Huang, H.Y.; Lan, J.; Yen, S.L.; et al. The clinicopathological significance of NAB2-STAT6 gene fusions in 52 cases of intrathoracic solitary fibrous tumors. Cancer Med. 2016, 5, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Ladanyi, M.; Lui, M.Y.; Antonescu, C.R.; Krause-Boehm, A.; Meindl, A.; Argani, P.; Healey, J.H.; Ueda, T.; Yoshikawa, H.; Meloni-Ehrig, A.; et al. The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene 2001, 20, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Antonescu, C.R.; Le Loarer, F.; Mosquera, J.; Sboner, A.; Zhang, L.; Chen, C.; Chen, H.; Pathan, N.; Krausz, T.; Dickson, B.C.; et al. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer 2013, 52, 775–784. [Google Scholar] [CrossRef]
- Argani, P.; Aulmann, S.; Illei, P.B.; Netto, G.J.; Ro, J.; Cho, H.-Y.; Dogan, S.; Ladanyi, M.; Martignoni, G.; Goldblum, J.R.; et al. A distinctive subset of PEComas harbors TFE3 gene fusions. Am. J. Surg. Pathol. 2010, 34, 1395–1406. [Google Scholar] [CrossRef] [PubMed]
- Bogan, J.S.; Hendon, N.; McKee, A.E.; Tsao, T.S.; Lodish, H.F. Functional cloning of TUG as a regulator of GLUT4 glucose transporter trafficking. Nature 2003, 425, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Bogan, J.S.; Rubin, B.R.; Yu, C.; Löffler, M.G.; Orme, C.M.; Belman, J.P.; McNally, L.J.; Hao, M.; Cresswell, J.A. Endoproteolytic cleavage of TUG protein regulates GLUT4 glucose transporter translocation. J. Biol. Chem. 2012, 287, 23932–23947. [Google Scholar] [CrossRef] [PubMed]
- Kobos, R.; Nagai, M.; Tsuda, M.; Merl, M.Y.; Saito, T.; Laé, M.; Mo, Q.; Olshen, A.; Lianoglou, S.; Leslie, C.; et al. Combining integrated genomics and functional genomics to dissect the biology of a cancer-associated, aberrant transcription factor, the ASPSCR1-TFE3 fusion oncoprotein. J. Pathol. 2013, 229, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, M.; Davis, I.J.; Argani, P.; Shukla, N.; McGill, G.G.; Nagai, M.; Saito, T.; Laé, M.; Fisher, D.E.; Ladanyi, M. TFE3 fusions activate MET signaling by transcriptional up-regulation, defining another class of tumors as candidates for therapeutic MET inhibition. Cancer Res. 2007, 67, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Tanas, M.R.; Sboner, A.; Oliveira, A.M.; Erickson-Johnson, M.R.; Hespelt, J.; Hanwright, P.J.; Flanagan, J.; Luo, Y.; Fenwick, K.; Natrajan, R.; et al. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci. Transl. Med. 2011, 3, 98ra82. [Google Scholar] [CrossRef] [PubMed]
- Suurmeijer, A.J.H.; Dickson, B.C.; Swanson, D.; Sung, Y.S.; Zhang, L.; Antonescu, C.R. Variant WWTR1 gene fusions in epithelioid hemangioendothelioma-A genetic subset associated with cardiac involvement. Genes Chromosomes Cancer 2020, 59, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Ye, X.; Yu, J.; Li, L.; Li, W.; Li, S.; Yu, J.; Lin, J.D.; Wang, C.-Y.; Chinnaiyan, A.M.; et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008, 22, 1962–1971. [Google Scholar] [CrossRef] [PubMed]
- Tanas, M.R.; Ma, S.; Jadaan, F.O.; Ng, C.K.Y.; Weigelt, B.; Reis-Filho, J.S.; Rubin, B.P. Mechanism of action of a WWTR1(TAZ)-CAMTA1 fusion oncoprotein. Oncogene 2016, 35, 929–938. [Google Scholar] [CrossRef]
- Szulzewsky, F.; Arora, S.; Hoellerbauer, P.; King, C.; Nathan, E.; Chan, M.; Cimino, P.J.; Ozawa, T.; Kawauchi, D.; Pajtler, K.W.; et al. Comparison of tumor-associated YAP1 fusions identifies a recurrent set of functions critical for oncogenesis. Genes Dev. 2020, 34, 1051–1064. [Google Scholar] [CrossRef]
- Merritt, N.; Garcia, K.; Rajendran, D.; Lin, Z.Y.; Zhang, X.; Mitchell, K.A.; Borcherding, N.; Fullenkamp, C.; Chimenti, M.S.; Gingras, A.C.; et al. TAZ-CAMTA1 and YAP-TFE3 alter the TAZ/YAP transcriptome by recruiting the ATAC histone acetyltransferase complex. Elife 2021, 10, e62857. [Google Scholar] [CrossRef]
- Dahlén, A.; Fletcher, C.D.; Mertens, F.; Fletcher, J.A.; Perez-Atayde, A.R.; Hicks, M.J.; Debiec-Rychter, M.; Sciot, R.; Wejde, J.; Wedin, R.; et al. Activation of the GLI oncogene through fusion with the beta-actin gene (ACTB) in a group of distinctive pericytic neoplasms: Pericytoma with t(7;12). Am. J. Pathol. 2004, 164, 1645–1653. [Google Scholar] [CrossRef]
- Antonescu, C.R.; Agaram, N.P.; Sung, Y.S.; Zhang, L.; Swanson, D.; Dickson, B.C. A Distinct Malignant Epithelioid Neoplasm with GLI1 Gene Rearrangements, Frequent S100 Protein Expression, and Metastatic Potential: Expanding the Spectrum of Pathologic Entities With ACTB/MALAT1/PTCH1-GLI1 Fusions. Am. J. Surg. Pathol. 2018, 42, 553–560. [Google Scholar] [CrossRef]
- Raffel, C.; Jenkins, R.B.; Frederick, L.; Hebrink, D.; Alderete, B.; Fults, D.W.; James, C.D. Sporadic medulloblastomas contain PTCH mutations. Cancer Res. 1997, 57, 842–845. [Google Scholar]
- Xie, J.; Murone, M.; Luoh, S.-M.; Ryan, A.; Gu, Q.; Zhang, C.; Bonifas, J.M.; Lam, C.-W.; Hynes, M.; Goddard, A.; et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature 1998, 391, 90–92. [Google Scholar] [CrossRef]
- Stransky, N.; Cerami, E.; Schalm, S.; Kim, J.L.; Lengauer, C. The landscape of kinase fusions in cancer. Nat. Commun. 2014, 5, 4846. [Google Scholar] [CrossRef]
- Delespaul, L.; Lesluyes, T.; Pérot, G.; Brulard, C.; Lartigue, L.; Baud, J.; Lagarde, P.; Le Guellec, S.; Neuville, A.; Terrier, P.; et al. Recurrent TRIO Fusion in Nontranslocation-Related Sarcomas. Clin. Cancer Res. 2017, 23, 857–867. [Google Scholar] [CrossRef]
- Akiyama, B.M.; Parks, J.W.; Stone, M.D. The telomerase essential N-terminal domain promotes DNA synthesis by stabilizing short RNA-DNA hybrids. Nucleic Acids Res. 2015, 43, 5537–5549. [Google Scholar] [CrossRef]
- Salhia, B.; Tran, N.L.; Chan, A.; Wolf, A.; Nakada, M.; Rutka, F.; Ennis, M.; McDonough, W.S.; Berens, M.E.; Symons, M.; et al. The guanine nucleotide exchange factors trio, Ect2, and Vav3 mediate the invasive behavior of glioblastoma. Am. J. Pathol. 2008, 173, 1828–1838. [Google Scholar] [CrossRef]
- Tirode, F.; Surdez, D.; Ma, X.; Parker, M.; Le Deley, M.C.; Bahrami, A.; Zhang, Z.; Lapouble, E.; Grossetête-Lalami, S.; Rusch, M.; et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014, 4, 1342–1353. [Google Scholar] [CrossRef]
- Chen, L.; Shern, J.F.; Wei, J.S.; Yohe, M.E.; Song, Y.K.; Hurd, L.; Liao, H.; Catchpoole, D.; Skapek, S.X.; Barr, F.G.; et al. Clonality and evolutionary history of rhabdomyosarcoma. PLoS Genet. 2015, 11, e1005075. [Google Scholar] [CrossRef]
- Bernasconi, M.; Remppis, A.; Fredericks, W.J.; Rauscher, F.J., 3rd; Schafer, B.W. Induction of apoptosis in rhabdomyosarcoma cells through down-regulation of PAX proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 13164–13169. [Google Scholar] [CrossRef] [PubMed]
- Chansky, H.A.; Barahmand-Pour, F.; Mei, Q.; Kahn-Farooqi, W.; Zielinska-Kwiatkowska, A.; Blackburn, M.; Chansky, K.; Conrad, E.U., III; Bruckner, J.D.; Greenlee, T.K.; et al. Targeting of EWS/FLI-1 by RNA interference attenuates the tumor phenotype of Ewing’s sarcoma cells in vitro. J. Orthop. Res. 2004, 22, 910–917. [Google Scholar] [CrossRef]
- Carmody Soni, E.E.; Schlottman, S.; Erkizan, H.V.; Uren, A.; Toretsky, J.A. Loss of SS18-SSX1 inhibits viability and induces apoptosis in synovial sarcoma. Clin. Orthop. Relat. Res. 2014, 472, 874–882. [Google Scholar] [CrossRef]
- Wachtel, M.; Schafer, B.W. PAX3-FOXO1: Zooming in on an “undruggable” target. Semin. Cancer Biol. 2018, 50, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559. [Google Scholar] [CrossRef] [PubMed]
- den Besten, W.; Lipford, J.R. Prospecting for molecular glues. Nat. Chem. Biol. 2020, 16, 1157–1158. [Google Scholar] [CrossRef]
- Danielli, S.G.; Porpiglia, E.; De Micheli, A.J.; Navarro, N.; Zellinger, M.J.; Bechtold, I.; Kisele, S.; Volken, L.; Marques, J.G.; Kasper, S.; et al. Single-cell profiling of alveolar rhabdomyosarcoma reveals RAS pathway inhibitors as cell-fate hijackers with therapeutic relevance. Sci. Adv. 2023, 9, eade9238. [Google Scholar] [CrossRef]
- Masuzawa, R.; Takahashi, K.; Takano, K.; Nishino, I.; Sakai, T.; Endo, T. DA-Raf and the MEK inhibitor trametinib reverse skeletal myocyte differentiation inhibition or muscle atrophy caused by myostatin and GDF11 through the non-Smad Ras-ERK pathway. J. Biochem. 2022, 171, 109–122. [Google Scholar] [CrossRef]
| Fusion Protein | Sarcoma Entity | Antibody Target | References |
|---|---|---|---|
| PAX3::FOXO1 | Alveolar Rhabdomyosarcoma | Breakpoint | [8] |
| SS18::SSX | Synovial Sarcoma | Breakpoint | [9,10,11,12] |
| NAB2::STAT6 | Solitary Fibrous Tumor | STAT6 | [13,14,15] |
| WWTR1::CAMTA1 | Epithelioid Hemangioendothelioma | CAMTA1 | [16,17] |
| FUS::DDIT3 | Myxoid Liposarcoma | DDIT3 | [18,19] |
| ALK-fusions | Inflammatory Myofibroblastic Tumor | ALK | [20,21] |
| Kinase | Sarcoma Entity | Potential Therapeutic Drugs |
|---|---|---|
| ALK | Inflammatory myofibroblastic tumor, Leiomyosarcoma | Crizotinib, Ceritinib, Alectinib, Brigatinib, Lorlatinib |
| ROS1 | Inflammatory myofibroblastic tumor | Crizotinib, Entrectinib |
| NTRK | Inflammatory myofibroblastic tumor, Congenital fibrosarcoma, Adult fibrosarcoma, Dermato- fibrosarcoma protuberans | Larotrectinib, Entrectinib |
| BRAF | Spindle cell sarcoma, Myxoinflammatory fibroblastic sarcoma, Myxofibrosarcoma | Dabrafenib, Vemurafenib |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wachtel, M.; Surdez, D.; Grünewald, T.G.P.; Schäfer, B.W. Functional Classification of Fusion Proteins in Sarcoma. Cancers 2024, 16, 1355. https://doi.org/10.3390/cancers16071355
Wachtel M, Surdez D, Grünewald TGP, Schäfer BW. Functional Classification of Fusion Proteins in Sarcoma. Cancers. 2024; 16(7):1355. https://doi.org/10.3390/cancers16071355
Chicago/Turabian StyleWachtel, Marco, Didier Surdez, Thomas G. P. Grünewald, and Beat W. Schäfer. 2024. "Functional Classification of Fusion Proteins in Sarcoma" Cancers 16, no. 7: 1355. https://doi.org/10.3390/cancers16071355
APA StyleWachtel, M., Surdez, D., Grünewald, T. G. P., & Schäfer, B. W. (2024). Functional Classification of Fusion Proteins in Sarcoma. Cancers, 16(7), 1355. https://doi.org/10.3390/cancers16071355






