Exosomes—Promising Carriers for Regulatory Therapy in Oncology
Abstract
Simple Summary
Abstract
1. Introduction
2. Exosomes—Variations and Biological Significance
3. Scientific Observations
3.1. Gene Expression and 5-Year Survival
3.2. Remodeling of the Tumor Microenvironment
EMT Regulators
3.3. Cancer-Associated Fibroblasts (CAFs)
3.4. Tumor-Associated Macrophages (TAMs)
3.5. Myeloid-Derived Suppressor Cells (MDSCs)
3.6. Mesenchymal Stem Cells (MSCs)
3.7. Cancer Stem Cells (CSCs)
3.8. Progression
3.8.1. Increase in Malignancy
3.8.2. EMT in Tumor Metastasis
3.9. Reprogramming of Metabolism
3.10. Impact on the Immune System
4. Exosome Technology: An Overview
4.1. EMT and Microenvironment
4.1.1. Cancer-Associated Fibroblasts (CAFs)
4.1.2. Tumor-Associated Macrophages (TAMs)
4.1.3. Bone Marrow Stem Cells (BMSCs)
4.2. Long Noncoding RNA (LncRNA)
4.3. Relationship with the Immune System
Tregs (T-Regulatory Cells)
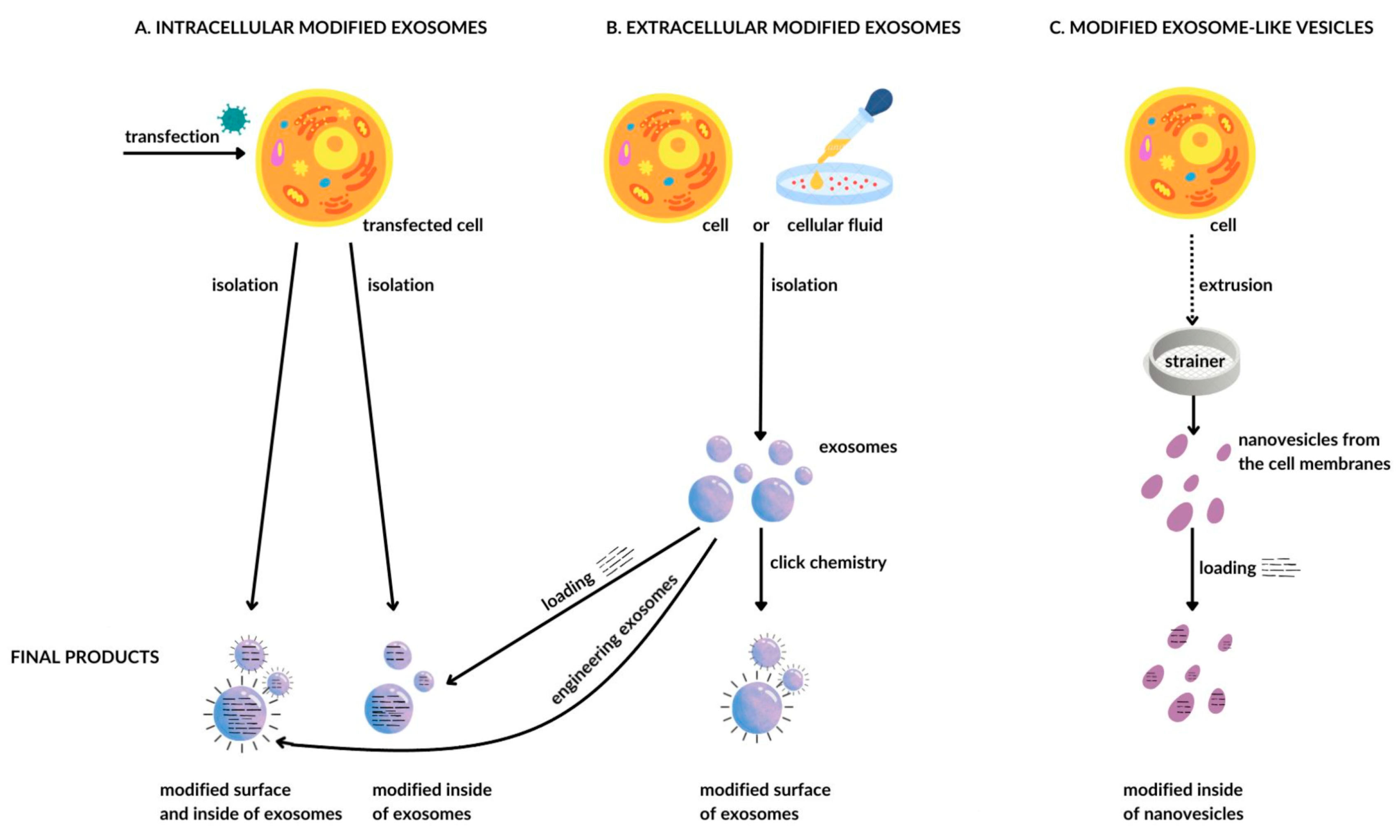
4.4. Technology of Obtaining and Modifying Exosomes
4.4.1. Exosomes Loaded with Chemotherapy Drugs
4.4.2. Exosomes for Delivery of Functional Proteins in Cancer Therapy
5. Discussion
6. Current Challenges and Future Outlook
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, Z.; Liu, X.; Chen, D.; Yu, J. Radiotherapy Combined with Immunotherapy: The Dawn of Cancer Treatment. Signal Transduct. Target. Ther. 2022, 7, 258. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, D. Exosomes in Cancer Development, Metastasis, and Immunity. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Maacha, S.; Bhat, A.A.; Jimenez, L.; Raza, A.; Haris, M.; Uddin, S.; Grivel, J.C. Extracellular Vesicles-Mediated Intercellular Communication: Roles in the Tumor Microenvironment and Anti-Cancer Drug Resistance. Mol. Cancer 2019, 18, 55. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular Mechanisms of Epithelial-Mesenchymal Transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Y.; Jastrzebska, B.; Golczak, M.; Gulati, S.; Tang, H.; Seibel, W.; Li, X.; Jin, H.; Han, Y.; et al. A Novel Small Molecule Chaperone of Rod Opsin and Its Potential Therapy for Retinal Degeneration. Nat. Commun. 2018, 9, 1976. [Google Scholar] [CrossRef] [PubMed]
- Vader, P.; Mol, E.A.; Pasterkamp, G.; Schiffelers, R.M. Extracellular Vesicles for Drug Delivery. Adv. Drug Deliv. Rev. 2016, 106, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Pascucci, B.; Spadaro, F.; Pietraforte, D.; De Nuccio, C.; Visentin, S.; Giglio, P.; Dogliotti, E.; D’errico, M. Drp1 Inhibition Rescues Mitochondrial Integrity and Excessive Apoptosis in Cs-a Disease Cell Models. Int. J. Mol. Sci. 2021, 22, 7123. [Google Scholar] [CrossRef]
- Wang, X.; Luo, G.; Zhang, K.; Cao, J.; Huang, C.; Jiang, T.; Liu, B.; Su, L.; Qiu, Z. Correction: Hypoxic Tumor-Derived Exosomal MiR-301a Mediates M2 Macrophage Polarization via PTEN/PI3Kγ to Promote Pancreatic Cancer Metastasis. Cancer Res. 2020, 80, 4586–4598. [Google Scholar] [CrossRef]
- Chang, W.H.; Cerione, R.A.; Antonyak, M.A. Extracellular Vesicles and Their Roles in Cancer Progression. Methods Mol. Biol. 2021, 2174, 143–170. [Google Scholar] [CrossRef]
- Nowak, M.; Górczyńska, J.; Kołodzińska, K.; Rubin, J.; Choromańska, A. Extracellular Vesicles as Drug Transporters. Int. J. Mol. Sci. 2023, 24, 267. [Google Scholar] [CrossRef]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef]
- Meng, W.; He, C.; Hao, Y.; Wang, L.; Li, L.; Zhu, G. Prospects and Challenges of Extracellular Vesicle-Based Drug Delivery System: Considering Cell Source. Drug Deliv. 2020, 27, 585–598. [Google Scholar] [CrossRef]
- Meldolesi, J. Extracellular Vesicles, News about Their Role in Immune Cells: Physiology, Pathology and Diseases. Clin. Exp. Immunol. 2019, 196, 318–327. [Google Scholar] [CrossRef]
- Popiołek, K.; Grzesiak, M. Exosomes as a New Approach into Cell-to-Cell Communication within the Mammalian Ovary. Postepy Biochem. 2019, 65, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Fernández, M.; de la Cuesta, F.; Tallón, A.; Cuesta, I.; Fernández-Fournier, M.; Laso-García, F.; Gómez-De Frutos, M.C.; Díez-Tejedor, E.; Otero-Ortega, L. Potential Roles of Extracellular Vesicles as Biomarkers and a Novel Treatment Approach in Multiple Sclerosis. Int. J. Mol. Sci. 2021, 22, 9011. [Google Scholar] [CrossRef] [PubMed]
- Isaac, R.; Reis, F.C.G.; Ying, W.; Olefsky, J.M. Exosomes as Mediators of Intercellular Crosstalk in Metabolism. Cell Metab. 2021, 33, 1744–1762. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering Exosomes for Targeted Drug Delivery. Theranostics 2021, 11, 3183–3195. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, Y.; Gong, H.; Luo, S.; Cui, Y. The Role of Exosomes and Their Applications in Cancer. Int. J. Mol. Sci. 2021, 22, 12204. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Gu, J.; Jiang, P.; Qian, H.; Xu, W.; Zhang, X. Exosomes in Gastric Cancer: Roles, Mechanisms, and Applications. Mol. Cancer 2019, 18, 41. [Google Scholar] [CrossRef] [PubMed]
- Gil, B.; Keshavarz, M.; Wales, D.; Darzi, A.; Yeatman, E. Orthogonal Surface-Enhanced Raman Scattering/Field-Effect Transistor Detection of Breast and Colorectal Cancer-Derived Exosomes Using Graphene as a Tag-Free Diagnostic Template. Adv. Nanobiomed Res. 2023, 3, 2300055. [Google Scholar] [CrossRef]
- Ding, Y.; Li, W.; Wang, K.; Xu, C.; Hao, M.; Ding, L. Perspectives of the Application of Liquid Biopsy in Colorectal Cancer. Biomed. Res. Int. 2020, 2020, 2300055. [Google Scholar] [CrossRef] [PubMed]
- Scavo, M.P.; Depalo, N.; Tutino, V.; De Nunzio, V.; Ingrosso, C.; Rizzi, F.; Notarnicola, M.; Curri, M.L.; Giannelli, G. Exosomes for Diagnosis and Therapy in Gastrointestinal Cancers. Int. J. Mol. Sci. 2020, 21, 367. [Google Scholar] [CrossRef] [PubMed]
- Khatami, S.H.; Karami, N.; Taheri-Anganeh, M.; Taghvimi, S.; Tondro, G.; Khorsand, M.; Soltani Fard, E.; Sedighimehr, N.; Kazemi, M.; Rahimi Jaberi, K.; et al. Exosomes: Promising Delivery Tools for Overcoming Blood-Brain Barrier and Glioblastoma Therapy. Mol. Neurobiol. 2023, 60, 4659–4678. [Google Scholar] [CrossRef]
- Lan, B.; Zeng, S.; Grützmann, R.; Pilarsky, C. The Role of Exosomes in Pancreatic Cancer. Int. J. Mol. Sci. 2019, 20, 4332. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A Pathology Atlas of the Human Cancer Transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Ye, Q.; Su, L.; Chen, D.; Zheng, W.; Liu, Y. Astragaloside IV Induced MIR-134 Expression Reduces EMT and Increases Chemotherapeutic Sensitivity by Suppressing CREB1 Signaling in Colorectal Cancer Cell Line SW-480. Cell. Physiol. Biochem. 2017, 43, 1617–1626. [Google Scholar] [CrossRef]
- Rezaei, R.; Baghaei, K.; Amani, D.; Piccin, A.; Hashemi, S.M.; Asadzadeh Aghdaei, H.; Zali, M.R. Exosome-Mediated Delivery of Functionally Active MiRNA-375-3p Mimic Regulate Epithelial Mesenchymal Transition (EMT) of Colon Cancer Cells. Life Sci. 2021, 269, 119035. [Google Scholar] [CrossRef]
- Hu, J.L.; Wang, W.; Lan, X.L.; Zeng, Z.C.; Liang, Y.S.; Yan, Y.R.; Song, F.Y.; Wang, F.F.; Zhu, X.H.; Liao, W.J.; et al. CAFs Secreted Exosomes Promote Metastasis and Chemotherapy Resistance by Enhancing Cell Stemness and Epithelial-Mesenchymal Transition in Colorectal Cancer. Mol. Cancer 2019, 18, 91. [Google Scholar] [CrossRef]
- Pakravan, K.; Babashah, S.; Sadeghizadeh, M.; Mowla, S.J.; Mossahebi-Mohammadi, M.; Ataei, F.; Dana, N.; Javan, M. MicroRNA-100 Shuttled by Mesenchymal Stem Cell-Derived Exosomes Suppresses in Vitro Angiogenesis through Modulating the MTOR/HIF-1α/VEGF Signaling Axis in Breast Cancer Cells. Cell. Oncol. 2017, 40, 457–470. [Google Scholar] [CrossRef]
- Chen, D.; Sun, Y.; Yuan, Y.; Han, Z.; Zhang, P.; Zhang, J.; You, M.J.; Teruya-Feldstein, J.; Wang, M.; Gupta, S.; et al. MiR-100 Induces Epithelial-Mesenchymal Transition but Suppresses Tumorigenesis, Migration and Invasion. PLoS Genet. 2014, 10, e1004177. [Google Scholar] [CrossRef]
- Kim, M.; Jang, K.; Miller, P.; Picon-Ruiz, M.; Yeasky, T.M.; El-Ashry, D.; Slingerland, J.M. VEGFA Links Self-Renewal and Metastasis by Inducing Sox2 to Repress MiR-452, Driving Slug. Oncogene 2017, 36, 5199–5211. [Google Scholar] [CrossRef]
- Chen, X.; Chen, R.X.; Wei, W.S.; Li, Y.H.; Feng, Z.H.; Tan, L.; Chen, J.W.; Yuan, G.J.; Chen, S.L.; Guo, S.J.; et al. PRMT5 Circular RNA Promotes Metastasis of Urothelial Carcinoma of the Bladder through Sponging MiR-30c to Induce Epithelial–Mesenchymal Transition. Clin. Cancer Res. 2018, 24, 6319–6330. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Mou, Z.; Xu, C.; Wu, S.; Dai, X.; Chen, X.; Ou, Y.; Chen, Y.; Yang, C.; Jiang, H. Autophagy-Associated Circular RNA Hsa_circ_0007813 Modulates Human Bladder Cancer Progression via Hsa-MiR-361-3p/IGF2R Regulation. Cell Death Dis. 2021, 12, 778. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Liu, B.; Jiang, H.; Li, Z.; Fan, C.; Zang, L. Bone Marrow Mesenchymal Stem Cells-Derived Exosomal MicroRNA-19b-1-5p Reduces Proliferation and Raises Apoptosis of Bladder Cancer Cells via Targeting ABL2. Genomics 2021, 113, 1338–1348. [Google Scholar] [CrossRef]
- Dong, P.; Xiong, Y.; Watari, H.; Hanley, S.J.B.; Konno, Y.; Ihira, K.; Yamada, T.; Kudo, M.; Yue, J.; Sakuragi, N. MiR-137 and MiR-34a Directly Target Snail and Inhibit EMT, Invasion and Sphere-Forming Ability of Ovarian Cancer Cells. J. Exp. Clin. Cancer Res. 2016, 35, 132. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Wan, Q.; Li, F.; Tang, C.E. MiR-363 Inhibits Cisplatin Chemoresistance of Epithelial Ovarian Cancer by Regulating Snail-Induced Epithelial-Mesenchymal Transition. BMB Rep. 2018, 51, 456–461. [Google Scholar] [CrossRef]
- Cai, J.; Gong, L.; Li, G.; Guo, J.; Yi, X.; Wang, Z. Exosomes in Ovarian Cancer Ascites Promote Epithelial–Mesenchymal Transition of Ovarian Cancer Cells by Delivery of MiR-6780b-5p. Cell Death Dis. 2021, 12, 210. [Google Scholar] [CrossRef]
- Zhou, X.; Men, X.; Zhao, R.; Han, J.; Fan, Z.; Wang, Y.; Lv, Y.; Zuo, J.; Zhao, L.; Sang, M.; et al. MiR-200c Inhibits TGF-β-Induced-EMT to Restore Trastuzumab Sensitivity by Targeting ZEB1 and ZEB2 in Gastric Cancer. Cancer Gene Ther. 2018, 25, 68–76. [Google Scholar] [CrossRef]
- Wang, L.; Yang, G.; Zhao, D.; Wang, J.; Bai, Y.; Peng, Q.; Wang, H.; Fang, R.; Chen, G.; Wang, Z.; et al. CD103-Positive CSC Exosome Promotes EMT of Clear Cell Renal Cell Carcinoma: Role of Remote MiR-19b-3p. Mol. Cancer 2019, 18, 86. [Google Scholar] [CrossRef]
- Yin, L.C.; Xiao, G.; Zhou, R.; Huang, X.P.; Li, N.L.; Tan, C.L.; Xie, F.J.; Weng, J.; Liu, L.X. MicroRNA-361-5p Inhibits Tumorigenesis and the EMT of HCC by Targeting Twist1. Biomed. Res. Int. 2020, 2020, 8891876. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Shi, Y.; Sun, L.; Liu, Z.; Song, T.; Liu, Q. Mir-4319 Induced an Inhibition of Epithelialmesenchymal Transition and Prevented Cancer Stemness of Hcc through Targeting Foxq1. Int. J. Biol. Sci. 2019, 15, 2936–2947. [Google Scholar] [CrossRef]
- Dong, F.; Lou, D. MicroRNA-34b/c Suppresses Uveal Melanoma Cell Proliferation and Migration through Multiple Targets. Mol. Vis. 2012, 18, 537–546. [Google Scholar] [PubMed]
- Debnath, P.; Huirem, R.S.; Dutta, P.; Palchaudhuri, S. Epithelial-Mesenchymal Transition and Its Transcription Factors. Biosci. Rep. 2022, 42, BSR20211754. [Google Scholar] [CrossRef]
- Elia, I.; Haigis, M.C. Metabolites and the Tumour Microenvironment: From Cellular Mechanisms to Systemic Metabolism. Nat. Metab. 2021, 3, 21–32. [Google Scholar] [CrossRef]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, Biogenesis, and Mechanisms in Cancer Metastasis and Drug Resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Guo, P.; He, Y.; Chen, Z.; Chen, L.; Luo, Y.; Qi, L.; Liu, Y.; Wu, Q.; Cui, Y.; et al. HCC-Derived Exosomes Elicit HCC Progression and Recurrence by Epithelial-Mesenchymal Transition through MAPK/ERK Signalling Pathway Article. Cell Death Dis. 2018, 9, 513. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.S.; Ho, J.Y.; Chiang, J.H.; Yu, C.P.; Yu, D.S. Exosome-Derived LINC00960 and LINC02470 Promote the Epithelial-Mesenchymal Transition and Aggressiveness of Bladder Cancer Cells. Cells 2020, 9, 1419. [Google Scholar] [CrossRef]
- Ning, X.; Zhang, H.; Wang, C.; Song, X. Exosomes Released by Gastric Cancer Cells Induce Transition of Pericytes into Cancer-Associated Fibroblasts. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 2350–2359. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Q.; Hosen, M.R.; Zietzer, A.; Flender, A.; Levermann, P.; Schmitz, T.; Frühwald, D.; Goody, P.; Nickenig, G.; et al. Atherosclerotic Conditions Promote the Packaging of Functional MicroRNA-92a-3p into Endothelial Microvesicles. Circ. Res. 2019, 124, 575–587. [Google Scholar] [CrossRef]
- Li, X.; Guo, S.; Min, L.; Guo, Q.; Zhang, S. MiR-92a-3p Promotes the Proliferation, Migration and Invasion of Esophageal Squamous Cell Cancer by Regulating PTEN. Int. J. Mol. Med. 2019, 44, 973–981. [Google Scholar] [CrossRef]
- Goulet, C.R.; Champagne, A.; Bernard, G.; Vandal, D.; Chabaud, S.; Pouliot, F.; Bolduc, S. Cancer-Associated Fibroblasts Induce Epithelial-Mesenchymal Transition of Bladder Cancer Cells through Paracrine IL-6 Signalling. BMC Cancer 2019, 19, 137. [Google Scholar] [CrossRef]
- Shin, N.; Mo Son, G.; Shin, D.H.; Kwon, M.S.; Park, B.S.; Kim, H.S.; Ryu, D.; Kang, C.D. Cancer-Associated Fibroblasts and Desmoplastic Reactions Related to Cancer Invasiveness in Patients with Colorectal Cancer. Ann. Coloproctol. 2019, 35, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Liu, T.; Chen, J.; Ni, H.; Li, W. Survivin in Breast Cancer-Derived Exosomes Activates Fibroblasts by up-Regulating SOD1, Whose Feedback Promotes Cancer Proliferation and Metastasis. J. Biol. Chem. 2020, 295, 13737–13752. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.; Zhou, L.; Sui, H.; Yang, L.; Wu, X.; Song, Q.; Jia, R.; Li, R.; Sun, J.; Wang, Z.; et al. Primary Tumors Release ITGBL1-Rich Extracellular Vesicles to Promote Distal Metastatic Tumor Growth through Fibroblast-Niche Formation. Nat. Commun. 2020, 11, 1211. [Google Scholar] [CrossRef]
- Wei, C.; Yang, C.; Wang, S.; Shi, D.; Zhang, C.; Lin, X.; Liu, Q.; Dou, R.; Xiong, B. Crosstalk between Cancer Cells and Tumor Associated Macrophages Is Required for Mesenchymal Circulating Tumor Cell-Mediated Colorectal Cancer Metastasis. Mol. Cancer 2019, 18, 64. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Yin, H.-B.; Li, X.Y.; Zhu, G.M.; He, W.Y.; Gou, X. Bladder Cancer Cell-Secreted Exosomal MiR-21 Activates the PI3K/AKT Pathway in Macrophages to Promote Cancer Progression. Int. J. Oncol. 2020, 56, 151–164. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Bronte, V. Coordinated Regulation of Myeloid Cells by Tumours. Nat. Rev. Immunol. 2012, 12, 253–268. [Google Scholar] [CrossRef]
- Kumar, V.; Cheng, P.; Condamine, T.; Mony, S.; Languino, L.R.; McCaffrey, J.C.; Hockstein, N.; Guarino, M.; Masters, G.; Penman, E.; et al. CD45 Phosphatase Inhibits STAT3 Transcription Factor Activity in Myeloid Cells and Promotes Tumor-Associated Macrophage Differentiation. Immunity 2016, 44, 303–315. [Google Scholar] [CrossRef]
- Bunt, S.K.; Sinha, P.; Clements, V.K.; Leips, J.; Ostrand-Rosenberg, S. Inflammation Induces Myeloid-Derived Suppressor Cells That Facilitate Tumor Progression. J. Immunol. 2006, 176, 284–290. [Google Scholar] [CrossRef]
- Gallina, G.; Dolcetti, L.; Serafini, P.; De Santo, C.; Marigo, I.; Colombo, M.P.; Basso, G.; Brombacher, F.; Borrello, I.; Zanovello, P.; et al. Tumors Induce a Subset of Inflammatory Monocytes with Immunosuppressive Activity on CD8+ T Cells. J. Clin. Investig. 2006, 116, 2777–2790. [Google Scholar] [CrossRef]
- Biswas, S.; Mandal, G.; Roy Chowdhury, S.; Purohit, S.; Payne, K.K.; Anadon, C.; Gupta, A.; Swanson, P.; Yu, X.; Conejo-Garcia, J.R.; et al. Exosomes Produced by Mesenchymal Stem Cells Drive Differentiation of Myeloid Cells into Immunosuppressive M2-Polarized Macrophages in Breast Cancer. J. Immunol. 2019, 203, 3447–3460. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhang, Y.; Zhang, Y.; Jia, Z.; Zhang, Z.; Yang, J. Cancer Derived Exosomes Induce Macrophages Immunosuppressive Polarization to Promote Bladder Cancer Progression. Cell Commun. Signal. 2021, 19, 93. [Google Scholar] [CrossRef]
- Li, W.; Zhang, X.; Wu, F.; Zhou, Y.; Bao, Z.; Li, H.; Zheng, P.; Zhao, S. Gastric Cancer-Derived Mesenchymal Stromal Cells Trigger M2 Macrophage Polarization That Promotes Metastasis and EMT in Gastric Cancer. Cell Death Dis. 2019, 10, 918. [Google Scholar] [CrossRef]
- Wei, X.; Yang, S.; Pu, X.; He, S.; Yang, Z.; Sheng, X.; Meng, X.; Chen, X.; Jin, L.; Chen, W.; et al. Tumor-Associated Macrophages Increase the Proportion of Cancer Stem Cells in Lymphoma by Secreting Pleiotrophin. Am. J. Transl. Res. 2019, 11, 6393–6402. [Google Scholar]
- Liguori, M.; Digifico, E.; Vacchini, A.; Avigni, R.; Colombo, F.S.; Borroni, E.M.; Farina, F.M.; Milanesi, S.; Castagna, A.; Mannarino, L.; et al. The Soluble Glycoprotein NMB (GPNMB) Produced by Macrophages Induces Cancer Stemness and Metastasis via CD44 and IL-33. Cell Mol. Immunol. 2021, 18, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Neal, M.L.; Boyle, A.M.; Budge, K.M.; Safadi, F.F.; Richardson, J.R. The Glycoprotein GPNMB Attenuates Astrocyte Inflammatory Responses through the CD44 Receptor. J. Neuroinflammation 2018, 15, 73. [Google Scholar] [CrossRef]
- Liu, J.; Ren, L.; Li, S.; Li, W.; Zheng, X.; Yang, Y.; Fu, W.; Yi, J.; Wang, J.; Du, G. The Biology, Function, and Applications of Exosomes in Cancer. Acta Pharm. Sin. B 2021, 11, 2783–2797. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, Y.; Zhang, Y.; Zhang, S.; Qiu, L.; Zhuang, Z.; Wei, M.; Deng, X.; Wang, Z.; Han, J. The Key Role of Exosomes on the Pre-Metastatic Niche Formation in Tumors. Front. Mol. Biosci. 2021, 8, 703640. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Zhou, X.; Fang, M.; Li, H.; Su, P.; Tu, Y.; Zhang, L.; Zhou, F. Extracellular Vesicles in Cancer Immune Microenvironment and Cancer Immunotherapy. Adv. Sci. 2019, 6, 1901779. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, D.W.; Cho, J.Y. Exploring the Key Communicator Role of Exosomes in Cancer Microenvironment through Proteomics. Proteome Sci. 2019, 17, 5. [Google Scholar] [CrossRef]
- Han, L.; Lam, E.W.F.; Sun, Y. Extracellular Vesicles in the Tumor Microenvironment: Old Stories, but New Tales. Mol. Cancer 2019, 18, 59. [Google Scholar] [CrossRef]
- Takenaga, K.; Koshikawa, N.; Nagase, H. Intercellular Transfer of Mitochondrial DNA Carrying Metastasis-Enhancing Pathogenic Mutations from High- to Low-Metastatic Tumor Cells and Stromal Cells via Extracellular Vesicles. BMC Mol. Cell Biol. 2021, 22, 52. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Peng, L.; Yang, J.; Sang, H.; Jin, D.; Li, X.; Chen, M.; Zhang, W.; Dang, Y.; Zhang, G. Exosomal Transfer of MiR-15b-3p Enhances Tumorigenesis and Malignant Transformation through the DYNLT1/Caspase-3/Caspase-9 Signaling Pathway in Gastric Cancer. J. Exp. Clin. Cancer Res. 2020, 39, 32. [Google Scholar] [CrossRef]
- Babaei, G.; Aziz, S.G.G.; Jaghi, N.Z.Z. EMT, Cancer Stem Cells and Autophagy; The Three Main Axes of Metastasis. Biomed. Pharmacother. 2021, 133, 110909. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhan, H. Communication between EMT and PD-L1 Signaling: New Insights into Tumor Immune Evasion. Cancer Lett. 2020, 468, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Xiao, P.; Long, X.; Zhang, L.; Ye, Y.; Guo, J.; Liu, P.; Zhang, R.; Ning, J.; Yu, W.; Wei, F.; et al. Neurotensin/IL-8 Pathway Orchestrates Local Inflammatory Response and Tumor Invasion by Inducing M2 Polarization of Tumor-Associated Macrophages and Epithelial-Mesenchymal Transition of Hepatocellular Carcinoma Cells. Oncoimmunology 2018, 7, e1440166. [Google Scholar] [CrossRef] [PubMed]
- Jafari, N.; Kolla, M.; Meshulam, T.; Shafran, J.S.; Qiu, Y.; Casey, A.N.; Pompa, I.R.; Ennis, C.S.; Mazzeo, C.S.; Rabhi, N.; et al. Adipocyte-Derived Exosomes May Promote Breast Cancer Progression in Type 2 Diabetes. Sci. Signal 2021, 14, eabj2807. [Google Scholar] [CrossRef] [PubMed]
- Gunaydin, G. CAFs Interacting With TAMs in Tumor Microenvironment to Enhance Tumorigenesis and Immune Evasion. Front. Oncol. 2021, 11, 668349. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Yang, J.; Liu, J.; Wang, Y.; Mu, J.; Zeng, Q.; Deng, S.; Zhou, H. Signaling Pathways in Cancer-Associated Fibroblasts and Targeted Therapy for Cancer. Signal Transduct. Target. Ther. 2021, 6, 218. [Google Scholar] [CrossRef]
- Ren, Y.; Cao, L.; Wang, L.; Zheng, S.; Zhang, Q.; Guo, X.; Li, X.; Chen, M.; Wu, X.; Furlong, F.; et al. Autophagic Secretion of HMGB1 from Cancer-Associated Fibroblasts Promotes Metastatic Potential of Non-Small Cell Lung Cancer Cells via NFκB Signaling. Cell Death Dis. 2021, 12, 858. [Google Scholar] [CrossRef]
- Liu, S.C.; Cao, Y.H.; Chen, L.B.; Kang, R.; Huang, Z.X.; Lu, X.S. BMSC-Derived Exosomal LncRNA PTENP1 Suppresses the Malignant Phenotypes of Bladder Cancer by Upregulating SCARA5 Expression. Cancer Biol. Ther. 2022, 23, 1–13. [Google Scholar] [CrossRef]
- Wang, S.; Su, X.; Xu, M.; Xiao, X.; Li, X.; Li, H.; Keating, A.; Zhao, R.C. Exosomes Secreted by Mesenchymal Stromal/Stem Cell-Derived Adipocytes Promote Breast Cancer Cell Growth via Activation of Hippo Signaling Pathway. Stem Cell Res. Ther. 2019, 10, 117. [Google Scholar] [CrossRef]
- Yang, E.; Wang, X.; Gong, Z.; Yu, M.; Wu, H.; Zhang, D. Exosome-Mediated Metabolic Reprogramming: The Emerging Role in Tumor Microenvironment Remodeling and Its Influence on Cancer Progression. Signal Transduct. Target. Ther. 2020, 5, 242. [Google Scholar] [CrossRef]
- Wang, B.; Wang, X.; Hou, D.; Huang, Q.; Zhan, W.; Chen, C.; Liu, J.; You, R.; Xie, J.; Chen, P.; et al. Exosomes Derived from Acute Myeloid Leukemia Cells Promote Chemoresistance by Enhancing Glycolysis-Mediated Vascular Remodeling. J. Cell Physiol. 2019, 234, 10602–10614. [Google Scholar] [CrossRef] [PubMed]
- Lopes, R.; Caetano, J.; Barahona, F.; Pestana, C.; Ferreira, B.V.; Lourenço, D.; Queirós, A.C.; Bilreiro, C.; Shemesh, N.; Beck, H.C.; et al. Multiple Myeloma-Derived Extracellular Vesicles Modulate the Bone Marrow Immune Microenvironment. Front. Immunol. 2022, 13, 909880. [Google Scholar] [CrossRef]
- Liu, H.; Yin, J.; Wang, H.; Jiang, G.; Deng, M.; Zhang, G.; Bu, X.; Cai, S.; Du, J.; He, Z. FOXO3a Modulates WNT/β-Catenin Signaling and Suppresses Epithelial-to-Mesenchymal Transition in Prostate Cancer Cells. Cell Signal 2015, 27, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zheng, L.; Huang, J.; Gao, F.; Lin, X.; He, L.; Li, D.; Li, Z.; Ding, Y.; Chen, L. MiR-124 Radiosensitizes Human Colorectal Cancer Cells by Targeting PRRX1. PLoS ONE 2014, 9, e93917. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Qiu, R.; Yu, S.; Xu, X.; Li, G.; Gu, R.; Tan, C.; Zhu, W.; Shen, B. Paclitaxel-Resistant Gastric Cancer MGC-803 Cells Promote Epithelial-to-Mesenchymal Transition and Chemoresistance in Paclitaxel-Sensitive Cells via Exosomal Delivery of MiR-155-5p. Int. J. Oncol. 2019, 54, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Feng, X.; Liu, H.; Tong, R.; Wu, J.; Li, C.; Yu, H.; Chen, Y.; Cheng, Q.; Chen, J.; et al. High-Metastatic Cancer Cells Derived Exosomal MiR92a-3p Promotes Epithelial-Mesenchymal Transition and Metastasis of Low-Metastatic Cancer Cells by Regulating PTEN/Akt Pathway in Hepatocellular Carcinoma. Oncogene 2020, 39, 6529–6543. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sai, B.; Wang, F.; Wang, L.; Wang, Y.; Zheng, L.; Li, G.; Tang, J.; Xiang, J. Hypoxic BMSC-Derived Exosomal MiRNAs Promote Metastasis of Lung Cancer Cells via STAT3-Induced EMT. Mol. Cancer 2019, 18, 40. [Google Scholar] [CrossRef] [PubMed]
- Hahn, S.; Jackstadt, R.; Siemens, H.; Hünten, S.; Hermeking, H. SNAIL and MiR-34a Feed-Forward Regulation of ZNF281/ZBP99 Promotes Epithelial-Mesenchymal Transition. EMBO J. 2013, 32, 3079–3095. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Fabbri, M.; Calin, G.A. MicroRNAs and Other Non-Coding RNAs as Targets for Anticancer Drug Development. Nat. Rev. Drug Discov. 2013, 12, 847–865. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.P.; Xu, K.; Cui, J.; Yuan, D.Y.; Zou, B.; Li, J.; Liu, J.L.; Li, K.Y.; Meng, Z.; Zhang, B. Cancer-Associated Fibroblast-Derived Exosomal MiR-382-5p Promotes the Migration and Invasion of Oral Squamous Cell Carcinoma. Oncol. Rep. 2019, 42, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.S.; Wu, T.M.; Ling, C.C.; Yu, F.; Zhang, J.; Cao, P.S.; Gu, L.P.; Wang, H.M.; Xu, H.; Li, L.; et al. M2 Macrophage-Derived Exosomal MicroRNA-155-5p Promotes the Immune Escape of Colon Cancer by Downregulating ZC3H12B. Mol. Ther. Oncolytics 2021, 20, 484–498. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, C.; Huang, X.; Yu, J.; Gui, Y. Circular RNA Hsa_circ_0075828 Promotes Bladder Cancer Cell Proliferation through Activation of CREB1. BMB Rep. 2020, 53, 82–87. [Google Scholar] [CrossRef]
- Jiang, S.; Mo, C.; Guo, S.; Zhuang, J.; Huang, B.; Mao, X. Human Bone Marrow Mesenchymal Stem Cells-Derived MicroRNA-205-Containing Exosomes Impede the Progression of Prostate Cancer through Suppression of RHPN2. J. Exp. Clin. Cancer Res. 2019, 38, 495. [Google Scholar] [CrossRef]
- Liu, D.; Zhong, L.; Yuan, Z.; Yao, J.; Zhong, P.; Liu, J.; Yao, S.; Zhao, Y.; Liu, L.; Chen, M.; et al. MiR-382-5p Modulates the ATRA-Induced Differentiation of Acute Promyelocytic Leukemia by Targeting Tumor Suppressor PTEN. Cell Signal. 2019, 54, 1–9. [Google Scholar] [CrossRef]
- Ho, J.Y.; Hsu, R.J.; Liu, J.M.; Chen, S.C.; Liao, G.S.; Gao, H.W.; Yu, C.P. MicroRNA-382-5p Aggravates Breast Cancer Progression by Regulating the RERG/Ras/ERK Signaling Axis. Oncotarget 2017, 8, 22443–22459. [Google Scholar] [CrossRef]
- Kok, V.C.; Yu, C.C. Cancer-Derived Exosomes: Their Role in Cancer Biology and Biomarker Development. Int. J. Nanomed. 2020, 15, 8019–8036. [Google Scholar] [CrossRef]
- Jiang, F.; Doudna, J.A. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef] [PubMed]
- Murugan, D.; Rangasamy, L. A Perspective to Weaponize MicroRNAs against Lung Cancer. Noncoding RNA Res. 2023, 8, 18–32. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Qin, P.; Zhang, D.; Cui, X.; Gao, J.; Yu, Z.; Chai, Y.; Wang, J.; Li, J. Long Non-Coding RNA PVT1 Encapsulated in Bone Marrow Mesenchymal Stem Cell-Derived Exosomes Promotes Osteosarcoma Growth and Metastasis by Stabilizing ERG and Sponging MiR-183-5p. Aging 2019, 11, 9581–9596. [Google Scholar] [CrossRef]
- Yao, Y.; Chen, R.; Wang, G.; Zhang, Y.; Liu, F. Exosomes Derived from Mesenchymal Stem Cells Reverse EMT via TGF-Β1/Smad Pathway and Promote Repair of Damaged Endometrium. Stem Cell Res. Ther. 2019, 10, 225. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal MiRNAs as Biomarkers for Prostate Cancer. Front. Genet. 2013, 4, 102860. [Google Scholar] [CrossRef]
- Zheng, R.; Du, M.; Wang, X.; Xu, W.; Liang, J.; Wang, W.; Lv, Q.; Qin, C.; Chu, H.; Wang, M.; et al. Exosome-Transmitted Long Non-Coding RNA PTENP1 Suppresses Bladder Cancer Progression. Mol. Cancer 2018, 17, 143. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Luo, Y.; He, W.; Zhao, Y.; Kong, Y.; Liu, H.; Zhong, G.; Li, Y.; Li, J.; Huang, J.; et al. Exosomal Long Noncoding RNA LNMAT2 Promotes Lymphatic Metastasis in Bladder Cancer. J. Clin. Investig. 2020, 130, 404–421. [Google Scholar] [CrossRef]
- Chen, C.; Zheng, H.; Luo, Y.; Kong, Y.; An, M.; Li, Y.; He, W.; Gao, B.; Zhao, Y.; Huang, H.; et al. SUMOylation Promotes Extracellular Vesicle-Mediated Transmission of LncRNA ELNAT1 and Lymph Node Metastasis in Bladder Cancer. J. Clin. Investig. 2021, 131, e146431. [Google Scholar] [CrossRef]
- Dou, D.; Ren, X.; Han, M.; Xu, X.; Ge, X.; Gu, Y.; Wang, X. Cancer-Associated Fibroblasts-Derived Exosomes Suppress Immune Cell Function in Breast Cancer via the MiR-92/PD-L1 Pathway. Front. Immunol. 2020, 11, 2026. [Google Scholar] [CrossRef]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 Contributes to Immunosuppression and Is Associated with Anti-PD-1 Response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef]
- Jóźwicki, W.; Brozyna, A.A.; Siekiera, J.; Slominski, A.T. Frequency of CD4+CD25+Foxp3+ Cells in Peripheral Blood in Relation to Urinary Bladder Cancer Malignancy Indicators before and after Surgical Removal. Oncotarget 2016, 7, 11450–11462. [Google Scholar] [CrossRef] [PubMed]
- Jóźwicki, W.; Brożyna, A.A.; Siekiera, J.; Slominski, A.T. Changes in Immunogenicity during the Development of Urinary Bladder Cancer: A Preliminary Study. Int. J. Mol. Sci. 2016, 17, 285. [Google Scholar] [CrossRef] [PubMed]
- Muller, L.; Simms, P.; Hong, C.S.; Nishimura, M.I.; Jackson, E.K.; Watkins, S.C.; Whiteside, T.L. Human Tumor-Derived Exosomes (TEX) Regulate Treg Functions via Cell Surface Signaling Rather than Uptake Mechanisms. Oncoimmunology 2017, 6, e1261243. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Camfield, R.; Gorski, S.M. The Interplay between Exosomes and Autophagy—Partners in Crime. J. Cell Sci. 2018, 131, jcs215210. [Google Scholar] [CrossRef] [PubMed]
- El Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J.A. Extracellular Vesicles: Biology and Emerging Therapeutic Opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Michael, I.J.; Park, J.; Granick, S.; Cho, Y.K. Cloaked Exosomes: Biocompatible, Durable, and Degradable Encapsulation. Small 2018, 14, 1802052. [Google Scholar] [CrossRef] [PubMed]
- Lara, P.; Huis In ‘T Veld, R.V.; Jorquera-Cordero, C.; Chan, A.B.; Ossendorp, F.; Cruz, L.J. Zinc-Phthalocyanine-Loaded Extracellular Vesicles Increase Efficacy and Selectivity of Photodynamic Therapy in Co-Culture and Preclinical Models of Colon Cancer. Pharmaceutics 2021, 13, 1547. [Google Scholar] [CrossRef]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of Exosome-Encapsulated Paclitaxel to Overcome MDR in Cancer Cells. Nanomedicine 2016, 12, 655–664. [Google Scholar] [CrossRef]
- Chinnappan, M.; Srivastava, A.; Amreddy, N.; Razaq, M.; Pareek, V.; Ahmed, R.; Mehta, M.; Peterson, J.E.; Munshi, A.; Ramesh, R. Exosomes as Drug Delivery Vehicle and Contributor of Resistance to Anticancer Drugs. Cancer Lett. 2020, 486, 18–28. [Google Scholar] [CrossRef]
- Lennaárd, A.J.; Mamand, D.R.; Wiklander, R.J.; Andaloussi, S.E.L.; Wiklander, O.P.B. Optimised Electroporation for Loading of Extracellular Vesicles with Doxorubicin. Pharmaceutics 2022, 14, 38. [Google Scholar] [CrossRef]
- Fuhrmann, G.; Serio, A.; Mazo, M.; Nair, R.; Stevens, M.M. Active Loading into Extracellular Vesicles Significantly Improves the Cellular Uptake and Photodynamic Effect of Porphyrins. J. Control. Release 2015, 205, 35–44. [Google Scholar] [CrossRef]
- Ebrahimian, M.; Hashemi, M.; Etemad, L.; Salmasi, Z. Thymoquinone-Loadedmesenchymalstemcell-Derivedexosome as an Efficient Nano-System against Breast Cancer Cells. Iran. J. Basic. Med. Sci. 2022, 25, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.T.; Umezaki, K.; Sawada, S.; Mukai, S.A.; Sasaki, Y.; Harada, N.; Shiku, H.; Akiyoshi, K. Engineering Hybrid Exosomes by Membrane Fusion with Liposomes. Sci. Rep. 2016, 6, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Jeyaram, A.; Lamichhane, T.N.; Wang, S.; Zou, L.; Dahal, E.; Kronstadt, S.M.; Levy, D.; Parajuli, B.; Knudsen, D.R.; Chao, W.; et al. Enhanced Loading of Functional MiRNA Cargo via PH Gradient Modification of Extracellular Vesicles. Mol. Ther. 2020, 28, 975–985. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Jiang, X.; Shi, J.; Gao, S.; Zhu, Y.; Gu, T.; Shi, E. Exosomes Derived from Bone Marrow Mesenchymal Stem Cells Overexpressing MicroRNA-25 Protect Spinal Cords against Transient Ischemia. J. Thorac. Cardiovasc. Surg. 2019, 157, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Cardarelli, F.; Digiacomo, L.; Marchini, C.; Amici, A.; Salomone, F.; Fiume, G.; Rossetta, A.; Gratton, E.; Pozzi, D.; Caracciolo, G. The Intracellular Trafficking Mechanism of Lipofectamine-Based Transfection Reagents and Its Implication for Gene Delivery. Sci. Rep. 2016, 6, 25879. [Google Scholar] [CrossRef] [PubMed]
- Melzer, C.; von der Ohe, J.; Hass, R. Anti-Tumor Effects of Exosomes Derived from Drug-Incubated Permanently Growing Human MSC. Int. J. Mol. Sci. 2020, 21, 7311. [Google Scholar] [CrossRef] [PubMed]
- Bellavia, D.; Raimondo, S.; Calabrese, G.; Forte, S.; Cristaldi, M.; Patinella, A.; Memeo, L.; Manno, M.; Raccosta, S.; Diana, P.; et al. Interleukin 3- Receptor Targeted Exosomes Inhibit in Vitro and in Vivo Chronic Myelogenous Leukemia Cell Growth. Theranostics 2017, 7, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Morishita, M.; Takahashi, Y.; Nishikawa, M.; Ariizumi, R.; Takakura, Y. Enhanced Class i Tumor Antigen Presentation via Cytosolic Delivery of Exosomal Cargos by Tumor-Cell-Derived Exosomes Displaying a PH-Sensitive Fusogenic Peptide. Mol. Pharm. 2017, 14, 4079–4086. [Google Scholar] [CrossRef]
- Smyth, T.; Petrova, K.; Payton, N.M.; Persaud, I.; Redzic, J.S.; Graner, M.W.; Smith-Jones, P.; Anchordoquy, T.J. Surface Functionalization of Exosomes Using Click Chemistry. Bioconjug Chem. 2014, 25, 1777–1784. [Google Scholar] [CrossRef]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Yuan, D.; Deygen, I.; Klyachko, N.L.; Kabanov, A.V.; Batrakova, E.V. Engineering Macrophage-Derived Exosomes for Targeted Paclitaxel Delivery to Pulmonary Metastases: In Vitro and in Vivo Evaluations. Nanomedicine 2018, 14, 195–204. [Google Scholar] [CrossRef]
- Jang, S.C.; Kim, O.Y.; Yoon, C.M.; Choi, D.S.; Roh, T.Y.; Park, J.; Nilsson, J.; Lötvall, J.; Kim, Y.K.; Gho, Y.S. Bioinspired Exosome-Mimetic Nanovesicles for Targeted Delivery of Chemotherapeutics to Malignant Tumors. ACS Nano 2013, 7, 7698–7710. [Google Scholar] [CrossRef]
- Smyth, T.J.; Redzic, J.S.; Graner, M.W.; Anchordoquy, T.J. Examination of the Specificity of Tumor Cell Derived Exosomes with Tumor Cells in Vitro. Biochim. Biophys. Acta Biomembr. 2014, 1838, 2954–2965. [Google Scholar] [CrossRef]
- Hadla, M.; Palazzolo, S.; Corona, G.; Caligiuri, I.; Canzonieri, V.; Toffoli, G.; Rizzolio, F. Exosomes Increase the Therapeutic Index of Doxorubicin in Breast and Ovarian Cancer Mouse Models. Nanomedicine 2016, 11, 2431–2441. [Google Scholar] [CrossRef]
- Aqil, F.; Kausar, H.; Agrawal, A.K.; Jeyabalan, J.; Kyakulaga, A.H.; Munagala, R.; Gupta, R. Exosomal Formulation Enhances Therapeutic Response of Celastrol against Lung Cancer. Exp. Mol. Pathol. 2016, 101, 12–21. [Google Scholar] [CrossRef]
- Gao, X.; Ran, N.; Dong, X.; Zuo, B.; Yang, R.; Zhou, Q.; Moulton, H.M.; Seow, Y.; Yin, H.F. Anchor Peptide Captures, Targets, and Loads Exosomes of Diverse Origins for Diagnostics and Therapy. Sci. Transl. Med. 2018, 10, eaat0195. [Google Scholar] [CrossRef]
- Illes, B.; Hirschle, P.; Barnert, S.; Cauda, V.; Wuttke, S.; Engelke, H. Exosome-Coated Metal-Organic Framework Nanoparticles: An Efficient Drug Delivery Platform. Chem. Mater. 2017, 29, 8042–8046. [Google Scholar] [CrossRef]
- Busatto, S.; Iannotta, D.; Walker, S.A.; Di Marzio, L.; Wolfram, J. A Simple and Quick Method for Loading Proteins in Extracellular Vesicles. Pharmaceuticals 2021, 14, 356. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Rich, J.; Hao, N.; Gu, Y.; Chen, C.; Yang, S.; Zhang, P.; Huang, T.J. Acoustofluidics for Simultaneous Nanoparticle-Based Drug Loading and Exosome Encapsulation. Microsyst. Nanoeng. 2022, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.M.J.; Fang, R.H.; Wang, K.C.; Luk, B.T.; Thamphiwatana, S.; Dehaini, D.; Nguyen, P.; Angsantikul, P.; Wen, C.H.; Kroll, A.V.; et al. Nanoparticle Biointerfacing by Platelet Membrane Cloaking. Nature 2015, 526, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wu, J.; Gu, W.; Huang, Y.; Tong, Z.; Huang, L.; Tan, J. Exosome–Liposome Hybrid Nanoparticles Deliver CRISPR/Cas9 System in MSCs. Adv. Sci. 2018, 5, 1700611. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.; Heuser, J.; Stahl, P. Receptor-Mediated Endocytosis of Transferrin and Recycling of the Transferrin Receptor in Rat Reticulocytes. J. Cell Biol. 1983, 97, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.T.; Johnstone, R.M. Fate of the Transferrin Receptor during Maturation of Sheep Reticulocytes in Vitro: Selective Externalization of the Receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron Microscopic Evidence for Externalization of the Transferrin Receptor in Vesicular Form in Sheep Reticulocytes. J. Cell Biol. 1985, 101, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Gerwing, M.; Kocman, V.; Stölting, M.; Helfen, A.; Masthoff, M.; Roth, J.; Barczyk-Kahlert, K.; Greune, L.; Schmidt, M.A.; Heindel, W.; et al. Tracking of Tumor Cell–Derived Extracellular Vesicles In Vivo Reveals a Specific Distribution Pattern with Consecutive Biological Effects on Target Sites of Metastasis. Mol. Imaging Biol. 2020, 22, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Buzas, E.I. The Roles of Extracellular Vesicles in the Immune System. Nat. Rev. Immunol. 2022, 23, 236–250. [Google Scholar] [CrossRef] [PubMed]
- Simeone, P.; Bologna, G.; Lanuti, P.; Pierdomenico, L.; Guagnano, M.T.; Pieragostino, D.; Del Boccio, P.; Vergara, D.; Marchisio, M.; Miscia, S.; et al. Extracellular Vesicles as Signaling Mediators and Disease Biomarkers across Biological Barriers. Int. J. Mol. Sci. 2020, 21, 2514. [Google Scholar] [CrossRef]
- Mahida, R.Y.; Matsumoto, S.; Matthay, M.A. Extracellular Vesicles in ARDS: New Insights into Pathogenesis with Novel Clinical Applications. In Annual Update in Intensive Care and Emergency Medicine 2020; Springer: Cham, Switzerland, 2020; pp. 53–65. [Google Scholar] [CrossRef]
- Hovhannisyan, L.; Czechowska, E.; Gutowska-Owsiak, D. The Role of Non-Immune Cell-Derived Extracellular Vesicles in Allergy. Front. Immunol. 2021, 12, 702381. [Google Scholar] [CrossRef]
- De Freitas, R.C.C.; Hirata, R.D.C.; Hirata, M.H.; Aikawa, E. Circulating Extracellular Vesicles as Biomarkers and Drug Delivery Vehicles in Cardiovascular Diseases. Biomolecules 2021, 11, 388. [Google Scholar] [CrossRef]
- Maia, J.; Caja, S.; Strano Moraes, M.C.; Couto, N.; Costa-Silva, B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol. 2018, 6, 18. [Google Scholar] [CrossRef]
- Gok Yavuz, B.; Gunaydin, G.; Gedik, M.E.; Kosemehmetoglu, K.; Karakoc, D.; Ozgur, F.; Guc, D. Cancer Associated Fibroblasts Sculpt Tumour Microenvironment by Recruiting Monocytes and Inducing Immunosuppressive PD-1 + TAMs. Sci. Rep. 2019, 9, 3172. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A Framework for Advancing Our Understanding of Cancer-Associated Fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.H.; Tai, S.K.; Yang, M.H. Snail-Overexpressing Cancer Cells Promote M2-Like Polarization of Tumor-Associated Macrophages by Delivering MiR-21-Abundant Exosomes. Neoplasia 2018, 20, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Gulati, S.; Cheng, T.M.K.; Bates, P.A. Cancer Networks and beyond: Interpreting Mutations Using the Human Interactome and Protein Structure. Semin. Cancer Biol. 2013, 23, 219–226. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, C.; Shang, Z.; Xing, R.; Shi, L.; Lv, Y. P42.3 Gene Expression in Gastric Cancer Cell and Its Protein Regulatory Network Analysis. Theor. Biol. Med. Model. 2012, 9, 53. [Google Scholar] [CrossRef]
- Sun, H.; Han, B.; Cheng, X.; Ma, K. Transcriptional Regulatory Network and Protein-Protein Interaction to Reveal the Mechanism of Pancreatic Cancer. Mol. Biol. Rep. 2014, 41, 387–395. [Google Scholar] [CrossRef]
| Potential Therapeutic Use | Coding Gene//Protein/Publication | Average/ Maximum FPKM Value | Best Expression Cut Off (FPKM) | 5-Year Survival High/Low [%] | p-Score |
|---|---|---|---|---|---|
| Colorectal cancer | CREB1//CAMP responsive element binding protein 1/ [27] | 3.9/27.0 | 3.2 | 66/49 | 0.033 |
| ZEB1//Zinc finger E-box binding homeobox 1/ [28] | 1.9/26.8 | 2.35 | 57/63 | 0.034 | |
| FBXW7//F-box/ [29] | 1.7/9.3 | 1.71 | 68/56 | 0.018 | |
| Breast cancer | VEGFB//Vascular endothelial growth factor B/ [30] | 48.5/296.1 | 44.57 | 85/79 | 0.049 |
| SMARCA5//SWI/SNF related/ [31] | 17.7/55.4 | 21.43 | 77/83 | 0.0092 | |
| SNAI2//Snail family transcriptional repressor 2/ [32] | 10.2/183.1 | 6.55 | 85/77 | 0.035 | |
| Urinary bladder carcinoma | SNAIL1//Snail family transcriptional repressor 1/ [33] | 3.1/57.1 | 0.77 | 38/56 | 0.019 |
| IGF2R//Insulin-like growth factor 2 receptor/ [34] | 9.1/27.9 | 8.89 | 30/50 | 0.00061 | |
| ABL2//ABL proto-oncogene 2/ [35] | 2,5/13,5 | 2.03 | 33/50 | 0.00084 | |
| Ovarian carcinoma | SNAIL1//Snail family transcriptional repressor 1/ [36,37] | 2.9/21.2 | 2.8 | 20/40 | 0.0098 |
| Notch-1//Notch receptor 1/ [38] | 7.1/ 61.1 | 10.2 | 25/34 | 0.0055 | |
| Gastric cancer | ZEB1//Zinc finger E-box binding homeobox 1/ [39] | 6.4/51.8 | 6.15 | 13/45 | 0.0056 |
| ZEB2//Zinc finger E-box binding homeobox 2/ [39] | 2.1/9.2 | 1.76 | 24/48 | 0.011 | |
| Renal cell carcinoma | PTEN//Phosphatase and tensin homolog / [40] | 8.7/37.8 | 6.05 | 66/77 | 0.00073 |
| Hepatic cell carcinoma | Twist1//Twist family bHLH transcription factor 1/ [41] | 0.4/45.1 | 0.14 | 37/52 | 0.018 |
| FOXQ1//Forkhead box Q1/ [42] | 4.2/79.4 | 2.6 | 39/52 | 0.033 | |
| Uveal melanoma | CDK4//Cyclin dependent kinase 4/ [43] | 38.9/176.2 | 34.4 | 0/57 * | 0.0032 |
| Method of Action | Potential Therapeutic Benefit | Y/CI * | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| miRNA | Action * | Effect of Modification of miRNA Expression | Clinical Application | ||||||||
| miRNA | Inhibition | CTH/RTH * Sensitivity | M0/M2 * | CM * | |||||||
| M/I * | P/A * | Meta * | EMT | ||||||||
| miR-134 | IR: CREB1 | + | +/+ | nd/nd | nd | + | + | nd | - | CRC [27] | 2017/52 |
| miR-100 | mTOR/HIF-1α/VEGF modulation | + | +/+ | nd/nd | nd | + | nd | nd | - | BC [30] | 2017/194 |
| IR: HOXA1 and SMARCA5 | + | +/+ | nd/nd | nd | - | nd | nd | - | BC [31] | 2014/101 | |
| miRNA-34b/c | IR: β-katenin | + | +/+ | nd/nd | nd | + | nd | nd | - | PC [87] | 2015/116 |
| miR-30c | IR: SNAIL1 | + | +/+ | nd/nd | nd | + | nd | nd | - | BC [33] | 2018/221 |
| miR-137 | IR: Snail | + | +/+ | nd/nd | nd | + | nd | nd | - | OC [36] | 2016/89 |
| MiR-363 | IR: Snail | + | nd/nd | nd/nd | nd | + | + | nd | - | OC [37] | 2018/29 |
| miR-452 | IR: SNAI2 | + | nd/+ | nd/nd | nd | + | nd | nd | - | BC [32] | 2017/58 |
| miR-200c | IR: Zeb1, Zeb2 | + | +/+ | nd/nd | nd | + | + | nd | - | GC [39] | 2018/56 |
| miR-361-5p | IR: Twist1 | + | +/+ | +/nd | nd | + | nd | nd | - | HCC [41] | 2020/9 |
| miR-124 | IR: PRRX1 | + | nd/nd | nd/nd | nd | + | + | nd | - | CRC [88] | 2014/93 |
| miR-155-5p | IR: GATA3 | - | nd/nd | nd/nd | nd | + | nd | nd | + | GC [89] | 2019/86 |
| miR-4319 | IR: FOXQ1 | + | nd/nd | +/+ | nd | + | nd | nd | - | HCC [42] | 2019/19 |
| miR-6780b-5p | RE: Notch/MAPK pathway | - | +/+ | +/nd | nd | + | nd | nd | + | OC [38] | 2021/20 |
| miR-19b-3p | RE: PTEN IR: E-cadherin, IN: N-cadherin, vimentin, Twist protein, CD103 | - | nd/nd | nd/nd | + | + | nd | nd | + | RCC [40] | 2019/98 |
| miR92a-3p | ID: Akt/Snail pathway | - | nd/nd | nd/nd | + | + | + | nd | + | HCC [90] | 2020/79 |
| miR-301a-3p | IR: PTEN, IN: PI3Kγ, p-AKT, p-mTOR | - | +/+ | nd/nd | + | + | nd | nd | + | PAC [8] | 2020/9 |
| IN: PTEN/ PI3Kγ pathway, Arg1, TGFβ, IL10 | - | +/+ | nd/nd | nd | nd | nd | + | + | PAC [8] | 2020/9 | |
| miR-1500, miR-210-3p, miR-193 | ID: STAT3, IR: E-cadherin, IN: snail, vimentin, slug, twist, fibronectin, ZEB1, N-cadherin | - | nd/+ | nd/nd | nd | + | nd | nd | + | LC [91] | 2019/246 |
| miR-34a | IR: Snail | + | nd/+ | nd/nd | nd | + | nd | nd | - | OC [36] | 2016/89 |
| IR: ZNF281 | + | +/+ | +/nd | + | + | nd | nd | - | CRC [92] | 2013/123 | |
| HCC [93] | 2013/1067 | ||||||||||
| miR-375-3p | IN: E-cadherin, IR: Snail, vimentin, ZEB-1, β-catenin | + | +/+ | nd/nd | nd | nd | nd | nd | - | CRC [28] | 2021/22 |
| miR-382-5p | IN: MMP-3, MMP-9, N-cadherin, β-catenin | - | +/+ | nd/nd | nd | + | nd | nd | + | OSCC [94] | 2019/81 |
| miR92a-3p | IN: Wnt/β-catenin pathway, IR: MOAP1, FBXW7 | - | +/+ | nd/- | nd | nd | + | nd | + | CRC [29] | 2019/299 |
| miR-21-5p | IN: PTEN, STAT3 pathway, IL-10, TGF-β | - | nd/nd | nd/nd | nd | nd | nd | + | + | BDC [57] | 2020/51 |
| miR-155-5p | IR: ZC3H12B, IN: IL-6 | - | nd/nd | +/- | nd | nd | nd | nd | + | CRC [95] | 2021/29 |
| miR-361-3p | IR: IGF2R | + | +/+ | +/nd | nd | nd | nd | nd | - | BDC [34] | 2021/9 |
| miR-1224-5p | IR: CREB1 | + | nd/nd | +/nd | nd | nd | nd | nd | - | BDC [96] | 2020/23 |
| miR-34b/c | IR: c-Met, (G1 cell cycle) | + | +/nd | +/nd | nd | nd | nd | + | - | Uveal MM [43] | 2012/58 |
| miR-205 | IR: RHPN2 | + | +/+ | +/- | nd | nd | nd | nd | - | PC [97] | 2019/45 |
| miR-19b-1-5p | IR: ABL2, Bcl-2, MMP2, MMP9, IN: Bax | + | +/+ | +/- | nd | nd | nd | nd | - | BDC [35] | 2021/7 |
| miR-382-5p | IR: PTEN, ATRA, | - | nd/nd | nd/nd | nd | nd | + | nd | + | APML [98] | 2019/23 |
| RE: cyclin D1 | |||||||||||
| RE: RERG | - | +/+ | nd/nd | nd | nd | nd | nd | + | BC [99] | 2017/55 | |
| Type of Action | Method | Description of the Method | Weaknesses * | Strengths * | Example of a Packaged Substance |
|---|---|---|---|---|---|
| Extracellular exosome loading | Incubation with target cargo | Temperature 37 °C for 1 hour with shaking | LE | SP and MEMI | Paclitaxel [118] |
| Sonication | UE–DEMI–LTC | ED, EA, and EF | HE and PCP | Paclitaxel [118] Catalase [119] | |
| Electroporation | ElF–DEMI–LTC | DEI, EA, EF, and ITL | SP, CP, and HE | Doxorubicin [120] Catalase [119] | |
| Extrusion | Exosomes and target cargo—FFDPS | ED | HE and PCP | Porphyrins [121] | |
| Freeze–thaw | METC—3 cycles of fast freezing and thawing | EA | HE and MF | Thymoquinone [122] | |
| Exosomes with liposomes—several freeze–thaw cycles | Membrane fusion [123] | ||||
| Modified CCM | METC—thermal shock | DNA and DEMI | SP and HE | miRNA [28] | |
| pH gradient-based method | Generation of pHG and incubation with target cargo | Unpredictable effect associated with DEMI | HE, possibility of reusing the cargo | miRNA packaging [124] | |
| Saponin | Increasing exosome lipid membrane permeabilization | Hemolytic activity | LHME | Porphyrins [121] | |
| Transfection | LmiRNAGV with exosomes—enhancement of miRNA expression | Limited rate of penetration through membranes | MGME and increasing the therapeutic effect | pre-microRNA [125] | |
| Transfection with lipofectamine | Increasing the efficiency of transfection of RNA or plasmid DNA into cell cultures in vitro | There is no known mechanism of action of lipofectamine | HE, “gold standard”, and low toxicity to cells | DNA [126] | |
| Intracellular loading during exosomes biogenesis | Bioengineering of exosome-producing cells | Cells with target cargo | TCM, DSM, and unwanted cellular content in exosomes | HE, preservation of native features of exosomes, and low toxicity to cells | Taxol [127] |
| Transfection into cells with miRNA/siRNA/pDNA/plasmid vector to increase gene product expression in the exosome | IL3-Lamp2b plasmid vector [128] and encoding the fusion protein [129] | ||||
| Surface-modified exosomes | Click chemistry | Attachment of molecules to the surface of exosomes through covalent bonds | Few scientific reports | HE, CP, no effect on size, adhesion, and internalization of exosomes | Copper-catalyzed azide–alkyne cycloaddition [130] |
| Combinations with pH-sensitive fusion peptides | Exosomes + fusion peptide = formation of pores in the lipid membrane due to lower pH | Complexity of the method, few scientific reports | Control of exosome movement, better presentation of tumor antigens | Exosomes from melanoma cells and mixing with GALA [129] | |
| Dual ligand engineering | Sonication and incubation of a mixture of exosomes, vector lipid molecules and target cargo | Identification of a vector molecule unique to a specific cancer | High ability to accumulate in cancer cells, the therapeutic effect | PTX-loaded exosomes with PEG-AA vector moiety [131] | |
| Exosome-mimetic | Nanovesicles from cell membranes | The result of cell disintegration using extrusion | Efficacy depends on the surface properties of the cells used | Features similar to exosomes but 100 times higher production efficiency | Nanovesicles from monocytes or macrophages with doxorubicin [132] |
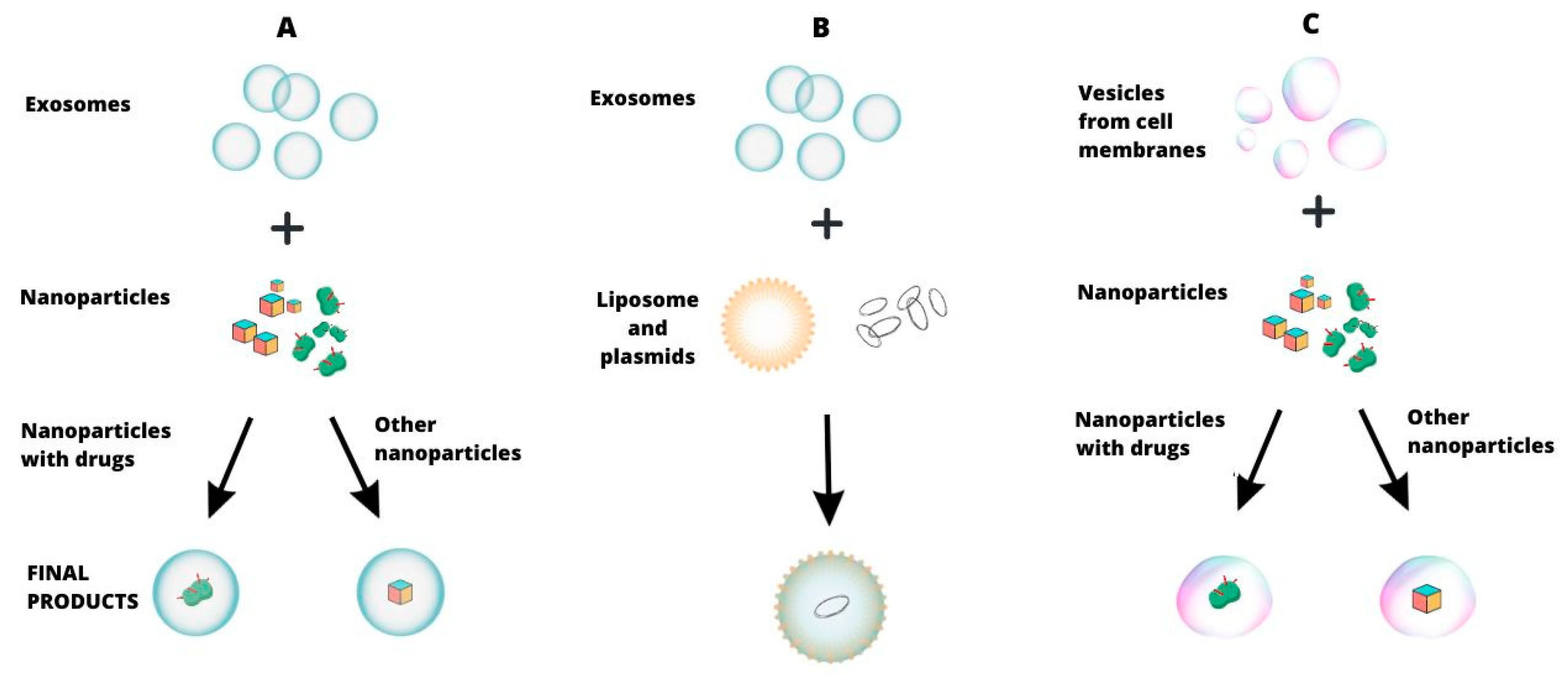
| Method | Particle Covered | Description of the Method | Weaknesses | Strengths | Application Examples |
|---|---|---|---|---|---|
| Covering a molecule with an exosomal membrane | Hybrid nanoparticles | Nanoparticles synthesized from inorganic and organic building block units, loaded with cargo, and coated with exosomes | The complexity of the method, few scientific reports | Exosomal transfer selectivity, preferential capture by specific cell types | Iron-based metal−organic framework nanoparticles with calcein or suberohydroxamic acid [137] |
| Cationic nanoparticles | Incubation of exosomes with a mixture of synthetic cationic nanoparticles using the interaction of their electric charges | High costs, little scientific reports | Maintaining native EV features, comparable loading efficiency, lower toxicity | Nanoparticles with Cas9 protein [138] | |
| Drug-loaded silica nanocarriers | Combination of exosomes, drugs, and porous silica nanoparticles using acoustofluidics | Complexity of the method, few scientific reports | Drug loading and encapsulation in minutes | Silica nanocarriers with doxorubicin [139] | |
| Covering a molecule with a cell membrane | Polymer nanoparticles | Combining membrane vesicles with PLGA (poly(lactic-co-glycolic acid)) particles using sonication | Complexity of the method, few scientific reports | Increased blood half-life and prolonged retention, decreased uptake by macrophages | Docetaxel-loaded PLGA nanoparticle [140] |
| Hybrids | Exosome–liposome | Incubation with the effect of fusion of exosomes with liposomes and encapsulation of plasmids | Few scientific reports | Effective encapsulation of large plasmids and drugs, reducing drug resistance | CRISPR/Cas9 vectors in an exosome–liposome hybrid [141] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jóźwicka, T.M.; Erdmańska, P.M.; Stachowicz-Karpińska, A.; Olkiewicz, M.; Jóźwicki, W. Exosomes—Promising Carriers for Regulatory Therapy in Oncology. Cancers 2024, 16, 923. https://doi.org/10.3390/cancers16050923
Jóźwicka TM, Erdmańska PM, Stachowicz-Karpińska A, Olkiewicz M, Jóźwicki W. Exosomes—Promising Carriers for Regulatory Therapy in Oncology. Cancers. 2024; 16(5):923. https://doi.org/10.3390/cancers16050923
Chicago/Turabian StyleJóźwicka, Teresa Maria, Patrycja Maria Erdmańska, Agnieszka Stachowicz-Karpińska, Magdalena Olkiewicz, and Wojciech Jóźwicki. 2024. "Exosomes—Promising Carriers for Regulatory Therapy in Oncology" Cancers 16, no. 5: 923. https://doi.org/10.3390/cancers16050923
APA StyleJóźwicka, T. M., Erdmańska, P. M., Stachowicz-Karpińska, A., Olkiewicz, M., & Jóźwicki, W. (2024). Exosomes—Promising Carriers for Regulatory Therapy in Oncology. Cancers, 16(5), 923. https://doi.org/10.3390/cancers16050923






