The Future of Targeted Therapy for Leiomyosarcoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Genomic Landscape of Leiomyosarcoma
3. Transcriptomic Profiling Has Revealed Distinct Molecular Subtypes of Leiomyosarcoma
4. Approved Targeted Therapies for Leiomyosarcoma
5. Pharmacogenomic Biomarkers in Leiomyosarcoma
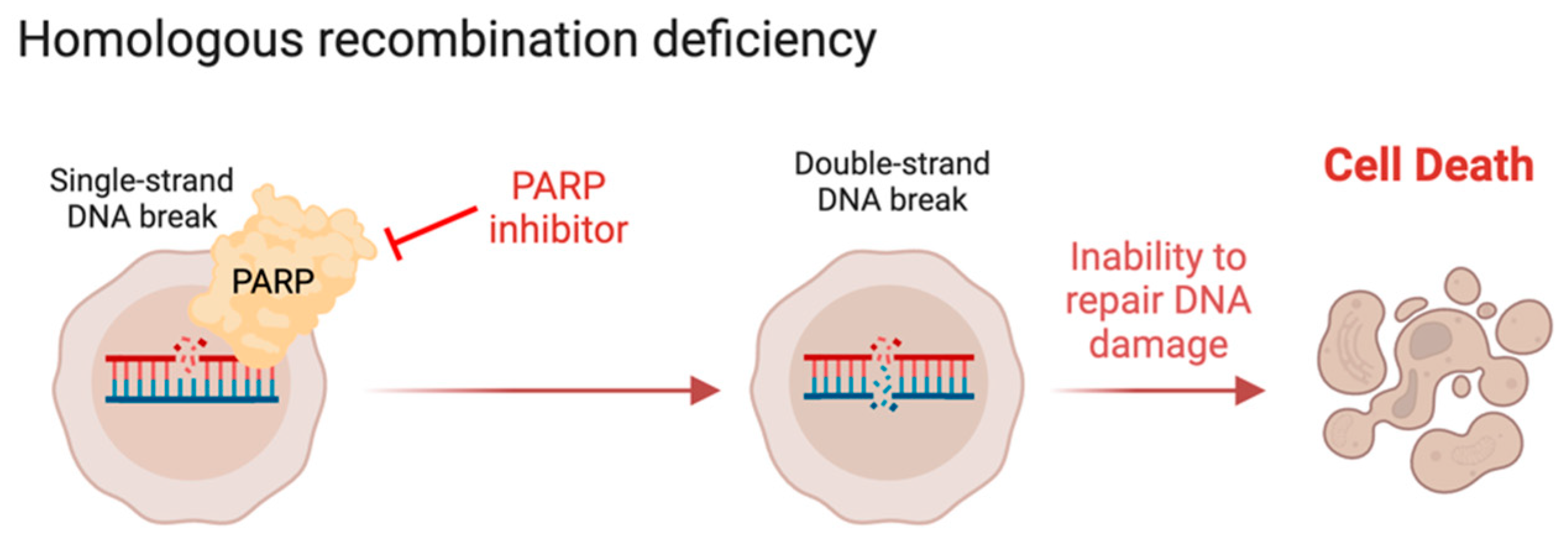
6. The DNA Damage Response in Leiomyosarcoma
7. The PI3K/PTEN/AKT/mTOR Signaling Pathway
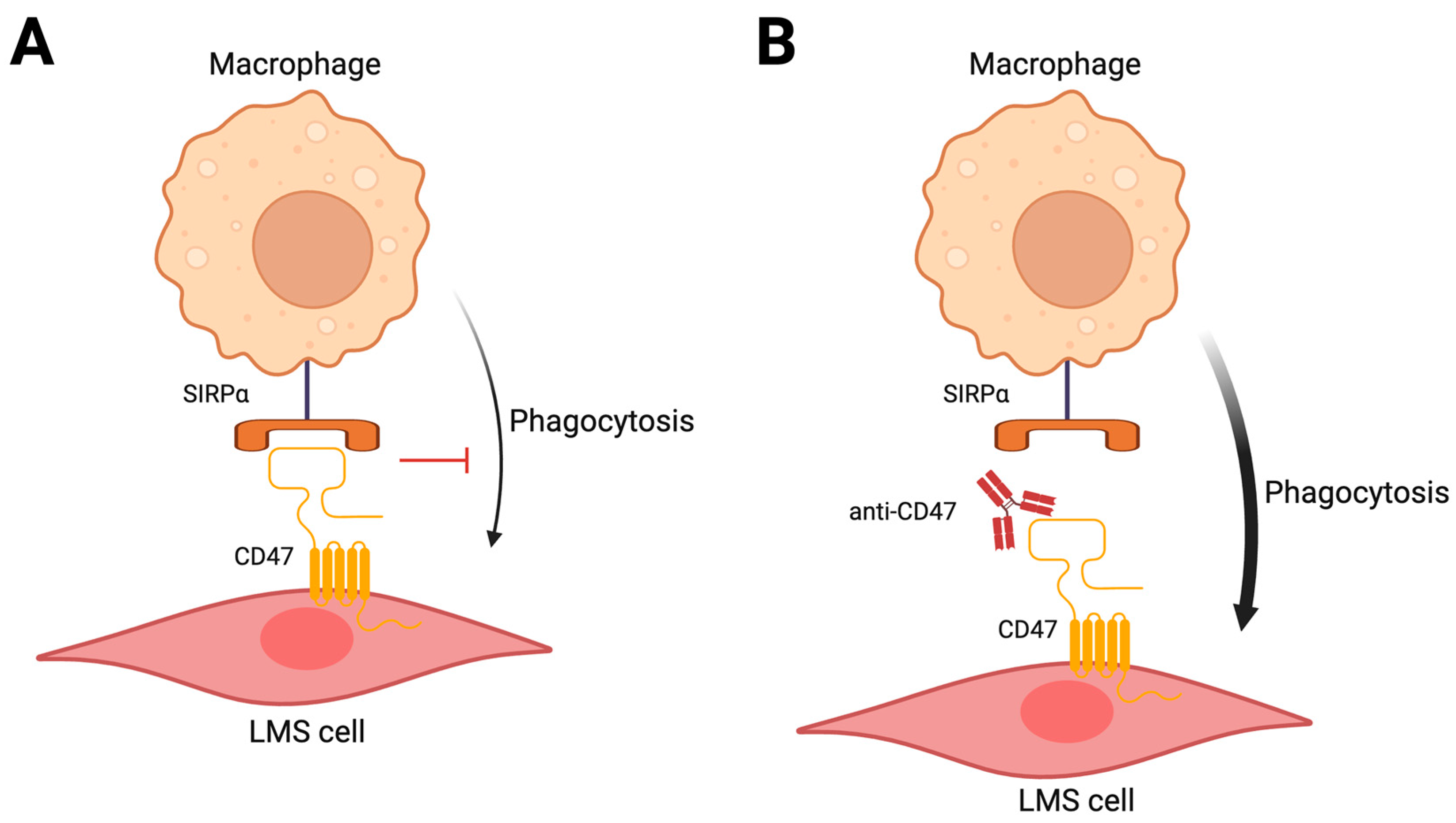
8. Targeting the Micro-Environment
9. The Leiomyosarcoma Epigenome
10. Telomere Biology
11. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Farshid, G.; Pradhan, M.; Goldblum, J.; Weiss, S.W. Leiomyosarcoma of Somatic Soft Tissues: A Tumor of Vascular Origin with Multivariate Analysis of Outcome in 42 Cases. Am. J. Surg. Pathol. 2002, 26, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Gladdy, R.A.; Qin, L.-X.; Moraco, N.; Agaram, N.P.; Brennan, M.F.; Singer, S. Predictors of Survival and Recurrence in Primary Leiomyosarcoma. Ann. Surg. Oncol. 2013, 20, 1851–1857. [Google Scholar] [CrossRef]
- Mbatani, N.; Olawaiye, A.B.; Prat, J. Uterine Sarcomas. Int. J. Gynaecol. Obstet. 2018, 143 (Suppl. 2), 51–58. [Google Scholar] [CrossRef]
- Gustafson, P.; Willén, H.; Baldetorp, B.; Fernö, M.; Akerman, M.; Rydholm, A. Soft Tissue Leiomyosarcoma. A Population-Based Epidemiologic and Prognostic Study of 48 Patients, Including Cellular DNA Content. Cancer 1992, 70, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Toro, J.R.; Travis, L.B.; Wu, H.J.; Zhu, K.; Fletcher, C.D.M.; Devesa, S.S. Incidence Patterns of Soft Tissue Sarcomas, Regardless of Primary Site, in the Surveillance, Epidemiology and End Results Program, 1978–2001: An Analysis of 26,758 Cases. Int. J. Cancer 2006, 119, 2922–2930. [Google Scholar] [CrossRef]
- Cope, B.M.; Traweek, R.S.; Lazcano, R.; Keung, E.Z.; Lazar, A.J.; Roland, C.L.; Nassif, E.F. Targeting the Molecular and Immunologic Features of Leiomyosarcoma. Cancers 2023, 15, 2099. [Google Scholar] [CrossRef] [PubMed]
- Pautier, P.; Italiano, A.; Piperno-Neumann, S.; Chevreau, C.; Penel, N.; Firmin, N.; Boudou-Rouquette, P.; Bertucci, F.; Balleyguier, C.; Lebrun-Ly, V.; et al. Doxorubicin Alone versus Doxorubicin with Trabectedin Followed by Trabectedin Alone as First-Line Therapy for Metastatic or Unresectable Leiomyosarcoma (LMS-04): A Randomised, Multicentre, Open-Label Phase 3 Trial. Lancet Oncol. 2022, 23, 1044–1054. [Google Scholar] [CrossRef]
- Judson, I.; Verweij, J.; Gelderblom, H.; Hartmann, J.T.; Schöffski, P.; Blay, J.-Y.; Kerst, J.M.; Sufliarsky, J.; Whelan, J.; Hohenberger, P.; et al. Doxorubicin Alone versus Intensified Doxorubicin plus Ifosfamide for First-Line Treatment of Advanced or Metastatic Soft-Tissue Sarcoma: A Randomised Controlled Phase 3 Trial. Lancet Oncol. 2014, 15, 415–423. [Google Scholar] [CrossRef]
- Maki, R.G.; Wathen, J.K.; Patel, S.R.; Priebat, D.A.; Okuno, S.H.; Samuels, B.; Fanucchi, M.; Harmon, D.C.; Schuetze, S.M.; Reinke, D.; et al. Randomized Phase II Study of Gemcitabine and Docetaxel Compared with Gemcitabine Alone in Patients with Metastatic Soft Tissue Sarcomas: Results of Sarcoma Alliance for Research through Collaboration Study 002 [Corrected]. J. Clin. Oncol. 2007, 25, 2755–2763. [Google Scholar] [CrossRef]
- Hensley, M.L.; Maki, R.; Venkatraman, E.; Geller, G.; Lovegren, M.; Aghajanian, C.; Sabbatini, P.; Tong, W.; Barakat, R.; Spriggs, D.R. Gemcitabine and Docetaxel in Patients with Unresectable Leiomyosarcoma: Results of a Phase II Trial. J. Clin. Oncol. 2002, 20, 2824–2831. [Google Scholar] [CrossRef]
- Patel, S.R.; Gandhi, V.; Jenkins, J.; Papadopolous, N.; Burgess, M.A.; Plager, C.; Plunkett, W.; Benjamin, R.S. Phase II Clinical Investigation of Gemcitabine in Advanced Soft Tissue Sarcomas and Window Evaluation of Dose Rate on Gemcitabine Triphosphate Accumulation. J. Clin. Oncol. 2001, 19, 3483–3489. [Google Scholar] [CrossRef]
- Verweij, J.; Lee, S.M.; Ruka, W.; Buesa, J.; Coleman, R.; van Hoessel, R.; Seynaeve, C.; di Paola, E.D.; van Glabbeke, M.; Tonelli, D.; et al. Randomized Phase II Study of Docetaxel versus Doxorubicin in First- and Second-Line Chemotherapy for Locally Advanced or Metastatic Soft Tissue Sarcomas in Adults: A Study of the European Organization for Research and Treatment of Cancer Soft Tissue and Bone Sarcoma Group. J. Clin. Oncol. 2000, 18, 2081–2086. [Google Scholar] [CrossRef] [PubMed]
- Guillou, L.; Aurias, A. Soft Tissue Sarcomas with Complex Genomic Profiles. Virchows Arch. 2010, 456, 201–217. [Google Scholar] [CrossRef] [PubMed]
- de Alava, E. Molecular Pathology in Sarcomas. Clin. Transl. Oncol. 2007, 9, 130–144. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Du, X.; Chen, K.; Ylipää, A.; Lazar, A.J.F.; Trent, J.; Lev, D.; Pollock, R.; Hao, X.; Hunt, K.; et al. Genetic Aberrations in Soft Tissue Leiomyosarcoma. Cancer Lett. 2009, 275, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Cancer Genome Atlas Research Network. Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell 2017, 171, 950–965.e28. [Google Scholar] [CrossRef] [PubMed]
- Agaram, N.P.; Zhang, L.; LeLoarer, F.; Silk, T.; Sung, Y.-S.; Scott, S.N.; Kuk, D.; Qin, L.-X.; Berger, M.F.; Antonescu, C.R.; et al. Targeted Exome Sequencing Profiles Genetic Alterations in Leiomyosarcoma. Genes. Chromosomes Cancer 2016, 55, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Movva, S.; Wen, W.; Chen, W.; Millis, S.Z.; Gatalica, Z.; Reddy, S.; von Mehren, M.; Van Tine, B.A. Multi-Platform Profiling of over 2000 Sarcomas: Identification of Biomarkers and Novel Therapeutic Targets. Oncotarget 2015, 6, 12234–12247. [Google Scholar] [CrossRef] [PubMed]
- Cuppens, T.; Moisse, M.; Depreeuw, J.; Annibali, D.; Colas, E.; Gil-Moreno, A.; Huvila, J.; Carpén, O.; Zikán, M.; Matias-Guiu, X.; et al. Integrated Genome Analysis of Uterine Leiomyosarcoma to Identify Novel Driver Genes and Targetable Pathways. Int. J. Cancer 2018, 142, 1230–1243. [Google Scholar] [CrossRef]
- Chudasama, P.; Mughal, S.S.; Sanders, M.A.; Hübschmann, D.; Chung, I.; Deeg, K.I.; Wong, S.-H.; Rabe, S.; Hlevnjak, M.; Zapatka, M.; et al. Integrative Genomic and Transcriptomic Analysis of Leiomyosarcoma. Nat. Commun. 2018, 9, 144. [Google Scholar] [CrossRef]
- Mäkinen, N.; Aavikko, M.; Heikkinen, T.; Taipale, M.; Taipale, J.; Koivisto-Korander, R.; Bützow, R.; Vahteristo, P. Exome Sequencing of Uterine Leiomyosarcomas Identifies Frequent Mutations in TP53, ATRX, and MED12. PLoS Genet. 2016, 12, e1005850. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.J.; Yoo, N.S.; Hagemann, I.S.; Pfeifer, J.D.; Cottrell, C.E.; Abel, H.J.; Duncavage, E.J. Spectrum of Mutations in Leiomyosarcomas Identified by Clinical Targeted Next-Generation Sequencing. Exp. Mol. Pathol. 2017, 102, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Hensley, M.L.; Chavan, S.S.; Solit, D.B.; Murali, R.; Soslow, R.; Chiang, S.; Jungbluth, A.A.; Bandlamudi, C.; Srinivasan, P.; Tap, W.D.; et al. Genomic Landscape of Uterine Sarcomas Defined Through Prospective Clinical Sequencing. Clin. Cancer Res. 2020, 26, 3881–3888. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.D.; Babichev, Y.; Fuligni, F.; Comitani, F.; Layeghifard, M.; Venier, R.E.; Dentro, S.C.; Maheshwari, A.; Guram, S.; Wunker, C.; et al. Lineage-Defined Leiomyosarcoma Subtypes Emerge Years before Diagnosis and Determine Patient Survival. Nat. Commun. 2021, 12, 4496. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Rao, U.N.M.; Jasani, S.; Khanna, V.; Yaw, K.; Surti, U. Loss of DNA Copy Number of 10q Is Associated with Aggressive Behavior of Leiomyosarcomas: A Comparative Genomic Hybridization Study. Cancer Genet. Cytogenet. 2005, 161, 20–27. [Google Scholar] [CrossRef]
- Larramendy, M.L.; Kaur, S.; Svarvar, C.; Böhling, T.; Knuutila, S. Gene Copy Number Profiling of Soft-Tissue Leiomyosarcomas by Array-Comparative Genomic Hybridization. Cancer Genet. Cytogenet. 2006, 169, 94–101. [Google Scholar] [CrossRef]
- El-Rifai, W.; Sarlomo-Rikala, M.; Knuutila, S.; Miettinen, M. DNA Copy Number Changes in Development and Progression in Leiomyosarcomas of Soft Tissues. Am. J. Pathol. 1998, 153, 985–990. [Google Scholar] [CrossRef][Green Version]
- Otaño-Joos, M.; Mechtersheimer, G.; Ohl, S.; Lehnert, T.; Willeke, F.; Möller, P.; Otto, H.F.; Lichter, P.; Joos, S. Analysis of chromosome copy number changes in leiomyosarcoma through molecular cytogenetic methods. Verh. Dtsch. Ges. Pathol. 1998, 82, 207–209. [Google Scholar]
- Parente, F.; Grosgeorge, J.; Coindre, J.M.; Terrier, P.; Vilain, O.; Turc-Carel, C. Comparative Genomic Hybridization Reveals Novel Chromosome Deletions in 90 Primary Soft Tissue Tumors. Cancer Genet. Cytogenet. 1999, 115, 89–95. [Google Scholar] [CrossRef]
- George, S.; Serrano, C.; Hensley, M.L.; Ray-Coquard, I. Soft Tissue and Uterine Leiomyosarcoma. J. Clin. Oncol. 2018, 36, 144–150. [Google Scholar] [CrossRef]
- Stephens, P.J.; Greenman, C.D.; Fu, B.; Yang, F.; Bignell, G.R.; Mudie, L.J.; Pleasance, E.D.; Lau, K.W.; Beare, D.; Stebbings, L.A.; et al. Massive Genomic Rearrangement Acquired in a Single Catastrophic Event during Cancer Development. Cell 2011, 144, 27–40. [Google Scholar] [CrossRef]
- Andreassen, P.R.; Lohez, O.D.; Lacroix, F.B.; Margolis, R.L. Tetraploid State Induces P53-Dependent Arrest of Nontransformed Mammalian Cells in G1. Mol. Biol. Cell 2001, 12, 1315–1328. [Google Scholar] [CrossRef] [PubMed]
- Storchova, Z.; Pellman, D. From Polyploidy to Aneuploidy, Genome Instability and Cancer. Nat. Rev. Mol. Cell Biol. 2004, 5, 45–54. [Google Scholar] [CrossRef]
- Storchova, Z.; Kuffer, C. The Consequences of Tetraploidy and Aneuploidy. J. Cell Sci. 2008, 121, 3859–3866. [Google Scholar] [CrossRef] [PubMed]
- Gounder, M.M.; Agaram, N.P.; Trabucco, S.E.; Robinson, V.; Ferraro, R.A.; Millis, S.Z.; Krishnan, A.; Lee, J.; Attia, S.; Abida, W.; et al. Clinical Genomic Profiling in the Management of Patients with Soft Tissue and Bone Sarcoma. Nat. Commun. 2022, 13, 3406. [Google Scholar] [CrossRef]
- Nacev, B.A.; Sanchez-Vega, F.; Smith, S.A.; Antonescu, C.R.; Rosenbaum, E.; Shi, H.; Tang, C.; Socci, N.D.; Rana, S.; Gularte-Mérida, R.; et al. Clinical Sequencing of Soft Tissue and Bone Sarcomas Delineates Diverse Genomic Landscapes and Potential Therapeutic Targets. Nat. Commun. 2022, 13, 3405. [Google Scholar] [CrossRef]
- AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017, 7, 818–831. [Google Scholar] [CrossRef]
- Pugh, T.J.; Bell, J.L.; Bruce, J.P.; Doherty, G.J.; Galvin, M.; Green, M.F.; Hunter-Zinck, H.; Kumari, P.; Lenoue-Newton, M.L.; Li, M.M.; et al. AACR Project GENIE: 100,000 Cases and Beyond. Cancer Discov. 2022, 12, 2044–2057. [Google Scholar] [CrossRef]
- Francis, P.; Namløs, H.M.; Müller, C.; Edén, P.; Fernebro, J.; Berner, J.-M.; Bjerkehagen, B.; Akerman, M.; Bendahl, P.-O.; Isinger, A.; et al. Diagnostic and Prognostic Gene Expression Signatures in 177 Soft Tissue Sarcomas: Hypoxia-Induced Transcription Profile Signifies Metastatic Potential. BMC Genom. 2007, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.H.; Lee, C.-H.; Witten, D.M.; Gleason, B.C.; Edris, B.; Espinosa, I.; Zhu, S.; Li, R.; Montgomery, K.D.; Marinelli, R.J.; et al. Discovery of Molecular Subtypes in Leiomyosarcoma through Integrative Molecular Profiling. Oncogene 2010, 29, 845–854. [Google Scholar] [CrossRef]
- Baird, K.; Davis, S.; Antonescu, C.R.; Harper, U.L.; Walker, R.L.; Chen, Y.; Glatfelter, A.A.; Duray, P.H.; Meltzer, P.S. Gene Expression Profiling of Human Sarcomas: Insights into Sarcoma Biology. Cancer Res. 2005, 65, 9226–9235. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Jo, V.Y.; Mills, A.M.; Zhu, S.X.; Lee, C.-H.; Espinosa, I.; Nucci, M.R.; Varma, S.; Forgó, E.; Hastie, T.; et al. Clinically Relevant Molecular Subtypes in Leiomyosarcoma. Clin. Cancer Res. 2015, 21, 3501–3511. [Google Scholar] [CrossRef] [PubMed]
- Schutz, F.A.B.; Choueiri, T.K.; Sternberg, C.N. Pazopanib: Clinical Development of a Potent Anti-Angiogenic Drug. Crit. Rev. Oncol. Hematol. 2011, 77, 163–171. [Google Scholar] [CrossRef] [PubMed]
- van der Graaf, W.T.A.; Blay, J.-Y.; Chawla, S.P.; Kim, D.-W.; Bui-Nguyen, B.; Casali, P.G.; Schöffski, P.; Aglietta, M.; Staddon, A.P.; Beppu, Y.; et al. Pazopanib for Metastatic Soft-Tissue Sarcoma (PALETTE): A Randomised, Double-Blind, Placebo-Controlled Phase 3 Trial. Lancet 2012, 379, 1879–1886. [Google Scholar] [CrossRef] [PubMed]
- Benson, C.; Ray-Coquard, I.; Sleijfer, S.; Litière, S.; Blay, J.-Y.; Le Cesne, A.; Papai, Z.; Judson, I.; Schöffski, P.; Chawla, S.; et al. Outcome of Uterine Sarcoma Patients Treated with Pazopanib: A Retrospective Analysis Based on Two European Organisation for Research and Treatment of Cancer (EORTC) Soft Tissue and Bone Sarcoma Group (STBSG) Clinical Trials 62043 and 62072. Gynecol. Oncol. 2016, 142, 89–94. [Google Scholar] [CrossRef]
- Marcus, L.; Lemery, S.J.; Keegan, P.; Pazdur, R. FDA Approval Summary: Pembrolizumab for the Treatment of Microsatellite Instability-High Solid Tumors. Clin. Cancer Res. 2019, 25, 3753–3758. [Google Scholar] [CrossRef] [PubMed]
- Marcus, L.; Fashoyin-Aje, L.A.; Donoghue, M.; Yuan, M.; Rodriguez, L.; Gallagher, P.S.; Philip, R.; Ghosh, S.; Theoret, M.R.; Beaver, J.A.; et al. FDA Approval Summary: Pembrolizumab for the Treatment of Tumor Mutational Burden-High Solid Tumors. Clin. Cancer Res. 2021, 27, 4685–4689. [Google Scholar] [CrossRef]
- Subbiah, V.; Wolf, J.; Konda, B.; Kang, H.; Spira, A.; Weiss, J.; Takeda, M.; Ohe, Y.; Khan, S.; Ohashi, K.; et al. Tumour-Agnostic Efficacy and Safety of Selpercatinib in Patients with RET Fusion-Positive Solid Tumours Other than Lung or Thyroid Tumours (LIBRETTO-001): A Phase 1/2, Open-Label, Basket Trial. Lancet Oncol. 2022, 23, 1261–1273. [Google Scholar] [CrossRef]
- Marcus, L.; Donoghue, M.; Aungst, S.; Myers, C.E.; Helms, W.S.; Shen, G.; Zhao, H.; Stephens, O.; Keegan, P.; Pazdur, R. FDA Approval Summary: Entrectinib for the Treatment of NTRK Gene Fusion Solid Tumors. Clin. Cancer Res. 2021, 27, 928–932. [Google Scholar] [CrossRef]
- Thanopoulou, E.; Thway, K.; Khabra, K.; Judson, I. Treatment of Hormone Positive Uterine Leiomyosarcoma with Aromatase Inhibitors. Clin. Sarcoma Res. 2014, 4, 5. [Google Scholar] [CrossRef]
- Maccaroni, E.; Lunerti, V.; Agostinelli, V.; Giampieri, R.; Zepponi, L.; Pagliacci, A.; Berardi, R. New Insights into Hormonal Therapies in Uterine Sarcomas. Cancers 2022, 14, 921. [Google Scholar] [CrossRef]
- Zang, Y.; Dong, M.; Zhang, K.; Gao, C.; Guo, F.; Wang, Y.; Xue, F. Hormonal Therapy in Uterine Sarcomas. Cancer Med. 2019, 8, 1339–1349. [Google Scholar] [CrossRef]
- Davidson, B.; Kjæreng, M.L.; Førsund, M.; Danielsen, H.E.; Kristensen, G.B.; Abeler, V.M. Progesterone Receptor Expression Is an Independent Prognosticator in FIGO Stage I Uterine Leiomyosarcoma. Am. J. Clin. Pathol. 2016, 145, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Akhan, S.E.; Yavuz, E.; Tecer, A.; Iyibozkurt, C.A.; Topuz, S.; Tuzlali, S.; Bengisu, E.; Berkman, S. The Expression of Ki-67, P53, Estrogen and Progesterone Receptors Affecting Survival in Uterine Leiomyosarcomas. A Clinicopathologic Study. Gynecol. Oncol. 2005, 99, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Bodner, K.; Bodner-Adler, B.; Kimberger, O.; Czerwenka, K.; Leodolter, S.; Mayerhofer, K. Estrogen and Progesterone Receptor Expression in Patients with Uterine Leiomyosarcoma and Correlation with Different Clinicopathological Parameters. Anticancer. Res. 2003, 23, 729–732. [Google Scholar]
- Kitaoka, Y.; Kitawaki, J.; Koshiba, H.; Inoue, S.; Ishihara, H.; Teramoto, M.; Honjo, H. Aromatase Cytochrome P450 and Estrogen and Progesterone Receptors in Uterine Sarcomas: Correlation with Clinical Parameters. J. Steroid Biochem. Mol. Biol. 2004, 88, 183–189. [Google Scholar] [CrossRef]
- Leitao, M.M.; Hensley, M.L.; Barakat, R.R.; Aghajanian, C.; Gardner, G.J.; Jewell, E.L.; O’Cearbhaill, R.; Soslow, R.A. Immunohistochemical Expression of Estrogen and Progesterone Receptors and Outcomes in Patients with Newly Diagnosed Uterine Leiomyosarcoma. Gynecol. Oncol. 2012, 124, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Ioffe, Y.J.; Li, A.J.; Walsh, C.S.; Karlan, B.Y.; Leuchter, R.; Forscher, C.; Cass, I. Hormone Receptor Expression in Uterine Sarcomas: Prognostic and Therapeutic Roles. Gynecol. Oncol. 2009, 115, 466–471. [Google Scholar] [CrossRef] [PubMed]
- George, S.; Feng, Y.; Manola, J.; Nucci, M.R.; Butrynski, J.E.; Morgan, J.A.; Ramaiya, N.; Quek, R.; Penson, R.T.; Wagner, A.J.; et al. Phase 2 Trial of Aromatase Inhibition with Letrozole in Patients with Uterine Leiomyosarcomas Expressing Estrogen and/or Progesterone Receptors. Cancer 2014, 120, 738–743. [Google Scholar] [CrossRef]
- Sanfilippo, R.; Sbaraglia, M.; Fabbroni, C.; Croce, S.; Ray-Coquard, I.; Guermazi, F.; Paolini, B.; Blanc-Durand, F.; Lecesne, A.; Chiappa, V.; et al. Low-Grade Uterine Leiomyosarcoma Is Highly Sensitive to Hormonal Treatment. Clin. Cancer Res. 2023, 29, 4679–4684. [Google Scholar] [CrossRef]
- Vincenzi, B.; Stacchiotti, S.; Collini, P.; Pantano, F.; Rabitti, C.; Perrone, G.; Iuliani, M.; Baldi, A.; Badalamenti, G.; Sanfilippo, R.; et al. Human Equilibrative Nucleoside Transporter 1 Gene Expression Is Associated with Gemcitabine Efficacy in Advanced Leiomyosarcoma and Angiosarcoma. Br. J. Cancer 2017, 117, 340–346. [Google Scholar] [CrossRef]
- Schöffski, P.; Taron, M.; Jimeno, J.; Grosso, F.; Sanfilipio, R.; Casali, P.G.; Le Cesne, A.; Jones, R.L.; Blay, J.-Y.; Poveda, A.; et al. Predictive Impact of DNA Repair Functionality on Clinical Outcome of Advanced Sarcoma Patients Treated with Trabectedin: A Retrospective Multicentric Study. Eur. J. Cancer 2011, 47, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Venkitaraman, A.R. Cancer Susceptibility and the Functions of BRCA1 and BRCA2. Cell 2002, 108, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Moynahan, M.E.; Chiu, J.W.; Koller, B.H.; Jasin, M. Brca1 Controls Homology-Directed DNA Repair. Mol. Cell 1999, 4, 511–518. [Google Scholar] [CrossRef]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.J.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA Repair Defect in BRCA Mutant Cells as a Therapeutic Strategy. Nature 2005, 434, 917–921. [Google Scholar] [CrossRef]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific Killing of BRCA2-Deficient Tumours with Inhibitors of Poly(ADP-Ribose) Polymerase. Nature 2005, 434, 913–917. [Google Scholar] [CrossRef]
- Cerrato, A.; Morra, F.; Celetti, A. Use of Poly ADP-Ribose Polymerase [PARP] Inhibitors in Cancer Cells Bearing DDR Defects: The Rationale for Their Inclusion in the Clinic. J. Exp. Clin. Cancer Res. 2016, 35, 179. [Google Scholar] [CrossRef]
- Helleday, T. The Underlying Mechanism for the PARP and BRCA Synthetic Lethality: Clearing up the Misunderstandings. Mol. Oncol. 2011, 5, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Teo, M.Y.; Bambury, R.M.; Zabor, E.C.; Jordan, E.; Al-Ahmadie, H.; Boyd, M.E.; Bouvier, N.; Mullane, S.A.; Cha, E.K.; Roper, N.; et al. DNA Damage Response and Repair Gene Alterations Are Associated with Improved Survival in Patients with Platinum-Treated Advanced Urothelial Carcinoma. Clin. Cancer Res. 2017, 23, 3610–3618. [Google Scholar] [CrossRef]
- Sehdev, A.; Gbolahan, O.; Hancock, B.A.; Stanley, M.; Shahda, S.; Wan, J.; Wu, H.H.; Radovich, M.; O’Neil, B.H. Germline and Somatic DNA Damage Repair Gene Mutations and Overall Survival in Metastatic Pancreatic Adenocarcinoma Patients Treated with FOLFIRINOX. Clin. Cancer Res. 2018, 24, 6204–6211. [Google Scholar] [CrossRef]
- Seligson, N.D.; Kautto, E.A.; Passen, E.N.; Stets, C.; Toland, A.E.; Millis, S.Z.; Meyer, C.F.; Hays, J.L.; Chen, J.L. BRCA1/2 Functional Loss Defines a Targetable Subset in Leiomyosarcoma. Oncologist 2019, 24, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, E.; Jonsson, P.; Seier, K.; Qin, L.-X.; Chi, P.; Dickson, M.; Gounder, M.; Kelly, C.; Keohan, M.L.; Nacev, B.; et al. Clinical Outcome of Leiomyosarcomas with Somatic Alteration in Homologous Recombination Pathway Genes. JCO Precis. Oncol. 2020, 4, PO.20.00122. [Google Scholar] [CrossRef]
- Dall, G.; Vandenberg, C.J.; Nesic, K.; Ratnayake, G.; Zhu, W.; Vissers, J.H.A.; Bedő, J.; Penington, J.; Wakefield, M.J.; Kee, D.; et al. Targeting Homologous Recombination Deficiency in Uterine Leiomyosarcoma. J. Exp. Clin. Cancer Res. 2023, 42, 112. [Google Scholar] [CrossRef] [PubMed]
- Dunlop, C.R.; Wallez, Y.; Johnson, T.I.; Bernaldo de Quirós Fernández, S.; Durant, S.T.; Cadogan, E.B.; Lau, A.; Richards, F.M.; Jodrell, D.I. Complete Loss of ATM Function Augments Replication Catastrophe Induced by ATR Inhibition and Gemcitabine in Pancreatic Cancer Models. Br. J. Cancer 2020, 123, 1424–1436. [Google Scholar] [CrossRef] [PubMed]
- Schofield, M.J.; Hsieh, P. DNA Mismatch Repair: Molecular Mechanisms and Biological Function. Annu. Rev. Microbiol. 2003, 57, 579–608. [Google Scholar] [CrossRef] [PubMed]
- Nilbert, M.; Therkildsen, C.; Nissen, A.; Akerman, M.; Bernstein, I. Sarcomas Associated with Hereditary Nonpolyposis Colorectal Cancer: Broad Anatomical and Morphological Spectrum. Fam. Cancer 2009, 8, 209–213. [Google Scholar] [CrossRef]
- Doyle, L.A.; Nowak, J.A.; Nathenson, M.J.; Thornton, K.; Wagner, A.J.; Johnson, J.M.; Albrayak, A.; George, S.; Sholl, L.M. Characteristics of Mismatch Repair Deficiency in Sarcomas. Mod. Pathol. 2019, 32, 977–987. [Google Scholar] [CrossRef]
- Grignani, G.; D’Ambrosio, L.; Pignochino, Y.; Palmerini, E.; Zucchetti, M.; Boccone, P.; Aliberti, S.; Stacchiotti, S.; Bertulli, R.; Piana, R.; et al. Trabectedin and Olaparib in Patients with Advanced and Non-Resectable Bone and Soft-Tissue Sarcomas (TOMAS): An Open-Label, Phase 1b Study from the Italian Sarcoma Group. Lancet Oncol. 2018, 19, 1360–1371. [Google Scholar] [CrossRef]
- D’Ambrosio, L.; Merlini, A.; Brunello, A.; Ferraresi, V. TOMAS2: A Randomized Phase 2 Study from the Italian Sarcoma Group (ISG) of Trabectedin plus Olaparib (T+O) or Trabectedin (T) in Advanced, Metastatic, or Unresectable Soft Tissue Sarcomas (STS) after Failure of Standard Treatments. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2023, 34 (Suppl. S2), S1281–S1282. [Google Scholar] [CrossRef]
- Ingham, M.; Allred, J.B.; Chen, L.; Das, B.; Kochupurakkal, B.; Gano, K.; George, S.; Attia, S.; Burgess, M.A.; Seetharam, M.; et al. Phase II Study of Olaparib and Temozolomide for Advanced Uterine Leiomyosarcoma (NCI Protocol 10250). J Clin Oncol 2023, 41, 4154–4163. [Google Scholar] [CrossRef]
- Vikas, P.; Borcherding, N.; Chennamadhavuni, A.; Garje, R. Therapeutic Potential of Combining PARP Inhibitor and Immunotherapy in Solid Tumors. Front. Oncol. 2020, 10, 570. [Google Scholar] [CrossRef]
- Yap, T.A.; Bardia, A.; Dvorkin, M.; Galsky, M.D.; Beck, J.T.; Wise, D.R.; Karyakin, O.; Rubovszky, G.; Kislov, N.; Rohrberg, K.; et al. Avelumab Plus Talazoparib in Patients with Advanced Solid Tumors: The JAVELIN PARP Medley Nonrandomized Controlled Trial. JAMA Oncol. 2023, 9, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Ayodele, O.; Wang, B.X.; Pfister, T.D.; Al-Ezzi, E.M. A Phase II, Open-Label, Randomized Trial of Durvalumab (D) with Olaparib (O) or Cediranib (C) in Patients (Pts) with Leiomyosarcoma (LMS). J. Clin. Oncol. 2021, 39, 11522. [Google Scholar] [CrossRef]
- Fresno Vara, J.A.; Casado, E.; de Castro, J.; Cejas, P.; Belda-Iniesta, C.; González-Barón, M. PI3K/Akt Signalling Pathway and Cancer. Cancer Treat. Rev. 2004, 30, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Weng, L.P.; Smith, W.M.; Dahia, P.L.; Ziebold, U.; Gil, E.; Lees, J.A.; Eng, C. PTEN Suppresses Breast Cancer Cell Growth by Phosphatase Activity-Dependent G1 Arrest Followed by Cell Death. Cancer Res. 1999, 59, 5808–5814. [Google Scholar]
- Zou, Z.; Tao, T.; Li, H.; Zhu, X. mTOR Signaling Pathway and mTOR Inhibitors in Cancer: Progress and Challenges. Cell Biosci. 2020, 10, 31. [Google Scholar] [CrossRef]
- Gibault, L.; Pérot, G.; Chibon, F.; Bonnin, S.; Lagarde, P.; Terrier, P.; Coindre, J.-M.; Aurias, A. New Insights in Sarcoma Oncogenesis: A Comprehensive Analysis of a Large Series of 160 Soft Tissue Sarcomas with Complex Genomics. J. Pathol. 2011, 223, 64–71. [Google Scholar] [CrossRef]
- Gibault, L.; Ferreira, C.; Pérot, G.; Audebourg, A.; Chibon, F.; Bonnin, S.; Lagarde, P.; Vacher-Lavenu, M.-C.; Terrier, P.; Coindre, J.-M.; et al. From PTEN Loss of Expression to RICTOR Role in Smooth Muscle Differentiation: Complex Involvement of the mTOR Pathway in Leiomyosarcomas and Pleomorphic Sarcomas. Mod. Pathol. 2012, 25, 197–211. [Google Scholar] [CrossRef]
- Cote, G.M.; He, J.; Choy, E. Next-Generation Sequencing for Patients with Sarcoma: A Single Center Experience. Oncologist 2018, 23, 234–242. [Google Scholar] [CrossRef]
- Hernando, E.; Charytonowicz, E.; Dudas, M.E.; Menendez, S.; Matushansky, I.; Mills, J.; Socci, N.D.; Behrendt, N.; Ma, L.; Maki, R.G.; et al. The AKT-mTOR Pathway Plays a Critical Role in the Development of Leiomyosarcomas. Nat. Med. 2007, 13, 748–753. [Google Scholar] [CrossRef]
- Li, J.; Dang, Y.; Gao, J.; Li, Y.; Zou, J.; Shen, L. PI3K/AKT/mTOR Pathway Is Activated after Imatinib Secondary Resistance in Gastrointestinal Stromal Tumors (GISTs). Med. Oncol. 2015, 32, 111. [Google Scholar] [CrossRef]
- George, S.; Miao, D.; Demetri, G.D.; Adeegbe, D.; Rodig, S.J.; Shukla, S.; Lipschitz, M.; Amin-Mansour, A.; Raut, C.P.; Carter, S.L.; et al. Loss of PTEN Is Associated with Resistance to Anti-PD-1 Checkpoint Blockade Therapy in Metastatic Uterine Leiomyosarcoma. Immunity 2017, 46, 197–204. [Google Scholar] [CrossRef]
- Peng, W.; Chen, J.Q.; Liu, C.; Malu, S.; Creasy, C.; Tetzlaff, M.T.; Xu, C.; McKenzie, J.A.; Zhang, C.; Liang, X.; et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov. 2016, 6, 202–216. [Google Scholar] [CrossRef]
- Ali, E.S.; Mitra, K.; Akter, S.; Ramproshad, S.; Mondal, B.; Khan, I.N.; Islam, M.T.; Sharifi-Rad, J.; Calina, D.; Cho, W.C. Recent Advances and Limitations of mTOR Inhibitors in the Treatment of Cancer. Cancer Cell Int. 2022, 22, 284. [Google Scholar] [CrossRef] [PubMed]
- Demetri, G.D.; Chawla, S.P.; Ray-Coquard, I.; Le Cesne, A.; Staddon, A.P.; Milhem, M.M.; Penel, N.; Riedel, R.F.; Bui-Nguyen, B.; Cranmer, L.D.; et al. Results of an International Randomized Phase III Trial of the Mammalian Target of Rapamycin Inhibitor Ridaforolimus versus Placebo to Control Metastatic Sarcomas in Patients after Benefit from Prior Chemotherapy. J. Clin. Oncol. 2013, 31, 2485–2492. [Google Scholar] [CrossRef]
- Wan, X.; Harkavy, B.; Shen, N.; Grohar, P.; Helman, L.J. Rapamycin Induces Feedback Activation of Akt Signaling through an IGF-1R-Dependent Mechanism. Oncogene 2007, 26, 1932–1940. [Google Scholar] [CrossRef] [PubMed]
- Fourneaux, B.; Chaire, V.; Lucchesi, C.; Karanian, M.; Pineau, R.; Laroche-Clary, A.; Italiano, A. Dual Inhibition of the PI3K/AKT/mTOR Pathway Suppresses the Growth of Leiomyosarcomas but Leads to ERK Activation through mTORC2: Biological and Clinical Implications. Oncotarget 2017, 8, 7878–7890. [Google Scholar] [CrossRef]
- Mendes-Pereira, A.M.; Lord, C.J.; Ashworth, A. NLK Is a Novel Therapeutic Target for PTEN Deficient Tumour Cells. PLoS ONE 2012, 7, e47249. [Google Scholar] [CrossRef] [PubMed]
- Kostine, M.; Briaire-de Bruijn, I.H.; Cleven, A.H.G.; Vervat, C.; Corver, W.E.; Schilham, M.W.; Van Beelen, E.; van Boven, H.; Haas, R.L.; Italiano, A.; et al. Increased Infiltration of M2-Macrophages, T-Cells and PD-L1 Expression in High Grade Leiomyosarcomas Supports Immunotherapeutic Strategies. Oncoimmunology 2018, 7, e1386828. [Google Scholar] [CrossRef]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage Polarization: Tumor-Associated Macrophages as a Paradigm for Polarized M2 Mononuclear Phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef]
- Lee, C.-H.; Espinosa, I.; Vrijaldenhoven, S.; Subramanian, S.; Montgomery, K.D.; Zhu, S.; Marinelli, R.J.; Peterse, J.L.; Poulin, N.; Nielsen, T.O.; et al. Prognostic Significance of Macrophage Infiltration in Leiomyosarcomas. Clin. Cancer Res. 2008, 14, 1423–1430. [Google Scholar] [CrossRef]
- Ben-Ami, E.; Barysauskas, C.M.; Solomon, S.; Tahlil, K.; Malley, R.; Hohos, M.; Polson, K.; Loucks, M.; Severgnini, M.; Patel, T.; et al. Immunotherapy with Single Agent Nivolumab for Advanced Leiomyosarcoma of the Uterus: Results of a Phase 2 Study. Cancer 2017, 123, 3285–3290. [Google Scholar] [CrossRef] [PubMed]
- Tawbi, H.A.; Burgess, M.; Bolejack, V.; Van Tine, B.A.; Schuetze, S.M.; Hu, J.; D’Angelo, S.; Attia, S.; Riedel, R.F.; Priebat, D.A.; et al. Pembrolizumab in Advanced Soft-Tissue Sarcoma and Bone Sarcoma (SARC028): A Multicentre, Two-Cohort, Single-Arm, Open-Label, Phase 2 Trial. Lancet Oncol. 2017, 18, 1493–1501. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, S.P.; Mahoney, M.R.; Van Tine, B.A.; Atkins, J.; Milhem, M.M.; Jahagirdar, B.N.; Antonescu, C.R.; Horvath, E.; Tap, W.D.; Schwartz, G.K.; et al. Nivolumab with or without Ipilimumab Treatment for Metastatic Sarcoma (Alliance A091401): Two Open-Label, Non-Comparative, Randomised, Phase 2 Trials. Lancet Oncol. 2018, 19, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Wilky, B.A.; Trent, J.C.; Gordon, M.; El-Khoueiry, A.; Bullock, B.S. Efficacy and Safety of Botensilimab plus Balstilimab in Patients with Refractory Metastatic Sarcoma. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2023, 34, S1032–S1033. [Google Scholar] [CrossRef]
- Smrke, A.; Ostler, A.; Napolitano, A.; Vergnano, M. 1526MO GEMMK: A Phase I Study of Gemcitabine (Gem) and Pembrolizumab (Pem) in Patients (Pts) with Leiomyosarcoma (LMS) and Undifferentiated Pleomorphic Sarcoma UPS). Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2021, 32, S1114. [Google Scholar] [CrossRef]
- Napolitano, A. Final Results of GEMMK: A Phase I Trial of Gemcitabine and Pembrolizumab in Patients with Leiomyosarcoma and Undifferentiated Pleomorphic Sarcoma. In Proceedings of the CTOS Annual Meeting, Vancouver, BC, Canada, 16–19 November 2022. [Google Scholar]
- Nathenson, M.; Choy, E.; Carr, N.D.; Hibbard, H.D. Phase II Study of Eribulin and Pembrolizumab in Patients (Pts) with Metastatic Soft Tissue Sarcomas (STS): Report of LMS Cohort. J. Clin. Oncol. 2020, 38, 11559. [Google Scholar] [CrossRef]
- Martin Broto, J.; Diaz Beveridge, R.; Moura, D.; Ramos, R.; Martinez-Trufero, J. ImmunoSarc2: A Spanish Sarcoma Group (GEIS) Phase Ib Trial of Doxorubicin and Dacarbazine plus Nivolumab in First Line Treatment of Advanced Leiomyosarcoma. J. Clin. Oncol. 2023, 41, 11502. [Google Scholar] [CrossRef]
- Rosenbaum, E.; Qin, L.; Dickson, M.A.; Keohan, M.L.; Gounder, M.M. Interim Results of a Phase II Trial of First Line Retifanlimab (R) plus Gemcitabine and Docetaxel (GD) in Patients (Pts) with Advanced Soft Tissue Sarcoma (STS). J. Clin. Oncol. 2023, 41, 11518. [Google Scholar] [CrossRef]
- Reichardt, P.; Andreou, D.; Florcken, A.; Gross, T.; Richter, S. Efficacy and Safety of Nivolumab and Trabectedin in Pretreated Patients with Advanced Soft Tissue Sarcomas (STS): Results of a Phase II Trial of the German Interdisciplinary Sarcoma Group (GISG-15, NitraSarc). J. Clin. Oncol. 2023, 41, 11500. [Google Scholar] [CrossRef]
- Wilky, B.A.; Maleddu, A.; Mailhot, A.; Cartwright, C.; Gao, D. A Single-Arm, Open-Label Phase 2 Trial of Doxorubicin plus Zalifrelimab, a CTLA-4 Inhibitor, with Balstilimab, a PD-1 Inhibitor, in Patients with Advanced/Metastatic Soft Tissue Sarcomas. J. Clin. Oncol. 2023, 41, 11501. [Google Scholar] [CrossRef]
- Wilky, B.A.; Trucco, M.M.; Subhawong, T.K.; Florou, V.; Park, W.; Kwon, D.; Wieder, E.D.; Kolonias, D.; Rosenberg, A.E.; Kerr, D.A.; et al. Axitinib plus Pembrolizumab in Patients with Advanced Sarcomas Including Alveolar Soft-Part Sarcoma: A Single-Centre, Single-Arm, Phase 2 Trial. Lancet Oncol. 2019, 20, 837–848. [Google Scholar] [CrossRef]
- Movva, S.; Avutu, V.; Chi, P.; Dickson, M.A.; Gounder, M.M. A Pilot Study of Lenvatinib plus Pembrolizumab in Patients with Advanced Sarcoma. J. Clin. Oncol. 2023, 41, 11517. [Google Scholar] [CrossRef]
- Van Tine, B.A.; Eulo, V.; Toeniskoetter, J.; Ruff, T.; Luo, J.; Kemp, L. Randomized Phase II Trial of Cabozantinib Combined with PD-1 and CTLA-4 Inhibition versus Cabozantinib in Metastatic Soft Tissue Sarcoma. J. Clin. Oncol. 2023, 41, LBA11504. [Google Scholar] [CrossRef]
- Sha, D.; Jin, Z.; Budczies, J.; Kluck, K.; Stenzinger, A.; Sinicrope, F.A. Tumor Mutational Burden as a Predictive Biomarker in Solid Tumors. Cancer Discov. 2020, 10, 1808–1825. [Google Scholar] [CrossRef]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 Human Cancer Genomes Reveals the Landscape of Tumor Mutational Burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.J.; Frazier, W.A. Integrin-Associated Protein (CD47) and Its Ligands. Trends Cell Biol. 2001, 11, 130–135. [Google Scholar] [CrossRef]
- Edris, B.; Weiskopf, K.; Volkmer, A.K.; Volkmer, J.-P.; Willingham, S.B.; Contreras-Trujillo, H.; Liu, J.; Majeti, R.; West, R.B.; Fletcher, J.A.; et al. Antibody Therapy Targeting the CD47 Protein Is Effective in a Model of Aggressive Metastatic Leiomyosarcoma. Proc. Natl. Acad. Sci. USA 2012, 109, 6656–6661. [Google Scholar] [CrossRef]
- Sant, C.; Kelly, C.M.; Gordon, E.M.; Quon, D. TTI-621-03: A Phase I/II Study of TTI-621 in Combination with Doxorubicin in Patients with Unresectable or Metastatic High-Grade Leiomyosarcoma (LMS). J. Clin. Oncol. 2022, 40, TPS11593. [Google Scholar]
- Movva, S.; Druta, M.; Davis, L.E.; Monga, V.; Milhem, M.M. Safety and Clinical Activity of TTI-621 in Combination with Doxorubicin in Patients with Unresectable or Metastatic High-Grade Leiomyosarcoma: Results from the Low-Dose Expansion Cohort. J. Clin. Oncol. 2023, 41, 11508. [Google Scholar] [CrossRef]
- Espinosa, I.; Beck, A.H.; Lee, C.-H.; Zhu, S.; Montgomery, K.D.; Marinelli, R.J.; Ganjoo, K.N.; Nielsen, T.O.; Gilks, C.B.; West, R.B.; et al. Coordinate Expression of Colony-Stimulating Factor-1 and Colony-Stimulating Factor-1-Related Proteins Is Associated with Poor Prognosis in Gynecological and Nongynecological Leiomyosarcoma. Am. J. Pathol. 2009, 174, 2347–2356. [Google Scholar] [CrossRef] [PubMed]
- Yonemitsu, K.; Pan, C.; Fujiwara, Y.; Miyasato, Y.; Shiota, T.; Yano, H.; Hosaka, S.; Tamada, K.; Yamamoto, Y.; Komohara, Y. GM-CSF Derived from the Inflammatory Microenvironment Potentially Enhanced PD-L1 Expression on Tumor-Associated Macrophages in Human Breast Cancer. Sci. Rep. 2022, 12, 12007. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Roca, C.; Cassier, P.; Zamarin, D.; Machiels, J.-P.; Perez Gracia, J.L.; Stephen Hodi, F.; Taus, A.; Martinez Garcia, M.; Boni, V.; Eder, J.P.; et al. Anti-CSF-1R Emactuzumab in Combination with Anti-PD-L1 Atezolizumab in Advanced Solid Tumor Patients Naïve or Experienced for Immune Checkpoint Blockade. J. Immunother. Cancer 2022, 10, e004076. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Wang, C.; Yang, L.; Wu, J.; Li, M.; Xiao, P.; Xu, Z.; Xu, Y.; Wang, K. Targeting Immune Checkpoints on Tumor-Associated Macrophages in Tumor Immunotherapy. Front. Immunol. 2023, 14, 1199631. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Maruyama, T.; Urade, Y.; Nagata, S. Immunosuppression via Adenosine Receptor Activation by Adenosine Monophosphate Released from Apoptotic Cells. eLife 2014, 3, e02172. [Google Scholar] [CrossRef] [PubMed]
- Gschwandtner, M.; Derler, R.; Midwood, K.S. More Than Just Attractive: How CCL2 Influences Myeloid Cell Behavior Beyond Chemotaxis. Front. Immunol. 2019, 10, 2759. [Google Scholar] [CrossRef] [PubMed]
- Abraham, R.T. Mammalian Target of Rapamycin: Immunosuppressive Drugs Uncover a Novel Pathway of Cytokine Receptor Signaling. Curr. Opin. Immunol. 1998, 10, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Sauer, S.; Bruno, L.; Hertweck, A.; Finlay, D.; Leleu, M.; Spivakov, M.; Knight, Z.A.; Cobb, B.S.; Cantrell, D.; O’Connor, E.; et al. T Cell Receptor Signaling Controls Foxp3 Expression via PI3K, Akt, and mTOR. Proc. Natl. Acad. Sci. USA 2008, 105, 7797–7802. [Google Scholar] [CrossRef]
- Jia, S.; Liu, Z.; Zhang, S.; Liu, P.; Zhang, L.; Lee, S.H.; Zhang, J.; Signoretti, S.; Loda, M.; Roberts, T.M.; et al. Essential Roles of PI(3)K-P110beta in Cell Growth, Metabolism and Tumorigenesis. Nature 2008, 454, 776–779. [Google Scholar] [CrossRef]
- Duan, Z.; Luo, Y. Targeting Macrophages in Cancer Immunotherapy. Signal Transduct. Target. Ther. 2021, 6, 127. [Google Scholar] [CrossRef]
- DeNardo, D.G.; Ruffell, B. Macrophages as Regulators of Tumour Immunity and Immunotherapy. Nat. Rev. Immunol. 2019, 19, 369–382. [Google Scholar] [CrossRef]
- Nacev, B.A.; Jones, K.B.; Intlekofer, A.M.; Yu, J.S.E.; Allis, C.D.; Tap, W.D.; Ladanyi, M.; Nielsen, T.O. The Epigenomics of Sarcoma. Nat. Rev. Cancer 2020, 20, 608–623. [Google Scholar] [CrossRef]
- Choi, J.; Manzano, A.; Dong, W.; Bellone, S.; Bonazzoli, E.; Zammataro, L.; Yao, X.; Deshpande, A.; Zaidi, S.; Guglielmi, A.; et al. Integrated Mutational Landscape Analysis of Uterine Leiomyosarcomas. Proc. Natl. Acad. Sci. USA 2021, 118, e2025182118. [Google Scholar] [CrossRef] [PubMed]
- Voon, H.P.J.; Hughes, J.R.; Rode, C.; De La Rosa-Velázquez, I.A.; Jenuwein, T.; Feil, R.; Higgs, D.R.; Gibbons, R.J. ATRX Plays a Key Role in Maintaining Silencing at Interstitial Heterochromatic Loci and Imprinted Genes. Cell Rep. 2015, 11, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Iwase, S.; Xiang, B.; Ghosh, S.; Ren, T.; Lewis, P.W.; Cochrane, J.C.; Allis, C.D.; Picketts, D.J.; Patel, D.J.; Li, H.; et al. ATRX ADD Domain Links an Atypical Histone Methylation Recognition Mechanism to Human Mental-Retardation Syndrome. Nat. Struct. Mol. Biol. 2011, 18, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Paeschke, K.; Capra, J.A.; Zakian, V.A. DNA Replication through G-Quadruplex Motifs Is Promoted by the Saccharomyces Cerevisiae Pif1 DNA Helicase. Cell 2011, 145, 678–691. [Google Scholar] [CrossRef] [PubMed]
- Garbarino, J.; Eckroate, J.; Sundaram, R.K.; Jensen, R.B.; Bindra, R.S. Loss of ATRX Confers DNA Repair Defects and PARP Inhibitor Sensitivity. Transl. Oncol. 2021, 14, 101147. [Google Scholar] [CrossRef] [PubMed]
- Floyd, W.; Pierpoint, M.; Su, C.; Patel, R.; Luo, L.; Deland, K.; Wisdom, A.J.; Zhu, D.; Ma, Y.; DeWitt, S.B.; et al. Atrx Deletion Impairs CGAS/STING Signaling and Increases Sarcoma Response to Radiation and Oncolytic Herpesvirus. J. Clin. Investig. 2023, 133, e149310. [Google Scholar] [CrossRef] [PubMed]
- Darmusey, L.; Pérot, G.; Thébault, N.; Le Guellec, S.; Desplat, N.; Gaston, L.; Delespaul, L.; Lesluyes, T.; Darbo, E.; Gomez-Brouchet, A.; et al. ATRX Alteration Contributes to Tumor Growth and Immune Escape in Pleomorphic Sarcomas. Cancers 2021, 13, 2151. [Google Scholar] [CrossRef]
- Mäkinen, N.; Mehine, M.; Tolvanen, J.; Kaasinen, E.; Li, Y.; Lehtonen, H.J.; Gentile, M.; Yan, J.; Enge, M.; Taipale, M.; et al. MED12, the Mediator Complex Subunit 12 Gene, Is Mutated at High Frequency in Uterine Leiomyomas. Science 2011, 334, 252–255. [Google Scholar] [CrossRef]
- Mittal, P.; Shin, Y.-H.; Yatsenko, S.A.; Castro, C.A.; Surti, U.; Rajkovic, A. Med12 Gain-of-Function Mutation Causes Leiomyomas and Genomic Instability. J. Clin. Investig. 2015, 125, 3280–3284. [Google Scholar] [CrossRef]
- Pérot, G.; Croce, S.; Ribeiro, A.; Lagarde, P.; Velasco, V.; Neuville, A.; Coindre, J.-M.; Stoeckle, E.; Floquet, A.; MacGrogan, G.; et al. MED12 Alterations in Both Human Benign and Malignant Uterine Soft Tissue Tumors. PLoS ONE 2012, 7, e40015. [Google Scholar] [CrossRef]
- Park, M.J.; Shen, H.; Spaeth, J.M.; Tolvanen, J.H.; Failor, C.; Knudtson, J.F.; McLaughlin, J.; Halder, S.K.; Yang, Q.; Bulun, S.E.; et al. Oncogenic Exon 2 Mutations in Mediator Subunit MED12 Disrupt Allosteric Activation of Cyclin C-CDK8/19. J. Biol. Chem. 2018, 293, 4870–4882. [Google Scholar] [CrossRef]
- Turunen, M.; Spaeth, J.M.; Keskitalo, S.; Park, M.J.; Kivioja, T.; Clark, A.D.; Mäkinen, N.; Gao, F.; Palin, K.; Nurkkala, H.; et al. Uterine Leiomyoma-Linked MED12 Mutations Disrupt Mediator-Associated CDK Activity. Cell Rep. 2014, 7, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Yasutake, N.; Iwasaki, T.; Yamamoto, H.; Sonoda, K.; Kodama, K.; Okugawa, K.; Asanoma, K.; Yahata, H.; Kato, K.; Oda, Y. Cyclin-Dependent Kinase 8 Is an Independent Prognosticator in Uterine Leiomyosarcoma. Pathol. Res. Pract. 2022, 235, 153920. [Google Scholar] [CrossRef] [PubMed]
- Buyukcelebi, K.; Chen, X.; Abdula, F.; Elkafas, H.; Duval, A.J.; Ozturk, H.; Seker-Polat, F.; Jin, Q.; Yin, P.; Feng, Y.; et al. Engineered MED12 Mutations Drive Leiomyoma-like Transcriptional and Metabolic Programs by Altering the 3D Genome Compartmentalization. Nat. Commun. 2023, 14, 4057. [Google Scholar] [CrossRef] [PubMed]
- Muralimanoharan, S.; Shamby, R.; Stansbury, N.; Schenken, R.; de la Pena Avalos, B.; Javanmardi, S.; Dray, E.; Sung, P.; Boyer, T.G. Aberrant R-Loop-Induced Replication Stress in MED12-Mutant Uterine Fibroids. Sci. Rep. 2022, 12, 6169. [Google Scholar] [CrossRef] [PubMed]
- Du, K.L.; Ip, H.S.; Li, J.; Chen, M.; Dandre, F.; Yu, W.; Lu, M.M.; Owens, G.K.; Parmacek, M.S. Myocardin Is a Critical Serum Response Factor Cofactor in the Transcriptional Program Regulating Smooth Muscle Cell Differentiation. Mol. Cell Biol. 2003, 23, 2425–2437. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, D.-Z.; Pipes, G.C.T.; Olson, E.N. Myocardin Is a Master Regulator of Smooth Muscle Gene Expression. Proc. Natl. Acad. Sci. USA 2003, 100, 7129–7134. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, D.-Z.; Hockemeyer, D.; McAnally, J.; Nordheim, A.; Olson, E.N. Myocardin and Ternary Complex Factors Compete for SRF to Control Smooth Muscle Gene Expression. Nature 2004, 428, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Pérot, G.; Derré, J.; Coindre, J.-M.; Tirode, F.; Lucchesi, C.; Mariani, O.; Gibault, L.; Guillou, L.; Terrier, P.; Aurias, A. Strong Smooth Muscle Differentiation Is Dependent on Myocardin Gene Amplification in Most Human Retroperitoneal Leiomyosarcomas. Cancer Res. 2009, 69, 2269–2278. [Google Scholar] [CrossRef]
- Guenther, M.G.; Barak, O.; Lazar, M.A. The SMRT and N-CoR Corepressors Are Activating Cofactors for Histone Deacetylase 3. Mol. Cell Biol. 2001, 21, 6091–6101. [Google Scholar] [CrossRef]
- Hasan, N.M.; Sharma, A.; Ruzgar, N.M.; Deshpande, H.; Olino, K.; Khan, S.; Ahuja, N. Epigenetic Signatures Differentiate Uterine and Soft Tissue Leiomyosarcoma. Oncotarget 2021, 12, 1566–1579. [Google Scholar] [CrossRef]
- Renner, M.; Wolf, T.; Meyer, H.; Hartmann, W.; Penzel, R.; Ulrich, A.; Lehner, B.; Hovestadt, V.; Czwan, E.; Egerer, G.; et al. Integrative DNA Methylation and Gene Expression Analysis in High-Grade Soft Tissue Sarcomas. Genome Biol. 2013, 14, r137. [Google Scholar] [CrossRef]
- Koelsche, C.; Schrimpf, D.; Stichel, D.; Sill, M.; Sahm, F.; Reuss, D.E.; Blattner, M.; Worst, B.; Heilig, C.E.; Beck, K.; et al. Sarcoma Classification by DNA Methylation Profiling. Nat. Commun. 2021, 12, 498. [Google Scholar] [CrossRef]
- Monga, V.; Dodd, R.; Scherer, A.; Gutierrez, W.R. Phase Ib Study of Decitabine in Combination with Gemcitabine in Treatment of Advanced Soft Tissue and Bone Sarcomas. J. Clin. Oncol. 2020, 38, 11550. [Google Scholar] [CrossRef]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres Shorten during Ageing of Human Fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef]
- Sobinoff, A.P.; Pickett, H.A. Mechanisms That Drive Telomere Maintenance and Recombination in Human Cancers. Curr. Opin. Genet. Dev. 2020, 60, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Henson, J.D.; Hannay, J.A.; McCarthy, S.W.; Royds, J.A.; Yeager, T.R.; Robinson, R.A.; Wharton, S.B.; Jellinek, D.A.; Arbuckle, S.M.; Yoo, J.; et al. A Robust Assay for Alternative Lengthening of Telomeres in Tumors Shows the Significance of Alternative Lengthening of Telomeres in Sarcomas and Astrocytomas. Clin. Cancer Res. 2005, 11, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Ulaner, G.A.; Huang, H.-Y.; Otero, J.; Zhao, Z.; Ben-Porat, L.; Satagopan, J.M.; Gorlick, R.; Meyers, P.; Healey, J.H.; Huvos, A.G.; et al. Absence of a Telomere Maintenance Mechanism as a Favorable Prognostic Factor in Patients with Osteosarcoma. Cancer Res. 2003, 63, 1759–1763. [Google Scholar]
- Liau, J.-Y.; Tsai, J.-H.; Jeng, Y.-M.; Lee, J.-C.; Hsu, H.-H.; Yang, C.-Y. Leiomyosarcoma with Alternative Lengthening of Telomeres Is Associated with Aggressive Histologic Features, Loss of ATRX Expression, and Poor Clinical Outcome. Am. J. Surg. Pathol. 2015, 39, 236–244. [Google Scholar] [CrossRef]
- Heaphy, C.M.; de Wilde, R.F.; Jiao, Y.; Klein, A.P.; Edil, B.H.; Shi, C.; Bettegowda, C.; Rodriguez, F.J.; Eberhart, C.G.; Hebbar, S.; et al. Altered Telomeres in Tumors with ATRX and DAXX Mutations. Science 2011, 333, 425. [Google Scholar] [CrossRef]
- Lafferty-Whyte, K.; Cairney, C.J.; Will, M.B.; Serakinci, N.; Daidone, M.-G.; Zaffaroni, N.; Bilsland, A.; Keith, W.N. A Gene Expression Signature Classifying Telomerase and ALT Immortalization Reveals an hTERT Regulatory Network and Suggests a Mesenchymal Stem Cell Origin for ALT. Oncogene 2009, 28, 3765–3774. [Google Scholar] [CrossRef]
- Amorim, J.P.; Santos, G.; Vinagre, J.; Soares, P. The Role of ATRX in the Alternative Lengthening of Telomeres (ALT) Phenotype. Genes 2016, 7, 66. [Google Scholar] [CrossRef]
- Lewis, P.W.; Elsaesser, S.J.; Noh, K.-M.; Stadler, S.C.; Allis, C.D. Daxx Is an H3.3-Specific Histone Chaperone and Cooperates with ATRX in Replication-Independent Chromatin Assembly at Telomeres. Proc. Natl. Acad. Sci. USA 2010, 107, 14075–14080. [Google Scholar] [CrossRef]
- Clynes, D.; Jelinska, C.; Xella, B.; Ayyub, H.; Scott, C.; Mitson, M.; Taylor, S.; Higgs, D.R.; Gibbons, R.J. Suppression of the Alternative Lengthening of Telomere Pathway by the Chromatin Remodelling Factor ATRX. Nat. Commun. 2015, 6, 7538. [Google Scholar] [CrossRef]
- Henson, J.D.; Cao, Y.; Huschtscha, L.I.; Chang, A.C.; Au, A.Y.M.; Pickett, H.A.; Reddel, R.R. DNA C-Circles Are Specific and Quantifiable Markers of Alternative-Lengthening-of-Telomeres Activity. Nat. Biotechnol. 2009, 27, 1181–1185. [Google Scholar] [CrossRef]
- Cesare, A.J.; Reddel, R.R. Alternative Lengthening of Telomeres: Models, Mechanisms and Implications. Nat. Rev. Genet. 2010, 11, 319–330. [Google Scholar] [CrossRef]
- Lawlor, R.T.; Veronese, N.; Pea, A.; Nottegar, A.; Smith, L.; Pilati, C.; Demurtas, J.; Fassan, M.; Cheng, L.; Luchini, C. Alternative Lengthening of Telomeres (ALT) Influences Survival in Soft Tissue Sarcomas: A Systematic Review with Meta-Analysis. BMC Cancer 2019, 19, 232. [Google Scholar] [CrossRef]
- Chiappori, A.A.; Kolevska, T.; Spigel, D.R.; Hager, S.; Rarick, M.; Gadgeel, S.; Blais, N.; Von Pawel, J.; Hart, L.; Reck, M.; et al. A Randomized Phase II Study of the Telomerase Inhibitor Imetelstat as Maintenance Therapy for Advanced Non-Small-Cell Lung Cancer. Ann. Oncol. 2015, 26, 354–362. [Google Scholar] [CrossRef]
- Flynn, R.L.; Cox, K.E.; Jeitany, M.; Wakimoto, H.; Bryll, A.R.; Ganem, N.J.; Bersani, F.; Pineda, J.R.; Suvà, M.L.; Benes, C.H.; et al. Alternative Lengthening of Telomeres Renders Cancer Cells Hypersensitive to ATR Inhibitors. Science 2015, 347, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Laroche-Clary, A.; Chaire, V.; Verbeke, S.; Algéo, M.-P.; Malykh, A.; Le Loarer, F.; Italiano, A. ATR Inhibition Broadly Sensitizes Soft-Tissue Sarcoma Cells to Chemotherapy Independent of Alternative Lengthening Telomere (ALT) Status. Sci. Rep. 2020, 10, 7488. [Google Scholar] [CrossRef] [PubMed]
- Deeg, K.I.; Chung, I.; Bauer, C.; Rippe, K. Cancer Cells with Alternative Lengthening of Telomeres Do Not Display a General Hypersensitivity to ATR Inhibition. Front. Oncol. 2016, 6, 186. [Google Scholar] [CrossRef] [PubMed]



| Ref. | Modality | Total n | STLMS | ULMS | Major Findings | TP53 | RB1 | ATRX | CDKN2A | CDKN2B | PTEN | Other |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [16] | WES | 80 | 53 | 27 | Predominantly chromosomal or arm-level deletions. | Mutation, 50%; deep deletion, 9% | Deep deletion, 14% | Deep deletion, 14% | Deep deletion, 13% | |||
| [17] | Targeted exome (230 genes) | 35 | 35 | 0 | Losses of chromosomal regions involving key tumor suppressor genes PTEN (10q), RB1 (13q), CDH1 (16q), and TP53 (17p) were the most frequent genetic events. Gains mainly involved chromosome regions 17p11.2 (MYOCD) and 15q25-26 (IGF1R). | Mutation, 37%; deletion, 43% | Mutation, 8.5%; deletion, 54% | Deletion, 60% | CDH1 deletion, 46% | |||
| [18] | Targeted exome (47 genes) | 751 | 350 | 401 | TP53 mutations in 42.2% of STLMS and 40.5% of ULMS, BRCA2 mutations in 11% of STLMS and 21.7% of ULMS. PTEN mutations in 6.3% of STLMS and 0% of ULMS. | Mutation, 41.7% | Mutation, 5.3% | Mutation, 4.4% | BRCA2 mutation, 17.1% | |||
| [19] | SNP arrays, RNA seq, WGS on subset | 84 | 0 | 84 | Alterations affecting TP53, RB1, PTEN, MED12, YWHAE and VIPR2 were present in the majority of ULMS. Enrichment in PI3K/AKT/mTOR, estrogen-mediated S phase entry, and DNA damage response signaling pathways. | Altered in 92%; mutation, 41.7%; deletion, 33% | 88% | 75% | Mutations in MED12, 12.5%; EIF3A, 16.7%; ABL1, 12.5%, IGF2R, 12.5%; ATR, 8.3%; RAD50, 8.3%. BRCA1, 8.3%. | |||
| [20] | WES | 49 | 39 | 10 | Notable mutational heterogeneity, near-universal loss of TP53 and RB1, widespread DNA copy number alterations with evidence of chromothripsis, and frequent whole-genome duplication. | 49% | 27% | 24% | 57% | |||
| [21] | WES | 19 | 0 | 19 | TP53, MED12, and ATRX mutations were prevalent. | 33% | 26% | MED12, 21% | ||||
| [22] | Targeted exome (151 genes) | 25 | 16 | 9 | CNVs were identified in 85% of cases. Most frequent losses in chromosomes 10 and 13 including PTEN and RB1. Most frequent gains in chromosomes 7 and 17. | 36% | 12% | 16% | ATM, 16%; EGFR, 12% | |||
| [23] | Targeted exome (341–468 genes) | 80 | 0 | 80 | Compared to ESS, STUMP. PTEN alteration frequency was higher in the metastases samples as compared with the primary samples. Genomes of low-grade tumors were largely silent, while 50.5% of high-grade tumors had whole-genome duplication. | 56% | 51% | 31% | ||||
| [24] | WGS | 53 samples (34 patients) | 23 | 11 | Mutational signatures highlight importance of DNA damage repair and homologous recombination deficiency. Dystrophin deletion associated with worse outcome. Whole-genome doubling was prevalent. Analysis of matched primary-metastatic samples suggested divergence 10–30 years prior to diagnosis. | Mutation, 82.3%; deletion, 14.7% | Mutation, 11.8%; deletion, 8.8% | Deletion, 8.8% |
| Ref. | Total n (LMS) | STLMS | ULMS | Modality | Major Findings |
|---|---|---|---|---|---|
| [16] | 80 | 53 | 27 | Bulk RNA sequencing | Identified three subgroups: ULMS group with poor prognosis and two STLMS clusters (C1, C2). C1 with hypermethylation, higher expression of IGF1R and cell cycle control genes, DNA replication, DNA repair, RB1 mutations, PTEN deletion. C2 with more inflammatory cells (NK and mast). |
| [20] | 49 | 39 | 10 | Bulk RNA sequencing | Identified three subgroups. Subgroup 1 with platelet degranulation, complement activation, and metabolism signatures. Subgroup 2 with muscle development and regulation of membrane potential signatures. Subgroup 3 with myofibril assembly, muscle filament function, and cell–cell signaling signatures. |
| [40] | 51 | 35 | 16 | Microarray | Identified three subgroups. Subgroup 1 with muscle contraction and actin cytoskeleton genes. Subgroup 2 with protein metabolism, cell proliferation, and organ development genes. Subgroup 3 with CSF1 response genes. |
| [42] | 99 | 50 | 49 | Bulk RNA sequencing | Identified three subtypes. Validated their findings from Beck et al. study using new cohort (n = 99) and TCGA data (n = 82). Identified IHC-compatible assays for different STLMS subtypes. |
| [19] | 24 | 0 | 24 | Bulk RNA sequencing | Enrichment in PI3K/AKT/mTOR, estrogen-mediated S phase entry, and DNA damage response signaling pathways. |
| [39] | 40 | Microarray | Identified a muscle gene-enriched group of 11. Remaining 29 were heterogeneous. | ||
| [41] | 17 | Microarray | No difference among anatomic site, tumor grade, or metastatic lesions. ULMS enriched for site-specific genes such as regulators of urogenital differentiation, development, and growth (ESR1, HOXA10, PBX1, and FAT) compared to STLMS. | ||
| [24] | 113 (130 samples, 51 newly sequenced, 79 from TCGA) | 23 | 11 | Bulk RNA sequencing | Identified three subtypes. Subtype 1 contained LMS from different anatomic sites, harbored higher TMB, and was associated with worse OS and DSS. Subtype 2 was mostly abdominal and was associated with better OS and DSS compared to the other subtypes. Subtype 3 was mostly uterine, harbored higher TMB, and was associated with worse OS and DSS. Matching primary-metastatic samples allowed for assessing tumor evolution; metastases maintained subtype. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Denu, R.A.; Dann, A.M.; Keung, E.Z.; Nakazawa, M.S.; Nassif Haddad, E.F. The Future of Targeted Therapy for Leiomyosarcoma. Cancers 2024, 16, 938. https://doi.org/10.3390/cancers16050938
Denu RA, Dann AM, Keung EZ, Nakazawa MS, Nassif Haddad EF. The Future of Targeted Therapy for Leiomyosarcoma. Cancers. 2024; 16(5):938. https://doi.org/10.3390/cancers16050938
Chicago/Turabian StyleDenu, Ryan A., Amanda M. Dann, Emily Z. Keung, Michael S. Nakazawa, and Elise F. Nassif Haddad. 2024. "The Future of Targeted Therapy for Leiomyosarcoma" Cancers 16, no. 5: 938. https://doi.org/10.3390/cancers16050938
APA StyleDenu, R. A., Dann, A. M., Keung, E. Z., Nakazawa, M. S., & Nassif Haddad, E. F. (2024). The Future of Targeted Therapy for Leiomyosarcoma. Cancers, 16(5), 938. https://doi.org/10.3390/cancers16050938






