Precision Medicine for Nasopharyngeal Cancer—A Review of Current Prognostic Strategies
Abstract
Simple Summary
Abstract
1. Introduction
2. Current Diagnosis, Staging, and Treatment Options
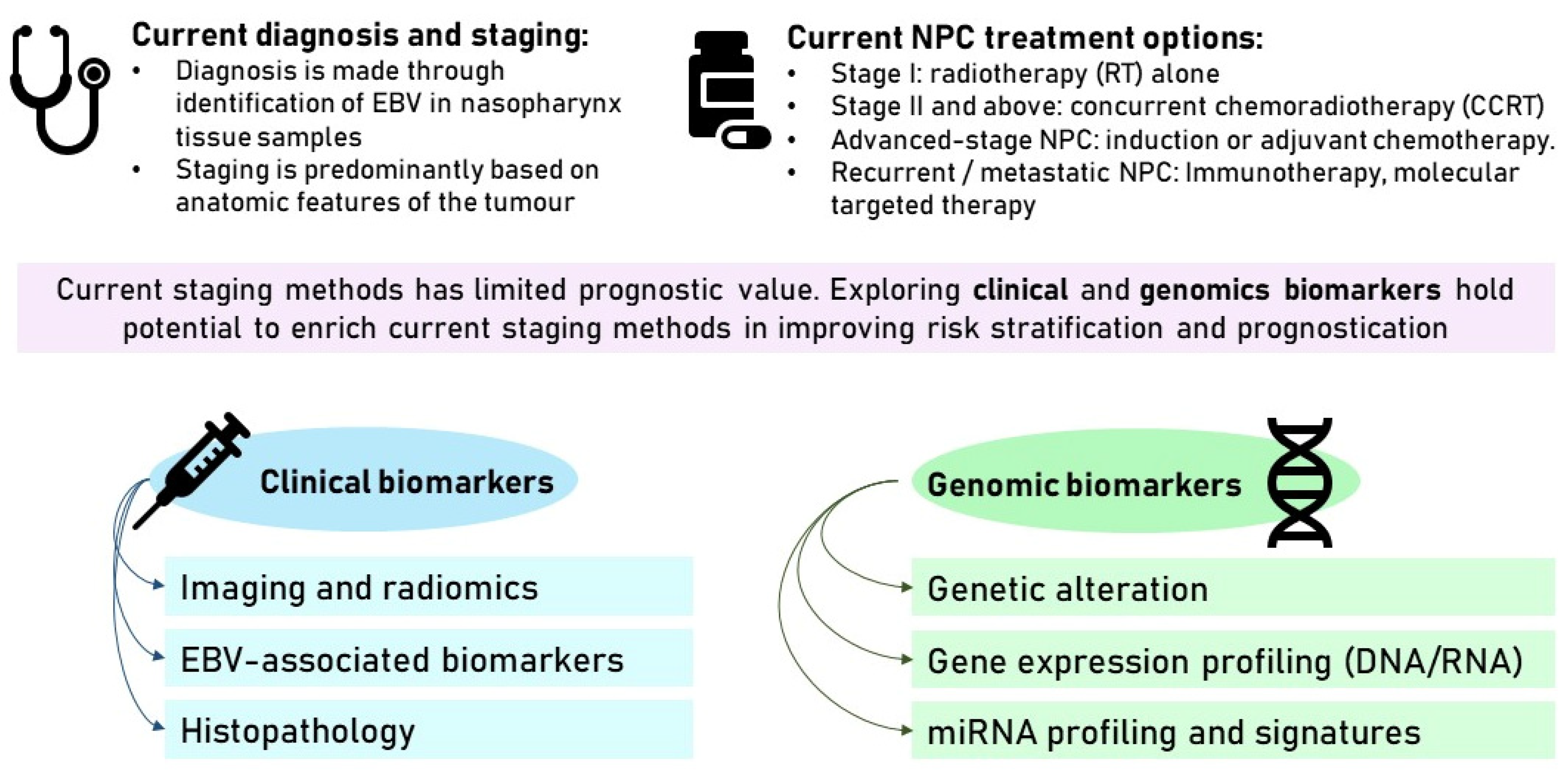
2.1. Diagnosis and Staging
2.2. Current Treatment Options
3. Limitations of the Current Staging System in Predicting Prognosis
4. Clinical Biomarkers for Risk Stratification
4.1. Radiomics
4.2. EBV-Associated Biomarkers
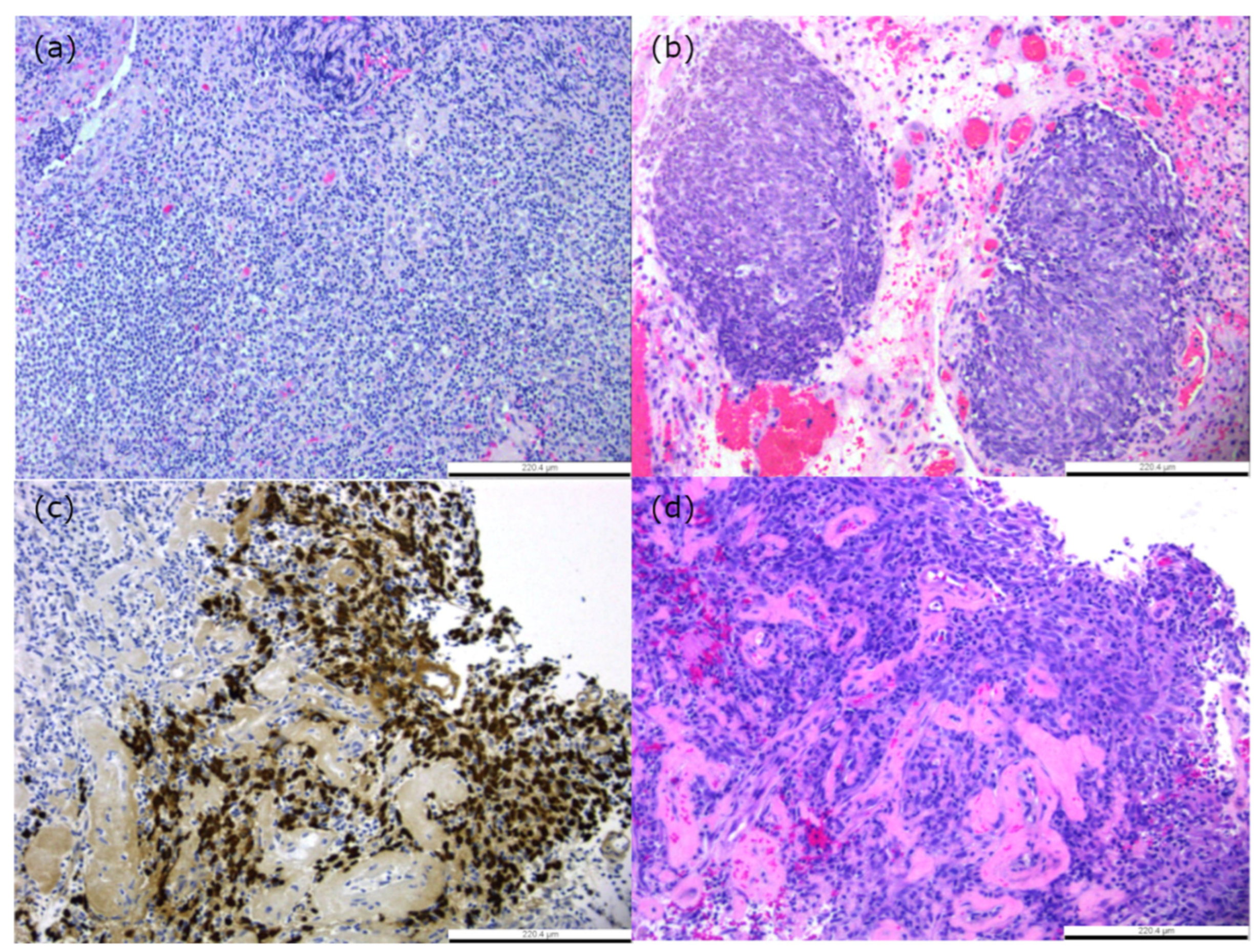
4.3. Histological Subtypes Associated with Prognosis
5. Genomic Biomarkers for Risk Stratification
5.1. Genetic Alteration in NPC Molecular Landscape
5.2. Gene Expression Profiling in NPC
5.3. miRNA Studies
6. Limitations Faced in Clinical Implementation of Predictors
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Petersson, F. Nasopharyngeal carcinoma: A review. Semin. Diagn. Pathol. 2015, 32, 54–73. [Google Scholar] [CrossRef]
- Glastonbury, C.M.; Salzman, K.L. Pitfalls in the staging of cancer of nasopharyngeal carcinoma. Neuroimaging Clin. 2013, 23, 9–25. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Zhang, R.; He, Y.; Wei, B.; Lu, Y.; Zhang, J.; Zhang, N.; He, R.; Xue, H.; Zhu, B. Nasopharyngeal Carcinoma Burden and Its Attributable Risk Factors in China: Estimates and Forecasts from 1990 to 2050. Int. J. Environ. Res. Public Health 2023, 20, 2926. [Google Scholar] [CrossRef]
- Yu, H.; Yin, X.; Mao, Y.; Chen, M.; Tang, Q.; Yan, S. The global burden of nasopharyngeal carcinoma from 2009 to 2019: An observational study based on the Global Burden of Disease Study 2019. Eur. Arch. Otorhinolaryngol. 2022, 279, 1519–1533. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.T.; Adami, H.O. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1765–1777. [Google Scholar] [CrossRef] [PubMed]
- Singapore Cancer Registry Annual Report 2020. 2022. Available online: https://nrdo.gov.sg/docs/librariesprovider3/default-document-library/scr-2020-annual-report_web-release.pdf (accessed on 1 February 2024).
- Xu, T.; Tang, J.; Gu, M.; Liu, L.; Wei, W.; Yang, H. Recurrent nasopharyngeal carcinoma: A clinical dilemma and challenge. Curr. Oncol. 2013, 20, 406–419. [Google Scholar] [CrossRef] [PubMed]
- Perri, F.; Della Vittoria Scarpati, G.; Caponigro, F.; Ionna, F.; Longo, F.; Buonopane, S.; Muto, P.; Di Marzo, M.; Pisconti, S.; Solla, R. Management of recurrent nasopharyngeal carcinoma: Current perspectives. Onco Targets Ther. 2019, 12, 1583–1591. [Google Scholar] [CrossRef] [PubMed]
- Lydiatt, W.M.; Patel, S.G.; O’Sullivan, B.; Brandwein, M.S.; Ridge, J.A.; Migliacci, J.C.; Loomis, A.M.; Shah, J.P. Head and Neck cancers-major changes in the American Joint Committee on cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017, 67, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.P.; Chan, A.T.C.; Le, Q.T.; Blanchard, P.; Sun, Y.; Ma, J. Nasopharyngeal carcinoma. Lancet 2019, 394, 64–80. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.W.; Foo, W.; Law, S.C.; Poon, Y.F.; Sze, W.M.; Tung, S.Y.; Lau, W.H. Nasopharyngeal carcinoma: Presenting symptoms and duration before diagnosis. Hong Kong Med. J. 1997, 3, 355–361. [Google Scholar]
- Tang, L.L.; Chen, Y.P.; Mao, Y.P.; Wang, Z.X.; Guo, R.; Chen, L.; Tian, L.; Lin, A.H.; Li, L.; Sun, Y.; et al. Validation of the 8th Edition of the UICC/AJCC Staging System for Nasopharyngeal Carcinoma From Endemic Areas in the Intensity-Modulated Radiotherapy Era. J. Natl. Compr. Cancer Netw. 2017, 15, 913–919. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Yan, R.N.; Chen, H.Y.; Zeng, Y.Y.; Xiang, Z.Z.; Liu, F.; Shao, B.F.; Ma, J.C.; Wang, X.R.; Liu, L. Comparing the 7th and 8th editions of UICC/AJCC staging system for nasopharyngeal carcinoma in the IMRT era. BMC Cancer 2021, 21, 327. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Zhou, P.; Li, G.; Yan, H.; Feng, G.; Liu, M.; Zhu, J.; Wang, R. Validation of the 8th edition of the UICC/AJCC staging system for nasopharyngeal carcinoma treated with intensity-modulated radiotherapy. Oncotarget 2017, 8, 70586–70594. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.R.; Zhang, X.M.; Xie, X.D.; Lu, Y.; Wu, J.F.; He, X. Validation of the 8th edition of AJCC/UICC staging system for nasopharyngeal carcinoma: Results from a non-endemic cohort with 10-year follow-up. Oral Oncol. 2019, 98, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.L.; Chen, Y.P.; Chen, C.B.; Chen, M.Y.; Chen, N.Y.; Chen, X.Z.; Du, X.J.; Fang, W.F.; Feng, M.; Gao, J.; et al. The Chinese Society of Clinical Oncology (CSCO) clinical guidelines for the diagnosis and treatment of nasopharyngeal carcinoma. Cancer Commun. 2021, 41, 1195–1227. [Google Scholar] [CrossRef] [PubMed]
- Rueda Domínguez, A.; Cirauqui, B.; García Castaño, A.; Alvarez Cabellos, R.; Carral Maseda, A.; Castelo Fernández, B.; Iglesias Rey, L.; Rubió-Casadevall, J.; Arrazubi, V.; Mesía, R. SEOM-TTCC clinical guideline in nasopharynx cancer (2021). Clin. Transl. Oncol. 2022, 24, 670–680. [Google Scholar] [CrossRef]
- Bossi, P.; Chan, A.T.; Licitra, L.; Trama, A.; Orlandi, E.; Hui, E.P.; Halámková, J.; Mattheis, S.; Baujat, B.; Hardillo, J.; et al. Nasopharyngeal carcinoma: ESMO-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up(†). Ann. Oncol. 2021, 32, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, E.J.; Luu, M.; David, J.M.; Kim, S.; Mita, A.; Scher, K.; Shiao, S.L.; Tighiouart, M.; Lee, N.Y.; Ho, A.S.; et al. Facility Volume and Survival in Nasopharyngeal Carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2018, 100, 408–417. [Google Scholar] [CrossRef]
- Liang, S.B.; Wang, Y.; Hu, X.F.; He, S.S.; Yang, X.L.; Liu, L.Z.; Cui, C.Y.; Chen, Y.; Fu, L.W. Survival and Toxicities of IMRT Based on the RTOG Protocols in Patients with Nasopharyngeal Carcinoma from the Endemic Regions of China. J. Cancer 2017, 8, 3718–3724. [Google Scholar] [CrossRef]
- Peng, G.; Wang, T.; Yang, K.Y.; Zhang, S.; Zhang, T.; Li, Q.; Han, J.; Wu, G. A prospective, randomized study comparing outcomes and toxicities of intensity-modulated radiotherapy vs. conventional two-dimensional radiotherapy for the treatment of nasopharyngeal carcinoma. Radiother Oncol. 2012, 104, 286–293. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, Y.; Hong, S.; Yang, Y.; Yu, G.; Jia, J.; Peng, P.; Wu, X.; Lin, Q.; Xi, X.; et al. Gemcitabine plus cisplatin versus fluorouracil plus cisplatin in recurrent or metastatic nasopharyngeal carcinoma: A multicentre, randomised, open-label, phase 3 trial. Lancet 2016, 388, 1883–1892. [Google Scholar] [CrossRef]
- Nakanishi, Y.; Wakisaka, N.; Kondo, S.; Endo, K.; Sugimoto, H.; Hatano, M.; Ueno, T.; Ishikawa, K.; Yoshizaki, T. Progression of understanding for the role of Epstein-Barr virus and management of nasopharyngeal carcinoma. Cancer Metastasis Rev. 2017, 36, 435–447. [Google Scholar] [CrossRef]
- Low, Y.H.; Loh, C.J.L.; Peh, D.Y.Y.; Chu, A.J.M.; Han, S.; Toh, H.C. Pathogenesis and therapeutic implications of EBV-associated epithelial cancers. Front. Oncol. 2023, 13, 1202117. [Google Scholar] [CrossRef]
- Jain, A.; Chia, W.K.; Toh, H.C. Immunotherapy for nasopharyngeal cancer-a review. Chin. Clin. Oncol. 2016, 5, 22. [Google Scholar] [CrossRef]
- Johnson, D.; Ma, B.B.Y. Targeting the PD-1/ PD-L1 interaction in nasopharyngeal carcinoma. Oral Oncol. 2021, 113, 105127. [Google Scholar] [CrossRef]
- Yu, J.; Pham, T.T.; Wandrey, N.; Daly, M.; Karam, S.D. Multimodality Management of EBV-Associated Nasopharyngeal Carcinoma. Cancers 2021, 13, 6078. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Li, H.; Ma, H.; Dong, A.; Xie, F.; Liang, S.; Li, L.; Zhou, J.; Xie, C.; Yan, Y.; et al. Staging of T2 and T3 nasopharyngeal carcinoma: Proposed modifications for improving the current AJCC staging system. Cancer Med. 2020, 9, 7572–7579. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Sun, X.; Huang, H.; Wang, Z.; Fang, X.; Chen, M.; Chen, Z.; Weng, H.; Guo, C.; Hong, H.; et al. Proposed prognostic subgroups and facilitated clinical decision-making for additional locoregional radiotherapy in de novo metastatic nasopharyngeal carcinoma: A retrospective study based on recursive partitioning analysis. Radiat. Oncol. 2023, 18, 15. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.K.; Lin, C.; Huang, S.H.; Chau, T.C.; Guo, Q.J.; O’Sullivan, B.; Lam, K.O.; Chau, S.C.; Chan, S.Y.; Tong, C.C.; et al. Refining TNM-8 M1 categories with anatomic subgroups for previously untreated de novo metastatic nasopharyngeal carcinoma. Oral Oncol. 2022, 126, 105736. [Google Scholar] [CrossRef] [PubMed]
- You, R.; Zou, X.; Wang, S.L.; Jiang, R.; Tang, L.Q.; Zhang, W.D.; Li, L.; Zhang, M.X.; Shen, G.P.; Guo, L.; et al. New surgical staging system for patients with recurrent nasopharyngeal carcinoma based on the AJCC/UICC rTNM classification system. Eur. J. Cancer 2015, 51, 1771–1779. [Google Scholar] [CrossRef]
- Goshtasbi, K.; Abiri, A.; Lehrich, B.M.; Haidar, Y.M.; Tjoa, T.; Kuan, E.C. The influence of facility volume on patient treatments and survival outcomes in nasopharyngeal carcinoma. Head Neck 2021, 43, 2755–2763. [Google Scholar] [CrossRef]
- Kong, F.; Zhou, J.; Du, C.; He, X.; Kong, L.; Hu, C.; Ying, H. Long-term survival and late complications of intensity-modulated radiotherapy for recurrent nasopharyngeal carcinoma. BMC Cancer 2018, 18, 1139. [Google Scholar] [CrossRef]
- Qian, J.; Yang, Y.; Xing, P.; Wang, C.; Tian, Y.; Lu, X. Differences in lower cranial nerve complications predicted by the NTCP model between RTOG and reduced-volume IMRT planning in radiotherapy for nasopharyngeal carcinoma. Transl. Cancer Res. 2020, 9, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Shim, H.J.; Kim, H.J.; Hwang, J.E.; Bae, W.K.; Chung, I.J.; Lee, D.H.; Mi, Y.T.; Lee, J.K.; Lim, S.C.; Chung, J.W.; et al. Long term complications and prognostic factors in locally advanced nasopharyngeal carcinoma treated with docetaxel, cisplatin, 5-fluorouracil induction chemotherapy followed by concurrent chemoradiotherapy: A retrospective cohort study. Medicine 2020, 99, e23173. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.M.; Yang, Q.; Guo, L.; Mai, H.Q.; Mo, H.Y.; Cao, K.J.; Qian, C.N.; Zhao, C.; Xiang, Y.Q.; Zhang, X.P.; et al. Neoadjuvant chemotherapy followed by concurrent chemoradiotherapy versus concurrent chemoradiotherapy alone in locoregionally advanced nasopharyngeal carcinoma: A phase III multicentre randomised controlled trial. Eur. J. Cancer 2017, 75, 14–23. [Google Scholar] [CrossRef]
- Yang, Q.; Cao, S.M.; Guo, L.; Hua, Y.J.; Huang, P.Y.; Zhang, X.L.; Lin, M.; You, R.; Zou, X.; Liu, Y.P.; et al. Induction chemotherapy followed by concurrent chemoradiotherapy versus concurrent chemoradiotherapy alone in locoregionally advanced nasopharyngeal carcinoma: Long-term results of a phase III multicentre randomised controlled trial. Eur. J. Cancer 2019, 119, 87–96. [Google Scholar] [CrossRef]
- Hong, R.L.; Hsiao, C.F.; Ting, L.L.; Ko, J.Y.; Wang, C.W.; Chang, J.T.C.; Lou, P.J.; Wang, H.M.; Tsai, M.H.; Lai, S.C.; et al. Final results of a randomized phase III trial of induction chemotherapy followed by concurrent chemoradiotherapy versus concurrent chemoradiotherapy alone in patients with stage IVA and IVB nasopharyngeal carcinoma-Taiwan Cooperative Oncology Group (TCOG) 1303 Study. Ann. Oncol. 2018, 29, 1972–1979. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, W.F.; Liu, X.; Chen, L.; Sun, R.; Sun, Y.; Liu, Q.; Ma, J. Nomogram to predict the benefit of additional induction chemotherapy to concurrent chemoradiotherapy in locoregionally advanced nasopharyngeal carcinoma: Analysis of a multicenter, phase III randomized trial. Radiother. Oncol. 2018, 129, 18–22. [Google Scholar] [CrossRef]
- Chiang, C.L.; Guo, Q.; Ng, W.T.; Lin, S.; Ma, T.S.W.; Xu, Z.; Xiao, Y.; Li, J.; Lu, T.; Choi, H.C.W.; et al. Prognostic Factors for Overall Survival in Nasopharyngeal Cancer and Implication for TNM Staging by UICC: A Systematic Review of the Literature. Front. Oncol. 2021, 11, 703995. [Google Scholar] [CrossRef]
- Cattell, R.; Chen, S.; Huang, C. Robustness of radiomic features in magnetic resonance imaging: Review and a phantom study. Vis. Comput. Ind. Biomed. Art 2019, 2, 19. [Google Scholar] [CrossRef]
- Ouyang, F.S.; Guo, B.L.; Zhang, B.; Dong, Y.H.; Zhang, L.; Mo, X.K.; Huang, W.H.; Zhang, S.X.; Hu, Q.G. Exploration and validation of radiomics signature as an independent prognostic biomarker in stage III-IVb nasopharyngeal carcinoma. Oncotarget 2017, 8, 74869–74879. [Google Scholar] [CrossRef][Green Version]
- Du, R.; Lee, V.H.; Yuan, H.; Lam, K.O.; Pang, H.H.; Chen, Y.; Lam, E.Y.; Khong, P.L.; Lee, A.W.; Kwong, D.L.; et al. Radiomics Model to Predict Early Progression of Nonmetastatic Nasopharyngeal Carcinoma after Intensity Modulation Radiation Therapy: A Multicenter Study. Radiol. Artif. Intell. 2019, 1, e180075. [Google Scholar] [CrossRef]
- Chan, S.C.; Chang, K.P.; Fang, Y.D.; Tsang, N.M.; Ng, S.H.; Hsu, C.L.; Liao, C.T.; Yen, T.C. Tumor heterogeneity measured on F-18 fluorodeoxyglucose positron emission tomography/computed tomography combined with plasma Epstein-Barr Virus load predicts prognosis in patients with primary nasopharyngeal carcinoma. Laryngoscope 2017, 127, E22–E28. [Google Scholar] [CrossRef] [PubMed]
- Intarak, S.; Chongpison, Y.; Vimolnoch, M.; Oonsiri, S.; Kitpanit, S.; Prayongrat, A.; Kannarunimit, D.; Chakkabat, C.; Sriswasdi, S.; Lertbutsayanukul, C.; et al. Tumor Prognostic Prediction of Nasopharyngeal Carcinoma Using CT-Based Radiomics in Non-Chinese Patients. Front. Oncol. 2022, 12, 775248. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Dong, D.; Fang, M.J.; Li, L.; Tang, L.L.; Chen, L.; Li, W.F.; Mao, Y.P.; Fan, W.; Liu, L.Z.; et al. Prognostic Value of Deep Learning PET/CT-Based Radiomics: Potential Role for Future Individual Induction Chemotherapy in Advanced Nasopharyngeal Carcinoma. Clin. Cancer Res. 2019, 25, 4271–4279. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Yuan, Q.; Wang, Q.; Ma, J.; Feng, Q.; Chen, W.; Rahmim, A.; Lu, L. Radiomics Analysis of PET and CT Components of PET/CT Imaging Integrated with Clinical Parameters: Application to Prognosis for Nasopharyngeal Carcinoma. Mol. Imaging Biol. 2019, 21, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wu, J.; Mai, W.; Li, H. Prediction of Changes in Tumor Regression during Radiotherapy for Nasopharyngeal Carcinoma by Using the Computed Tomography-Based Radiomics. Contrast Media Mol. Imaging 2022, 2022, 3417480. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.; Xiong, B.; Tian, T.; Zou, X.; He, Z.; Zhang, L. Radiomics in Nasopharyngeal Carcinoma. Clin. Med. Insights Oncol. 2022, 16, 11795549221079186. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Limkin, E.J.; Vakalopoulou, M.; Dercle, L.; Champiat, S.; Han, S.R.; Verlingue, L.; Brandao, D.; Lancia, A.; Ammari, S.; et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: An imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018, 19, 1180–1191. [Google Scholar] [CrossRef]
- Gao, Y.; Mao, Y.; Lu, S.; Tan, L.; Li, G.; Chen, J.; Huang, D.; Zhang, X.; Qiu, Y.; Liu, Y. Magnetic resonance imaging-based radiogenomics analysis for predicting prognosis and gene expression profile in advanced nasopharyngeal carcinoma. Head Neck 2021, 43, 3730–3742. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Shen, D.S.; Chen, X.B.; Su, D.K.; Liang, Z.G.; Chen, K.H.; Li, L.; Liang, X.; Liao, H.; Zhu, X.D. CT-Based Radiomics Nomogram for Prediction of Progression-Free Survival in Locoregionally Advanced Nasopharyngeal Carcinoma. Cancer Manag. Res. 2021, 13, 6911–6923. [Google Scholar] [CrossRef]
- Li, Q.; Yu, Q.; Gong, B.; Ning, Y.; Chen, X.; Gu, J.; Lv, F.; Peng, J.; Luo, T. The Effect of Magnetic Resonance Imaging Based Radiomics Models in Discriminating stage I-II and III-IVa Nasopharyngeal Carcinoma. Diagnostics 2023, 13, 300. [Google Scholar] [CrossRef] [PubMed]
- Poh, S.S.; Chua, M.L.; Wee, J.T. Carcinogenesis of nasopharyngeal carcinoma: An alternate hypothetical mechanism. Chin. J. Cancer 2016, 35, 9. [Google Scholar] [CrossRef]
- Tsao, S.W.; Tsang, C.M.; Lo, K.W. Epstein-Barr virus infection and nasopharyngeal carcinoma. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160270. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Li, Z.; Long, Y.; Li, J.; Liu, Z.; Zhou, R. Clinical value of a plasma Epstein-Barr virus DNA assay in the diagnosis of recurrent or metastatic nasopharyngeal carcinoma: A meta-analysis. Biosci. Rep. 2019, 39, BSR20190691. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Song, Y.L.; Huang, S.M.; Tao, H.X.; Zhao, Y.Q.; Yan, N.; Xu, D.Y. The clinical significance of EBV DNA analysis in nasopharyngeal carcinoma screening. J. Clin. Otorhinolaryngol. Head Neck Surg. 2018, 32, 298–301. [Google Scholar] [CrossRef]
- Xie, X.; Ren, Y.; Wang, K.; Yi, B. Molecular Prognostic Value of Circulating Epstein-Barr Viral DNA in Nasopharyngeal Carcinoma: A Meta-Analysis of 27,235 Cases in the Endemic Area of Southeast Asia. Genet. Test. Mol. Biomark. 2019, 23, 448–459. [Google Scholar] [CrossRef]
- Li, W.; Chen, J.; Liang, B.; Li, Z.; Li, J.; Yuan, X.; Wu, S.; Zeng, F.; Peng, X.; Li, Y.; et al. Long-term monitoring of dynamic changes in plasma EBV DNA for improved prognosis prediction of nasopharyngeal carcinoma. Cancer Med. 2021, 10, 883–894. [Google Scholar] [CrossRef]
- Chan, K.C.A.; Woo, J.K.S.; King, A.; Zee, B.C.Y.; Lam, W.K.J.; Chan, S.L.; Chu, S.W.I.; Mak, C.; Tse, I.O.L.; Leung, S.Y.M.; et al. Analysis of Plasma Epstein-Barr Virus DNA to Screen for Nasopharyngeal Cancer. N. Engl. J. Med. 2017, 377, 513–522. [Google Scholar] [CrossRef]
- He, S.S.; Wang, Y.; Bao, Y.; Cai, X.Y.; Yang, X.L.; Chen, D.M.; Chen, Y.; Lu, L.X. Dynamic changes in plasma Epstein-Barr virus DNA load during treatment have prognostic value in nasopharyngeal carcinoma: A retrospective study. Cancer Med. 2018, 7, 1110–1117. [Google Scholar] [CrossRef]
- Lee, V.H.; Kwong, D.L.; Leung, T.W.; Choi, C.W.; Lai, V.; Ng, L.; Lam, K.O.; Ng, S.C.; Sze, C.K.; Tong, C.C.; et al. Prognostication of serial post-intensity-modulated radiation therapy undetectable plasma EBV DNA for nasopharyngeal carcinoma. Oncotarget 2017, 8, 5292–5308. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.Y.; Li, Y.H.; Gao, H.Y.; Wu, Q.L.; Cui, N.J.; Zhang, L.; Cheng, G.; Hu, L.F.; Ernberg, I.; Zeng, Y.X. Comparison of plasma Epstein-Barr virus (EBV) DNA levels and serum EBV immunoglobulin A/virus capsid antigen antibody titers in patients with nasopharyngeal carcinoma. Cancer 2004, 100, 1162–1170. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.H.; Kwong, D.L.; Leung, T.W.; Choi, C.W.; O’Sullivan, B.; Lam, K.O.; Lai, V.; Khong, P.L.; Chan, S.K.; Ng, C.Y.; et al. The addition of pretreatment plasma Epstein-Barr virus DNA into the eighth edition of nasopharyngeal cancer TNM stage classification. Int. J. Cancer 2019, 144, 1713–1722. [Google Scholar] [CrossRef] [PubMed]
- Leung, S.F.; Zee, B.; Ma, B.B.; Hui, E.P.; Mo, F.; Lai, M.; Chan, K.C.; Chan, L.Y.; Kwan, W.H.; Lo, Y.M.; et al. Plasma Epstein-Barr viral deoxyribonucleic acid quantitation complements tumor-node-metastasis staging prognostication in nasopharyngeal carcinoma. J. Clin. Oncol. 2006, 24, 5414–5418. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.; Chan, A.T.; Chan, L.Y.; Leung, S.F.; Lam, C.W.; Huang, D.P.; Johnson, P.J. Molecular prognostication of nasopharyngeal carcinoma by quantitative analysis of circulating Epstein-Barr virus DNA. Cancer Res. 2000, 60, 6878–6881. [Google Scholar] [PubMed]
- Yang, L.; Hong, S.; Wang, Y.; Chen, H.; Liang, S.; Peng, P.; Chen, Y. Development and External Validation of Nomograms for Predicting Survival in Nasopharyngeal Carcinoma Patients after Definitive Radiotherapy. Sci. Rep. 2015, 5, 15638. [Google Scholar] [CrossRef]
- Chen, F.P.; Luo, Y.S.; Chen, K.; Li, J.Y.; Huo, L.Q.; Shi, L.; Ou-Yang, Y.; Cao, X.P. Circulating Epstein-Barr virus DNA level post induction chemotherapy contributes to prognostication in advanced-stage nasopharyngeal carcinoma. Eur. J. Cancer 2021, 151, 63–71. [Google Scholar] [CrossRef]
- Chan, A.T.C.; Hui, E.P.; Ngan, R.K.C.; Tung, S.Y.; Cheng, A.C.K.; Ng, W.T.; Lee, V.H.F.; Ma, B.B.Y.; Cheng, H.C.; Wong, F.C.S.; et al. Analysis of Plasma Epstein-Barr Virus DNA in Nasopharyngeal Cancer After Chemoradiation to Identify High-Risk Patients for Adjuvant Chemotherapy: A Randomized Controlled Trial. J. Clin. Oncol. 2018, 36, 3091–3100. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, Y.; Chen, L.; Guo, R.; Zhou, G.; Tang, L.; Mao, Y.; Li, W.; Liu, X.; Du, X.; et al. The clinical utility of plasma Epstein-Barr virus DNA assays in nasopharyngeal carcinoma: The dawn of a new era?: A systematic review and meta-analysis of 7836 cases. Medicine 2015, 94, e845. [Google Scholar] [CrossRef]
- Zhang, Q.; Peng, Z.W.; Gu, Z.S.; Wang, Y.; He, F.; Zhao, W.B.; Luo, W.; Mei, Y.Y. Can Epstein-Barr virus-deoxyribonucleic acid load after induction chemotherapy combined with American Joint Committee on Cancer stage determine the chemotherapy intensity of locally advanced nasopharyngeal carcinoma? Cancer Med. 2023, 12, 223–235. [Google Scholar] [CrossRef]
- Twu, C.W.; Wang, W.Y.; Chen, C.C.; Liang, K.L.; Jiang, R.S.; Wu, C.T.; Shih, Y.T.; Lin, P.J.; Liu, Y.C.; Lin, J.C. Metronomic adjuvant chemotherapy improves treatment outcome in nasopharyngeal carcinoma patients with postradiation persistently detectable plasma Epstein-Barr virus deoxyribonucleic acid. Int. J. Radiat. Oncol. Biol. Phys. 2014, 89, 21–29. [Google Scholar] [CrossRef]
- Sun, X.S.; Liu, S.L.; Liang, Y.J.; Chen, Q.Y.; Li, X.Y.; Tang, L.Q.; Mai, H.Q. The role of capecitabine as maintenance therapy in de novo metastatic nasopharyngeal carcinoma: A propensity score matching study. Cancer Commun. 2020, 40, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.Y.; Wong, S.T. The role of plasma Epstein-Barr virus DNA in the management of recurrent nasopharyngeal carcinoma. Laryngoscope 2014, 124, 126–130. [Google Scholar] [CrossRef]
- Kim, K.Y.; Le, Q.T.; Yom, S.S.; Ng, R.H.W.; Chan, K.C.A.; Bratman, S.V.; Welch, J.J.; Divi, R.L.; Petryshyn, R.A.; Conley, B.A. Clinical Utility of Epstein-Barr Virus DNA Testing in the Treatment of Nasopharyngeal Carcinoma Patients. Int. J. Radiat. Oncol. Biol. Phys. 2017, 98, 996–1001. [Google Scholar] [CrossRef]
- Guo, R.; Tang, L.L.; Mao, Y.P.; Du, X.J.; Chen, L.; Zhang, Z.C.; Liu, L.Z.; Tian, L.; Luo, X.T.; Xie, Y.B.; et al. Proposed modifications and incorporation of plasma Epstein-Barr virus DNA improve the TNM staging system for Epstein-Barr virus-related nasopharyngeal carcinoma. Cancer 2019, 125, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, R.; Arora, S.; Sharma, P.; Biswas, A.; Nayak, B.; Thakar, A.; Sharma, A.; Ghose, S. Cell-free EBV DNA as a biomarker during clinical management of nasopharyngeal carcinoma in a nonendemic region. J. Med. Virol. 2022, 94, 720–728. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Zhou, Y.; Zhou, J.; Zhao, D.; Li, L.; Li, X.; Huang, Y.; Wang, Q.; Zou, H.; Zhang, K.; et al. Clinical relevance of plasma EBV DNA as a biomarker for nasopharyngeal carcinoma in non-endemic areas: A multicenter study in southwestern China. Clin. Chim. Acta 2023, 541, 117244. [Google Scholar] [CrossRef]
- Du, J.L.; Chen, S.H.; Huang, Q.H.; Xie, S.H.; Ye, Y.F.; Gao, R.; Guo, J.; Yang, M.J.; Liu, Q.; Hong, M.H.; et al. Subtype distribution and long-term titer fluctuation patterns of serum anti-Epstein-Barr virus antibodies in a non-nasopharyngeal carcinoma population from an endemic area in South China: A cohort study. Chin. J. Cancer 2016, 35, 78. [Google Scholar] [CrossRef]
- Tan, L.P.; Tan, G.W.; Sivanesan, V.M.; Goh, S.L.; Ng, X.J.; Lim, C.S.; Kim, W.R.; Mohidin, T.; Mohd Dali, N.S.; Ong, S.H.; et al. Systematic comparison of plasma EBV DNA, anti-EBV antibodies and miRNA levels for early detection and prognosis of nasopharyngeal carcinoma. Int. J. Cancer 2020, 146, 2336–2347. [Google Scholar] [CrossRef]
- Liang, T.; Liu, W.; Xie, J.; Wang, Y.; Chen, G.; Liao, W.; Song, L.; Zhang, X. Serum EA-IgA and D-dimer, but not VCA-IgA, are associated with prognosis in patients with nasopharyngeal carcinoma: A meta-analysis. Cancer Cell Int. 2021, 21, 329. [Google Scholar] [CrossRef]
- Yao, J.J.; Lin, L.; Jin, Y.N.; Wang, S.Y.; Zhang, W.J.; Zhang, F.; Zhou, G.Q.; Cheng, Z.B.; Qi, Z.Y.; Sun, Y. Prognostic value of serum Epstein-Barr virus antibodies in patients with nasopharyngeal carcinoma and undetectable pretreatment Epstein-Barr virus DNA. Cancer Sci. 2017, 108, 1640–1647. [Google Scholar] [CrossRef]
- Gurtsevitch, V.E.; Senyuta, N.B.; Ignatova, A.V.; Lomaya, M.V.; Kondratova, V.N.; Pavlovskaya, A.I.; Dushenkina, T.E.; Maximovich, D.M.; Smirnova, K.V.; Mudunov, A.M.; et al. Epstein-Barr virus biomarkers for nasopharyngeal carcinoma in non-endemic regions. J. Gen. Virol. 2017, 98, 2118–2127. [Google Scholar] [CrossRef]
- Ding, R.-B.; Chen, P.; Rajendran, B.K.; Lyu, X.; Wang, H.; Bao, J.; Zeng, J.; Hao, W.; Sun, H.; Wong, A.H.-H.; et al. Molecular landscape and subtype-specific therapeutic response of nasopharyngeal carcinoma revealed by integrative pharmacogenomics. Nat. Commun. 2021, 12, 3046. [Google Scholar] [CrossRef]
- Stelow, E.B.; Wenig, B.M. Update From The 4th Edition of the World Health Organization Classification of Head and Neck Tumours: Nasopharynx. Head Neck Pathol. 2017, 11, 16–22. [Google Scholar] [CrossRef]
- Brennan, B. Nasopharyngeal carcinoma. Orphanet J. Rare Dis. 2006, 1, 23. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Yao, K. Molecular characterization and clinical implications of spindle cells in nasopharyngeal carcinoma: A novel molecule-morphology model of tumor progression proposed. PLoS ONE 2013, 8, e83135. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.G.; Lian, C.L.; Wang, J.; Zhang, W.W.; Sun, J.Y.; Lin, Q.; He, Z.Y. The effect of histological subtypes on survival outcome in nasopharyngeal carcinoma after extensive follow up. Ann. Transl. Med. 2019, 7, 768. [Google Scholar] [CrossRef]
- Wang, H.Y.; Chang, Y.L.; To, K.F.; Hwang, J.S.; Mai, H.Q.; Feng, Y.F.; Chang, E.T.; Wang, C.P.; Kam, M.K.; Cheah, S.L.; et al. A new prognostic histopathologic classification of nasopharyngeal carcinoma. Chin. J. Cancer 2016, 35, 41. [Google Scholar] [CrossRef] [PubMed]
- Prasetyo, A.; Budiman, J.; Sadhana, U. The Relationship between Tumor-infiltrating Lymphocytes (TILs) and Nasopharyngeal Carcinoma (NPC): A Systematic Review. Iran. J. Otorhinolaryngol. 2021, 33, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.T.; Sheen, T.S.; Chen, C.L.; Lu, J.; Chang, Y.; Chen, J.Y.; Tsai, C.H. Profile of cytokine expression in nasopharyngeal carcinomas: A distinct expression of interleukin 1 in tumor and CD4+ T cells. Cancer Res. 1999, 59, 1599–1605. [Google Scholar]
- Liu, S.-X.; Zhao, G.-X.; Lin, R.-B.; Zeng, M.-S.; Zhong, Q. Classifying the tumor microenvironment to stratify nasopharyngeal carcinoma patients. Ann. Nasopharynx Cancer 2022, 6, 1–16. [Google Scholar] [CrossRef]
- Almangush, A.; Ruuskanen, M.; Hagström, J.; Hirvikoski, P.; Tommola, S.; Kosma, V.M.; Nieminen, P.; Mäkitie, A.; Leivo, I. Tumor-infiltrating lymphocytes associate with outcome in nonendemic nasopharyngeal carcinoma: A multicenter study. Hum. Pathol. 2018, 81, 211–219. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Chen, Y.P.; Zhang, Y.; Jiang, W.; Liu, N.; Yun, J.P.; Sun, Y.; He, Q.M.; Tang, X.R.; Wen, X.; et al. Prognostic significance of tumor-infiltrating lymphocytes in nondisseminated nasopharyngeal carcinoma: A large-scale cohort study. Int. J. Cancer 2018, 142, 2558–2566. [Google Scholar] [CrossRef]
- Zhang, L.; MacIsaac, K.D.; Zhou, T.; Huang, P.-Y.; Xin, C.; Dobson, J.R.; Yu, K.; Chiang, D.Y.; Fan, Y.; Pelletier, M.; et al. Genomic Analysis of Nasopharyngeal Carcinoma Reveals TME-Based Subtypes. Mol. Cancer Res. 2017, 15, 1722–1732. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Li, J.; Mo, H.Y.; Qiu, F.; Zheng, L.M.; Qian, C.N.; Zeng, Y.X. Different subsets of tumor infiltrating lymphocytes correlate with NPC progression in different ways. Mol. Cancer 2010, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Azuma, K.; Kawahara, A.; Sasada, T.; Matsuo, N.; Kakuma, T.; Kamimura, H.; Maeda, R.; Hattori, C.; On, K.; et al. Prognostic stratification of patients with nasopharyngeal carcinoma based on tumor immune microenvironment. Head Neck 2018, 40, 2007–2019. [Google Scholar] [CrossRef] [PubMed]
- Ooft, M.L.; van Ipenburg, J.A.; Braunius, W.W.; Zuur, C.I.; Koljenović, S.; Willems, S.M. Prognostic role of tumor infiltrating lymphocytes in EBV positive and EBV negative nasopharyngeal carcinoma. Oral Oncol. 2017, 71, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Tessari, A.; Palmieri, D.; Di Cosimo, S. Overview of diagnostic/targeted treatment combinations in personalized medicine for breast cancer patients. Pharmgenom. Pers. Med. 2013, 7, 1–19. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hensing, T.; Chawla, A.; Batra, R.; Salgia, R. A personalized treatment for lung cancer: Molecular pathways, targeted therapies, and genomic characterization. Adv. Exp. Med. Biol. 2014, 799, 85–117. [Google Scholar] [CrossRef] [PubMed]
- Lo, K.W.; Huang, D.P. Genetic and epigenetic changes in nasopharyngeal carcinoma. Semin. Cancer Biol. 2002, 12, 451–462. [Google Scholar] [CrossRef]
- Lee, A.W.; Ma, B.B.; Ng, W.T.; Chan, A.T. Management of Nasopharyngeal Carcinoma: Current Practice and Future Perspective. J. Clin. Oncol. 2015, 33, 3356–3364. [Google Scholar] [CrossRef] [PubMed]
- Hui, A.B.-Y.; Lo, K.-W.; Leung, S.-F.; Teo, P.; Fung, M.K.F.; To, K.F.; Wong, N.; Choi, P.H.K.; Lee, J.C.K.; Huang, D.P. Detection of recurrent chromosomal gains and losses in primary nasopharyngeal carcinoma by comparative genomic hybridisation. Int. J. Cancer 1999, 82, 498–503. [Google Scholar] [CrossRef]
- Shao, J.Y.; Huang, X.M.; Yu, X.J.; Huang, L.X.; Wu, Q.L.; Xia, J.C.; Wang, H.Y.; Feng, Q.S.; Ren, Z.F.; Ernberg, I.; et al. Loss of heterozygosity and its correlation with clinical outcome and Epstein-Barr virus infection in nasopharyngeal carcinoma. Anticancer Res. 2001, 21, 3021–3029. [Google Scholar]
- Lin, D.-C.; Meng, X.; Hazawa, M.; Nagata, Y.; Varela, A.M.; Xu, L.; Sato, Y.; Liu, L.-Z.; Ding, L.-W.; Sharma, A.; et al. The genomic landscape of nasopharyngeal carcinoma. Nat. Genet. 2014, 46, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Guan, B.; Wang, T.L.; Shih Ie, M. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011, 71, 6718–6727. [Google Scholar] [CrossRef]
- Guan, B.; Gao, M.; Wu, C.H.; Wang, T.L.; Shih Ie, M. Functional analysis of in-frame indel ARID1A mutations reveals new regulatory mechanisms of its tumor suppressor functions. Neoplasia 2012, 14, 986–993. [Google Scholar] [CrossRef]
- Zhang, J.-W.; Qin, T.; Hong, S.-D.; Zhang, J.; Fang, W.-F.; Zhao, Y.-Y.; Yang, Y.-P.; Xue, C.; Huang, Y.; Zhao, H.-Y.; et al. Multiple oncogenic mutations related to targeted therapy in nasopharyngeal carcinoma. Chin. J. Cancer 2015, 34, 10. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, P.; Zhang, X.; Xu, J.; Xu, J.; Yu, S.; Wang, D.; Dong, W.; Cao, X.; Yan, H.; et al. Mutational landscape of nasopharyngeal carcinoma based on targeted next-generation sequencing: Implications for predicting clinical outcomes. Mol. Med. 2022, 28, 55. [Google Scholar] [CrossRef]
- Ma, B.B.; Goh, B.C.; Lim, W.T.; Hui, E.P.; Tan, E.H.; Lopes Gde, L.; Lo, K.W.; Li, L.; Loong, H.; Foster, N.R.; et al. Multicenter phase II study of the AKT inhibitor MK-2206 in recurrent or metastatic nasopharyngeal carcinoma from patients in the mayo phase II consortium and the cancer therapeutics research group (MC1079). Investig. New Drugs 2015, 33, 985–991. [Google Scholar] [CrossRef]
- Li, Y.Y.; Chung, G.T.; Lui, V.W.; To, K.F.; Ma, B.B.; Chow, C.; Woo, J.K.; Yip, K.Y.; Seo, J.; Hui, E.P.; et al. Exome and genome sequencing of nasopharynx cancer identifies NF-κB pathway activating mutations. Nat. Commun. 2017, 8, 14121. [Google Scholar] [CrossRef]
- Bruce, J.P.; To, K.F.; Lui, V.W.Y.; Chung, G.T.Y.; Chan, Y.Y.; Tsang, C.M.; Yip, K.Y.; Ma, B.B.Y.; Woo, J.K.S.; Hui, E.P.; et al. Whole-genome profiling of nasopharyngeal carcinoma reveals viral-host co-operation in inflammatory NF-κB activation and immune escape. Nat. Commun. 2021, 12, 4193. [Google Scholar] [CrossRef] [PubMed]
- Chow, Y.P.; Tan, L.P.; Chai, S.J.; Abdul Aziz, N.; Choo, S.W.; Lim, P.V.H.; Pathmanathan, R.; Mohd Kornain, N.K.; Lum, C.L.; Pua, K.C.; et al. Exome Sequencing Identifies Potentially Druggable Mutations in Nasopharyngeal Carcinoma. Sci. Rep. 2017, 7, 42980. [Google Scholar] [CrossRef] [PubMed]
- DiDonato, J.A.; Mercurio, F.; Karin, M. NF-κB and the link between inflammation and cancer. Immunol. Rev. 2012, 246, 379–400. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Tarca, A.L.; Romero, R.; Draghici, S. Analysis of microarray experiments of gene expression profiling. Am. J. Obstet. Gynecol. 2006, 195, 373–388. [Google Scholar] [CrossRef] [PubMed]
- Zaatar, A.M.; Lim, C.R.; Bong, C.W.; Lee, M.M.L.; Ooi, J.J.; Suria, D.; Raman, R.; Chao, S.; Yang, H.; Neoh, S.B.; et al. Whole blood transcriptome correlates with treatment response in nasopharyngeal carcinoma. J. Exp. Clin. Cancer Res. 2012, 31, 76. [Google Scholar] [CrossRef]
- Tang, X.R.; Li, Y.Q.; Liang, S.B.; Jiang, W.; Liu, F.; Ge, W.X.; Tang, L.L.; Mao, Y.P.; He, Q.M.; Yang, X.J.; et al. Development and validation of a gene expression-based signature to predict distant metastasis in locoregionally advanced nasopharyngeal carcinoma: A retrospective, multicentre, cohort study. Lancet Oncol. 2018, 19, 382–393. [Google Scholar] [CrossRef]
- Zhao, S.; Dong, X.; Ni, X.; Li, L.; Lu, X.; Zhang, K.; Gao, Y. Exploration of a Novel Prognostic Risk Signature and Its Effect on the Immune Response in Nasopharyngeal Carcinoma. Front. Oncol. 2021, 11, 709931. [Google Scholar] [CrossRef]
- Liu, S.-L.; Sun, X.-S.; Chen, Q.-Y.; Liu, Z.-X.; Bian, L.-J.; Yuan, L.; Xiao, B.-B.; Lu, Z.-J.; Li, X.-Y.; Yan, J.-J.; et al. Development and validation of a transcriptomics-based gene signature to predict distant metastasis and guide induction chemotherapy in locoregionally advanced nasopharyngeal carcinoma. Eur. J. Cancer 2022, 163, 26–34. [Google Scholar] [CrossRef]
- Luo, M.S.; Huang, G.J.; Liu, B.X. Immune infiltration in nasopharyngeal carcinoma based on gene expression. Medicine 2019, 98, e17311. [Google Scholar] [CrossRef]
- Chen, Y.-P.; Yin, J.-H.; Li, W.-F.; Li, H.-J.; Chen, D.-P.; Zhang, C.-J.; Lv, J.-W.; Wang, Y.-Q.; Li, X.-M.; Li, J.-Y.; et al. Single-cell transcriptomics reveals regulators underlying immune cell diversity and immune subtypes associated with prognosis in nasopharyngeal carcinoma. Cell Res. 2020, 30, 1024–1042. [Google Scholar] [CrossRef]
- Liu, Y.; He, S.; Wang, X.-L.; Peng, W.; Chen, Q.-Y.; Chi, D.-M.; Chen, J.-R.; Han, B.-W.; Lin, G.-W.; Li, Y.-Q.; et al. Tumour heterogeneity and intercellular networks of nasopharyngeal carcinoma at single cell resolution. Nat. Commun. 2021, 12, 741. [Google Scholar] [CrossRef]
- Li, S.; Hang, L.; Ma, Y.; Wu, C. Distinctive microRNA expression in early stage nasopharyngeal carcinoma patients. J. Cell. Mol. Med. 2016, 20, 2259–2268. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Chen, N.Y.; Cui, R.X.; Li, W.F.; Li, Y.; Wei, R.R.; Zhang, M.Y.; Sun, Y.; Huang, B.J.; Chen, M.; et al. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: A microRNA expression analysis. Lancet Oncol. 2012, 13, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Bruce, J.P.; Hui, A.B.; Shi, W.; Perez-Ordonez, B.; Weinreb, I.; Xu, W.; Haibe-Kains, B.; Waggott, D.M.; Boutros, P.C.; O’Sullivan, B.; et al. Identification of a microRNA signature associated with risk of distant metastasis in nasopharyngeal carcinoma. Oncotarget 2015, 6, 4537–4550. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Mai, S.-J.; Lin, H.-X.; Zhang, M.-Y.; Huang, J.-L.; Hua, X.; Lin, C.; Long, Z.-Q.; Lu, Z.-J.; Sun, X.-Q.; et al. Identification of two microRNA signatures in whole blood as novel biomarkers for diagnosis of nasopharyngeal carcinoma. J. Transl. Med. 2019, 17, 186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zou, X.; Wu, L.; Zhang, S.; Wang, T.; Liu, P.; Zhu, W.; Zhu, J. Identification of a 7-microRNA signature in plasma as promising biomarker for nasopharyngeal carcinoma detection. Cancer Med. 2020, 9, 1230–1241. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, B.; Zhang, X.; Wang, C.; Xu, Y. Identification of a 3-miRNA Signature Associated With the Prediction of Prognosis in Nasopharyngeal Carcinoma. Front. Oncol. 2022, 11, 823603. [Google Scholar] [CrossRef]
| Year | Imaging Type | Cohort (Training, Validation) | Number of Radiomic Features Extracted | Endpoint | Reference |
|---|---|---|---|---|---|
| 2016 | 18F-FDG PET/CT | 101 primary NPC patients pre-treatment (101, 0) | 15 (4 histogram-based heterogeneity parameters, 6 second-order texture features, 5 higher-order features) | OS, recurrence-free survival (RFS) | Chan et al. [45] |
| 2017 | CE-T1WI, T2WI | 100 NPC patients pre-treatment (stage III-IVb) (70, 30) | 970 (tumour intensity, shape, texture, and wavelet features) | PFS | Ouyang et al. [43] |
| 2019 | CE-T1WI, T2WI | 277 non-metastatic NPC (217, 60) | 525 (11 first-order intensity features, 41 texture features, 5 shape features, with 4 subbands of Coiflet wavelet transforms per MRI sequence) | 3-year disease progression | Du et al. [44] |
| 2019 | 18F-FDG PET/CT | 707 patients with advanced (Stage III-IVA) NPC (470, 237) | 269 (136 deep learning features, 133 handcrafted features—divided into shape, histogram, grey-level co-occurrence matrix, and grey-level run-length matrix features) | Disease-free survival (DFS), distant metastasis-free survival (DMFS), OS, locoregional relapse-free survival (LRRFS) | Peng et al. [47] |
| 2021 | CT | 311 patients with locally-advanced NPC (stage III-IVa) (218, 93) | 1409 (shape, first-order, texture features) | PFS | Yan et al. [53] |
| 2022 | CT | 197 non-Chinese non-metastatic NPC patients (158, 39) | 842 (shape, first-order intensity, texture, and wavelet-based class) | 3-year OS, PFS, DMFS | Intarak et al. [46] |
| 2023 | CE-T1WI, T1WI, T2WI | 329 primary NPC patients pre-treatment (229, 100) | 3669 (shape, first-order, grey-level co-occurrence matrix (GLCM), grey-level dependence matrix, grey-level run length matrix, grey-level size zone matrix, neighbouring grey tone difference matrix; 3 directions of wavelet decomposition applied) | Staging classification | Li et al. [54] |
| Year | Cohort | Methods Used | Sample Source | Gene Expression Signatures | Outcome Measure | Reference |
|---|---|---|---|---|---|---|
| 2012 | 66 NPC patients and 33 healthy controls | Microarray | Peripheral whole blood samples | Primary genes: LDLRAP1, PHF20, and LUC7L3 Associated secondary suppressor genes: EZH1, IFI35, and UQCRH | Differentiating NPC from control and other diseases. The average ROC AUC was 0.98 (95% C.I. 0.98–0.99) for the combination of the three primary genes together with their associated suppressor genes. | Zaatar, A.M. et al. [118] |
| 2018 | 937 patients with locoregionally advanced NPC | Microarray | Paraffin-embedded tissues—patients with stage III–IVa locoregionally advanced nasopharyngeal carcinoma | YBX3, CBR3, CXCL10, CLASP1, DCTN1, FNDC3B, WSB2, LR1G1, GRM4, ANXA1, WNK1, HDLBP, POLR2M | Stratifying patients into high risk and low risk; patients with high-risk scores had shorter DMFS (HR 4.93, 95% CI 2.99–8.16; p < 0.0001), DFS (HR 3.51, 95% CI 2.43–5.07; p < 0.0001), and OS (HR 3.22, 95% CI 2.18–4.76; p < 0.0001) patients with low-risk scores. | Tang, X.R., et al. [119] |
| 2021 | 60 NPC tumour | RNAseq | Paired tumour tissue and normal tissue | U2AF1L5, TMEM265, GLB1L, and MLF1 | Stratifying patients into high risk and low risk; both OS (HR 2.72, 95% CI (1.679–4.400) and PFS (HR 1.92, 95% CI (1.446–2.563) of the patients in the high-risk group were significantly shorter (p < 0.05) | Zhao, S., et al. [120] |
| 2022 | 12 pairs of NPC patients with similar clinical characteristics NPC, but different metastasis risk | RNAseq | NPC biopsy tissue | AK4, CPAMD8, DDAH1, and CRTR1 | Stratifying patients into high risk and low risk; patients in the high-risk group had a significantly lower DMFS (88.4 versus 73.9%; p = 0.00057) and PFS (75.1 versus 60.4%; p = 0.0058) than the low-risk group. Low-risk groups could benefit from IC + CCRT, but not those identified as high risk. | Liu et al. [121] |
| Year | Cohort | Sample Source | MiRNA Signatures | Outcome Measure | References |
|---|---|---|---|---|---|
| 2012 | 312 NPC and 18 non-cancer specimens | Paraffin-embedded tissue—primary nasopharyngeal carcinoma and non-cancer nasopharyngitis biopsy specimens | miR-142-3p, miR-29c, miR-26a, and miR-30e | The high-risk patient group had shorter DFS (HR 2.73, 95% CI 1.46–5.11; p = 0·0019), DMFS (HR 3.48, 95% CI 1.57–7.75; p = 0·0020), and OS (HR 2.48, 95% CI 1.24–4.96; p = 0·010). Concurrent chemotherapy did not benefit advanced-stage patients with high-risk scores. | Liu et al. [126] |
| 2015 | 734 NPC specimens taken from the training and validation cohort | Paraffin-embedded tissue—primary nasopharyngeal carcinoma | miR-154-5p, miR-449b-5p, miR-140-5Pe, and miR34c-5p | High-risk patients had a higher possibility of distant metastasis (DM) (HR 8.25; p < 0.001 in the training data set and HR 3.2; p = 0.01 in the independent validation set). The high-risk group could benefit from the administration of combined chemoradiation therapy since radiation alone was associated with a 45% risk of developing DM, compared to a 20% risk when treated with combined treatment. | Bruce et al. [127] |
| 2016 | Eight patients in stage I–IV and two normal samples | Paraffin-embedded tissue—taken from poorly differentiated squamous NPC patients | Downregulation of miR-203, miR-199b-5p and miR-4324 and upregulation of miR-2117, miR-4502, miR-4494 in stage I NPC | Down-regulation and upregulation of the miRNAs listed might promote the formation of NPC in stage 1 patients. The upregulated miRNAs were found to suppress apoptosis pathways. | Li et al. [125] |
| 2019 | 120 patients with NPC, 30 patients with head-neck tumours (HNT), and 30 healthy subjects (HS) | Whole blood samples | 8 miRNA signature: miR-188-5p, miR-1908, miR-3196, miR-3935, miR-4284, miR-4433-5p, miR-4665-3p, and miR-513b 16 miRNA signature: miR-1224-3p, miR-1280, miR-155-5p, miR-1908, miR-1973, miR-296-5p, miR-361-3p, miR-425-5p, miR-4284, miR-4436b-5p, miR-4439, miR-4665-3p, miR-4706, miR-4740-3p, miR-5091, and miR-513b | The 8-miRNA signature is used to diagnose NPC in training group 1 (96.43% sensitivity and 100% specificity, AUC = 0.995) and validation group 1 (86.11% sensitivity and 88.89% specificity (AUC = 0.941)). 16-miRNA signature is used to differentiate NPC from HNT and HS with 100% accuracy (AUC = 1.000) in training group 2 and 87.04% (AUC = 0.924) in validation group 2. | Wen et al. [128] |
| 2020 | 200 NPC patients and 189 healthy donors 48 NPC patients and 32 healthy donors | Plasma samples Frozen NPC tissue specimens and paraffin-embedded nasal mucosa tissue specimens from healthy donors | let-7b-5p, miR-140-3p, miR-144-3p, miR-17-5p, miR-20a-5p, miR-20b-5p, and miR-205-5p | None of the seven miRNAs seemed to be associated with NPC prognosis. 7 miRNA signature classified NPC patients from healthy control (AUC 0.807). All of these miRNAs from plasma samples have shown an upregulation trend in NPC compared to healthy patients. Upregulation of miR-144-3p, miR-17-5p, miR-20a-5p, and miR-205-5p and downregulation of let-7b-5p and miR-140-3p in NPC tissue compared to healthy control | Zhang et al. [129] |
| 2022 | 62 cases of NPC and six cases of non-cancerous tissues | Clinical data set (GSE36682) | ebv-miR-BART19-3p, hsa-miR-135b, and hsa-miR-141 | The high-risk patient group has lower OS than the low-risk group (HR 3.98, 95% CI (2.13, 7.42)) | Zhou et al. [130] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suryani, L.; Lee, H.P.Y.; Teo, W.K.; Chin, Z.K.; Loh, K.S.; Tay, J.K. Precision Medicine for Nasopharyngeal Cancer—A Review of Current Prognostic Strategies. Cancers 2024, 16, 918. https://doi.org/10.3390/cancers16050918
Suryani L, Lee HPY, Teo WK, Chin ZK, Loh KS, Tay JK. Precision Medicine for Nasopharyngeal Cancer—A Review of Current Prognostic Strategies. Cancers. 2024; 16(5):918. https://doi.org/10.3390/cancers16050918
Chicago/Turabian StyleSuryani, Luvita, Hazel P. Y. Lee, Wei Keat Teo, Zhi Kang Chin, Kwok Seng Loh, and Joshua K. Tay. 2024. "Precision Medicine for Nasopharyngeal Cancer—A Review of Current Prognostic Strategies" Cancers 16, no. 5: 918. https://doi.org/10.3390/cancers16050918
APA StyleSuryani, L., Lee, H. P. Y., Teo, W. K., Chin, Z. K., Loh, K. S., & Tay, J. K. (2024). Precision Medicine for Nasopharyngeal Cancer—A Review of Current Prognostic Strategies. Cancers, 16(5), 918. https://doi.org/10.3390/cancers16050918






