Machine Learning-Based Prediction of Glioma IDH Gene Mutation Status Using Physio-Metabolic MRI of Oxygen Metabolism and Neovascularization (A Bicenter Study)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. MRI Data Acquisition
2.2.1. The MRI Study Protocol at the University Clinic St. Pölten
2.2.2. The MRI Study Protocol at the FAU Erlangen-Nürnberg
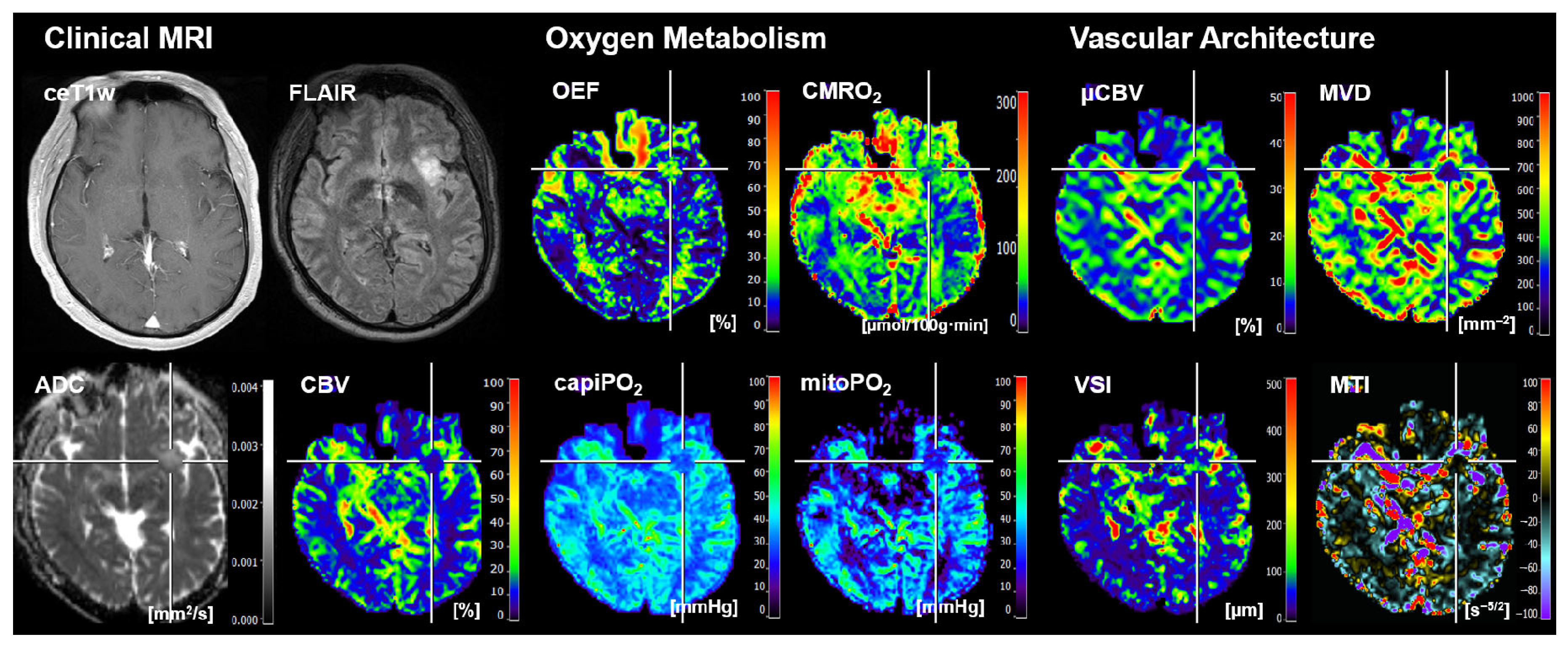
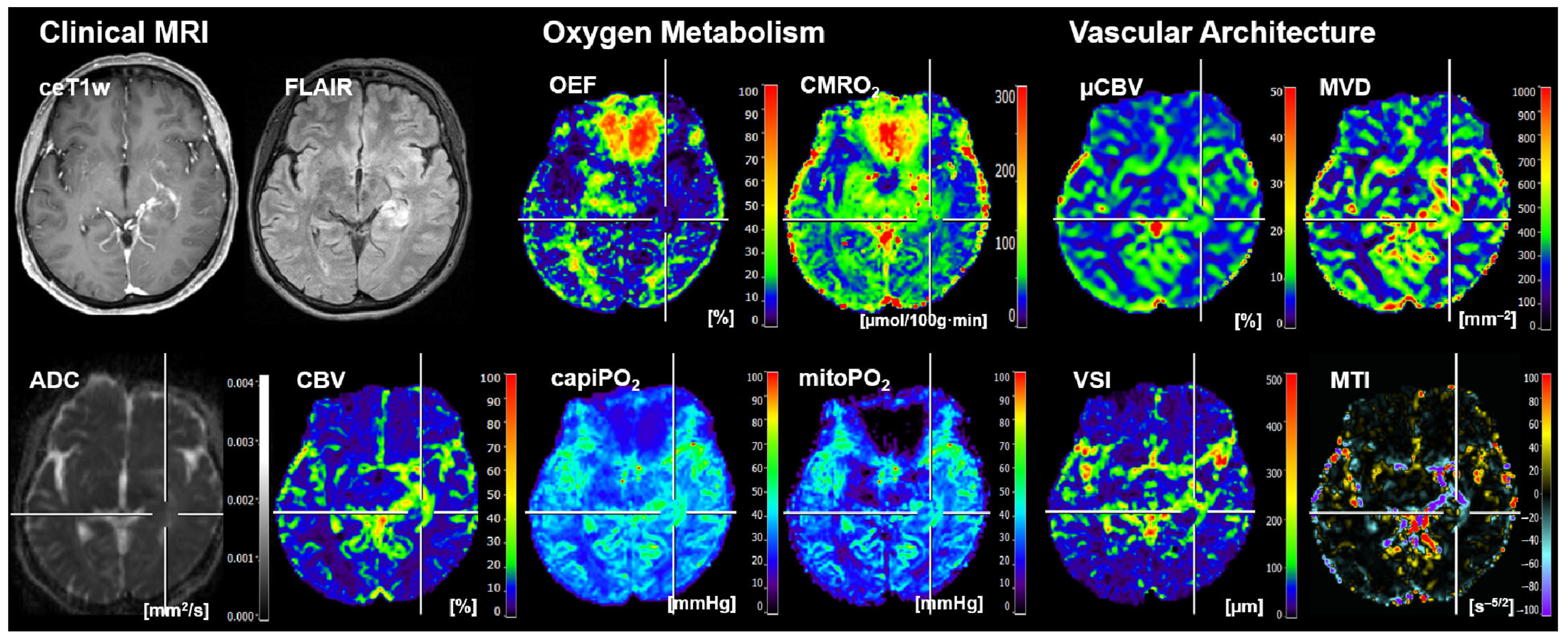
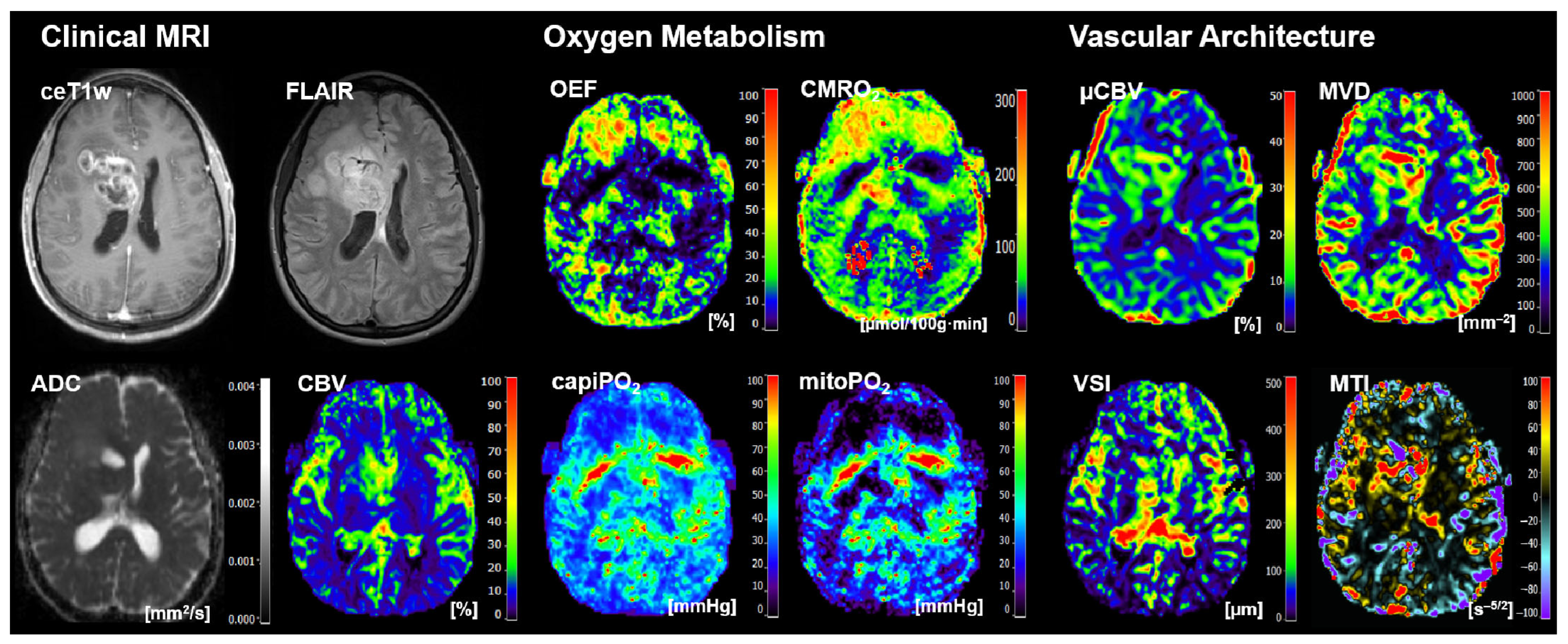
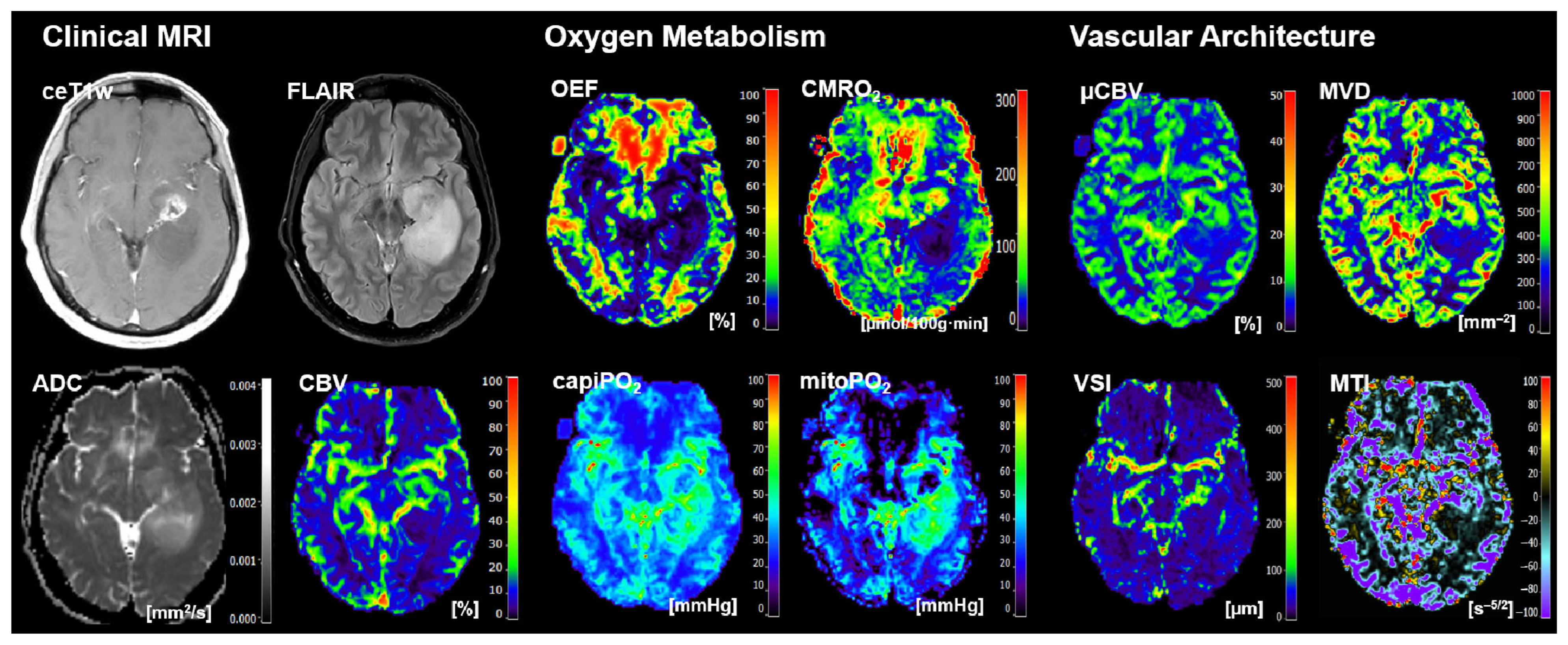
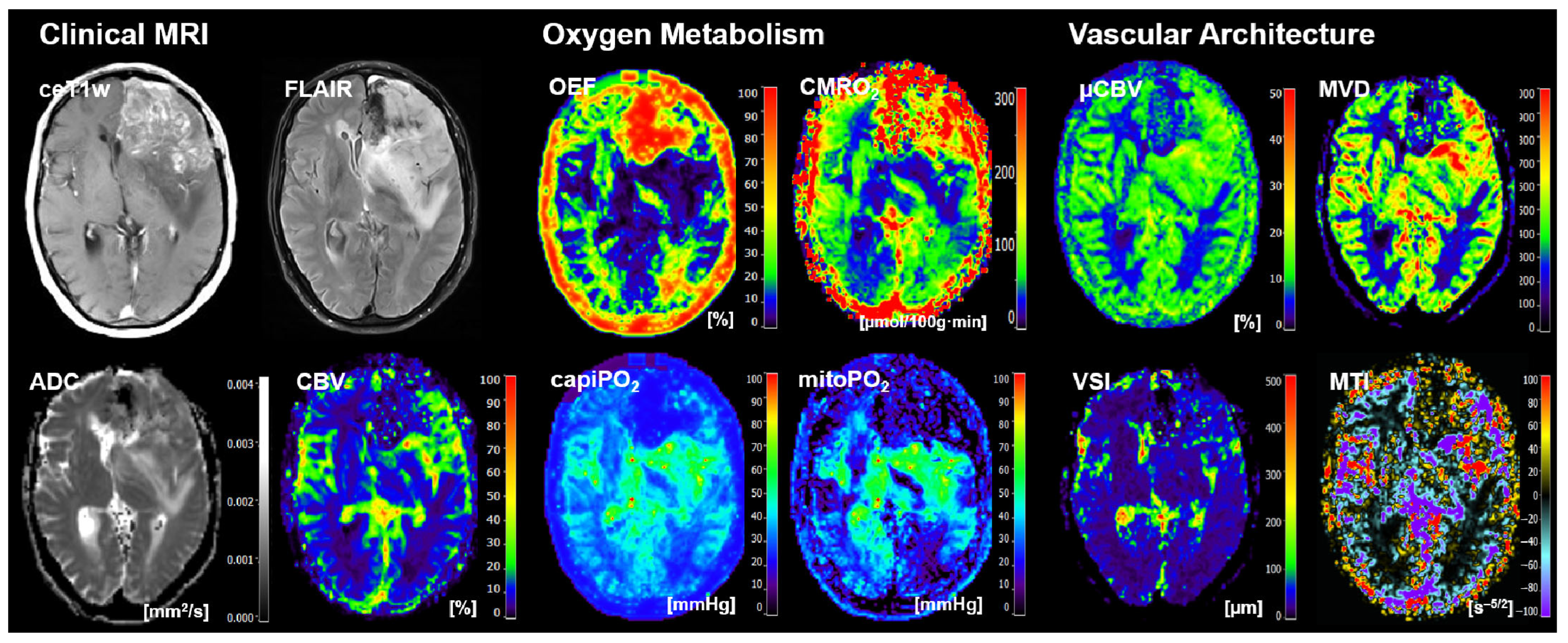
2.3. MRI Data Processing and Calculation of MRI Biomarker Maps
- Four cMRI data sets: FLAIR and ceT1w MRI data as well as the ADC maps for microstructural density and the CBV maps for macrovascular perfusion.
- Four biomarker maps for oxygen metabolism: MRI-based tissue oxygen metabolism (OEF and CMRO2) as well as MRI-based capillary oxygen tension (capiPO2) and mitochondrial (tissue) oxygen tension (mitoPO2).
- Four biomarker maps for microvascular architecture and neovascularization activity: microvascular density (MVD), microvascular diameter (VSI), microvascular perfusion (µCBV), and microvascular type (MTI).
2.4. Radiomic Feature Extraction
2.5. Traditional Machine Learning
- A multilayer perceptron (MLP) with one hidden layer and number of neurons = number of features + number of classes;
- Adaptive boosting (ABoost) using decision tree “J48” as classifier; and
- Random forest (RF).
- cMRI (ceT1w, FLAIR, ADC, CBV);
- MRI-based oxygen metabolism (CMRO2, OEF, capiPO2, mitoPO2);
- MRI-based vascular architecture (µCBV, MVD, MTI, VSI); and
- The combination of MRI-based oxygen metabolism and vascular architecture.
2.6. Deep Learning
2.7. Model Performance Testing
3. Results
3.1. The Selected Radiomic Features
3.2. Testing with an Independent Internal Cohort
3.3. Testing with an Independent External Cohort
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Molinaro, A.M.; Taylor, J.W.; Wiencke, J.K.; Wrensch, M.R. Genetic and molecular epidemiology of adult diffuse glioma. Nat. Rev. Neurol. 2019, 15, 405–417. [Google Scholar] [CrossRef]
- Vigneswaran, K.; Neill, S.; Hadjipanayis, C.G. Beyond the World Health Organization grading of infiltrating gliomas: Advances in the molecular genetics of glioma classification. Ann. Transl. Med. 2015, 3, 95. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.-M.; Gallia, G.L.; et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-R.; Yao, Y.; Xu, H.-Z.; Qin, Z.-Y. Isocitrate Dehydrogenase (IDH)1/2 Mutations as Prognostic Markers in Patients with Glioblastomas. Medicine 2016, 95, e2583. [Google Scholar] [CrossRef]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009, 462, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Tateishi, K.; Wakimoto, H.; Iafrate, A.J.; Tanaka, S.; Loebel, F.; Lelic, N.; Wiederschain, D.; Bedel, O.; Deng, G.; Zhang, B.; et al. Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell 2015, 28, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Reitman, Z.J.; Duncan, C.G.; Poteet, E.; Winters, A.; Yan, L.-J.; Gooden, D.M.; Spasojevic, I.; Boros, L.G.; Yang, S.-H.; Yan, H. Cancer-associated isocitrate dehydrogenase 1 (IDH1) R132H mutation and d-2-hydroxyglutarate stimulate glutamine metabolism under hypoxia. J. Biol. Chem. 2014, 289, 23318–23328. [Google Scholar] [CrossRef]
- Grassian, A.R.; Parker, S.J.; Davidson, S.M.; Divakaruni, A.S.; Green, C.R.; Zhang, X.; Slocum, K.L.; Pu, M.; Lin, F.; Vickers, C.; et al. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res. 2014, 74, 3317–3331. [Google Scholar] [CrossRef]
- Yao, J.; Chakhoyan, A.; Nathanson, D.A.; Yong, W.H.; Salamon, N.; Raymond, C.; Mareninov, S.; Lai, A.; Nghiemphu, P.L.; Prins, R.M.; et al. Metabolic characterization of human IDH mutant and wild type gliomas using simultaneous pH- and oxygen-sensitive molecular MRI. Neuro. Oncol. 2019, 21, 1184–1196. [Google Scholar] [CrossRef]
- Wu, B.; Warnock, G.; Zaiss, M.; Lin, C.; Chen, M.; Zhou, Z.; Mu, L.; Nanz, D.; Tuura, R.; Delso, G. An overview of CEST MRI for non-MR physicists. EJNMMI Phys. 2016, 3, 19. [Google Scholar] [CrossRef]
- Hardee, M.E.; Zagzag, D. Mechanisms of Glioma-Associated Neovascularization. Am. J. Pathol. 2012, 181, 1126–1141. [Google Scholar] [CrossRef]
- Stadlbauer, A.; Zimmermann, M.; Bennani-Baiti, B.; Helbich, T.H.; Baltzer, P.; Clauser, P.; Kapetas, P.; Bago-Horvath, Z.; Pinker, K. Development of a Non-invasive Assessment of Hypoxia and Neovascularization with Magnetic Resonance Imaging in Benign and Malignant Breast Tumors: Initial Results. Mol. Imaging Biol. 2019, 21, 758–770. [Google Scholar] [CrossRef]
- Stadlbauer, A.; Zimmermann, M.; Doerfler, A.; Oberndorfer, S.; Buchfelder, M.; Coras, R.; Kitzwögerer, M.; Roessler, K. Intratumoral heterogeneity of oxygen metabolism and neovascularization uncovers 2 survival-relevant subgroups of IDH1 wild-type glioblastoma. Neuro Oncol. 2018, 20, 1536–1546. [Google Scholar] [CrossRef] [PubMed]
- Stadlbauer, A.; Kinfe, T.M.; Eyüpoglu, I.; Zimmermann, M.; Kitzwögerer, M.; Podar, K.; Buchfelder, M.; Heinz, G.; Oberndorfer, S.; Marhold, F. Tissue Hypoxia and Alterations in Microvascular Architecture Predict Glioblastoma Recurrence in Humans. Clin. Cancer Res. 2021, 27, 1641–1649. [Google Scholar] [CrossRef]
- Christen, T.; Schmiedeskamp, H.; Straka, M.; Bammer, R.; Zaharchuk, G. Measuring brain oxygenation in humans using a multiparametric quantitative blood oxygenation level dependent MRI approach. Magn. Reson. Med. 2012, 68, 905–911. [Google Scholar] [CrossRef]
- Stadlbauer, A.; Zimmermann, M.; Heinz, G.; Oberndorfer, S.; Doerfler, A.; Buchfelder, M.; Rössler, K. Magnetic resonance imaging biomarkers for clinical routine assessment of microvascular architecture in glioma. J. Cereb. Blood Flow Metab. 2017, 37, 632–643. [Google Scholar] [CrossRef]
- Boxerman, J.L.; Hamberg, L.M.; Rosen, B.R.; Weisskoff, R.M. Mr contrast due to intravascular magnetic susceptibility perturbations. Magn. Reson. Med. 1995, 34, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- SchmIDHuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Radenovic, F.; Tolias, G.; Chum, O. Fine-Tuning CNN Image Retrieval with No Human Annotation. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 41, 1655–1668. [Google Scholar] [CrossRef]
- Kong, W.; Dong, Z.Y.; Jia, Y.; Hill, D.J.; Xu, Y.; Zhang, Y. Short-Term Residential Load Forecasting Based on LSTM Recurrent Neural Network. IEEE Trans. Smart Grid 2019, 10, 841–851. [Google Scholar] [CrossRef]
- Choi, E.; Schuetz, A.; Stewart, W.F.; Sun, J. Using recurrent neural network models for early detection of heart failure onset. J. Am. Med. Inform. Assoc. 2017, 24, 361–370. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhao, K.; Xia, K.; Xue, J.; Zhou, L.; Ding, Y.; Qian, P. A Novel Distributed Multitask Fuzzy Clustering Algorithm for Automatic MR Brain Image Segmentation. J. Med. Syst. 2019, 43, 118. [Google Scholar] [CrossRef] [PubMed]
- Hanson, J.; Yang, Y.; Paliwal, K.; Zhou, Y. Improving protein disorder prediction by deep bidirectional long short-term memory recurrent neural networks. Bioinformatics 2017, 33, 685–692. [Google Scholar] [CrossRef]
- Jing, X.; Dorrius, M.D.; Wielema, M.; Sijens, P.E.; Oudkerk, M.; van Ooijen, P. Breast Tumor Identification in Ultrafast MRI Using Temporal and Spatial Information. Cancers 2022, 14, 2042. [Google Scholar] [CrossRef]
- Antropova, N.; Huynh, B.; Li, H.; Giger, M.L. Breast lesion classification based on dynamic contrast-enhanced magnetic resonance images sequences with long short-term memory networks. J. Med. Imaging 2019, 6, 011002. [Google Scholar] [CrossRef]
- Zou, J.; Balter, J.M.; Cao, Y. Estimation of pharmacokinetic parameters from DCE-MRI by extracting long and short time-dependent features using an LSTM network. Med. Phys. 2020, 47, 3447–3457. [Google Scholar] [CrossRef]
- Boxerman, J.L.; Schmainda, K.M.; Weisskoff, R.M. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am. J. Neuroradiol. 2006, 27, 859–867. [Google Scholar] [PubMed]
- Boxerman, J.L.; Prah, D.E.; Paulson, E.S.; Machan, J.T.; Bedekar, D.; Schmainda, K.M. The Role of Preload and Leakage Correction in Gadolinium-Based Cerebral Blood Volume Estimation Determined by Comparison with MION as a Criterion Standard. Am. J. Neuroradiol. 2012, 33, 1081–1087. [Google Scholar] [CrossRef]
- Smith, A.M.; Grandin, C.B.; Duprez, T.; Mataigne, F.; Cosnard, G. Whole brain quantitative CBF, CBV, and MTT measurements using MRI bolus tracking: Implementation and application to data acquired from hyperacute stroke patients. J. Magn. Reson. Imaging 2000, 12, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Bjørnerud, A.; Emblem, K.E. A Fully Automated Method for Quantitative Cerebral Hemodynamic Analysis Using DSC–MRI. J. Cereb. Blood Flow Metab. 2010, 30, 1066–1078. [Google Scholar] [CrossRef] [PubMed]
- Preibisch, C.; Volz, S.; Anti, S.; Deichmann, R. Exponential excitation pulses for improved water content mapping in the presence of background gradients. Magn. Reson. Med. 2008, 60, 908–916. [Google Scholar] [CrossRef]
- Prasloski, T.; Mädler, B.; Xiang, Q.-S.; MacKay, A.; Jones, C. Applications of stimulated echo correction to multicomponent T 2 analysis. Magn. Reson. Med. 2012, 67, 1803–1814. [Google Scholar] [CrossRef]
- Stadlbauer, A.; Zimmermann, M.; Kitzwögerer, M.; Oberndorfer, S.; Rössler, K.; Dörfler, A.; Buchfelder, M.; Heinz, G. MR Imaging–derived Oxygen Metabolism and Neovascularization Characterization for Grading and IDH Gene Mutation Detection of Gliomas. Radiology 2017, 283, 799–809. [Google Scholar] [CrossRef]
- Kennan, R.P.; Zhong, J.; Gore, J.C. Intravascular susceptibility contrast mechanisms in tissues. Magn. Reson. Med. 1994, 31, 9–21. [Google Scholar] [CrossRef]
- Vafaee, M.S.; Vang, K.; Bergersen, L.H.; Gjedde, A. Oxygen Consumption and Blood Flow Coupling in Human Motor Cortex during Intense Finger Tapping: Implication for a Role of Lactate. J. Cereb. Blood Flow Metab. 2012, 32, 1859–1868. [Google Scholar] [CrossRef]
- Gjedde, A. Cerebral Blood Flow Change in Arterial Hypoxemia Is Consistent with Negligible Oxygen Tension in Brain Mitochondria. Neuroimage 2002, 17, 1876–1881. [Google Scholar] [CrossRef]
- Vafaee, M.S.; Gjedde, A. Model of Blood–Brain Transfer of Oxygen Explains Nonlinear Flow-Metabolism Coupling During Stimulation of Visual Cortex. J. Cereb. Blood Flow Metab. 2000, 20, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Ducreux, D.; Buvat, I.; Meder, J.F.; Mikulis, D.; Crawley, A.; Fredy, D.; TerBrugge, K.; Lasjaunias, P.; Bittoun, J. Perfusion-weighted MR imaging studies in brain hypervascular diseases: Comparison of arterial input function extractions for perfusion measurement. AJNR Am. J. Neuroradiol. 2006, 27, 1059–1069. [Google Scholar]
- Xu, C.; Kiselev, V.G.; Möller, H.E.; Fiebach, J.B. Dynamic hysteresis between gradient echo and spin echo attenuations in dynamic susceptibility contrast imaging. Magn. Reson. Med. 2013, 69, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.H.; Lu, H.; Inglese, M. Microvessel density estimation in the human brain by means of dynamic contrast-enhanced echo-planar imaging. Magn. Reson. Med. 2006, 56, 1145–1150. [Google Scholar] [CrossRef]
- Emblem, K.E.; Mouridsen, K.; Bjornerud, A.; Farrar, C.T.; Jennings, D.; Borra, R.J.H.; Wen, P.Y.; Ivy, P.; Batchelor, T.T.; Rosen, B.R.; et al. Vessel architectural imaging identifies cancer patient responders to anti-angiogenic therapy. Nat. Med. 2013, 19, 1178–1183. [Google Scholar] [CrossRef]
- Stadlbauer, A.; Marhold, F.; Oberndorfer, S.; Heinz, G.; Buchfelder, M.; Kinfe, T.M.; Meyer-Bäse, A. Radiophysiomics: Brain Tumors Classification by Machine Learning and Physiological MRI Data. Cancers 2022, 14, 2363. [Google Scholar] [CrossRef]
- Carré, A.; Klausner, G.; Edjlali, M.; Lerousseau, M.; Briend-Diop, J.; Sun, R.; Ammari, S.; Reuzé, S.; Alvarez Andres, E.; Estienne, T.; et al. Standardization of brain MR images across machines and protocols: Bridging the gap for MRI-based radiomics. Sci. Rep. 2020, 10, 12340. [Google Scholar] [CrossRef]
- Collewet, G.; Strzelecki, M.; Mariette, F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn. Reson. Imaging 2004, 22, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Shafiq-Ul-Hassan, M.; Zhang, G.G.; Latifi, K.; Ullah, G.; Hunt, D.C.; Balagurunathan, Y.; Abdalah, M.A.; Schabath, M.B.; Goldgof, D.G.; Mackin, D.; et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med. Phys. 2017, 44, 1050–1062. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Zacharaki, E.I.; Wang, S.; Chawla, S.; Soo Yoo, D.; Wolf, R.; Melhem, E.R.; Davatzikos, C. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn. Reson. Med. 2009, 62, 1609–1618. [Google Scholar] [CrossRef] [PubMed]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- An, J.Y.; Seo, H.; Kim, Y.-G.; Lee, K.E.; Kim, S.; Kong, H.-J. Codeless Deep Learning of COVID-19 Chest X-Ray Image Dataset with KNIME Analytics Platform. Healthc. Inform. Res. 2021, 27, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Berthold, M.R.; Cebron, N.; Dill, F.; Gabriel, T.R.; Kötter, T.; Meinl, T.; Ohl, P.; Thiel, K.; Wiswedel, B. KNIME-the Konstanz information miner: Version 2.0 and beyond. ACM SIGKDD Explor. Newsl. 2009, 11, 26–31. [Google Scholar] [CrossRef]
- Weigert, M.; Schmidt, U.; Boothe, T.; Müller, A.; Dibrov, A.; Jain, A.; Wilhelm, B.; Schmidt, D.; Broaddus, C.; Culley, S.; et al. Content-aware image restoration: Pushing the limits of fluorescence microscopy. Nat. Methods 2018, 15, 1090–1097. [Google Scholar] [CrossRef] [PubMed]
- Kiranyaz, S.; Avci, O.; Abdeljaber, O.; Ince, T.; Gabbouj, M.; Inman, D.J. 1D convolutional neural networks and applications: A survey. Mech. Syst. Signal Process. 2021, 151, 107398. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Ng, W.; Minasny, B.; Montazerolghaem, M.; Padarian, J.; Ferguson, R.; Bailey, S.; McBratney, A.B. Convolutional neural network for simultaneous prediction of several soil properties using visible/near-infrared, mid-infrared, and their combined spectra. Geoderma 2019, 352, 251–267. [Google Scholar] [CrossRef]
- Hochreiter, S.; SchmIDHuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Bradley, A.P. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997, 30, 1145–1159. [Google Scholar] [CrossRef]
- Ozturk-Isik, E.; Cengiz, S.; Ozcan, A.; Yakicier, C.; Ersen Danyeli, A.; Pamir, M.N.; Özduman, K.; Dincer, A. Identification of IDH and TERTp mutation status using 1 H-MRS in 112 hemispheric diffuse gliomas. J. Magn. Reson. Imaging 2020, 51, 1799–1809. [Google Scholar] [CrossRef]
- Tatekawa, H.; Hagiwara, A.; Uetani, H.; Bahri, S.; Raymond, C.; Lai, A.; Cloughesy, T.F.; Nghiemphu, P.L.; Liau, L.M.; Pope, W.B.; et al. Differentiating IDH status in human gliomas using machine learning and multiparametric MR/PET. Cancer Imaging 2021, 21, 27. [Google Scholar] [CrossRef] [PubMed]
- Kesler, S.R.; Noll, K.; Cahill, D.P.; Rao, G.; Wefel, J.S. The effect of IDH1 mutation on the structural connectome in malignant astrocytoma. J. Neurooncol. 2017, 131, 565–574. [Google Scholar] [CrossRef]
- Kesler, S.R.; Harrison, R.A.; Petersen, M.L.; Rao, V.; Dyson, H.; Alfaro-Munoz, K.; Weathers, S.-P.; de Groot, J. Pre-surgical connectome features predict IDH status in diffuse gliomas. Oncotarget 2019, 10, 6484–6493. [Google Scholar] [CrossRef] [PubMed]
- Karami, G.; Pascuzzo, R.; Figini, M.; Del Gratta, C.; Zhang, H.; Bizzi, A. Combining Multi-Shell Diffusion with Conventional MRI Improves Molecular Diagnosis of Diffuse Gliomas with Deep Learning. Cancers 2023, 15, 482. [Google Scholar] [CrossRef]
- Choi, K.S.; Choi, S.H.; Jeong, B. Prediction of IDH genotype in gliomas with dynamic susceptibility contrast perfusion MR imaging using an explainable recurrent neural network. Neuro Oncol. 2019, 21, 1197–1209. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Scarpace, L.; Flanders, A.; Jain, R.; Mikkelsen, T.; Andrews, D. Data from REMBRANDT. Version 1. Cancer Imaging Arch. 2015. [Google Scholar] [CrossRef]
- National Cancer Institute Clinical Proteomic Tumor Analysis Consortium. (CPTAC) Radiology Data from the Clinical Proteomic Tumor Analysis Consortium Glioblastoma Multiforme CPTAC-GBM collection. Version 6. Cancer Imaging Arch. 2019. [Google Scholar] [CrossRef]
- Puchalski, R.B.; Shah, N.; Miller, J.; Dalley, R.; Nomura, S.R.; Yoon, J.-G.; Smith, K.A.; Lankerovich, M.; Bertagnolli, D.; Bickley, K.; et al. An anatomic transcriptional atlas of human glioblastoma. Science 2018, 360, 660–663. [Google Scholar] [CrossRef]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef] [PubMed]
- van der Voort, S.R.; Incekara, F.; Wijnenga, M.M.J.; Kapsas, G.; Gahrmann, R.; Schouten, J.W.; Dubbink, H.J.; Vincent, A.J.P.E.; van den Bent, M.J.; French, P.J.; et al. The Erasmus Glioma Database (EGD): Structural MRI scans, WHO 2016 subtypes, and segmentations of 774 patients with glioma. Data Br. 2021, 37, 107191. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, S.; LaMontagne, P.; Shimony, J.; Marcus, D.S.; Sotiras, A. MRI-based classification of IDH mutation and 1p/19q codeletion status of gliomas using a 2.5D hybrid multi-task convolutional neural network. Neuro-Oncol. Adv. 2023, 5, vdad023. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.S.; Bae, S.; Chang, J.H.; Kang, S.-G.; Kim, S.H.; Kim, J.; Rim, T.H.; Choi, S.H.; Jain, R.; Lee, S.-K. Fully automated hybrid approach to predict the IDH mutation status of gliomas via deep learning and radiomics. Neuro. Oncol. 2021, 23, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Bangalore Yogananda, C.G.; Wagner, B.C.; Truong, N.C.D.; Holcomb, J.M.; Reddy, D.D.; Saadat, N.; Hatanpaa, K.J.; Patel, T.R.; Fei, B.; Lee, M.D.; et al. MRI-Based Deep Learning Method for Classification of IDH Mutation Status. Bioengineering 2023, 10, 1045. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- van der Voort, S.R.; Incekara, F.; Wijnenga, M.M.J.; Kapsas, G.; Gahrmann, R.; Schouten, J.W.; Nandoe Tewarie, R.; Lycklama, G.J.; De Witt Hamer, P.C.; Eijgelaar, R.S.; et al. Combined molecular subtyping, grading, and segmentation of glioma using multi-task deep learning. Neuro Oncol. 2023, 25, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Liu, J.; Kuang, H.; Wang, J. A Fully Automated Multimodal MRI-Based Multi-Task Learning for Glioma Segmentation and IDH Genotyping. IEEE Trans. Med. Imaging 2022, 41, 1520–1532. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.; Bai, H.X.; Zhou, H.; Su, C.; Bi, W.L.; Agbodza, E.; Kavouridis, V.K.; Senders, J.T.; Boaro, A.; Beers, A.; et al. Residual Convolutional Neural Network for the Determination of IDH Status in Low- and High-Grade Gliomas from MR Imaging. Clin. Cancer Res. 2018, 24, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Nalawade, S.S.; Yu, F.F.; Bangalore Yogananda, C.G.; Murugesan, G.K.; Shah, B.R.; Pinho, M.C.; Wagner, B.C.; Xi, Y.; Mickey, B.; Patel, T.R.; et al. Brain tumor IDH, 1p/19q, and MGMT molecular classification using MRI-based deep learning: An initial study on the effect of motion and motion correction. J. Med. Imaging 2022, 9, 016001. [Google Scholar] [CrossRef] [PubMed]
- Manikis, G.C.; Ioannidis, G.S.; Siakallis, L.; Nikiforaki, K.; Iv, M.; Vozlic, D.; Surlan-Popovic, K.; Wintermark, M.; Bisdas, S.; Marias, K. Multicenter DSC-MRI-Based Radiomics Predict IDH Mutation in Gliomas. Cancers 2021, 13, 3965. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, G.S.; Pigott, L.E.; Iv, M.; Surlan-Popovic, K.; Wintermark, M.; Bisdas, S.; Marias, K. Investigating the value of radiomics stemming from DSC quantitative biomarkers in IDH mutation prediction in gliomas. Front. Neurol. 2023, 14, 1249452. [Google Scholar] [CrossRef] [PubMed]
- Wiggins, W.F.; Tejani, A.S. On the Opportunities and Risks of Foundation Models for Natural Language Processing in Radiology. Radiol. Artif. Intell. 2022, 4, e220119. [Google Scholar] [CrossRef] [PubMed]
- Moor, M.; Banerjee, O.; Abad, Z.S.H.; Krumholz, H.M.; Leskovec, J.; Topol, E.J.; Rajpurkar, P. Foundation models for generalist medical artificial intelligence. Nature 2023, 616, 259–265. [Google Scholar] [CrossRef]
- Krishnan, R.; Rajpurkar, P.; Topol, E.J. Self-supervised learning in medicine and healthcare. Nat. Biomed. Eng. 2022, 6, 1346–1352. [Google Scholar] [CrossRef]
- Zhou, Y.; Chia, M.A.; Wagner, S.K.; Ayhan, M.S.; Williamson, D.J.; Struyven, R.R.; Liu, T.; Xu, M.; Lozano, M.G.; Woodward-Court, P.; et al. A foundation model for generalizable disease detection from retinal images. Nature 2023, 622, 156–163. [Google Scholar] [CrossRef]
- Beebe-Wang, N.; Celik, S.; Weinberger, E.; Sturmfels, P.; De Jager, P.L.; Mostafavi, S.; Lee, S.-I. Unified AI framework to uncover deep interrelationships between gene expression and Alzheimer’s disease neuropathologies. Nat. Commun. 2021, 12, 5369. [Google Scholar] [CrossRef]
- Panizza, E. DeepOmicsAE: Representing Signaling Modules in Alzheimer’s Disease with Deep Learning Analysis of Proteomics, Metabolomics, and Clinical Data. J. Vis. Exp. 2023. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man. Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Sun, C.; Wee, W.G. Neighboring gray level dependence matrix for texture classification. Comput. Vision Graph. Image Process. 1983, 23, 341–352. [Google Scholar] [CrossRef]
- Galloway, M.M. Texture analysis using gray level run lengths. Comput. Graph. Image Process. 1975, 4, 172–179. [Google Scholar] [CrossRef]
- Thibault, G.; Fertil, B.; Navarro, C.; Pereira, S.; Cau, P.; Levy, N.; Sequeira, J.; Mari, J.-L. Shape and texture indexes application tocell nuclei classification. Int. J. Pattern Recognit. Artif. Intell. 2013, 27, 1357002. [Google Scholar] [CrossRef]
- Amadasun, M.; King, R. Textural features corresponding to textural properties. IEEE Trans. Syst. Man. Cybern. 1989, 19, 1264–1274. [Google Scholar] [CrossRef]
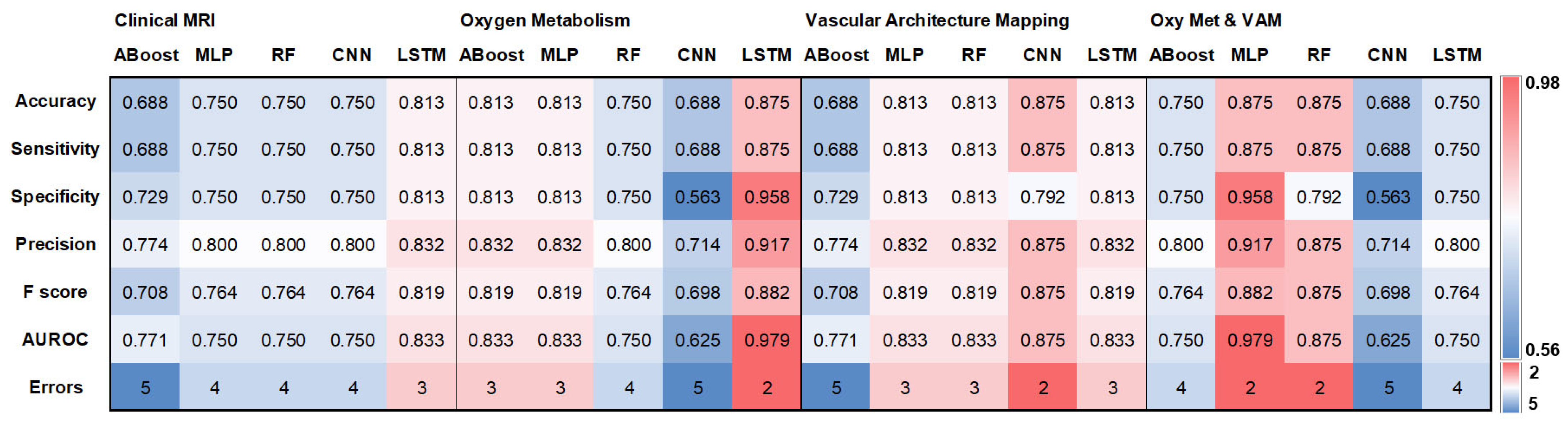

| ABoost | MLP | RF | CNN | LSTM | |
|---|---|---|---|---|---|
| Clinical MRI | 0.858 | 0.866 | 0.907 | 0.891 | 0.94 |
| Oxygen Metabolism | 0.87 | 0.907 | 0.902 | 0.855 | 0.88 |
| Vascular Architecture Mapping | 0.85 | 0.902 | 0.907 | 0.891 | 0.88 |
| Oxy Met & VAM | 0.829 | 0.902 | 0.898 | 0.873 | 0.86 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stadlbauer, A.; Nikolic, K.; Oberndorfer, S.; Marhold, F.; Kinfe, T.M.; Meyer-Bäse, A.; Bistrian, D.A.; Schnell, O.; Doerfler, A. Machine Learning-Based Prediction of Glioma IDH Gene Mutation Status Using Physio-Metabolic MRI of Oxygen Metabolism and Neovascularization (A Bicenter Study). Cancers 2024, 16, 1102. https://doi.org/10.3390/cancers16061102
Stadlbauer A, Nikolic K, Oberndorfer S, Marhold F, Kinfe TM, Meyer-Bäse A, Bistrian DA, Schnell O, Doerfler A. Machine Learning-Based Prediction of Glioma IDH Gene Mutation Status Using Physio-Metabolic MRI of Oxygen Metabolism and Neovascularization (A Bicenter Study). Cancers. 2024; 16(6):1102. https://doi.org/10.3390/cancers16061102
Chicago/Turabian StyleStadlbauer, Andreas, Katarina Nikolic, Stefan Oberndorfer, Franz Marhold, Thomas M. Kinfe, Anke Meyer-Bäse, Diana Alina Bistrian, Oliver Schnell, and Arnd Doerfler. 2024. "Machine Learning-Based Prediction of Glioma IDH Gene Mutation Status Using Physio-Metabolic MRI of Oxygen Metabolism and Neovascularization (A Bicenter Study)" Cancers 16, no. 6: 1102. https://doi.org/10.3390/cancers16061102
APA StyleStadlbauer, A., Nikolic, K., Oberndorfer, S., Marhold, F., Kinfe, T. M., Meyer-Bäse, A., Bistrian, D. A., Schnell, O., & Doerfler, A. (2024). Machine Learning-Based Prediction of Glioma IDH Gene Mutation Status Using Physio-Metabolic MRI of Oxygen Metabolism and Neovascularization (A Bicenter Study). Cancers, 16(6), 1102. https://doi.org/10.3390/cancers16061102








