Exploring Genetic Determinants: A Comprehensive Analysis of Serpin B Family SNPs and Prognosis in Glioblastoma Multiforme Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. DNA Extraction
2.3. Bioinformatics Analysis
2.4. Statistical Analysis
3. Results
3.1. Primary Cohort
3.2. Serpin B 5-Gene Risk Score
3.3. Bioinformatics Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mattiuzzi, C.; Lippi, G. Current Cancer Epidemiology. J. Epidemiol. Glob. Health 2019, 9, 217–222. [Google Scholar] [CrossRef]
- Schaff, L.R.; Mellinghoff, I.K. Glioblastoma and Other Primary Brain Malignancies in Adults: A Review. JAMA 2023, 329, 574–587. [Google Scholar] [CrossRef]
- Grochans, S.; Cybulska, A.M.; Simińska, D.; Korbecki, J.; Kojder, K.; Chlubek, D.; Baranowska-Bosiacka, I. Epidemiology of Glioblastoma Multiforme—Literature Review. Cancers 2022, 14, 2412. [Google Scholar] [CrossRef]
- Koshy, M.; Villano, J.L.; Dolecek, T.A.; Howard, A.; Mahmood, U.; Chmura, S.J.; Weichselbaum, R.R.; McCarthy, B.J. Improved survival time trends for glioblastoma using the SEER 17 population-based registries. J. Neuro-Oncol. 2012, 107, 207–212. [Google Scholar] [CrossRef]
- Mohammadi, E.; Moghaddam, S.S.; Azadnajafabad, S.; Maroufi, S.F.; Rashidi, M.M.; Naderian, M.; Jafari, A.; Sharifi, G.; Ghasemi, E.; Rezaei, N.; et al. Epidemiology of Brain and Other Central Nervous System Cancers in the North Africa and Middle East Region: A Systematic Analysis of the Global Burden of Disease Study 1990–2019. World Neurosurg. 2023, 171, e796–e819. [Google Scholar] [CrossRef]
- Falzone, L.; Bordonaro, R.; Libra, M. SnapShot: Cancer chemotherapy. Cell 2023, 186, 1816. [Google Scholar] [CrossRef]
- Rodríguez-Camacho, A.; Flores-Vázquez, J.G.; Moscardini-Martelli, J.; Torres-Ríos, J.A.; Olmos-Guzmán, A.; Ortiz-Arce, C.S.; Cid-Sánchez, D.R.; Pérez, S.R.; Macías-González, M.D.S.; Hernández-Sánchez, L.C.; et al. Glioblastoma Treatment: State-of-the-Art and Future Perspectives. Int. J. Mol. Sci. 2022, 23, 7207. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, C.; Costa, A.; Osório, L.; Lago, R.C.; Linhares, P.; Carvalho, B.; Caeiro, C. Current Standards of Care in Glioblastoma Therapy. In Glioblastoma; Codon Publications: Brisbane, Australia, 2017; pp. 197–241. [Google Scholar]
- Lowe, S.; Bhat, K.P.; Olar, A. Current clinical management of patients with glioblastoma. Cancer Rep. 2019, 2, e1216. [Google Scholar] [CrossRef] [PubMed]
- Silantyev, A.S.; Falzone, L.; Libra, M.; Gurina, O.I.; Kardashova, K.S.; Nikolouzakis, T.K.; Nosyrev, A.E.; Sutton, C.W.; Panayioti, M.; Tsatsakis, A. Current and Future Trends on Diagnosis and Prognosis of Glioblastoma: From Molecular Biology to Proteomics. Cells 2019, 8, 863. [Google Scholar] [CrossRef] [PubMed]
- Gilard, V.; Tebani, A.; Dabaj, I.; Laquerrière, A.; Fontanilles, M.; Derrey, S.; Marret, S.; Bekri, S. Diagnosis and management of glioblastoma: A comprehensive perspective. J. Pers. Med. 2021, 11, 258. [Google Scholar] [CrossRef] [PubMed]
- Bikfalvi, A.; da Costa, C.A.; Avril, T.; Barnier, J.V.; Bauchet, L.; Brisson, L.; Cartron, P.F.; Castel, H.; Chevet, E.; Chneiweiss, H.; et al. Challenges in glioblastoma research: Focus on the tumor microenvironment. Trends Cancer 2023, 9, 9–27. [Google Scholar] [CrossRef] [PubMed]
- Kaiserman, D.; Bird, P.I. Analysis of vertebrate genomes suggests a new model for clade B serpin evolution. BMC Genom. 2005, 6, 1–10. [Google Scholar] [CrossRef]
- Askew, D.J.; Cataltepe, S.; Kumar, V.; Edwards, C.; Pace, S.M.; Howarth, R.N.; Pak, S.C.; Askew, Y.S.; Brömme, D.; Luke, C.J.; et al. SERPINB11 is a new noninhibitory intracellular serpin: Common single nucleotide polymorphisms in the scaffold impair conformational change. J. Biol. Chem. 2007, 282, 24948–24960. [Google Scholar] [CrossRef]
- Wesseling, P.; Capper, D. WHO 2016 Classification of gliomas. Neuropathol. Appl. Neurobiol. 2018, 44, 139–150. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Program (TCGA)—NCI. Available online: https://www.cancer.gov/ccg/research/genome-sequencing/tcga (accessed on 14 November 2023).
- de Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2013, 83, 3861–3867. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling tumor infiltrating immune cells with, CIBERSORT. Methods Mol. Biol. 2018, 1711, 243. [Google Scholar]
- Yalamarty, S.S.K.; Filipczak, N.; Li, X.; Subhan, M.A.; Parveen, F.; Ataide, J.A.; Rajmalani, B.A.; Torchilin, V.P. Mechanisms of Resistance and Current Treatment Options for Glioblastoma Multiforme (GBM). Cancers 2023, 15, 2116. [Google Scholar] [CrossRef]
- Tamimi, A.F.; Juweid, M. Epidemiology and Outcome of Glioblastoma. In Glioblastoma; Codon Publications: Brisbane, Australia, 2017; pp. 143–153. [Google Scholar]
- Izuhara, K.; Ohta, S.; Kanaji, S.; Shiraishi, H.; Arima, K. Recent progress in understanding the diversity of the human ov-serpin/clade B serpin family. Cell. Mol. Life Sci. 2008, 65, 2541–2553. [Google Scholar] [CrossRef]
- Kryvalap, Y.; Czyzyk, J. The Role of Proteases and Serpin Protease Inhibitors in β-Cell Biology and Diabetes. Biomolecules 2022, 12, 67. [Google Scholar] [CrossRef] [PubMed]
- Al-Hadyan, K.S.; Al-Harbi, N.M.; Al-Qahtani, S.S.; Alsbeih, G.A. Involvement of Single-Nucleotide Polymorphisms in Predisposition to Head and Neck Cancer in Saudi Arabia. Genet. Test Mol. Biomark. 2012, 16, 95. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Chen, H.; Davids, J.; Bryant, M.; Lucas, A. Serpins for diagnosis and therapy in cancer. Cardiovasc. Hematol. Disord. Drug Targets 2013, 13, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Bae, H.; Yang, C.; Park, S.; Youn, B.S.; Kim, H.S.; Song, G.; Lim, W. Eupatilin Promotes Cell Death by Calcium Influx through ER-Mitochondria Axis with SERPINB11 Inhibition in Epithelial Ovarian Cancer. Cancers 2020, 12, 1459. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hou, H.; Li, M.; Yang, Y.; Sun, L. Anticancer effect of eupatilin on glioma cells through inhibition of the Notch-1 signaling pathway. Mol. Med. Rep. 2016, 13, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.H.; Lee, J.G.; Yang, Y.I.; Kim, J.H.; Ahn, J.H.; Baek, N.I.; Lee, K.T.; Choi, J.H. Eupatilin, a dietary flavonoid, induces G2/M cell cycle arrest in human endometrial cancer cells. Food Chem. Toxicol. 2011, 49, 1737–1744. [Google Scholar] [CrossRef] [PubMed]
- Park, B.B.; Yoon, J.S.; Kim, E.S.; Choi, J.; Won, Y.W.; Choi, J.H.; Lee, Y.Y. Inhibitory effects of eupatilin on tumor invasion of human gastric cancer MKN-1 cells. Tumour Biol. 2013, 34, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Lim, W.; Mun, J.; Paik, H.; Park, S.; Lim, H.; Kim, J.; Lee, E.J.; Yim, G.W.; Lee, N.; et al. SERPINB11 Expression Is Associated with Prognosis of High-grade Serous and Clear Cell Carcinoma of the Ovary. In Vivo 2021, 35, 2647–2653. [Google Scholar] [CrossRef]
- Heit, C.; Jackson, B.C.; McAndrews, M.; Wright, M.W.; Thompson, D.C.; Silverman, G.A.; Nebert, D.W.; Vasiliou, V. Update of the human and mouse SERPIN gene superfamily. Hum. Genom. 2013, 7, 1–14. [Google Scholar] [CrossRef]
- Snoeren, N.; Emmink, B.L.; Koerkamp, M.J.; van Hooff, S.R.; Goos, J.A.; van Houdt, W.J.; de Wit, M.; Prins, A.M.; Piersma, S.R.; Pham, T.V.; et al. Maspin is a marker for early recurrence in primary stage III and IV colorectal cancer. Br. J. Cancer 2013, 109, 1636–1647. [Google Scholar] [CrossRef]
- Zou, Z.; Anisowicz, A.; Hendrix, M.J.C.; Thor, A.; Neveu, M.; Sheng, S.; Rafidi, K.; Seftor, E.; Sager, R. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science 1994, 263, 526–529. [Google Scholar] [CrossRef]
- Ma, S.; Pang, C.; Song, L.; Guo, F.; Sun, H. Activating transcription factor 3 is overexpressed in human glioma and its knockdown in glioblastoma cells causes growth inhibition both in vitro and in vivo. Int. J. Mol. Med. 2015, 35, 1561–1573. [Google Scholar] [CrossRef]
- Lin, K.; Yang, R.; Zheng, Z.; Zhou, Y.; Geng, Y.; Hu, Y.; Wu, S.; Wu, W. Sulforaphane-cysteine-induced apoptosis via phosphorylated ERK1/2-mediated maspin pathway in human non-small cell lung cancer cells. Cell Death Discov. 2017, 3, 1–8. [Google Scholar] [CrossRef][Green Version]
- He, X.; Ma, Y.; Huang, Z.; Wang, G.; Wang, W.; Zhang, R.; Guo, G.; Zhang, X.; Wen, Y.; Zhang, L. SERPINB5 is a prognostic biomarker and promotes proliferation, metastasis and epithelial-mesenchymal transition (EMT) in lung adenocarcinoma. Thorac. Cancer 2023, 14, 2275–2287. [Google Scholar] [CrossRef] [PubMed]
- Scott, F.L.; Hirst, C.E.; Sun, J.; Bird, C.H.; Bottomley, S.P.; Bird, P.I. The Intracellular Serpin Proteinase Inhibitor 6 Is Expressed in Monocytes and Granulocytes and Is a Potent Inhibitor of the Azurophilic Granule Protease, Cathepsin G. Blood 1999, 93, 2089–2097. [Google Scholar] [CrossRef]
- Scarff, K.L.; Ung, K.S.; Nandurkar, H.; Crack, P.J.; Bird, C.H.; Bird, P.I. Targeted Disruption of SPI3/Serpinb6 Does Not Result in Developmental or Growth Defects, Leukocyte Dysfunction, or Susceptibility to Stroke. Mol. Cell Biol. 2004, 24, 4075. [Google Scholar] [CrossRef] [PubMed]
- Burgener, S.S.; Leborgne, N.G.F.; Snipas, S.J.; Salvesen, G.S.; Bird, P.I.; Benarafa, C. Cathepsin G Inhibition by Serpinb1 and Serpinb6 Prevents Programmed Necrosis in Neutrophils and Monocytes and Reduces GSDMD-Driven Inflammation. Cell. Rep. 2019, 27, 3646–3656.e5. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Huang, F.M.; Chen, L.; Feng, K.Y.; Jian, F.; Huang, T.; Cai, Y.D. Identification of methylation signatures associated with CAR T cell in B-cell acute lymphoblastic leukemia and non-hodgkin’s lymphoma. Front. Oncol. 2022, 12, 976262. [Google Scholar] [CrossRef]
- Lauko, A.; Volovetz, J.; Turaga, S.M.; Bayik, D.; Silver, D.J.; Mitchell, K.; Mulkearns-Hubert, E.E.; Watson, D.C.; Desai, K.; Midha, M.; et al. SerpinB3 drives cancer stem cell survival in glioblastoma. Cell Rep. 2022, 40, 111348. [Google Scholar] [CrossRef] [PubMed]
- Niehaus, J.Z.; Good, M.; Jackson, L.E.; Ozolek, J.A.; Silverman, G.A.; Luke, C.J. Human SERPINB12 Is an Abundant Intracellular Serpin Expressed in Most Surface and Glandular Epithelia. J. Histochem. Cytochem. 2015, 63, 854–865. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Li, J.; Li, X.; Yang, X.; Zhang, S.; Wang, X.; Wang, N.; Xu, K.; Jiang, X.; Zhang, Y. A Combined RNA Signature Predicts Recurrence Risk of Stage I-IIIA Lung Squamous Cell Carcinoma. Front. Genet. 2021, 12, 676464. [Google Scholar] [CrossRef] [PubMed]
- Jo, G.; Lim, W.; Bae, S.M.; Bazer, F.W.; Song, G. Avian SERPINB12 expression in the avian oviduct is regulated by estrogen and up-regulated in epithelial cell-derived ovarian carcinomas of laying hens. PLoS ONE 2014, 9, e99792. [Google Scholar] [CrossRef][Green Version]

| SNP-ID | Gene | Chr ^ | bp * | Primer Forward | Primer Reverse |
|---|---|---|---|---|---|
| rs4940595 | Serpinb11 | 18 | 63,712,604 | ACGTTGGATGCTGGAAGAATTCATTCCGAG | ACGTTGGATGTACAGTTAGAGTCTGGCTGG |
| Variable | GBM (n = 63) |
|---|---|
| Age at diagnosis, Mean (SD) | 50.1 (18.4) |
| Sex, n (%) | |
| Females | 26 (41.3%) |
| Males | 37.0 (58.7%) |
| Survival Status, n (%) | |
| Alive | 33 (52.4%) |
| Dead | 30 (47.6%) |
| Overall survival (months), Median (Q1, Q3) | 2.8 (0.5, 9.9) |
| Serum LDH (U/L), Mean (SD) | 34.0 (179.0) |
| Total protein (g/L), Mean (SD) | 47.3 (33.1) |
| Monocytes (×109/L), Mean (SD) | 3.7 (4.0) |
| Lymphocytes (×109/L), Mean (SD) | 9.2 (10.0) |
| Platelets (×103/μL), Mean (SD) | 281.9 (96.4) |
| Tumor size (mm), Mean (SD) | 126.7 (96.9) |
| Tumor laterality, n (%) | |
| Right | 31 (49.2%) |
| Left | 29 (46.0%) |
| Bilateral | 3 (4.8%) |
| Necrosis, n (%) | |
| Coagulative | 7 (11.7%) |
| Geographic | 1 (1.7%) |
| Liquefactive | 48 (80.0%) |
| None | 4 (6.7%) |
| Degree of necrosis, n (%) | |
| Foci of palisading necrosis | 34 (57.6%) |
| Whole tumor | 21 (35.6%) |
| None | 4 (6.8%) |
| Radiotherapy, n (%) | 9 (14.3%) |
| Chemotherapy, n (%) | 6 (19.4%) |
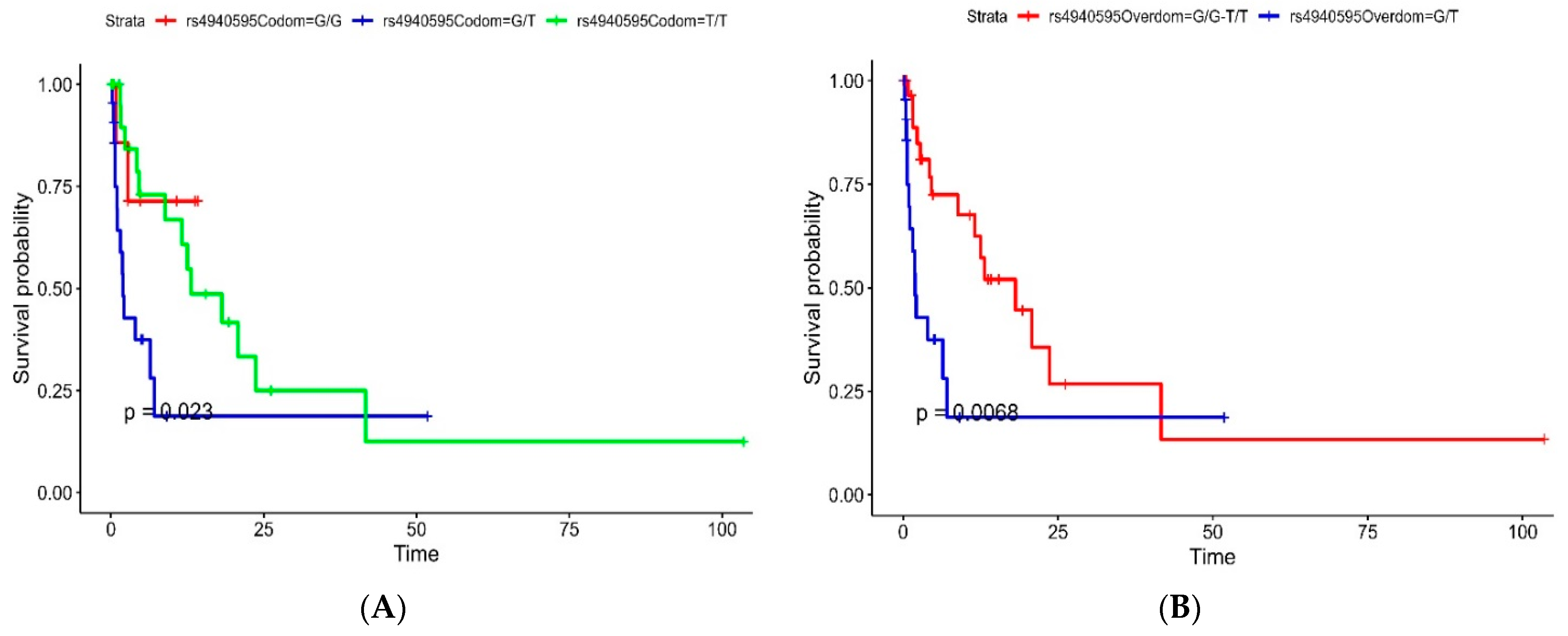
| SNP ID | Model | Genotype | HR (95% CI, p-Value) |
|---|---|---|---|
| rs4940595 | Codominant | G/G | - |
| G/T | 3.87 (0.87–17.26, p = 0.076) | ||
| T/T | 1.51 (0.34–6.79, p = 0.592) | ||
| Overdominant | G/G-T/T | - | |
| G/T | 2.75 (1.29–5.88, p = 0.009) | ||
| Dominant | G/G | - | |
| G/T-T/T | 2.25 (0.53–9.56, p = 0.271) | ||
| Recessive | G/G-G/T | - | |
| T/T | 0.53 (0.25–1.14, p = 0.106) |
| Characteristic | High, n = 80 1 | Low, n = 80 1 | p-Value 2 |
|---|---|---|---|
| Sex | 0.4 | ||
| Female | 16 (36%) | 28 (44%) | |
| Male | 28 (64%) | 35 (56%) | |
| Sample Type | >0.9 | ||
| Primary | 76 (95%) | 77 (96%) | |
| Recurrence | 4 (5.0%) | 3 (3.8%) | |
| Subtype | 0.9 | ||
| IDHmut | 4 (6.3%) | 3 (4.7%) | |
| IDHwt | 57 (90%) | 59 (92%) | |
| Fraction Genome Altered | 0.20 (0.13) | 0.23 (0.14) | 0.13 |
| MSIsensor Score | 0.31 (1.02) | 0.28 (0.32) | <0.001 |
| Mutation Count | 57 (64) | 216 (1367) | 0.2 |
| OS Time (Months) | 14 (12) | 14 (13) | 0.8 |
| OS Status | 68 (86%) | 59 (74%) | 0.053 |
| PFS Time (Months) | 8 (10) | 9 (8) | 0.082 |
| PFS Status | 69 (87%) | 57 (71%) | 0.012 |
| TMB (nonsynonymous) | 1.87 (2.14) | 7.15 (45.51) | 0.2 |
| Factor | OS Univariable | OS Multivariable |
|---|---|---|
| HR (95% CI, p-value) | HR (95% CI, p-value) | |
| SERPINB11 | 0.92 (0.18–4.70, p = 0.920) | 0.75 (0.13–4.19, p = 0.741) |
| SERPINB12 | 0.86 (0.41–1.81, p = 0.690) | 0.93 (0.43–2.01, p = 0.852) |
| SERPINB3 | 1.13 (0.60–2.10, p = 0.705) | 1.10 (0.57–2.11, p = 0.776) |
| SERPINB5 | 1.05 (0.69–1.59, p = 0.817) | 1.02 (0.67–1.56, p = 0.925) |
| SERPINB6 | 1.23 (0.91–1.67, p = 0.172) | 1.22 (0.89–1.66, p = 0.212) |
| SERPINB9 | 1.07 (0.85–1.35, p = 0.571) | 1.04 (0.67–1.62, p = 0.854) |
| Risk Score | 1.11 (0.88–1.40, p = 0.384) | NA (NA-NA, p = NA) |
| Risk Group | ||
| High | Reference | Reference |
| Low | 0.91 (0.64–1.30, p = 0.607) | 0.98 (0.51–1.89, p = 0.951) |
| Factor | PFS Univariable | PFS Multivariable |
| HR (95% CI, p-value) | HR (95% CI, p-value) | |
| SERPINB11 | 1.61 (0.40–6.48, p = 0.505) | 1.30 (0.29–5.79, p = 0.728) |
| SERPINB12 | 0.48 (0.16–1.46, p = 0.196) | 0.49 (0.16–1.57, p = 0.232) |
| SERPINB3 | 1.24 (0.70–2.17, p = 0.461) | 1.03 (0.56–1.90, p = 0.925) |
| SERPINB5 | 1.67 (1.15–2.43, p = 0.007) | 1.62 (1.12–2.35, p = 0.010) |
| SERPINB6 | 1.44 (1.06–1.96, p = 0.021) | 1.30 (0.94–1.79, p = 0.107) |
| SERPINB9 | 1.19 (0.94–1.52, p = 0.149) | 0.94 (0.61–1.46, p = 0.789) |
| Risk Score | 1.27 (1.00–1.61, p = 0.052) | NA (NA-NA, p = NA) |
| Risk Group | ||
| High | Reference | Reference |
| Low | 0.72 (0.51–1.03, p = 0.073) | 0.72 (0.38–1.37, p = 0.311) |
| Cells | High, n = 80 | Low, n = 80 | p-Value |
|---|---|---|---|
| B cells naive | 0.006 (0.012) | 0.004 (0.007) | 0.9 |
| B cells memory | 0.012 (0.018) | 0.013 (0.017) | 0.8 |
| Plasma cells | 0.001 (0.003) | 0.002 (0.007) | 0.6 |
| T cells CD8 | 0.04 (0.03) | 0.05 (0.04) | 0.1 |
| T cells CD4 naive | 0.0000 (0.0002) | 0.0020 (0.0110) | 0.2 |
| T cells CD4 memory resting | 0.08 (0.05) | 0.08 (0.06) | >0.9 |
| T cells CD4 memory activated | 0.0020 (0.0086) | 0.0001 (0.0007) | 0.061 |
| T cells follicular helper | 0.023 (0.019) | 0.034 (0.035) | 0.11 |
| T cells regulatory Tregs | 0.009 (0.012) | 0.008 (0.011) | 0.4 |
| T cells gamma delta | 0.002 (0.009) | 0.005 (0.015) | 0.2 |
| NK cells resting | 0.04 (0.04) | 0.04 (0.05) | 0.5 |
| NK cells activated | 0.017 (0.021) | 0.021 (0.025) | 0.3 |
| Monocytes | 0.10 (0.06) | 0.07 (0.06) | <0.001 |
| Macrophages M0 | 0.03 (0.07) | 0.06 (0.11) | 0.15 |
| Macrophages M1 | 0.015 (0.019) | 0.011 (0.016) | 0.043 |
| Macrophages M2 | 0.52 (0.11) | 0.48 (0.12) | 0.021 |
| Dendritic cells resting | 0.0010 (0.0036) | 0.0001 (0.0006) | 0.082 |
| Dendritic cells activated | 0.0013 (0.0024) | 0.0018 (0.0042) | >0.9 |
| Mast cells resting | 0.02 (0.04) | 0.06 (0.07) | <0.001 |
| Mast cells activated | 0.04 (0.06) | 0.02 (0.04) | 0.006 |
| Eosinophils | 0.003 (0.010) | 0.004 (0.012) | 0.5 |
| Neutrophils | 0.028 (0.020) | 0.023 (0.019) | 0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Khatib, S.M.; Al-Bzour, A.N.; Al-Majali, M.N.; Sa’d, L.M.; Alramadneh, J.A.; Othman, N.R.; Al-Mistarehi, A.-H.; Alomari, S. Exploring Genetic Determinants: A Comprehensive Analysis of Serpin B Family SNPs and Prognosis in Glioblastoma Multiforme Patients. Cancers 2024, 16, 1112. https://doi.org/10.3390/cancers16061112
Al-Khatib SM, Al-Bzour AN, Al-Majali MN, Sa’d LM, Alramadneh JA, Othman NR, Al-Mistarehi A-H, Alomari S. Exploring Genetic Determinants: A Comprehensive Analysis of Serpin B Family SNPs and Prognosis in Glioblastoma Multiforme Patients. Cancers. 2024; 16(6):1112. https://doi.org/10.3390/cancers16061112
Chicago/Turabian StyleAl-Khatib, Sohaib M., Ayah N. Al-Bzour, Mohammad N. Al-Majali, Laila M. Sa’d, Joud A. Alramadneh, Nour R. Othman, Abdel-Hameed Al-Mistarehi, and Safwan Alomari. 2024. "Exploring Genetic Determinants: A Comprehensive Analysis of Serpin B Family SNPs and Prognosis in Glioblastoma Multiforme Patients" Cancers 16, no. 6: 1112. https://doi.org/10.3390/cancers16061112
APA StyleAl-Khatib, S. M., Al-Bzour, A. N., Al-Majali, M. N., Sa’d, L. M., Alramadneh, J. A., Othman, N. R., Al-Mistarehi, A.-H., & Alomari, S. (2024). Exploring Genetic Determinants: A Comprehensive Analysis of Serpin B Family SNPs and Prognosis in Glioblastoma Multiforme Patients. Cancers, 16(6), 1112. https://doi.org/10.3390/cancers16061112






