Hepatocellular Carcinoma: Old and Emerging Therapeutic Targets
Simple Summary
Abstract
1. Introduction
2. Targeted Therapy: State of the Art
2.1. General Standard of Care
2.2. Immunotherapy
2.3. Antibody–Drug Conjugates (ADCs)
2.4. Chimeric Antigen Receptor T-Cells (CAR-Ts)
3. Review of Established and Novel Potential Targets in HCC
3.1. Receptor Tyrosine Kinases (RTKs)
3.1.1. Epidermal Growth Factor Receptors (EGFRs)
3.1.2. Platelet-Derived Growth Factor Receptors (PDGFRs)
3.1.3. Fibroblast Growth Factor Receptors (FGFRs)
3.1.4. Vascular Endothelial Growth Factor Receptors (VEGFRs)
3.1.5. Mesenchymal–Epithelial Transition Factor (c-Met)
3.2. Toll-Like Receptors
3.3. Chemokine Receptors
3.4. RAS/MAPK Pathway
3.5. JAK/STAT Pathway
3.6. PI3K/AKT/mTOR
3.7. Wnt/β-Catenin
3.8. p53
3.9. Cyclins and Cyclin-Dependent Kinases
3.10. TGFβ Signaling
4. Conclusions
| Target Class | Molecule | References | Clinical Trial IDs |
|---|---|---|---|
| VEGF/VEGFRs | VEGFR2 | [64,164,165] | NCT02435433 (Phase III) NCT01140347 (Phase III) |
| VEGF | [64,157,162,163,164,166,167,168] | NCT04487067 (Phase III) NCT04732286 (Phase III) NCT04102098 (Phase III) NCT03434379 (Phase III) NCT05904886 (Phase III) | |
| HGF/c-Met | c-MET | [24,187,194,195,196] | NCT01755767 (Phase III) NCT01908426 (Phase III) NCT03755791 (Phase III) |
| Target Class | Molecule | References | Clinical Trial IDs |
|---|---|---|---|
| EGF/EGFRs | EGFR | [66,72,74,75,76,77,78] | |
| ERRfI1 | [64,83] | ||
| TGFα | [70,90,91] | ||
| EGF | [70,92] | ||
| ADAM17 | [70] | ||
| ERBB2 | [93] | ||
| ERBB3 | [93,94] | ||
| NRG1 | [95] | ||
| PDGF/PDGFRs | PDGFRα | [93,116,118,123,125,126] | |
| PDGFRβ | [93,117,123,126] | ||
| FGF/FGFRs | FGFR3 | [129,130] | |
| FGFR4 | [93,131,140,142,146] | NCT04194801 (Phase I/II) NCT02508467 (Phase I) | |
| FGF1 | [132,133] | ||
| FGF2 | [136,137,138] | ||
| FGF8 | [128,139] | ||
| FGF19 | [140,142,143,144,148,149,150] | ||
| VEGF/VEGFRs | VEGFR1 | [64,164,165] | |
| HGF/c-Met | c-MET | [24,187,194,195,196] | NCT01988493 (Phase I/II) NCT01737827 (Phase II) NCT03672305 (Phase I) |
| HGF | [188,193] | ||
| Toll-like Receptors | TLR2 | [219,220,221,222] | NCT05937295 (Phase I) |
| TLR4 | [211,226,227,228,229,230,231,263] | ||
| Chemokine Receptors and Ligands | CXCL12/CXCR7 axis | [239,240] | |
| CXCL12/CXCR4 axis | [238,241,242,243,244,245] | ||
| CXCL9–CXCL10/CXCR3 axis | [248] | ||
| CXCL1–CXCL2/ CXCR2 axis | [249] | ||
| CXCL5/CXCR2 axis | [250] | ||
| CXCR6 | [251] | ||
| CCL2/CCR2 | [252,253] | ||
| CCL5/CCR5 | [254,255] | ||
| CCL20/CCR6 | [257,258] | ||
| CCR10 | [256] | ||
| RAS/MAPK | CRAF | [64,267,268] | |
| BRAF | [64,267] | ||
| MEK1 and MEK2 | [266,269] | NCT00604721 (Phase II) NCT02292173 (Phase I) | |
| JAK/STAT | JAK1 | [283,296] | |
| JAK2 | [64,292,294,295] | ||
| STAT3 | [297,298,299,301,307,308,313,317] | NCT03195699 (Phase I) | |
| IL-6 | [309,310,311,312,314,315,316] | ||
| PI3K/AKT/mTOR | PTEN | [263,333] | |
| PI3K | [263,333,334] | NCT03735628 (Phase I) | |
| SYK | [64] | ||
| RHEB | [64] | ||
| AKT | [64,263,333] | NCT01239355 (Phase II) | |
| mTOR | [263,333,335,336] | NCT01239355 (Phase II) NCT03591965 (Phase II) NCT02575339 (Phase I/II) | |
| Wnt/β-catenin | β-catenin | [263,283,340] | NCT04008797 (Phase I) NCT05091346 (Phase I/II) |
| APC | [64,263,283,340] | ||
| AXIN1/AXIN2 | [263,283,340] | ||
| DKK1 | [341,342] | NCT03645980 (Phase I/II) | |
| TERT | [343,344] | ||
| PORCN | [283] | NCT02675946 (Phase I) | |
| FZD8/Wnt complex | [283] | NCT02069145 (Phase I) | |
| P53 and Cell Cycle | P53 | [64,283,317,357,358,359,362,381] | |
| Cyclin D1 | [283,333] | ||
| Cyclin E1 | [283,333] | ||
| CDKN2A, CDKN2B | [283,333] | ||
| CDK1 | [64] | ||
| WEE1 | [64] | ||
| CDK4/6 | [379,380,381,382,383] | NCT01356628 (Phase II) NCT02524119 (Phase II) NCT03781960 (Phase II) | |
| Tumor Microenvironment | TGFβ | [243,387,388,391,392,398,399,401,402,403] | NCT01246986 (Phase II) NCT02906397 (Phase I) NCT02423343 (Phase I/II) NCT02699515 (Phase I) |
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Llovet, J.M.; Zucman-Rossi, J.; Pikarsky, E.; Sangro, B.; Schwartz, M.; Sherman, M.; Gores, G. Hepatocellular Carcinoma. Nat. Rev. Dis. Prim. 2016, 2, 16018. [Google Scholar] [CrossRef]
- Russo, F.P.; Zanetto, A.; Pinto, E.; Battistella, S.; Penzo, B.; Burra, P.; Farinati, F. Hepatocellular Carcinoma in Chronic Viral Hepatitis: Where Do We Stand? Int. J. Mol. Sci. 2022, 23, 500. [Google Scholar] [CrossRef] [PubMed]
- Janevska, D.; Chaloska-Ivanova, V.; Janevski, V. Hepatocellular Carcinoma: Risk Factors, Diagnosis and Treatment. Open Access Maced. J. Med. Sci. 2015, 3, 732–736. [Google Scholar] [CrossRef]
- Bogentoft, C.; Ericsson, O.; Kvist, M.; Danielsson, B. Studies on the Medicinal Chemistry of Oxoquinazolines. XI. Synthesis of 1-Methyl-3-Substituted 1,2,3,4-Tetrahydro-4-Oxoquinazolines. Acta Pharm. Suec. 1971, 8, 667–670. [Google Scholar] [PubMed]
- Gosalia, A.J.; Martin, P.; Jones, P.D. Advances and Future Directions in the Treatment of Hepatocellular Carcinoma. Gastroenterol. Hepatol. 2017, 13, 398–410. [Google Scholar]
- Ju, M.R.; Yopp, A.C. Evolving Thresholds for Liver Transplantation in Hepatocellular Carcinoma: A Western Experience. Ann. Gastroenterol. Surg. 2020, 4, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Morizane, C.; Ueno, M.; Okusaka, T.; Ishii, H.; Furuse, J. Chemotherapy for Hepatocellular Carcinoma: Current Status and Future Perspectives. Jpn. J. Clin. Oncol. 2018, 48, 103–114. [Google Scholar] [CrossRef]
- Chen, C.; Wang, Z.; Ding, Y.; Qin, Y. Tumor Microenvironment-Mediated Immune Evasion in Hepatocellular Carcinoma. Front. Immunol. 2023, 14, 1133308. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Y.; Gao, P.; Ding, J. Immune Checkpoint Inhibitor Resistance in Hepatocellular Carcinoma. Cancer Lett. 2023, 555, 216038. [Google Scholar] [CrossRef]
- Makary, M.S.; Khandpur, U.; Cloyd, J.M.; Mumtaz, K.; Dowell, J.D. Locoregional Therapy Approaches for Hepatocellular Carcinoma: Recent Advances and Management Strategies. Cancers 2020, 12, 1914. [Google Scholar] [CrossRef]
- Iyer, R.; Fetterly, G.; Lugade, A.; Thanavala, Y. Sorafenib: A Clinical and Pharmacologic Review. Expert Opin. Pharmacother. 2010, 11, 1943–1955. [Google Scholar] [CrossRef]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.-F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.-L.; Forner, A.; et al. Sorafenib in Advanced Hepatocellular Carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Parikh, N.D.; Marshall, V.D.; Singal, A.G.; Nathan, H.; Lok, A.S.; Balkrishnan, R.; Shahinian, V. Survival and Cost-Effectiveness of Sorafenib Therapy in Advanced Hepatocellular Carcinoma: An Analysis of the SEER-Medicare Database. Hepatology 2017, 65, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cai, L.; Fang, J.; Chen, F.; Pan, F.; Zhang, K.; Huang, Q.; Huang, Y.; Li, D.; Lv, L.; et al. Efficacy and Safety of Transarterial Chemoembolization plus Sorafenib in Patients with Recurrent Hepatocellular Carcinoma after Liver Transplantation. Front. Oncol. 2022, 12, 1101351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hu, P.; Chen, X.; Bie, P. Transarterial Chemoembolization (TACE) plus Sorafenib versus TACE for Intermediate or Advanced Stage Hepatocellular Carcinoma: A Meta-Analysis. PLoS ONE 2014, 9, e100305. [Google Scholar] [CrossRef] [PubMed]
- Lencioni, R.; Llovet, J.M.; Han, G.; Tak, W.Y.; Yang, J.; Guglielmi, A.; Paik, S.W.; Reig, M.; Kim, D.Y.; Chau, G.Y.; et al. Sorafenib or Placebo plus TACE with Doxorubicin-Eluting Beads for Intermediate Stage HCC: The SPACE Trial. J. Hepatol. 2016, 64, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Alturki, N.A. Review of the Immune Checkpoint Inhibitors in the Context of Cancer Treatment. J. Clin. Med. 2023, 12, 4301. [Google Scholar] [CrossRef]
- Herbst, R.S.; Soria, J.C.; Kowanetz, M.; Fine, G.D.; Hamid, O.; Gordon, M.S.; Sosman, J.A.; McDermott, D.F.; Powderly, J.D.; Gettinger, S.N.; et al. Predictive Correlates of Response to the Anti-PD-L1 Antibody MPDL3280A in Cancer Patients. Nature 2014, 515, 563–567. [Google Scholar] [CrossRef]
- Kudo, M. Durvalumab plus Tremelimumab in Unresectable Hepatocellular Carcinoma. Hepatobiliary Surg. Nutr. 2022, 11, 592–596. [Google Scholar] [CrossRef]
- Finn, R.S.; Ryoo, B.Y.; Merle, P.; Kudo, M.; Bouattour, M.; Lim, H.Y.; Breder, V.; Edeline, J.; Chao, Y.; Ogasawara, S.; et al. Pembrolizumab as Second-Line Therapy in Patients with Advanced Hepatocellular Carcinoma in KEYNOTE-240: A Randomized, Double-Blind, Phase III Trial. J. Clin. Oncol. 2020, 38, 193–202. [Google Scholar] [CrossRef]
- Yau, T.; Park, J.W.; Finn, R.S.; Cheng, A.L.; Mathurin, P.; Edeline, J.; Kudo, M.; Harding, J.J.; Merle, P.; Rosmorduc, O.; et al. Nivolumab versus Sorafenib in Advanced Hepatocellular Carcinoma (CheckMate 459): A Randomised, Multicentre, Open-Label, Phase 3 Trial. Lancet Oncol. 2022, 23, 77–90. [Google Scholar] [CrossRef]
- Sun, H.; Song, X.; Li, C.; Li, Q.; Liu, S.; Deng, N. Humanized Disulfide-Stabilized Diabody against Fibroblast Growth Factor-2 Inhibits PD-L1 Expression and Epithelial-Mesenchymal Transition in Hepatoma Cells through STAT3. IUBMB Life 2023, 75, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Liang, Q.; Sun, Z.; Yuan, X.; Hou, W.; Wang, Y.; Wang, H.; Yu, M. Development of Bispecific Anti-c-Met/PD-1 Diabodies for the Treatment of Solid Tumors and the Effect of c-Met Binding Affinity on Efficacy. Oncoimmunology 2021, 10, 1914954. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Li, T.; Guo, J.; Wang, J.; Jia, L.; Shi, X.; Yang, T.; Jiao, R.; Wei, X.; Feng, Z.; et al. Bispecific C-Met/PD-L1 CAR-T Cells Have Enhanced Therapeutic Effects on Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 546586. [Google Scholar] [CrossRef]
- Dahlgren, D.; Lennernäs, H. Antibody-Drug Conjugates and Targeted Treatment Strategies for Hepatocellular Carcinoma: A Drug-Delivery Perspective. Molecules 2020, 25, 2861. [Google Scholar] [CrossRef]
- Huang, L.R.; Hsu, H.C. Cloning and Expression of CD24 Gene in Human Hepatocellular Carcinoma: A Potential Early Tumor Marker Gene Correlates with P53 Mutation and Tumor Differentiation. Cancer Res. 1995, 55, 4717–4721. [Google Scholar] [PubMed]
- Zhang, Y.-F.; Ho, M. Humanization of High-Affinity Antibodies Targeting Glypican-3 in Hepatocellular Carcinoma. Sci. Rep. 2016, 6, 33878. [Google Scholar] [CrossRef]
- Fu, Y.; Urban, D.J.; Nani, R.R.; Zhang, Y.F.; Li, N.; Fu, H.; Shah, H.; Gorka, A.P.; Guha, R.; Chen, L.; et al. Glypican-3-Specific Antibody Drug Conjugates Targeting Hepatocellular Carcinoma. Hepatology 2019, 70, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; He, H.; Sun, F.; Xu, Y.; Huang, X.; Ma, Y.; Zhao, H.; Wang, Y.; Wang, M.; Zhang, J. Selective Targeted Delivery of Doxorubicin via Conjugating to Anti-CD24 Antibody Results in Enhanced Antitumor Potency for Hepatocellular Carcinoma Both in Vitro and in Vivo. J. Cancer Res. Clin. Oncol. 2017, 143, 1929–1940. [Google Scholar] [CrossRef]
- Sun, F.; Wang, T.; Jiang, J.; Wang, Y.; Ma, Z.; Li, Z.; Han, Y.; Pan, M.; Cai, J.; Wang, M.; et al. Engineering a High-Affinity Humanized Anti-CD24 Antibody to Target Hepatocellular Carcinoma by a Novel CDR Grafting Design. Oncotarget 2017, 8, 51238–51252. [Google Scholar] [CrossRef]
- Mullard, A. FDA Approves First CAR T Therapy. Nat. Rev. Drug Discov. 2017, 16, 669. [Google Scholar] [CrossRef]
- Ozer, M.; Goksu, S.Y.; Akagunduz, B.; George, A.; Sahin, I. Adoptive Cell Therapy in Hepatocellular Carcinoma: A Review of Clinical Trials. Cancers 2023, 15, 1808. [Google Scholar] [CrossRef]
- Tseng, H.-C.; Xiong, W.; Badeti, S.; Yang, Y.; Ma, M.; Liu, T.; Ramos, C.A.; Dotti, G.; Fritzky, L.; Jiang, J.-G.; et al. Efficacy of Anti-CD147 Chimeric Antigen Receptors Targeting Hepatocellular Carcinoma. Nat. Commun. 2020, 11, 4810. [Google Scholar] [CrossRef]
- Lian, C.; Guo, Y.; Zhang, J.; Chen, X.; Peng, C. Targeting CD147 Is a Novel Strategy for Antitumor Therapy. Curr. Pharm. Des. 2017, 23, 4410–4421. [Google Scholar] [CrossRef] [PubMed]
- Landras, A.; de Moura, C.R.; Jouenne, F.; Lebbe, C.; Menashi, S.; Mourah, S. CD147 Is a Promising Target of Tumor Progression and a Prognostic Biomarker. Cancers 2019, 11, 1803. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Yazdanifar, M.; Das Roy, L.; Whilding, L.M.; Gavrill, A.; Maher, J.; Mukherjee, P. CAR T Cells Targeting the Tumor MUC1 Glycoprotein Reduce Triple-Negative Breast Cancer Growth. Front. Immunol. 2019, 10, 1149. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Tong, C.; Shi, D.; Chen, M.; Guo, Y.; Chen, D.; Han, X.; Wang, H.; Wang, Y.; Shen, P. Efficacy and Biomarker Analysis of CD133-Directed CAR T Cells in Advanced Hepatocellular Carcinoma: A Single-Arm, Open-Label, Phase II Trial. Oncoimmunology 2020, 9, 1846926. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Jiang, X.; Chen, S.; Lai, Y.; Wei, X.; Li, B.; Lin, S.; Wang, S.; Wu, Q.; Liang, Q.; et al. Anti-GPC3-CAR T Cells Suppress the Growth of Tumor Cells in Patient-Derived Xenografts of Hepatocellular Carcinoma. Front. Immunol. 2016, 7, 690. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yang, C.; Lu, W.; Zeng, Y. Prognostic Significance of Glypican-3 Expression in Hepatocellular Carcinoma: A Meta-Analysis. Medicine 2018, 97, e9702. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Ke, C.; Guo, X.; Ren, P.; Tong, Y.; Luo, S.; He, Y.; Wei, Z.; Cheng, B.; Li, R.; et al. Both Glypican-3/Wnt/β-Catenin Signaling Pathway and Autophagy Contributed to the Inhibitory Effect of Curcumin on Hepatocellular Carcinoma. Dig. Liver Dis. 2019, 51, 120–126. [Google Scholar] [CrossRef]
- Shi, D.; Shi, Y.; Kaseb, A.O.; Qi, X.; Zhang, Y.; Chi, J.; Lu, Q.; Gao, H.; Jiang, H.; Wang, H.; et al. Chimeric Antigen Receptor-Glypican-3 T-Cell Therapy for Advanced Hepatocellular Carcinoma: Results of Phase I Trials. Clin. Cancer Res. 2020, 26, 3979–3989. [Google Scholar] [CrossRef]
- Li, W.; Guo, L.; Rathi, P.; Marinova, E.; Gao, X.; Wu, M.F.; Liu, H.; Dotti, G.; Gottschalk, S.; Metelitsa, L.S.; et al. Redirecting T Cells to Glypican-3 with 4-1BB Zeta Chimeric Antigen Receptors Results in Th1 Polarization and Potent Antitumor Activity. Hum. Gene Ther. 2017, 28, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Pang, N.; Shi, J.; Qin, L.; Chen, A.; Tang, Y.; Yang, H.; Huang, Y.; Wu, Q.; Li, X.; He, B.; et al. IL-7 and CCL19-Secreting CAR-T Cell Therapy for Tumors with Positive Glypican-3 or Mesothelin. J. Hematol. Oncol. 2021, 14, 118. [Google Scholar] [CrossRef]
- Liu, H.; Xu, Y.; Xiang, J.; Long, L.; Green, S.; Yang, Z.; Zimdahl, B.; Lu, J.; Cheng, N.; Horan, L.H.; et al. Targeting Alpha-Fetoprotein (AFP)-MHC Complex with CAR T-Cell Therapy for Liver Cancer. Clin. Cancer Res. 2017, 23, 478–488. [Google Scholar] [CrossRef]
- Xue, J.; Cao, Z.; Cheng, Y.; Wang, J.; Liu, Y.; Yang, R.; Li, H.; Jiang, W.; Li, G.; Zhao, W.; et al. Acetylation of Alpha-Fetoprotein Promotes Hepatocellular Carcinoma Progression. Cancer Lett. 2020, 471, 12–26. [Google Scholar] [CrossRef]
- Li, W.; Liu, K.; Chen, Y.; Zhu, M.; Li, M. Role of Alpha-Fetoprotein in Hepatocellular Carcinoma Drug Resistance. Curr. Med. Chem. 2021, 28, 1126–1142. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Zhao, J.; Tan, A.T.; Hu, W.; Wang, S.Y.; Jin, J.; Wu, J.; Li, Y.; Shi, L.; Fu, J.L.; et al. Immunotherapy of HBV-Related Advanced Hepatocellular Carcinoma with Short-Term HBV-Specific TCR Expressed T Cells: Results of Dose Escalation, Phase I Trial. Hepatol. Int. 2021, 15, 1402–1412. [Google Scholar] [CrossRef]
- Tan, A.T.; Yang, N.; Lee Krishnamoorthy, T.; Oei, V.; Chua, A.; Zhao, X.; Tan, H.S.; Chia, A.; Le Bert, N.; Low, D.; et al. Use of Expression Profiles of HBV-DNA Integrated Into Genomes of Hepatocellular Carcinoma Cells to Select T Cells for Immunotherapy. Gastroenterology 2019, 156, 1862–1876.e9. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Yang, D.; Dai, H.; Liu, X.; Jia, R.; Cui, X.; Li, W.; Cai, C.; Xu, J.; Zhao, X. Eradication of Hepatocellular Carcinoma by NKG2D-Based CAR-T Cells. Cancer Immunol. Res. 2019, 7, 1813–1823. [Google Scholar] [CrossRef]
- Zhang, J.; Basher, F.; Wu, J.D. NKG2D Ligands in Tumor Immunity: Two Sides of a Coin. Front. Immunol. 2015, 6, 97. [Google Scholar] [CrossRef]
- Oliviero, B.; Varchetta, S.; Mele, D.; Pessino, G.; Maiello, R.; Falleni, M.; Tosi, D.; Donadon, M.; Soldani, C.; Franceschini, B.; et al. MICA/B-Targeted Antibody Promotes NK Cell-Driven Tumor Immunity in Patients with Intrahepatic Cholangiocarcinoma. Oncoimmunology 2022, 11, 2035919. [Google Scholar] [CrossRef]
- Mendelsohn, C.L.; Wimmer, E.; Racaniello, V.R. Cellular Receptor for Poliovirus: Molecular Cloning, Nucleotide Sequence, and Expression of a New Member of the Immunoglobulin Superfamily. Cell 1989, 56, 855–865. [Google Scholar] [CrossRef]
- Molfetta, R.; Zitti, B.; Lecce, M.; Milito, N.D.; Stabile, H.; Fionda, C.; Cippitelli, M.; Gismondi, A.; Santoni, A.; Paolini, R. CD155: A Multi-Functional Molecule in Tumor Progression. Int. J. Mol. Sci. 2020, 21, 922. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, S.; Varchetta, S.; Mele, D.; Maiello, R.; Donadon, M.; Soldani, C.; Franceschini, B.; Torzilli, G.; Tartaglia, G.; Maestri, M.; et al. Defective DNAM-1 Dependent Cytotoxicity in Hepatocellular Carcinoma-Infiltrating NK Cells. Cancers 2022, 14, 4060. [Google Scholar] [CrossRef] [PubMed]
- Freed-Pastor, W.A.; Lambert, L.J.; Ely, Z.A.; Pattada, N.B.; Bhutkar, A.; Eng, G.; Mercer, K.L.; Garcia, A.P.; Lin, L.; Rideout, W.M.; et al. The CD155/TIGIT Axis Promotes and Maintains Immune Evasion in Neoantigen-Expressing Pancreatic Cancer. Cancer Cell 2021, 39, 1342–1360.e14. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhang, F.; Ji, F.; Chen, J.; Li, J.; Chen, Z.; Hu, Z.; Guo, Z. Self-Delivery of TIGIT-Blocking ScFv Enhances CAR-T Immunotherapy in Solid Tumors. Front. Immunol. 2023, 14, 1175920. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Jiang, L.; Guan, X.Y. The Genetic and Epigenetic Alterations in Human Hepatocellular Carcinoma: A Recent Update. Protein Cell 2014, 5, 673–691. [Google Scholar] [CrossRef] [PubMed]
- Dimri, M.; Satyanarayana, A. Molecular Signaling Pathways and Therapeutic Targets in Hepatocellular Carcinoma. Cancers 2020, 12, 491. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Nishioka, K. Therapeutic Approaches Targeting Tumor Vasculature in Gastrointestinal Cancers. Front. Biosci. 2011, 3, 541–548. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chuma, M.; Terashita, K.; Sakamoto, N. New Molecularly Targeted Therapies against Advanced Hepatocellular Carcinoma: From Molecular Pathogenesis to Clinical Trials and Future Directions. Hepatol. Res. 2015, 45, E1–E11. [Google Scholar] [CrossRef]
- Lee, Y.H.; Seo, D.; Choi, K.J.; Andersen, J.B.; Won, M.A.; Kitade, M.; Gómez-Quiroz, L.E.; Judge, A.D.; Marquardt, J.U.; Raggi, C.; et al. Antitumor Effects in Hepatocarcinoma of Isoform-Selective Inhibition of HDAC2. Cancer Res. 2014, 74, 4752–4761. [Google Scholar] [CrossRef]
- Dimri, M.; Bilogan, C.; Pierce, L.X.; Naegele, G.; Vasanji, A.; Gibson, I.; McClendon, A.; Tae, K.; Sakaguchi, T.F. Three-Dimensional Structural Analysis Reveals a Cdk5-Mediated Kinase Cascade Regulating Hepatic Biliary Network Branching in Zebrafish. Development 2017, 144, 2595–2605. [Google Scholar] [CrossRef]
- Elattar, S.; Dimri, M.; Satyanarayana, A. The Tumor Secretory Factor ZAG Promotes White Adipose Tissue Browning and Energy Wasting. FASEB J. 2018, 32, 4727–4743. [Google Scholar] [CrossRef]
- Fujita, M.; Chen, M.J.M.; Siwak, D.R.; Sasagawa, S.; Oosawa-Tatsuguchi, A.; Arihiro, K.; Ono, A.; Miura, R.; Maejima, K.; Aikata, H.; et al. Proteo-Genomic Characterization of Virus-Associated Liver Cancers Reveals Potential Subtypes and Therapeutic Targets. Nat. Commun. 2022, 13, 6481. [Google Scholar] [CrossRef] [PubMed]
- Blume-Jensen, P.; Hunter, T. Oncogenic Kinase Signalling. Nature 2001, 411, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Takeda, T.; Sakon, M.; Tsujimoto, M.; Higashiyama, S.; Noda, K.; Miyoshi, E.; Monden, M.; Matsuura, N. Expression and Clinical Significance of Erb-B Receptor Family in Hepatocellular Carcinoma. Br. J. Cancer 2001, 84, 1377–1383. [Google Scholar] [CrossRef] [PubMed]
- Jorissen, R.N.; Walker, F.; Pouliot, N.; Garrett, T.P.J.; Ward, C.W.; Burgess, A.W. Epidermal Growth Factor Receptor: Mechanisms of Activation and Signalling. Exp. Cell Res. 2003, 284, 31–53. [Google Scholar] [CrossRef]
- Citri, A.; Yarden, Y. EGF-ERBB Signalling: Towards the Systems Level. Nat. Rev. Mol. Cell Biol. 2006, 7, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.R.; Wolf, E. The Epidermal Growth Factor Receptor Ligands at a Glance. J. Cell. Physiol. 2009, 218, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Komposch, K.; Sibilia, M. EGFR Signaling in Liver Diseases. Int. J. Mol. Sci. 2015, 17, 30. [Google Scholar] [CrossRef] [PubMed]
- Yarden, Y.; Shilo, B.-Z. SnapShot: EGFR Signaling Pathway. Cell 2007, 131, 1018. [Google Scholar] [CrossRef] [PubMed]
- Buckley, A.F.; Burgart, L.J.; Sahai, V.; Kakar, S. Epidermal Growth Factor Receptor Expression and Gene Copy Number in Conventional Hepatocellular Carcinoma. Am. J. Clin. Pathol. 2008, 129, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Moon, W.S.; Park, H.S.; Yu, K.H.; Park, M.Y.; Kim, K.R.; Jang, K.Y.; Kim, J.S.; Cho, B.H. Expression of Betacellulin and Epidermal Growth Factor Receptor in Hepatocellular Carcinoma: Implications for Angiogenesis. Hum. Pathol. 2006, 37, 1324–1332. [Google Scholar] [CrossRef] [PubMed]
- Lanaya, H.; Natarajan, A.; Komposch, K.; Li, L.; Amberg, N.; Chen, L.; Wculek, S.K.; Hammer, M.; Zenz, R.; Peck-Radosavljevic, M.; et al. EGFR Has a Tumour-Promoting Role in Liver Macrophages during Hepatocellular Carcinoma Formation. Nat. Cell Biol. 2014, 16, 972–981. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Huo, Z.; Song, X.; Shao, Q.; Ren, W.; Huang, X.; Zhou, S.; Tang, X. EGFR Mediates Epithelial-mesenchymal Transition through the Akt/GSK-3β/Snail Signaling Pathway to Promote Liver Cancer Proliferation and Migration. Oncol. Lett. 2024, 27, 59. [Google Scholar] [CrossRef] [PubMed]
- Schiffer, E.; Housset, C.; Cacheux, W.; Wendum, D.; Desbois-Mouthon, C.; Rey, C.; Clergue, F.; Poupon, R.; Barbu, V.; Rosmorduc, O. Gefitinib, an EGFR Inhibitor, Prevents Hepatocellular Carcinoma Development in the Rat Liver with Cirrhosis. Hepatology 2005, 41, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, B.C.; Hoshida, Y.; Fujii, T.; Wei, L.; Yamada, S.; Lauwers, G.Y.; Mcginn, C.M.; Deperalta, D.K.; Chen, X.; Kuroda, T.; et al. Epidermal Growth Factor Receptor Inhibition Attenuates Liver Fibrosis and Development of Hepatocellular Carcinoma. Hepatology 2014, 59, 1577–1590. [Google Scholar] [CrossRef] [PubMed]
- López-Luque, J.; Caballero-Díaz, D.; Martinez-Palacián, A.; Roncero, C.; Moreno-Càceres, J.; García-Bravo, M.; Grueso, E.; Fernández, A.; Crosas-Molist, E.; García-Álvaro, M.; et al. Dissecting the Role of Epidermal Growth Factor Receptor Catalytic Activity during Liver Regeneration and Hepatocarcinogenesis. Hepatology 2016, 63, 604–619. [Google Scholar] [CrossRef]
- Sanoff, H.K.; Bernard, S.; Goldberg, R.M.; Morse, M.A.; Garcia, R.; Woods, L.; Moore, D.T.; O’Neil, B.H. Phase II Study of Capecitabine, Oxaliplatin, and Cetuximab for Advanced Hepatocellular Carcinoma. Gastrointest. Cancer Res. 2011, 4, 78–83. [Google Scholar]
- Zhu, A.X.; Stuart, K.; Blaszkowsky, L.S.; Muzikansky, A.; Reitberg, D.P.; Clark, J.W.; Enzinger, P.C.; Bhargava, P.; Meyerhardt, J.A.; Horgan, K.; et al. Phase 2 Study of Cetuximab in Patients with Advanced Hepatocellular Carcinoma. Cancer 2007, 110, 581–589. [Google Scholar] [CrossRef]
- Berasain, C.; Ujue Latasa, M.; Urtasun, R.; Goñi, S.; Elizalde, M.; Garcia-Irigoyen, O.; Azcona, M.; Prieto, J.; Avila, M.A. Epidermal Growth Factor Receptor (EGFR) Crosstalks in Liver Cancer. Cancers 2011, 3, 2444–2461. [Google Scholar] [CrossRef]
- Chen, L.; Hodges, R.R.; Funaki, C.; Zoukhri, D.; Gaivin, R.J.; Perez, D.M.; Dartt, D.A. Effects of Alpha1D-Adrenergic Receptors on Shedding of Biologically Active EGF in Freshly Isolated Lacrimal Gland Epithelial Cells. Am. J. Physiol. Cell Physiol. 2006, 291, C946–C956. [Google Scholar] [CrossRef] [PubMed]
- Ally, A.; Balasundaram, M.; Carlsen, R.; Chuah, E.; Clarke, A.; Dhalla, N.; Holt, R.A.; Jones, S.J.M.; Lee, D.; Ma, Y.; et al. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341.e23. [Google Scholar] [CrossRef]
- Dubuisson, L.; Monvoisin, A.; Nielsen, B.S.; Le Bail, B.; Bioulac-Sage, P.; Rosenbaum, J. Expression and Cellular Localization of the Urokinase-Type Plasminogen Activator and Its Receptor in Human Hepatocellular Carcinoma. J. Pathol. 2000, 190, 190–195. [Google Scholar] [CrossRef]
- Zheng, Q.; Tang, Z.; Wu, Z.; Shi, D.; Song, H. Inhibitor of Plasminogen Activator 1 (PAI-1) in Hepatocellular Carcinoma. Zhonghua Wai Ke Za Zhi 1998, 36, 474–476. [Google Scholar]
- Zheng, Q.; Tang, Z.; Wu, Z. Urokinase-Type Plasminogen Activator (UPA), UPA Receptor (UPA-R) and Inhibitors (PA I -1) Expression in Hepatocellular Carcinoma in Relation to Cancer Invasion/Metastasis and Prognosis. Zhonghua Zhong Liu Za Zhi 1998, 20, 57–59. [Google Scholar]
- Zhai, B.T.; Tian, H.; Sun, J.; Zou, J.B.; Zhang, X.F.; Cheng, J.X.; Shi, Y.J.; Fan, Y.; Guo, D.Y. Urokinase-Type Plasminogen Activator Receptor (UPAR) as a Therapeutic Target in Cancer. J. Transl. Med. 2022, 20, 135. [Google Scholar] [CrossRef] [PubMed]
- Baart, V.M.; van der Horst, G.; Deken, M.M.; Bhairosingh, S.S.; Schomann, T.; Sier, V.Q.; van der Mark, M.H.; Iamele, L.; de Jonge, H.; Resnati, M.; et al. A Multimodal Molecular Imaging Approach Targeting Urokinase Plasminogen Activator Receptor for the Diagnosis, Resection and Surveillance of Urothelial Cell Carcinoma. Eur. J. Cancer 2021, 146, 11–20. [Google Scholar] [CrossRef]
- Cuesta, Á.M.; Palao, N.; Bragado, P.; Gutierrez-Uzquiza, A.; Herrera, B.; Sánchez, A.; Porras, A. New and Old Key Players in Liver Cancer. Int. J. Mol. Sci. 2023, 24, 17152. [Google Scholar] [CrossRef]
- Lee, G.H.; Fausto, N. Development of Liver Tumors in Transforming Growth Factor a Transgenic Mice. Cancer Res. 1992, 52, 5162–5170. [Google Scholar]
- Zambreg, I.; Assouline, B.; Housset, C.; Schiffer, E. Overexpression of TGF-α and EGFR, a Key Event in Liver Carcinogenesis, Is Induced by Hypoxia Specifically in Hepatocytes. Gastroenterol. Hepatol. Endosc. 2019, 4, 1–4. [Google Scholar] [CrossRef]
- Caja, L.; Sancho, P.; Bertran, E.; Fabregat, I. Dissecting the Effect of Targeting the Epidermal Growth Factor Receptor on TGF-β-Induced-Apoptosis in Human Hepatocellular Carcinoma Cells. J. Hepatol. 2011, 55, 351–358. [Google Scholar] [CrossRef]
- Elmas, A.; Lujambio, A.; Huang, K.-L. Proteomic Analyses Identify Therapeutic Targets in Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 814120. [Google Scholar] [CrossRef]
- Chen, J.Y.; Chen, Y.J.; Yen, C.J.; Chen, W.S.; Huang, W.C. HBx Sensitizes Hepatocellular Carcinoma Cells to Lapatinib by Up-Regulating ErbB3. Oncotarget 2016, 7, 473–489. [Google Scholar] [CrossRef]
- Shi, D.M.; Li, L.X.; Bian, X.Y.; Shi, X.J.; Lu, L.L.; Zhou, H.X.; Pan, T.J.; Zhou, J.; Fan, J.; Wu, W.Z. MiR-296-5p Suppresses EMT of Hepatocellular Carcinoma via Attenuating NRG1/ERBB2/ERBB3 Signaling. J. Exp. Clin. Cancer Res. 2018, 37, 294. [Google Scholar] [CrossRef]
- Andrae, J.; Gallini, R.; Betsholtz, C. Role of Platelet-Derived Growth Factors in Physiology and Medicine. Genes Dev. 2008, 22, 1276–1312. [Google Scholar] [CrossRef]
- Heldin, C.H.; Östman, A.; Rönnstrand, L. Signal Transduction via Platelet-Derived Growth Factor Receptors. Biochim. Biophys. Acta 1998, 1378, F79–F113. [Google Scholar] [CrossRef]
- Birge, R.B.; Kalodimos, C.; Inagaki, F.; Tanaka, S. Crk and CrkL Adaptor Proteins: Networks for Physiological and Pathological Signaling. Cell Commun. Signal. 2009, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, A.; Nånberg, E.; Rönnstrand, L.; Engström, U.; Hellman, U.; Rupp, E.; Carpenter, G.; Heldin, C.H.; Claesson-Welsh, L. Demonstration of Functionally Different Interactions between Phospholipase C-Gamma and the Two Types of Platelet-Derived Growth Factor Receptors. J. Biol. Chem. 1995, 270, 7773–7781. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Lin, G.; Fang, W.; Zhu, H.; Chu, K. Increased Expression of Annexin A1 Predicts Poor Prognosis in Human Hepatocellular Carcinoma and Enhances Cell Malignant Phenotype. Med. Oncol. 2014, 31, 327. [Google Scholar] [CrossRef] [PubMed]
- Bishayee, S.; Ross, A.H.; Womer, R.; Scher, C.D. Purified Human Platelet-Derived Growth Factor Receptor Has Ligand-Stimulated Tyrosine Kinase Activity. Proc. Natl. Acad. Sci. USA 1986, 83, 6756–6760. [Google Scholar] [CrossRef]
- Tallquist, M.; Kazlauskas, A. PDGF Signaling in Cells and Mice. Cytokine Growth Factor Rev. 2004, 15, 205–213. [Google Scholar] [CrossRef]
- Ekman, S.; Thuresson, E.R.; Heldin, C.H.; Rönnstrand, L. Increased Mitogenicity of an Alphabeta Heterodimeric PDGF Receptor Complex Correlates with Lack of RasGAP Binding. Oncogene 1999, 18, 2481–2488. [Google Scholar] [CrossRef]
- Papadopoulos, N.; Lennartsson, J. The PDGF/PDGFR Pathway as a Drug Target. Mol. Asp. Med. 2018, 62, 75–88. [Google Scholar] [CrossRef]
- Ng, F.; Boucher, S.; Koh, S.; Sastry, K.S.R.; Chase, L.; Lakshmipathy, U.; Choong, C.; Yang, Z.; Vemuri, M.C.; Rao, M.S.; et al. PDGF, TGF-Beta, and FGF Signaling Is Important for Differentiation and Growth of Mesenchymal Stem Cells (MSCs): Transcriptional Profiling Can Identify Markers and Signaling Pathways Important in Differentiation of MSCs into Adipogenic, Chondrogenic, and Osteogenic Lineages. Blood 2008, 112, 295–307. [Google Scholar] [CrossRef]
- Heldin, C.H.; Lennartsson, J. Structural and Functional Properties of Platelet-Derived Growth Factor and Stem Cell Factor Receptors. Cold Spring Harb. Perspect. Biol. 2013, 5, a009100. [Google Scholar] [CrossRef]
- Raica, M.; Cimpean, A.M. Platelet-Derived Growth Factor (PDGF)/PDGF Receptors (PDGFR) Axis as Target for Antitumor and Antiangiogenic Therapy. Pharmaceuticals 2010, 3, 572–599. [Google Scholar] [CrossRef]
- Heldin, C.H. Autocrine PDGF Stimulation in Malignancies. Ups. J. Med. Sci. 2012, 117, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Coussens, L.M. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Pietras, K.; Östman, A. Hallmarks of Cancer: Interactions with the Tumor Stroma. Exp. Cell Res. 2010, 316, 1324–1331. [Google Scholar] [CrossRef] [PubMed]
- Bataller, R.; Brenner, D.A. Liver Fibrosis. J. Clin. Investig. 2005, 115, 209–218. [Google Scholar] [CrossRef]
- Hernandez-Gea, V.; Friedman, S.L. Pathogenesis of Liver Fibrosis. Annu. Rev. Pathol. 2011, 6, 425–456. [Google Scholar] [CrossRef]
- Mogler, C.; König, C.; Wieland, M.; Runge, A.; Besemfelder, E.; Komljenovic, D.; Longerich, T.; Schirmacher, P.; Augustin, H.G. Hepatic Stellate Cells Limit Hepatocellular Carcinoma Progression through the Orphan Receptor Endosialin. EMBO Mol. Med. 2017, 9, 741–749. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, L.; Xu, Y.; Zhang, Z.; Ren, G.; Tang, K.; Kuang, P.; Zhao, B.; Yin, Z.; Wang, X. Hepatic Stellate Cells Promote Tumor Progression by Enhancement of Immunosuppressive Cells in an Orthotopic Liver Tumor Mouse Model. Lab. Investig. 2014, 94, 182–191. [Google Scholar] [CrossRef]
- Xu, Y.; Zhao, W.; Xu, J.; Li, J.; Hong, Z.; Yin, Z.; Wang, X. Activated Hepatic Stellate Cells Promote Liver Cancer by Induction of Myeloid-Derived Suppressor Cells through Cyclooxygenase-2. Oncotarget 2016, 7, 8866–8878. [Google Scholar] [CrossRef]
- Yu, J.-H.; Kim, J.M.; Kim, J.K.; Choi, S.J.; Lee, K.S.; Lee, J.-W.; Chang, H.Y.; Lee, J. Il Platelet-Derived Growth Factor Receptor α in Hepatocellular Carcinoma Is a Prognostic Marker Independent of Underlying Liver Cirrhosis. Oncotarget 2017, 8, 39534–39546. [Google Scholar] [CrossRef] [PubMed]
- Borkham-Kamphorst, E.; Herrmann, J.; Stoll, D.; Treptau, J.; Gressner, A.M.; Weiskirchen, R. Dominant-Negative Soluble PDGF-Beta Receptor Inhibits Hepatic Stellate Cell Activation and Attenuates Liver Fibrosis. Lab. Invest. 2004, 84, 766–777. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Pan, Q.; Zhang, X.; Kong, L.Q.; Fan, J.; Dai, Z.; Wang, L.; Yang, X.R.; Hu, J.; Wan, J.L.; et al. MiR-146a Enhances Angiogenic Activity of Endothelial Cells in Hepatocellular Carcinoma by Promoting PDGFRA Expression. Carcinogenesis 2013, 34, 2071–2079. [Google Scholar] [CrossRef] [PubMed]
- von Marschall, Z.; Scholz, A.; Cramer, T.; Schäfer, G.; Schirner, M.; Öberg, K.; Wiedenmann, B.; Höcker, M.; Rosewicz, S. Effects of Interferon Alpha on Vascular Endothelial Growth Factor Gene Transcription and Tumor Angiogenesis. J. Natl. Cancer Inst. 2003, 95, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Ball, S.G.; Shuttleworth, C.A.; Kielty, C.M. Vascular Endothelial Growth Factor Can Signal through Platelet-Derived Growth Factor Receptors. J. Cell Biol. 2007, 177, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.F.; Tan, X.; Zeng, G.; Misse, A.; Singh, S.; Kim, Y.; Klaunig, J.E.; Monga, S.P.S. Conditional Beta-Catenin Loss in Mice Promotes Chemical Hepatocarcinogenesis: Role of Oxidative Stress and Platelet-Derived Growth Factor Receptor Alpha/Phosphoinositide 3-Kinase Signaling. Hepatology 2010, 52, 954–965. [Google Scholar] [CrossRef]
- Awuah, P.K.; Rhieu, B.H.; Singh, S.; Misse, A.; Monga, S.P.S. β-Catenin Loss in Hepatocytes Promotes Hepatocellular Cancer after Diethylnitrosamine and Phenobarbital Administration to Mice. PLoS ONE 2012, 7, e39771. [Google Scholar] [CrossRef]
- Fischer, A.N.M.; Fuchs, E.; Mikula, M.; Huber, H.; Beug, H.; Mikulits, W. PDGF Essentially Links TGF-Beta Signaling to Nuclear Beta-Catenin Accumulation in Hepatocellular Carcinoma Progression. Oncogene 2007, 26, 3395–3405. [Google Scholar] [CrossRef]
- Zhou, L.; An, N.; Haydon, R.C.; Zhou, Q.; Cheng, H.; Peng, Y.; Jiang, W.; Luu, H.H.; Vanichakarn, P.; Szatkowski, J.P.; et al. Tyrosine Kinase Inhibitor STI-571/Gleevec down-Regulates the β-Catenin Signaling Activity. Cancer Lett. 2003, 193, 161–170. [Google Scholar] [CrossRef]
- Kikuchi, A.; Monga, S.P. PDGFRα in Liver Pathophysiology: Emerging Roles in Development, Regeneration, Fibrosis, and Cancer. Gene Expr. 2015, 16, 109–127. [Google Scholar] [CrossRef]
- Hernandez-Gea, V.; Toffanin, S.; Friedman, S.L.; Llovet, J.M. Role of the Microenvironment in the Pathogenesis and Treatment of Hepatocellular Carcinoma. Gastroenterology 2013, 144, 512–527. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, D.; Zhang, T.; Xia, L. FGF/FGFR Signaling in Hepatocellular Carcinoma: From Carcinogenesis to Recent Therapeutic Intervention. Cancers 2021, 13, 1360. [Google Scholar] [CrossRef]
- Gauglhofer, C.; Sagmeister, S.; Schrottmaier, W.; Fischer, C.; Rodgarkia-Dara, C.; Mohr, T.; Stättner, S.; Bichler, C.; Kandioler, D.; Wrba, F.; et al. Up-Regulation of the Fibroblast Growth Factor 8 Subfamily in Human Hepatocellular Carcinoma for Cell Survival and Neoangiogenesis. Hepatology 2011, 53, 854–864. [Google Scholar] [CrossRef]
- Qiu, W.-H.; Zhou, B.-S.; Chu, P.-G.; Chen, W.-G.; Chung, C.; Shih, J.; Hwu, P.; Yeh, C.; Lopez, R.; Yen, Y. Over-Expression of Fibroblast Growth Factor Receptor 3 in Human Hepatocellular Carcinoma. World J. Gastroenterol. 2005, 11, 5266–5272. [Google Scholar] [CrossRef] [PubMed]
- Paur, J.; Nika, L.; Maier, C.; Moscu-Gregor, A.; Kostka, J.; Huber, D.; Mohr, T.; Heffeter, P.; Schrottmaier, W.C.; Kappel, S.; et al. Fibroblast Growth Factor Receptor 3 Isoforms: Novel Therapeutic Targets for Hepatocellular Carcinoma? Hepatology 2015, 62, 1767–1778. [Google Scholar] [CrossRef] [PubMed]
- Raja, A.; Park, I.; Haq, F.; Ahn, S.-M. FGF19-FGFR4 Signaling in Hepatocellular Carcinoma. Cells 2019, 8, 536. [Google Scholar] [CrossRef]
- Wang, H.; Yang, J.; Zhang, K.; Liu, J.; Li, Y.; Su, W.; Song, N. Advances of Fibroblast Growth Factor/Receptor Signaling Pathway in Hepatocellular Carcinoma and Its Pharmacotherapeutic Targets. Front. Pharmacol. 2021, 12, 650388. [Google Scholar] [CrossRef]
- Asada, N.; Tanaka, Y.; Hayashido, Y.; Toratani, S.; Kan, M.; Kitamoto, M.; Nakanishi, T.; Kajiyama, G.; Chayama, K.; Okamoto, T. Expression of Fibroblast Growth Factor Receptor Genes in Human Hepatoma-Derived Cell Lines. Vitr. Cell. Dev. Biol. Anim. 2003, 39, 321–328. [Google Scholar] [CrossRef]
- Sandhu, D.S.; Baichoo, E.; Roberts, L.R. Fibroblast Growth Factor Signaling in Liver Carcinogenesis. Hepatology 2014, 59, 1166–1173. [Google Scholar] [CrossRef]
- Cheng, A.L.; Shen, Y.C.; Zhu, A.X. Targeting Fibroblast Growth Factor Receptor Signaling in Hepatocellular Carcinoma. Oncology 2011, 81, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Poon, R.T.P.; Ng, I.O.L.; Lau, C.; Yu, W.C.; Fan, S.T.; Wong, J. Correlation of Serum Basic Fibroblast Growth Factor Levels with Clinicopathologic Features and Postoperative Recurrence in Hepatocellular Carcinoma. Am. J. Surg. 2001, 182, 298–304. [Google Scholar] [CrossRef]
- Midorikawa, Y.; Ishikawa, S.; Iwanari, H.; Imamura, T.; Sakamoto, H.; Miyazono, K.; Kodama, T.; Makuuchi, M.; Aburatani, H. Glypican-3, Overexpressed in Hepatocellular Carcinoma, Modulates FGF2 and BMP-7 Signaling. Int. J. Cancer 2003, 103, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Eun, J.W.; Cho, H.J.; Lee, H.Y.; Seo, C.W.; Noh, C.K.; Shin, S.J.; Lee, K.M.; Cho, S.W.; Cheong, J.Y. Effect of Fibroblast Growth Factor-2 and Its Receptor Gene Polymorphisms on the Survival of Patients with Hepatitis B Virus-Associated Hepatocellular Carcinoma. Anticancer Res. 2019, 39, 2217–2226. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Sun, X.; Guo, X.; Yin, H.; Wang, L.; Tian, F.; Jing, H.; Liang, X.; Xu, J.; Shi, P. FGF8 Promotes Cell Proliferation and Resistance to EGFR Inhibitors via Upregulation of EGFR in Human Hepatocellular Carcinoma Cells. Oncol. Rep. 2017, 38, 2205–2210. [Google Scholar] [CrossRef]
- Ho, H.K.; Pok, S.; Streit, S.; Ruhe, J.E.; Hart, S.; Lim, K.S.; Loo, H.L.; Aung, M.O.; Lim, S.G.; Ullrich, A. Fibroblast Growth Factor Receptor 4 Regulates Proliferation, Anti-Apoptosis and Alpha-Fetoprotein Secretion during Hepatocellular Carcinoma Progression and Represents a Potential Target for Therapeutic Intervention. J. Hepatol. 2009, 50, 118–127. [Google Scholar] [CrossRef]
- Yang, H.; Fang, F.; Chang, R.; Yang, L. MicroRNA-140-5p Suppresses Tumor Growth and Metastasis by Targeting Transforming Growth Factor β Receptor 1 and Fibroblast Growth Factor 9 in Hepatocellular Carcinoma. Hepatology 2013, 58, 205–217. [Google Scholar] [CrossRef]
- French, D.M.; Lin, B.C.; Wang, M.; Adams, C.; Shek, T.; Hötzel, K.; Bolon, B.; Ferrando, R.; Blackmore, C.; Schroeder, K.; et al. Targeting FGFR4 Inhibits Hepatocellular Carcinoma in Preclinical Mouse Models. PLoS ONE 2012, 7, e36713. [Google Scholar] [CrossRef]
- Lin, B.C.; Desnoyers, L.R. FGF19 and Cancer. Adv. Exp. Med. Biol. 2012, 728, 183–194. [Google Scholar] [CrossRef]
- Liu, W.Y.; Xie, D.M.; Zhu, G.Q.; Huang, G.Q.; Lin, Y.Q.; Wang, L.R.; Shi, K.Q.; Hu, B.; Braddock, M.; Chen, Y.P.; et al. Targeting Fibroblast Growth Factor 19 in Liver Disease: A Potential Biomarker and Therapeutic Target. Expert Opin. Ther. Targets 2015, 19, 675–685. [Google Scholar] [CrossRef]
- Gu, Q.; Zhang, B.; Sun, H.; Xu, Q.; Tan, Y.; Wang, G.; Luo, Q.; Xu, W.; Yang, S.; Li, J.; et al. Genomic Characterization of a Large Panel of Patient-Derived Hepatocellular Carcinoma Xenograft Tumor Models for Preclinical Development. Oncotarget 2015, 6, 20160–20176. [Google Scholar] [CrossRef]
- Sheu, M.J.; Hsieh, M.J.; Chiang, W.L.; Yang, S.F.; Lee, H.L.; Lee, L.M.; Yeh, C. Bin Fibroblast Growth Factor Receptor 4 Polymorphism Is Associated with Liver Cirrhosis in Hepatocarcinoma. PLoS ONE 2015, 10, e0122961. [Google Scholar] [CrossRef]
- Lin, Z.Z.; Hsu, C.; Jeng, Y.M.; Hu, F.C.; Pan, H.W.; Wu, Y.M.; Hsu, H.C.; Cheng, A.L. Klotho-Beta and Fibroblast Growth Factor 19 Expression Correlates with Early Recurrence of Resectable Hepatocellular Carcinoma. Liver Int. 2019, 39, 1682–1691. [Google Scholar] [CrossRef]
- Naugler, W.E.; Tarlow, B.D.; Fedorov, L.M.; Taylor, M.; Pelz, C.; Li, B.; Darnell, J.; Grompe, M. Fibroblast Growth Factor Signaling Controls Liver Size in Mice with Humanized Livers. Gastroenterology 2015, 149, 728–740.e15. [Google Scholar] [CrossRef] [PubMed]
- Sawey, E.T.; Chanrion, M.; Cai, C.; Wu, G.; Zhang, J.; Zender, L.; Zhao, A.; Busuttil, R.W.; Yee, H.; Stein, L.; et al. Identification of a Therapeutic Strategy Targeting Amplified FGF19 in Liver Cancer by Oncogenomic Screening. Cancer Cell 2011, 19, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Miura, S.; Mitsuhashi, N.; Shimizu, H.; Kimura, F.; Yoshidome, H.; Otsuka, M.; Kato, A.; Shida, T.; Okamura, D.; Miyazaki, M. Fibroblast Growth Factor 19 Expression Correlates with Tumor Progression and Poorer Prognosis of Hepatocellular Carcinoma. BMC Cancer 2012, 12, 56. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Shibuya, M. The Vascular Endothelial Growth Factor (VEGF)/ VEGF Receptor System and Its Role under Physiological and Pathological Conditions. Clin. Sci. 2005, 109, 227–241. [Google Scholar] [CrossRef]
- Apte, R.S.; Chen, D.S.; Ferrara, N. Leading Edge Review VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264. [Google Scholar] [CrossRef]
- Cannito, S.; Foglia, B.; Villano, G.; Turato, C.; Delgado, T.C.; Morello, E.; Pin, F.; Novo, E.; Napione, L.; Quarta, S.; et al. SerpinB3 Differently Up-Regulates Hypoxia Inducible Factors-1α and -2α in Hepatocellular Carcinoma: Mechanisms Revealing Novel Potential Therapeutic Targets. Cancers 2019, 11, 1933. [Google Scholar] [CrossRef]
- Germain, S.; Monnot, C.; Muller, L.; Eichmann, A. Hypoxia-Driven Angiogenesis: Role of Tip Cells and Extracellular Matrix Scaffolding. Curr. Opin. Hematol. 2010, 17, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, M.; Heymach, J.V. Vascular Endothelial Growth Factor (VEGF) Pathway. J. Thorac. Oncol. 2006, 1, 768–770. [Google Scholar] [CrossRef] [PubMed]
- Kerbel, R.S. Tumor Angiogenesis. N. Engl. J. Med. 2008, 358, 2039. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Qin, S.; Zheng, Y.; Han, L.; Zhang, M.; Luo, N.; Liu, Z.; Gu, N.; Gu, X.; Yin, X. Molecular Targeting of VEGF/VEGFR Signaling by the Anti-VEGF Monoclonal Antibody BD0801 Inhibits the Growth and Induces Apoptosis of Human Hepatocellular Carcinoma Cells in Vitro and in Vivo. Cancer Biol. Ther. 2017, 18, 166–176. [Google Scholar] [CrossRef]
- Ferrara, N. Vascular Endothelial Growth Factor and Age-Related Macular Degeneration: From Basic Science to Therapy. Nat. Med. 2010, 16, 1107–1111. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.K. Molecular Regulation of Vessel Maturation. Nat. Med. 2003, 9, 685–693. [Google Scholar] [CrossRef]
- Nagy, J.A.; Chang, S.H.; Dvorak, A.M.; Dvorak, H.F. Why Are Tumour Blood Vessels Abnormal and Why Is It Important to Know? Br. J. Cancer 2009, 100, 865–869. [Google Scholar] [CrossRef]
- Ferrara, N. Vascular Endothelial Growth Factor: Basic Science and Clinical Progress. Endocr. Rev. 2004, 25, 581–611. [Google Scholar] [CrossRef]
- Tseng, P.L.; Tai, M.H.; Huang, C.C.; Wang, C.C.; Lin, J.W.; Hung, C.H.; Chen, C.H.; Wang, J.H.; Lu, S.N.; Lee, C.M.; et al. Overexpression of VEGF Is Associated with Positive P53 Immunostaining in Hepatocellular Carcinoma (HCC) and Adverse Outcome of HCC Patients. J. Surg. Oncol. 2008, 98, 349–357. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, J.N.; Tang, J.M.; Kong, X.; Yang, J.Y.; Zheng, F.; Guo, L.Y.; Huang, Y.Z.; Zhang, L.; Tian, L.; et al. VEGF Is Essential for the Growth and Migration of Human Hepatocellular Carcinoma Cells. Mol. Biol. Rep. 2012, 39, 5085–5093. [Google Scholar] [CrossRef]
- Liu, M.; Yang, S.; Zhang, D.; Shui, P.; Song, S.; Yao, J.; Dai, Y.; Sun, Q. Fructopyrano-(1→4)-Glucopyranose Inhibits the Proliferation of Liver Cancer Cells and Angiogenesis in a VEGF/VEGFR Dependent Manner. Int. J. Clin. Exp. Med. 2014, 7, 3859. [Google Scholar] [PubMed]
- Peng, S.; Wang, Y.; Peng, H.; Chen, D.; Shen, S.; Peng, B.; Chen, M.; Lencioni, R.; Kuang, M. Autocrine Vascular Endothelial Growth Factor Signaling Promotes Cell Proliferation and Modulates Sorafenib Treatment Efficacy in Hepatocellular Carcinoma. Hepatology 2014, 60, 1264–1277. [Google Scholar] [CrossRef] [PubMed]
- Mise, M.; Arii, S.; Higashituji, H.; Furutani, M.; Niwano, M.; Harada, T.; Ishigami, S.; Toda, Y.; Nakayama, H.; Fukumoto, M.; et al. Clinical Significance of Vascular Endothelial Growth Factor and Basic Fibroblast Growth Factor Gene Expression in Liver Tumor. Hepatology 1996, 23, 455–464. [Google Scholar] [CrossRef]
- Zhan, P.; Qian, Q.; Yu, L.-K. Prognostic Significance of Vascular Endothelial Growth Factor Expression in Hepatocellular Carcinoma Tissue: A Meta-Analysis. Hepatobiliary Surg. Nutr. 2013, 2, 148. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Luo, L.; Chen, W.; Chen, L.Z.; Zeng, W.T.; Li, W.; Huang, X.H. Significance of the Vascular Endothelial Growth Factor and the Macrophage Migration Inhibitory Factor in the Progression of Hepatocellular Carcinoma. Oncol. Rep. 2014, 31, 1199–1204. [Google Scholar] [CrossRef]
- Chinnasamy, D.; Yu, Z.; Theoret, M.R.; Zhao, Y.; Shrimali, R.K.; Morgan, R.A.; Feldman, S.A.; Restifo, N.P.; Rosenberg, S.A. Gene Therapy Using Genetically Modified Lymphocytes Targeting VEGFR-2 Inhibits the Growth of Vascularized Syngenic Tumors in Mice. J. Clin. Investig. 2010, 120, 3953–3968. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ma, Y.; Li, J.; Shi, H.S.; Wang, L.Q.; Guo, F.C.; Zhang, J.; Li, D.; Mo, B.H.; Wen, F.; et al. Specificity Redirection by CAR with Human VEGFR-1 Affinity Endows T Lymphocytes with Tumor-Killing Ability and Anti-Angiogenic Potency. Gene Ther. 2013, 20, 970–978. [Google Scholar] [CrossRef]
- Hajari Taheri, F.; Hassani, M.; Sharifzadeh, Z.; Behdani, M.; Arashkia, A.; Abolhassani, M. T Cell Engineered with a Novel Nanobody-Based Chimeric Antigen Receptor against VEGFR2 as a Candidate for Tumor Immunotherapy. IUBMB Life 2019, 71, 1259–1267. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Yang, X.; Xu, Y.; Tang, K.; Tian, Z.; Chen, Z.; Zhang, Y.; Xue, Z.; Rao, Q.; Wang, M.; et al. Anti-Tumor Effects of Vascular Endothelial Growth Factor/Vascular Endothelial Growth Factor Receptor Binding Domain-Modified Chimeric Antigen Receptor T Cells. Cytotherapy 2021, 23, 810–819. [Google Scholar] [CrossRef]
- Akbari, P.; Katsarou, A.; Daghighian, R.; Van Mil, L.W.H.G.; Huijbers, E.J.M.; Griffioen, A.W.; Van Beijnum, J.R. Directing CAR T Cells towards the Tumor Vasculature for the Treatment of Solid Tumors. BBA Rev. Cancer 2022, 1877, 188701. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Rao, B.; Lou, J.; Li, J.; Liu, Z.; Li, A.; Cui, G.; Ren, Z.; Yu, Z. The Function of the HGF/c-Met Axis in Hepatocellular Carcinoma. Front. Cell Dev. Biol. 2020, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Nawa, K.; Ichihara, A. Partial Purification and Characterization of Hepatocyte Growth Factor from Serum of Hepatectomized Rats. Biochem. Biophys. Res. Commun. 1984, 122, 1450–1459. [Google Scholar] [CrossRef]
- Gherardi, E.; Stoker, M. Hepatocytes and Scatter Factor. Nature 1990, 346, 228. [Google Scholar] [CrossRef] [PubMed]
- García-Vilas, J.A.; Medina, M.Á. Updates on the Hepatocyte Growth Factor/c-Met Axis in Hepatocellular Carcinoma and Its Therapeutic Implications. World J. Gastroenterol. 2018, 24, 3695. [Google Scholar] [CrossRef] [PubMed]
- Basilico, C.; Hultberg, A.; Blanchetot, C.; De Jonge, N.; Festjens, E.; Hanssens, V.; Osepa, S.I.; De Boeck, G.; Mira, A.; Cazzanti, M.; et al. Four Individually Druggable MET Hotspots Mediate HGF-Driven Tumor Progression. J. Clin. Investig. 2014, 124, 3172–3186. [Google Scholar] [CrossRef]
- González, M.N.; de Mello, W.; Butler-Browne, G.S.; Silva-Barbosa, S.D.; Mouly, V.; Savino, W.; Riederer, I. HGF Potentiates Extracellular Matrix-Driven Migration of Human Myoblasts: Involvement of Matrix Metalloproteinases and MAPK/ERK Pathway. Skelet. Muscle 2017, 7, 20. [Google Scholar] [CrossRef]
- Pascale, R.M.; Feo, F.; Calvisi, D.F. An Infernal Cross-Talk between Oncogenic β-Catenin and c-Met in Hepatocellular Carcinoma: Evidence from Mouse Modeling. Hepatology 2016, 64, 1421–1423. [Google Scholar] [CrossRef]
- Scagliotti, G.V.; Novello, S.; von Pawel, J. The Emerging Role of MET/HGF Inhibitors in Oncology. Cancer Treat. Rev. 2013, 39, 793–801. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Chu, J.H.; Cui, S.X.; Song, Z.Y.; Qu, X.J. Des-γ-Carboxy Prothrombin (DCP) as a Potential Autologous Growth Factor for the Development of Hepatocellular Carcinoma. Cell. Physiol. Biochem. 2014, 34, 903–915. [Google Scholar] [CrossRef]
- Bozkaya, G.; Korhan, P.; Çokakli, M.; Erdal, E.; Saǧol, Ö.; Karademir, S.; Korch, C.; Atabey, N. Cooperative Interaction of MUC1 with the HGF/c-Met Pathway during Hepatocarcinogenesis. Mol. Cancer 2012, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Xu, E.; Zhao, Y.; Singh, S.; Li, X.; Couchy, G.; Chen, X.; Zucman-Rossi, J.; Chikina, M.; Monga, S.P.S. Modeling a Human Hepatocellular Carcinoma Subset in Mice through Coexpression of Met and Point-Mutant β-Catenin. Hepatology 2016, 64, 1587–1605. [Google Scholar] [CrossRef]
- Corso, S.; Giordano, S. Cell-Autonomous and Non-Cell-Autonomous Mechanisms of HGF/MET-Driven Resistance to Targeted Therapies: From Basic Research to a Clinical Perspective. Cancer Discov. 2013, 3, 978–992. [Google Scholar] [CrossRef] [PubMed]
- Bouattour, M.; Raymond, E.; Qin, S.; Cheng, A.L.; Stammberger, U.; Locatelli, G.; Faivre, S. Recent Developments of C-Met as a Therapeutic Target in Hepatocellular Carcinoma. Hepatology 2018, 67, 1132–1149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Z.; Pan, F.Y.; Xu, J.F.; Yuan, J.; Guo, S.Y.; Dai, G.; Xue, B.; Shen, W.G.; Wen, C.J.; Zhao, D.H.; et al. Knockdown of C-Met by Adenovirus-Delivered Small Interfering RNA Inhibits Hepatocellular Carcinoma Growth in Vitro and in Vivo. Mol. Cancer Ther. 2005, 4, 1577–1584. [Google Scholar] [CrossRef]
- Wang, S.W.; Pan, S.L.; Peng, C.Y.; Huang, D.Y.; Tsai, A.C.; Chang, Y.L.; Guh, J.H.; Kuo, S.C.; Lee, K.H.; Teng, C.M. CHM-1 Inhibits Hepatocyte Growth Factor-Induced Invasion of SK-Hep-1 Human Hepatocellular Carcinoma Cells by Suppressing Matrix Metalloproteinase-9 Expression. Cancer Lett. 2007, 257, 87–96. [Google Scholar] [CrossRef]
- Ding, W.; You, H.; Dang, H.; LeBlanc, F.; Galicia, V.; Lu, S.C.; Stiles, B.; Rountree, C.B. Epithelial-to-Mesenchymal Transition of Murine Liver Tumor Cells Promotes Invasion. Hepatology 2010, 52, 945–953. [Google Scholar] [CrossRef]
- He, M.; Peng, A.; Huang, X.Z.; Shi, D.C.; Wang, J.C.; Zhao, Q.; Lin, H.; Kuang, D.M.; Ke, P.F.; Lao, X.M. Peritumoral Stromal Neutrophils Are Essential for C-Met-Elicited Metastasis in Human Hepatocellular Carcinoma. Oncoimmunology 2016, 5, e1219828. [Google Scholar] [CrossRef]
- Jia, C.C.; Wang, T.T.; Liu, W.; Fu, B.S.; Hua, X.F.; Wang, G.Y.; Li, T.J.; Li, X.; Wu, X.Y.; Tai, Y.; et al. Cancer-Associated Fibroblasts from Hepatocellular Carcinoma Promote Malignant Cell Proliferation by HGF Secretion. PLoS ONE 2013, 8, e63243. [Google Scholar] [CrossRef] [PubMed]
- Karabulut, S.; Tas, F.; Akyüz, F.; Ormeci, A.C.; Serilmez, M.; Soydinç, H.O.; Vatansever, S.; Yasasever, V. Clinical Significance of Serum Hepatocyte Growth Factor (HGF) Levels in Hepatocellular Carcinoma. Tumor Biol. 2014, 35, 2327–2333. [Google Scholar] [CrossRef] [PubMed]
- Unić, A.; Derek, L.; Duvnjak, M.; Patrlj, L.; Rakić, M.; Kujundžić, M.; Renjić, V.; Štoković, N.; Dinjar, P.; Jukic, A.; et al. Diagnostic Specificity and Sensitivity of PIVKAII, GP3, CSTB, SCCA1 and HGF for the Diagnosis of Hepatocellular Carcinoma in Patients with Alcoholic Liver Cirrhosis. Ann. Clin. Biochem. 2018, 55, 355–362. [Google Scholar] [CrossRef] [PubMed]
- You, H.; Ding, W.; Dang, H.; Jiang, Y.; Rountree, C.B. C-Met Represents a Potential Therapeutic Target for Personalized Treatment in Hepatocellular Carcinoma. Hepatology 2011, 54, 879–889. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Caenepeel, S.; Shen, Y.; Rex, K.; Zhang, Y.; He, Y.; Tang, E.T.; Wang, O.; Zhong, W.; Zhou, H.; et al. Preclinical Evaluation of AMG 337, a Highly Selective Small Molecule MET Inhibitor, in Hepatocellular Carcinoma. Mol. Cancer Ther. 2016, 15, 1227–1237. [Google Scholar] [CrossRef]
- Luo, T.; Zhang, S.G.; Zhu, L.F.; Zhang, F.X.; Li, W.; Zhao, K.; Wen, X.X.; Yu, M.; Zhan, Y.Q.; Chen, H.; et al. A Selective C-Met and Trks Inhibitor Indo5 Suppresses Hepatocellular Carcinoma Growth. J. Exp. Clin. Cancer Res. 2019, 38, 130. [Google Scholar] [CrossRef]
- Goyal, L.; Muzumdar, M.D.; Zhu, A.X. Targeting the HGF/c-MET Pathway in Hepatocellular Carcinoma. Clin. Cancer Res. 2013, 19, 2310–2318. [Google Scholar] [CrossRef]
- Santoro, A.; Rimassa, L.; Borbath, I.; Daniele, B.; Salvagni, S.; Van Laethem, J.L.; Van Vlierberghe, H.; Trojan, J.; Kolligs, F.T.; Weiss, A.; et al. Tivantinib for Second-Line Treatment of Advanced Hepatocellular Carcinoma: A Randomised, Placebo-Controlled Phase 2 Study. Lancet. Oncol. 2013, 14, 55–63. [Google Scholar] [CrossRef]
- Liu, J.J.; Li, Y.; Chen, W.S.; Liang, Y.; Wang, G.; Zong, M.; Kaneko, K.; Xu, R.; Karin, M.; Feng, G.S. Shp2 Deletion in Hepatocytes Suppresses Hepatocarcinogenesis Driven by Oncogenic β-Catenin, PIK3CA and MET. J. Hepatol. 2018, 69, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Karagonlar, Z.F.; Korhan, P.; Atabey, N. Targeting C-Met in Cancer by MicroRNAs: Potential Therapeutic Applications in Hepatocellular Carcinoma. Drug Dev. Res. 2015, 76, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Zhou, H.; Miao, Y.; Li, N.; Zhao, L.; Jia, L. MiRNA Expression Profiles Reveal the Involvement of MiR-26a, MiR-548l and MiR-34a in Hepatocellular Carcinoma Progression through Regulation of ST3GAL5. Lab. Invest. 2017, 97, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Tao, J.; Li, X.; Zhang, T.; Zhao, L.; Wang, Y.; Zhang, L.; Xiong, J.; Zeng, Z.; Zhan, N.; et al. MicroRNA-206 Prevents the Pathogenesis of Hepatocellular Carcinoma by Modulating Expression of Met Proto-Oncogene and Cyclin-Dependent Kinase 6 in Mice. Hepatology 2017, 66, 1952–1967. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Shan, X.; Chen, K.; Liu, Y.; Yu, G.; Chen, Q.; Zeng, T.; Zhu, L.; Dang, H.; Chen, F.; et al. LINC00052/MiR-101-3p Axis Inhibits Cell Proliferation and Metastasis by Targeting SOX9 in Hepatocellular Carcinoma. Gene 2018, 679, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Wang, X.; Zhao, Y.; Hu, R.; Qin, L. Exosomal MiR-93 Promotes Proliferation and Invasion in Hepatocellular Carcinoma by Directly Inhibiting TIMP2/TP53INP1/CDKN1A. Biochem. Biophys. Res. Commun. 2018, 502, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tan, J.; Ou, S.; Chen, J.; Chen, L. MicroRNA-101-3p Suppresses Proliferation and Migration in Hepatocellular Carcinoma by Targeting the HGF/c-Met Pathway. Investig. New Drugs 2020, 38, 60–69. [Google Scholar] [CrossRef]
- Guizhen, Z.; Guanchang, J.; Liwen, L.; Huifen, W.; Zhigang, R.; Ranran, S.; Zujiang, Y. The Tumor Microenvironment of Hepatocellular Carcinoma and Its Targeting Strategy by CAR-T Cell Immunotherapy. Front. Endocrinol. 2022, 13, 918869. [Google Scholar] [CrossRef]
- Tchou, J.; Zhao, Y.; Levine, B.L.; Zhang, P.J.; Davis, M.M.; Melenhorst, J.J.; Kulikovskaya, I.; Brennan, A.L.; Liu, X.; Lacey, S.F.; et al. Safety and Efficacy of Intratumoral Injections of Chimeric Antigen Receptor (CAR) T Cells in Metastatic Breast Cancer. Cancer Immunol. Res. 2017, 5, 1152–1161. [Google Scholar] [CrossRef]
- Mori, J.; Adachi, K.; Sakoda, Y.; Sasaki, T.; Goto, S.; Matsumoto, H.; Nagashima, Y.; Matsuyama, H.; Tamada, K. Anti-tumor Efficacy of Human Anti-c-met CAR-T Cells against Papillary Renal Cell Carcinoma in an Orthotopic Model. Cancer Sci. 2021, 112, 1417. [Google Scholar] [CrossRef]
- Kang, C.H.; Kim, Y.; Lee, D.Y.; Choi, S.U.; Lee, H.K.; Park, C.H. C-Met-Specific Chimeric Antigen Receptor T Cells Demonstrate Anti-Tumor Effect in c-Met Positive Gastric Cancer. Cancers 2021, 13, 5738. [Google Scholar] [CrossRef]
- Liu, L.; Liu, B.; Yu, J.; Zhang, D.; Shi, J.; Liang, P. Development of a Toll-Like Receptor-Based Gene Signature That Can Predict Prognosis, Tumor Microenvironment, and Chemotherapy Response for Hepatocellular Carcinoma. Front. Mol. Biosci. 2021, 8, 729789. [Google Scholar] [CrossRef]
- Soares, J.B.; Pimentel-Nunes, P.; Afonso, L.; Rolanda, C.; Lopes, P.; Roncon-Albuquerque, R.; Gonçalves, N.; Boal-Carvalho, I.; Pardal, F.; Lopes, S.; et al. Increased Hepatic Expression of TLR2 and TLR4 in the Hepatic Inflammation-Fibrosis-Carcinoma Sequence. Innate Immun. 2012, 18, 700–708. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Takeda, K. Toll-like Receptor Signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Fukata, M.; Chen, A.; Klepper, A.; Krishnareddy, S.; Vamadevan, A.S.; Thomas, L.S.; Xu, R.; Inoue, H.; Arditi, M.; Dannenberg, A.J.; et al. Cox-2 Is Regulated by Toll-like Receptor-4 (TLR4) Signaling: Role in Proliferation and Apoptosis in the Intestine. Gastroenterology 2006, 131, 862–877. [Google Scholar] [CrossRef]
- Spitzer, J.A.; Zheng, M.; Kolls, J.K.; Vande Stouwe, C.; Spitzer, J.J. Ethanol and LPS Modulate NF-KappaB Activation, Inducible NO Synthase and COX-2 Gene Expression in Rat Liver Cells in Vivo. Front. Biosci. 2002, 7, a99–a108. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.T.; Lee, C.W.; Tung, W.H.; Wang, S.W.; Lin, C.C.; Shu, J.C.; Yang, C.M. Cooperation of TLR2 with MyD88, PI3K, and Rac1 in Lipoteichoic Acid-Induced CPLA2/COX-2-Dependent Airway Inflammatory Responses. Am. J. Pathol. 2010, 176, 1671–1684. [Google Scholar] [CrossRef]
- Szabo, G.; Dolganiuc, A.; Mandrekar, P. Pattern Recognition Receptors: A Contemporary View on Liver Diseases. Hepatology 2006, 44, 287–298. [Google Scholar] [CrossRef]
- Gao, B.; Jeong, W.-I.; Tian, Z. Liver: An Organ with Predominant Innate Immunity. Hepatology 2008, 47, 729–736. [Google Scholar] [CrossRef]
- Roh, Y.S.; Zhang, B.; Loomba, R.; Seki, E. TLR2 and TLR9 Contribute to Alcohol-Mediated Liver Injury through Induction of CXCL1 and Neutrophil Infiltration. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 309, 30–41. [Google Scholar] [CrossRef]
- Li, S.; Sun, R.; Chen, Y.; Wei, H.; Tian, Z. TLR2 Limits Development of Hepatocellular Carcinoma by Reducing IL18-Mediated Immunosuppression. Cancer Res. 2015, 75, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Cai, B.; Xu, M.; Qiu, Z.; Tao, Y.; Zhang, Y.; Wang, J.; Xu, Y.; Zhou, Y.; Yang, J.; et al. Gene Silencing of Toll-like Receptor 2 Inhibits Proliferation of Human Liver Cancer Cells and Secretion of Inflammatory Cytokines. PLoS ONE 2012, 7, e38890. [Google Scholar] [CrossRef] [PubMed]
- Nischalke, H.D.; Coenen, M.; Berger, C.; Aldenhoff, K.; Müller, T.; Berg, T.; Krämer, B.; Körner, C.; Odenthal, M.; Schulze, F.; et al. The Toll-like Receptor 2 (TLR2) -196 to -174 Del/Ins Polymorphism Affects Viral Loads and Susceptibility to Hepatocellular Carcinoma in Chronic Hepatitis C. Int. J. Cancer 2012, 130, 1470–1475. [Google Scholar] [CrossRef]
- Mohamed, F.E.Z.A.; Hammad, S.; Luong, T.V.; Dewidar, B.; Al-Jehani, R.; Davies, N.; Dooley, S.; Jalan, R. Expression of TLR-2 in Hepatocellular Carcinoma Is Associated with Tumour Proliferation, Angiogenesis and Caspase-3 Expression. Pathol. Res. Pract. 2020, 216, 152980. [Google Scholar] [CrossRef] [PubMed]
- Rakoff-Nahoum, S.; Medzhitov, R. Toll-like Receptors and Cancer. Nat. Rev. Cancer 2009, 9, 57–63. [Google Scholar] [CrossRef]
- Chen, R.; Alvero, A.B.; Silasi, D.A.; Steffensen, K.D.; Mor, G. Cancers Take Their Toll--the Function and Regulation of Toll-like Receptors in Cancer Cells. Oncogene 2008, 27, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Pimentel-Nunes, P.; Soares, J.B.; Roncon-Albuquerque, R.; Dinis-Ribeiro, M.; Leite-Moreira, A.F. Toll-like Receptors as Therapeutic Targets in Gastrointestinal Diseases. Expert Opin. Ther. Targets 2010, 14, 347–368. [Google Scholar] [CrossRef]
- Eiró, N.; Altadill, A.; Juárez, L.M.; Rodríguez, M.; González, L.O.; Atienza, S.; Bermúdez, S.; Fernandez-Garcia, B.; Fresno-Forcelledo, M.F.; Rodrigo, L.; et al. Toll-like Receptors 3, 4 and 9 in Hepatocellular Carcinoma: Relationship with Clinicopathological Characteristics and Prognosis. Hepatol. Res. 2014, 44, 769–778. [Google Scholar] [CrossRef]
- Wang, Z.; Lin, H.; Hua, F.; Hu, Z.W. Repairing DNA Damage by XRCC6/KU70 Reverses TLR4-Deficiency-Worsened HCC Development via Restoring Senescence and Autophagic Flux. Autophagy 2013, 9, 925–927. [Google Scholar] [CrossRef]
- Papadakos, S.P.; Arvanitakis, K.; Stergiou, I.E.; Lekakis, V.; Davakis, S.; Christodoulou, M.I.; Germanidis, G.; Theocharis, S. The Role of TLR4 in the Immunotherapy of Hepatocellular Carcinoma: Can We Teach an Old Dog New Tricks? Cancers 2023, 15, 2795. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, J.X.; Wang, H.; Wang, G.L.; Hu, Q.G.; Zheng, Q.C. Hepatocellular Carcinoma and Macrophage Interaction Induced Tumor Immunosuppression via Treg Requires TLR4 Signaling. World J. Gastroenterol. 2012, 18, 2938–2947. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Q.; Lou, Y.; Fu, Q.; Chen, Q.; Wei, T.; Yang, J.; Tang, J.; Wang, J.; Chen, Y.; et al. Hypoxia-Inducible Factor-1α/Interleukin-1β Signaling Enhances Hepatoma Epithelial-Mesenchymal Transition through Macrophages in a Hypoxic-Inflammatory Microenvironment. Hepatology 2018, 67, 1872–1889. [Google Scholar] [CrossRef]
- Zhou, S.; Du, R.; Wang, Z.; Shen, W.; Gao, R.; Jiang, S.; Fang, Y.; Shi, Y.; Chang, A.; Liu, L.; et al. TLR4 Increases the Stemness and Is Highly Expressed in Relapsed Human Hepatocellular Carcinoma. Cancer Med. 2019, 8, 2325–2337. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, N.; Tan, H.Y.; Guo, W.; Chen, F.; Zhong, Z.; Man, K.; Tsao, S.W.; Lao, L.; Feng, Y. Direct Inhibition of the TLR4/MyD88 Pathway by Geniposide Suppresses HIF-1α-Independent VEGF Expression and Angiogenesis in Hepatocellular Carcinoma. Br. J. Pharmacol. 2020, 177, 3240–3257. [Google Scholar] [CrossRef]
- Zlotnik, A.; Yoshie, O. The Chemokine Superfamily Revisited. Immunity 2012, 36, 705–716. [Google Scholar] [CrossRef]
- Molon, B.; Gri, G.; Bettella, M.; Gómez-Moutón, C.; Lanzavecchia, A.; Martínez-A, C.; Mañes, S.; Viola, A. T Cell Costimulation by Chemokine Receptors. Nat. Immunol. 2005, 6, 465–471. [Google Scholar] [CrossRef]
- Kim, J.W.; Ferris, R.L.; Whiteside, T.L. Chemokine C Receptor 7 Expression and Protection of Circulating CD8+ T Lymphocytes from Apoptosis. Clin. Cancer Res. 2005, 11, 7901–7910. [Google Scholar] [CrossRef]
- Coghill, J.M.; Fowler, K.A.; West, M.L.; Fulton, L.S.M.; Van Deventer, H.; McKinnon, K.P.; Vincent, B.G.; Lin, K.; Panoskaltsis-Mortari, A.; Cook, D.N.; et al. CC Chemokine Receptor 8 Potentiates Donor Treg Survival and Is Critical for the Prevention of Murine Graft-versus-Host Disease. Blood 2013, 122, 825–836. [Google Scholar] [CrossRef]
- Zhang, Z.; Mao, M.; Wang, F.; Zhang, Y.; Shi, J.; Chang, L.; Wu, X.; Zhang, Z.; Xu, P.; Lu, S. Comprehensive Analysis and Immune Landscape of Chemokines- and Chemokine Receptors-Based Signature in Hepatocellular Carcinoma. Front. Immunol. 2023, 14, 1164669. [Google Scholar] [CrossRef]
- Shibuta, K.; Mori, M.; Shimoda, K.; Inoue, H.; Mitra, P.; Barnard, G.F. Regional Expression of CXCL12/CXCR4 in Liver and Hepatocellular Carcinoma and Cell-Cycle Variation during in Vitro Differentiation. Jpn. J. Cancer Res. 2002, 93, 789–797. [Google Scholar] [CrossRef]
- Zheng, K.; Li, H.-Y.; Su, X.-L.; Wang, X.-Y.; Tian, T.; Li, F.; Ren, G.-S. Chemokine Receptor CXCR7 Regulates the Invasion, Angiogenesis and Tumor Growth of Human Hepatocellular Carcinoma Cells. J. Exp. Clin. Cancer Res. 2010, 29, 31. [Google Scholar] [CrossRef] [PubMed]
- Monnier, J.; Boissan, M.; L’Helgoualc’H, A.; Lacombe, M.L.; Turlin, B.; Zucman-Rossi, J.; Théret, N.; Piquet-Pellorce, C.; Samson, M. CXCR7 Is Up-Regulated in Human and Murine Hepatocellular Carcinoma and Is Specifically Expressed by Endothelial Cells. Eur. J. Cancer 2012, 48, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Kaemmerer, D.; Schindler, R.; Mußbach, F.; Dahmen, U.; Altendorf-Hofmann, A.; Dirsch, O.; Sänger, J.; Schulz, S.; Lupp, A. Somatostatin and CXCR4 Chemokine Receptor Expression in Hepatocellular and Cholangiocellular Carcinomas: Tumor Capillaries as Promising Targets. BMC Cancer 2017, 17, 896. [Google Scholar] [CrossRef]
- Liu, H.; Pan, Z.; Li, A.; Fu, S.; Lei, Y.; Sun, H.; Wu, M.; Zhou, W. Roles of Chemokine Receptor 4 (CXCR4) and Chemokine Ligand 12 (CXCL12) in Metastasis of Hepatocellular Carcinoma Cells. Cell. Mol. Immunol. 2008, 5, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Bertran, E.; Crosas-Molist, E.; Sancho, P.; Caja, L.; Lopez-Luque, J.; Navarro, E.; Egea, G.; Lastra, R.; Serrano, T.; Ramos, E.; et al. Overactivation of the TGF-β Pathway Confers a Mesenchymal-like Phenotype and CXCR4-Dependent Migratory Properties to Liver Tumor Cells. Hepatology 2013, 58, 2032–2044. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Pan, X.; Huang, Z.; Weber, G.F.; Zhang, G. Osteopontin Enhances the Expression and Activity of MMP-2 via the SDF-1/CXCR4 Axis in Hepatocellular Carcinoma Cell Lines. PLoS ONE 2011, 6, e23831. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Deng, H.; Yan, W.; Luo, M.; Tu, W.; Xia, Y.; He, J.; Han, P.; Fu, Y.; Tian, D. AEG-1 Promotes Anoikis Resistance and Orientation Chemotaxis in Hepatocellular Carcinoma Cells. PLoS ONE 2014, 9, e100372. [Google Scholar] [CrossRef] [PubMed]
- Jahanban-Esfahlan, R.; de la Guardia, M.; Ahmadi, D.; Yousefi, B. Modulating Tumor Hypoxia by Nanomedicine for Effective Cancer Therapy. J. Cell. Physiol. 2018, 233, 2019–2031. [Google Scholar] [CrossRef] [PubMed]
- Deol, A.; Abrams, J.; Masood, A.; Al-Kadhimi, Z.; Abidi, M.H.; Ayash, L.; Lum, L.G.; Ratanatharathorn, V.; Uberti, J.P. Long-Term Follow up of Patients Proceeding to Transplant Using Plerixafor Mobilized Stem Cells and Incidence of Secondary Myelodysplastic Syndrome/AML. Bone Marrow Transpl. 2013, 48, 1112–1116. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hirano, S.; Iwashita, Y.; Sasaki, A.; Kai, S.; Ohta, M.; Kitano, S. Increased MRNA Expression of Chemokines in Hepatocellular Carcinoma with Tumor-Infiltrating Lymphocytes. J. Gastroenterol. Hepatol. 2007, 22, 690–696. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yang, L.; Xu, J.; Zhang, X.; Wang, B. Enhanced Expression and Clinical Significance of Chemokine Receptor CXCR2 in Hepatocellular Carcinoma. J. Surg. Res. 2011, 166, 241–246. [Google Scholar] [CrossRef]
- Zhou, S.L.; Zhou, Z.J.; Hu, Z.Q.; Li, X.; Huang, X.W.; Wang, Z.; Fan, J.; Dai, Z.; Zhou, J. CXCR2/CXCL5 Axis Contributes to Epithelial-Mesenchymal Transition of HCC Cells through Activating PI3K/Akt/GSK-3β/Snail Signaling. Cancer Lett. 2015, 358, 124–135. [Google Scholar] [CrossRef]
- Gao, Q.; Zhao, Y.J.; Wang, X.Y.; Qiu, S.J.; Shi, Y.H.; Sun, J.; Yi, Y.; Shi, J.Y.; Shi, G.M.; Ding, Z.-B.; et al. CXCR6 Upregulation Contributes to a Proinflammatory Tumor Microenvironment That Drives Metastasis and Poor Patient Outcomes in Hepatocellular Carcinoma. Cancer Res. 2012, 72, 3546–3556. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.-B.; Tsai, H.T.; Chen, Y.C.; Kuo, W.H.; Chen, T.Y.; Hsieh, Y.H.; Chou, M.C.; Yang, S.F. Genetic Polymorphism of CCR2-64I Increased the Susceptibility of Hepatocellular Carcinoma. J. Surg. Oncol. 2010, 102, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.J.; Zhao, L.H.; Zhou, X.; Zhang, H.L.; Wen, W.; Tang, L.; Zeng, M.; Wang, M.-D.; Fu, G.B.; Huang, S.; et al. Inhibition of Dipeptidyl Peptidase IV Prevents High Fat Diet-Induced Liver Cancer Angiogenesis by Downregulating Chemokine Ligand 2. Cancer Lett. 2018, 420, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Nahon, P.; Sutton, A.; Rufat, P.; Simon, C.; Trinchet, J.C.; Gattegno, L.; Beaugrand, M.; Charnaux, N. Chemokine System Polymorphisms, Survival and Hepatocellular Carcinoma Occurrence in Patients with Hepatitis C Virus-Related Cirrhosis. World J. Gastroenterol. 2008, 14, 713–719. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ochoa-Callejero, L.; Pérez-Martínez, L.; Rubio-Mediavilla, S.; Oteo, J.A.; Martínez, A.; Blanco, J.R. Maraviroc, a CCR5 Antagonist, Prevents Development of Hepatocellular Carcinoma in a Mouse Model. PLoS ONE 2013, 8, e53992. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Chen, J.X.; Chen, Y.; Cai, L.L.; Wang, X.Z.; Guo, W.H.; Zheng, J.F. The Chemokine Receptor CCR10 Promotes Inflammation-Driven Hepatocarcinogenesis via PI3K/Akt Pathway Activation. Cell Death Dis. 2018, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Liu, Y.; Qian, H.; Zhang, B.; Tang, X.; Zhang, T.; Liu, W. The Effects of the CCR6/CCL20 Biological Axis on the Invasion and Metastasis of Hepatocellular Carcinoma. Int. J. Mol. Sci. 2014, 15, 6441–6452. [Google Scholar] [CrossRef]
- Hippe, A.; Braun, S.A.; Oláh, P.; Gerber, P.A.; Schorr, A.; Seeliger, S.; Holtz, S.; Jannasch, K.; Pivarcsi, A.; Buhren, B.; et al. EGFR/Ras-Induced CCL20 Production Modulates the Tumour Microenvironment. Br. J. Cancer 2020, 123, 942–954. [Google Scholar] [CrossRef]
- Wei, Z.; Liu, H.T. MAPK Signal Pathways in the Regulation of Cell Proliferation in Mammalian Cells. Cell Res. 2002, 12, 9–18. [Google Scholar] [CrossRef]
- Guo, Y.; Pan, W.; Liu, S.; Shen, Z.; Xu, Y.; Hu, L. ERK/MAPK Signalling Pathway and Tumorigenesis. Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef]
- Pudewell, S.; Wittich, C.; Kazemein Jasemi, N.S.; Bazgir, F.; Ahmadian, M.R. Accessory Proteins of the RAS-MAPK Pathway: Moving from the Side Line to the Front Line. Commun. Biol. 2021, 4, 696. [Google Scholar] [CrossRef]
- Burotto, M.; Chiou, V.L.; Lee, J.M.; Kohn, E.C. The MAPK Pathway across Different Malignancies: A New Perspective. Cancer 2014, 120, 3446. [Google Scholar] [CrossRef]
- Campbell, P.J.; Getz, G.; Korbel, J.O.; Stuart, J.M.; Jennings, J.L.; Stein, L.D.; Perry, M.D.; Nahal-Bose, H.K.; Ouellette, B.F.F.; Li, C.H.; et al. Pan-Cancer Analysis of Whole Genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef]
- Campbell, B.B.; Galati, M.A.; Stone, S.C.; Riemenschneider, A.N.; Edwards, M.; Sudhaman, S.; Siddaway, R.; Komosa, M.; Nunes, N.M.; Nobre, L.; et al. Mutations in the RAS/MAPK Pathway Drive Replication Repair-Deficient Hypermutated Tumors and Confer Sensitivity to MEK Inhibition. Cancer Discov. 2021, 11, 1454–1467. [Google Scholar] [CrossRef]
- Delire, B.; Stärkel, P. The Ras/MAPK Pathway and Hepatocarcinoma: Pathogenesis and Therapeutic Implications. Eur. J. Clin. Investig. 2015, 45, 609–623. [Google Scholar] [CrossRef]
- Roberts, L.R.; Gores, G.J. Hepatocellular Carcinoma: Molecular Pathways and New Therapeutic Targets. Semin. Liver Dis. 2005, 25, 212–225. [Google Scholar] [CrossRef] [PubMed]
- McCubrey, J.A.; Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Wong, E.W.T.; Chang, F.; Lehmann, B.; Terrian, D.M.; Milella, M.; Tafuri, A.; et al. Roles of the Raf/MEK/ERK Pathway in Cell Growth, Malignant Transformation and Drug Resistance. Biochim. Biophys. Acta Mol. Cell Res. 2007, 1773, 1263–1284. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shi, Y.; Jiang, C.Y.; Wei, L.X.; Wang, Y.L.; Dai, G.H. Expression and Prognostic Role of Pan-Ras, Raf-1, PMEK1 and PERK1/2 in Patients with Hepatocellular Carcinoma. Eur. J. Surg. Oncol. 2011, 37, 513–520. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, B.H.; Goff, L.W.; Kauh, J.S.W.; Strosberg, J.R.; Bekaii-Saab, T.S.; Lee, R.M.; Kazi, A.; Moore, D.T.; Learoyd, M.; Lush, R.M.; et al. Phase II Study of the Mitogen-Activated Protein Kinase 1/2 Inhibitor Selumetinib in Patients with Advanced Hepatocellular Carcinoma. J. Clin. Oncol. 2011, 29, 2350–2356. [Google Scholar] [CrossRef]
- Kim, R.; Tan, E.; Wang, E.; Mahipal, A.; Chen, D.-T.; Cao, B.; Masawi, F.; Machado, C.; Yu, J.; Kim, D.W. A Phase I Trial of Trametinib in Combination with Sorafenib in Patients with Advanced Hepatocellular Cancer. Oncologist 2020, 25, e1893–e1899. [Google Scholar] [CrossRef]
- Harrison, D.A. The JAK/STAT Pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011205. [Google Scholar] [CrossRef]
- Schindler, C.; Levy, D.E.; Decker, T. JAK-STAT Signaling: From Interferons to Cytokines. J. Biol. Chem. 2007, 282, 20059–20063. [Google Scholar] [CrossRef]
- Hu, X.; li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT Signaling Pathway: From Bench to Clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.-N.; Baik, E.J. JAK-STAT Pathway and Myogenic Differentiation. JAK-STAT 2013, 2, e23282. [Google Scholar] [CrossRef]
- Herrera, S.C.; Bach, E.A. JAK/STAT Signaling in Stem Cells and Regeneration: From Drosophila to Vertebrates. Development 2019, 146, dev167643. [Google Scholar] [CrossRef]
- Al Zaid Siddiquee, K.; Turkson, J. STAT3 as a Target for Inducing Apoptosis in Solid and Hematological Tumors. Cell Res. 2008, 18, 254. [Google Scholar] [CrossRef]
- Banerjee, S.; Biehl, A.; Gadina, M.; Hasni, S.; Schwartz, D.M. JAK–STAT Signaling as a Target for Inflammatory and Autoimmune Diseases: Current and Future Prospects. Drugs 2017, 77, 521. [Google Scholar] [CrossRef] [PubMed]
- Gurzov, E.N.; Stanley, W.J.; Pappas, E.G.; Thomas, H.E.; Gough, D.J. The JAK/STAT Pathway in Obesity and Diabetes. FEBS J. 2016, 283, 3002–3015. [Google Scholar] [CrossRef] [PubMed]
- Lokau, J.; Garbers, C. Activating Mutations of the Gp130/JAK/STAT Pathway in Human Diseases. Adv. Protein Chem. Struct. Biol. 2019, 116, 283–309. [Google Scholar] [CrossRef]
- Shahmarvand, N.; Nagy, A.; Shahryari, J.; Ohgami, R.S. Mutations in the Signal Transducer and Activator of Transcription Family of Genes in Cancer. Cancer Sci. 2018, 109, 926. [Google Scholar] [CrossRef]
- Hornakova, T.; Springuel, L.; Devreux, J.; Dusa, A.; Constantinescu, S.N.; Knoops, L.; Renauld, J.C. Oncogenic JAK1 and JAK2-Activating Mutations Resistant to ATP-Competitive Inhibitors. Haematologica 2011, 96, 845–853. [Google Scholar] [CrossRef][Green Version]
- Brooks, A.J.; Putoczki, T. JAK-STAT Signalling Pathway in Cancer. Cancers 2020, 12, 1971. [Google Scholar] [CrossRef] [PubMed]
- Kan, Z.; Zheng, H.; Liu, X.; Li, S.; Barber, T.D.; Gong, Z.; Gao, H.; Hao, K.; Willard, M.D.; Xu, J.; et al. Whole-Genome Sequencing Identifies Recurrent Mutations in Hepatocellular Carcinoma. Genome Res. 2013, 23, 1422–1433. [Google Scholar] [CrossRef]
- Dhillon, S. Tofacitinib: A Review in Rheumatoid Arthritis. Drugs 2017, 77, 1987–2001. [Google Scholar] [CrossRef] [PubMed]
- Al-Salama, Z.T.; Scott, L.J. Baricitinib: A Review in Rheumatoid Arthritis. Drugs 2018, 78, 761–772. [Google Scholar] [CrossRef]
- Tanaka, Y. A Review of Upadacitinib in Rheumatoid Arthritis. Mod. Rheumatol. 2020, 30, 779–787. [Google Scholar] [CrossRef]
- Harrington, R.; Al Nokhatha, S.A.; Conway, R. JAK Inhibitors in Rheumatoid Arthritis: An Evidence-Based Review on the Emerging Clinical Data. J. Inflamm. Res. 2020, 13, 519. [Google Scholar] [CrossRef] [PubMed]
- Qureshy, Z.; Johnson, D.E.; Grandis, J.R. Targeting the JAK/STAT Pathway in Solid Tumors. J. Cancer Metastasis Treat. 2020, 6, 27. [Google Scholar] [CrossRef]
- Hosseini, A.; Gharibi, T.; Marofi, F.; Javadian, M.; Babaloo, Z.; Baradaran, B. Janus Kinase Inhibitors: A Therapeutic Strategy for Cancer and Autoimmune Diseases. J. Cell. Physiol. 2020, 235, 5903–5924. [Google Scholar] [CrossRef]
- Vainchenker, W.; Leroy, E.; Gilles, L.; Marty, C.; Plo, I.; Constantinescu, S.N. JAK Inhibitors for the Treatment of Myeloproliferative Neoplasms and Other Disorders. F1000Research 2018, 7, 82. [Google Scholar] [CrossRef]
- Kirito, K. Recent Progress of JAK Inhibitors for Hematological Disorders. Immunol. Med. 2023, 46, 131–142. [Google Scholar] [CrossRef]
- Zhou, Q.; Jiang, H.; Zhang, J.; Yu, W.; Zhou, Z.; Huang, P.; Wang, J.; Xiao, Z. Uridine-Cytidine Kinase 2 Promotes Metastasis of Hepatocellular Carcinoma Cells via the Stat3 Pathway. Cancer Manag. Res. 2018, 10, 6339–6355. [Google Scholar] [CrossRef] [PubMed]
- Al-Fayoumi, S.; Hashiguchi, T.; Shirakata, Y.; Mascarenhas, J.; Singer, J.W. Pilot Study of the Antifibrotic Effects of the Multikinase Inhibitor Pacritinib in a Mouse Model of Liver Fibrosis. J. Exp. Pharmacol. 2018, 10, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Liu, S.; Lin, H.; Trivett, A.L.; Hannifin, S.; Yang, D.; Oppenheim, J.J. Inhibition of Murine Hepatoma Tumor Growth by Cryptotanshinone Involves TLR7-Dependent Activation of Macrophages and Induction of Adaptive Antitumor Immune Defenses. Cancer Immunol. Immunother. 2019, 68, 1073–1085. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.S.; Tian, A.; Hebbard, L.; Duan, W.; George, J.; Li, X.; Qiao, L. Tumoricidal Effects of the JAK Inhibitor Ruxolitinib (INC424) on Hepatocellular Carcinoma in Vitro. Cancer Lett. 2013, 341, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Luo, C.; Gu, Q.; Xu, Q.; Wang, G.; Sun, H.; Qian, Z.; Tan, Y.; Qin, Y.; Shen, Y.; et al. Activating JAK1 Mutation May Predict the Sensitivity of JAK-STAT Inhibition in Hepatocellular Carcinoma. Oncotarget 2016, 7, 5461. [Google Scholar] [CrossRef]
- Lee, C.; Cheung, S.T. STAT3: An Emerging Therapeutic Target for Hepatocellular Carcinoma. Cancers 2019, 11, 1646. [Google Scholar] [CrossRef] [PubMed]
- Tolomeo, M.; Cascio, A. The Multifaced Role of STAT3 in Cancer and Its Implication for Anticancer Therapy. Int. J. Mol. Sci. 2021, 22, 603. [Google Scholar] [CrossRef]
- Okusaka, T.; Morimoto, M.; Eguchi, Y.; Nakamura, S.; Iino, S.; Kageyama, R. A Phase I Study to Investigate the Safety, Tolerability and Pharmacokinetics of Napabucasin Combined with Sorafenib in Japanese Patients with Unresectable Hepatocellular Carcinoma. Drugs R&D 2023, 23, 99–107. [Google Scholar] [CrossRef]
- Ren, Z.; Xu, J.; Bai, Y.; Xu, A.; Cang, S.; Du, C.; Li, Q.; Lu, Y.; Chen, Y.; Guo, Y.; et al. Sintilimab plus a Bevacizumab Biosimilar (IBI305) versus Sorafenib in Unresectable Hepatocellular Carcinoma (ORIENT-32): A Randomised, Open-Label, Phase 2-3 Study. Lancet. Oncol. 2021, 22, 977–990. [Google Scholar] [CrossRef]
- Yoo, C.; Kang, J.; Lim, H.Y.; Kim, J.H.; Lee, M.A.; Lee, K.H.; Kim, T.Y.; Ryoo, B.Y. Phase I Dose-Finding Study of OPB-111077, a Novel STAT3 Inhibitor, in Patients with Advanced Hepatocellular Carcinoma. Cancer Res. Treat. 2019, 51, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Tolcher, A.; Flaherty, K.; Shapiro, G.I.; Berlin, J.; Witzig, T.; Habermann, T.; Bullock, A.; Rock, E.; Elekes, A.; Lin, C.; et al. A First-in-Human Phase I Study of OPB-111077, a Small-Molecule STAT3 and Oxidative Phosphorylation Inhibitor, in Patients with Advanced Cancers. Oncologist 2018, 23, 658-e72. [Google Scholar] [CrossRef] [PubMed]
- Bendell, J.C.; Hong, D.S.; Burris, H.A.; Naing, A.; Jones, S.F.; Falchook, G.; Bricmont, P.; Elekes, A.; Rock, E.P.; Kurzrock, R. Phase 1, Open-Label, Dose-Escalation, and Pharmacokinetic Study of STAT3 Inhibitor OPB-31121 in Subjects with Advanced Solid Tumors. Cancer Chemother. Pharmacol. 2014, 74, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.Y.; Lee, S.H.; Han, S.W.; Kim, M.J.; Kim, T.M.; Kim, T.Y.; Heo, D.S.; Yuasa, M.; Yanagihara, Y.; Bang, Y.J. Phase I Study of OPB-31121, an Oral STAT3 Inhibitor, in Patients with Advanced Solid Tumors. Cancer Res. Treat. 2015, 47, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Nishina, T.; Fujita, T.; Yoshizuka, N.; Sugibayashi, K.; Murayama, K.; Kuboki, Y. Safety, Tolerability, Pharmacokinetics and Preliminary Antitumour Activity of an Antisense Oligonucleotide Targeting STAT3 (Danvatirsen) as Monotherapy and in Combination with Durvalumab in Japanese Patients with Advanced Solid Malignancies: A Phase 1 Study. BMJ Open 2022, 12, e055718. [Google Scholar] [CrossRef] [PubMed]
- Roschewski, M.; Patel, M.R.; Reagan, P.M.; Saba, N.S.; Collins, G.P.; Arkenau, H.T.; de Vos, S.; Nuttall, B.; Acar, M.; Burke, K.; et al. Phase I Study of Acalabrutinib Plus Danvatirsen (AZD9150) in Relapsed/Refractory Diffuse Large B-Cell Lymphoma Including Circulating Tumor DNA Biomarker Assessment. Clin. Cancer Res. 2023, 29, 3301–3312. [Google Scholar] [CrossRef]
- Mohan, C.D.; Bharathkumar, H.; Bulusu, K.C.; Pandey, V.; Rangappa, S.; Fuchs, J.E.; Shanmugam, M.K.; Dai, X.; Li, F.; Deivasigamani, A.; et al. Development of a Novel Azaspirane That Targets the Janus Kinase-Signal Transducer and Activator of Transcription (STAT) Pathway in Hepatocellular Carcinoma in Vitro and in Vivo. J. Biol. Chem. 2014, 289, 34296–34307. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.F.; Tai, W.T.; Liu, T.H.; Huang, H.P.; Lin, Y.C.; Shiau, C.W.; Li, P.K.; Chen, P.J.; Cheng, A.L. Sorafenib Overcomes TRAIL Resistance of Hepatocellular Carcinoma Cells through the Inhibition of STAT3. Clin. Cancer Res. 2010, 16, 5189–5199. [Google Scholar] [CrossRef]
- Bergmann, J.; Müller, M.; Baumann, N.; Reichert, M.; Heneweer, C.; Bolik, J.; Lücke, K.; Gruber, S.; Carambia, A.; Boretius, S.; et al. IL-6 Trans-Signaling Is Essential for the Development of Hepatocellular Carcinoma in Mice. Hepatology 2017, 65, 89–103. [Google Scholar] [CrossRef]
- Xu, J.; Lin, H.; Wu, G.; Zhu, M.; Li, M. IL-6/STAT3 Is a Promising Therapeutic Target for Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 760971. [Google Scholar] [CrossRef]
- Kao, J.T.; Feng, C.L.; Yu, C.J.; Tsai, S.M.; Hsu, P.N.; Chen, Y.L.; Wu, Y.Y. IL-6, through p-STAT3 Rather than p-STAT1, Activates Hepatocarcinogenesis and Affects Survival of Hepatocellular Carcinoma Patients: A Cohort Study. BMC Gastroenterol. 2015, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Mao, R.; Yang, J. NF-ΚB and STAT3 Signaling Pathways Collaboratively Link Inflammation to Cancer. Protein Cell 2013, 4, 176–185. [Google Scholar] [CrossRef]
- Ghoshal, S.; Fuchs, B.C.; Tanabe, K.K. STAT3 Is a Key Transcriptional Regulator of Cancer Stem Cell Marker CD133 in HCC. Hepatobiliary Surg. Nutr. 2016, 5, 201–203. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Xu, M.; Yao, B.; Wang, C.; Jia, Y.; Liu, Q. IL-6/STAT3 Axis Initiated CAFs via up-Regulating TIMP-1 Which Was Attenuated by Acetylation of STAT3 Induced by PCAF in HCC Microenvironment. Cell. Signal. 2016, 28, 1314–1324. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Park, B. Saffron Carotenoids Inhibit STAT3 Activation and Promote Apoptotic Progression in IL-6-Stimulated Liver Cancer Cells. Oncol. Rep. 2018, 39, 1883–1891. [Google Scholar] [CrossRef]
- Yang, J.; Wang, J.; Luo, J. Decreased IL-6 Induces Sensitivity of Hepatocellular Carcinoma Cells to Sorafenib. Pathol. Res. Pract. 2019, 215, 152565. [Google Scholar] [CrossRef]
- Pham, T.H.; Park, H.M.; Kim, J.; Hong, J.T.; Yoon, D.Y. STAT3 and P53: Dual Target for Cancer Therapy. Biomedicines 2020, 8, 637. [Google Scholar] [CrossRef]
- Chen, R.; Chen, B. Siltuximab (CNTO 328): A Promising Option for Human Malignancies. Drug Des. Devel. Ther. 2015, 9, 3455–3458. [Google Scholar] [CrossRef]
- Rovin, B.H.; van Vollenhoven, R.F.; Aranow, C.; Wagner, C.; Gordon, R.; Zhuang, Y.; Belkowski, S.; Hsu, B. A Multicenter, Randomized, Double-Blind, Placebo-Controlled Study to Evaluate the Efficacy and Safety of Treatment with Sirukumab (CNTO 136) in Patients with Active Lupus Nephritis. Arthritis Rheumatol. 2016, 68, 2174–2183. [Google Scholar] [CrossRef]
- Bayliss, T.J.; Smith, J.T.; Schuster, M.; Dragnev, K.H.; Rigas, J.R. A Humanized Anti-IL-6 Antibody (ALD518) in Non-Small Cell Lung Cancer. Expert Opin. Biol. Ther. 2011, 11, 1663–1668. [Google Scholar] [CrossRef]
- Dijkgraaf, E.M.; Santegoets, S.J.A.M.; Reyners, A.K.L.; Goedemans, R.; Wouters, M.C.A.; Kenter, G.G.; van Erkel, A.R.; van Poelgeest, M.I.E.; Nijman, H.W.; van der Hoeven, J.J.M.; et al. A Phase I Trial Combining Carboplatin/Doxorubicin with Tocilizumab, an Anti-IL-6R Monoclonal Antibody, and Interferon-A2b in Patients with Recurrent Epithelial Ovarian Cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015, 26, 2141–2149. [Google Scholar] [CrossRef] [PubMed]
- Campo, S.; Serlupi-Crescenzi, O.; Arseni, B.; Rossi, S.; Saggio, I.; Salone, B.; Cherubini, G.; Carminati, P.; De Santis, R. Comparative Activity of Sant7 and Anti-IL-6, IL-6R Monoclonal Antibodies in a Murine Model of B-Cell Lymphoma. Cytokine 2005, 31, 368–374. [Google Scholar] [CrossRef]
- Oguro, T.; Ishibashi, K.; Sugino, T.; Hashimoto, K.; Tomita, S.; Takahashi, N.; Yanagida, T.; Haga, N.; Aikawa, K.; Suzutani, T.; et al. Humanised Antihuman IL-6R Antibody with Interferon Inhibits Renal Cell Carcinoma Cell Growth in Vitro and in Vivo through Suppressed SOCS3 Expression. Eur. J. Cancer 2013, 49, 1715–1724. [Google Scholar] [CrossRef] [PubMed]
- Dhar, D.; Baglieri, J.; Kisseleva, T.; Brenner, D.A. Mechanisms of Liver Fibrosis and Its Role in Liver Cancer. Exp. Biol. Med. 2020, 245, 96. [Google Scholar] [CrossRef]
- Zhao, J.; Qi, Y.F.; Yu, Y.R. STAT3: A Key Regulator in Liver Fibrosis. Ann. Hepatol. 2021, 21, 100224. [Google Scholar] [CrossRef]
- Bi, Y.H.; Han, W.Q.; Li, R.F.; Wang, Y.J.; Du, Z.S.; Wang, X.J.; Jiang, Y. Signal Transducer and Activator of Transcription 3 Promotes the Warburg Effect Possibly by Inducing Pyruvate Kinase M2 Phosphorylation in Liver Precancerous Lesions. World J. Gastroenterol. 2019, 25, 1936. [Google Scholar] [CrossRef]
- Park, H.; Lee, S.; Lee, J.; Moon, H.; Ro, S.W. Exploring the JAK/STAT Signaling Pathway in Hepatocellular Carcinoma: Unraveling Signaling Complexity and Therapeutic Implications. Int. J. Mol. Sci. 2023, 24, 13764. [Google Scholar] [CrossRef]
- Hin Tang, J.J.; Hao Thng, D.K.; Lim, J.J.; Toh, T.B. JAK/STAT Signaling in Hepatocellular Carcinoma. Hepatic Oncol. 2020, 7, HEP18. [Google Scholar] [CrossRef]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605. [Google Scholar] [CrossRef] [PubMed]
- Glaviano, A.; Foo, A.S.C.; Lam, H.Y.; Yap, K.C.H.; Jacot, W.; Jones, R.H.; Eng, H.; Nair, M.G.; Makvandi, P.; Geoerger, B.; et al. PI3K/AKT/MTOR Signaling Transduction Pathway and Targeted Therapies in Cancer. Mol. Cancer 2023, 22, 138. [Google Scholar] [CrossRef]
- Terracciano, L.M.; Piscuoglio, S.; Ng, C.K.Y. Hepatocellular Carcinoma: Pathology and Genetics. Encycl. Cancer 2019, 2019, 198–210. [Google Scholar] [CrossRef]
- Ligresti, G.; Militello, L.; Steelman, L.S.; Cavallaro, A.; Basile, F.; Nicoletti, F.; Stivala, F.; McCubrey, J.A.; Libra, M. PIK3CA Mutations in Human Solid Tumors: Role in Sensitivity to Various Therapeutic Approaches. Cell Cycle 2009, 8, 1352. [Google Scholar] [CrossRef] [PubMed]
- Schulze, K.; Imbeaud, S.; Letouzé, E.; Alexandrov, L.B.; Calderaro, J.; Rebouissou, S.; Couchy, G.; Meiller, C.; Shinde, J.; Soysouvanh, F.; et al. Exome Sequencing of Hepatocellular Carcinomas Identifies New Mutational Signatures and Potential Therapeutic Targets. Nat. Genet. 2015, 47, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Mayerle, J.; Ziesch, A.; Reiter, F.P.; Gerbes, A.L.; De Toni, E.N. The PI3K Inhibitor Copanlisib Synergizes with Sorafenib to Induce Cell Death in Hepatocellular Carcinoma. Cell Death Discov. 2019, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.X.; Abrams, T.A.; Miksad, R.; Blaszkowsky, L.S.; Meyerhardt, J.A.; Zheng, H.; Muzikansky, A.; Clark, J.W.; Kwak, E.L.; Schrag, D.; et al. Phase 1/2 Study of Everolimus in Advanced Hepatocellular Carcinoma. Cancer 2011, 117, 5094–5102. [Google Scholar] [CrossRef] [PubMed]
- Koeberle, D.; Dufour, J.F.; Demeter, G.; Li, Q.; Ribi, K.; Samaras, P.; Saletti, P.; Roth, A.D.; Horber, D.; Buehlmann, M.; et al. Sorafenib with or without Everolimus in Patients with Advanced Hepatocellular Carcinoma (HCC): A Randomized Multicenter, Multinational Phase II Trial (SAKK 77/08 and SASL 29). Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2016, 27, 856–861. [Google Scholar] [CrossRef] [PubMed]
- Sun, E.J.; Wankell, M.; Palamuthusingam, P.; McFarlane, C.; Hebbard, L. Targeting the PI3K/Akt/MTOR Pathway in Hepatocellular Carcinoma. Biomedicines 2021, 9, 1639. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/β-Catenin Signalling: Function, Biological Mechanisms, and Therapeutic Opportunities. Signal Transduct. Target. Ther. 2022, 7, 3. [Google Scholar] [CrossRef]
- Gajos-Michniewicz, A.; Czyz, M. WNT/β-Catenin Signaling in Hepatocellular Carcinoma: The Aberrant Activation, Pathogenic Roles, and Therapeutic Opportunities. Genes Dis. 2024, 11, 727. [Google Scholar] [CrossRef]
- Tian, L.Y.; Smit, D.J.; Jücker, M. The Role of PI3K/AKT/MTOR Signaling in Hepatocellular Carcinoma Metabolism. Int. J. Mol. Sci. 2023, 24, 2652. [Google Scholar] [CrossRef]
- Jang, E.S.; Jeong, S.H.; Kim, J.W.; Choi, Y.S.; Leissner, P.; Brechot, C. Diagnostic Performance of Alpha-Fetoprotein, Protein Induced by Vitamin K Absence, Osteopontin, Dickkopf-1 and Its Combinations for Hepatocellular Carcinoma. PLoS ONE 2016, 11, e0151069. [Google Scholar] [CrossRef]
- Suda, T.; Yamashita, T.; Sunagozaka, H.; Okada, H.; Nio, K.; Sakai, Y.; Yamashita, T.; Mizukoshi, E.; Honda, M.; Kaneko, S. Dickkopf-1 Promotes Angiogenesis and Is a Biomarker for Hepatic Stem Cell-like Hepatocellular Carcinoma. Int. J. Mol. Sci. 2022, 23, 2801. [Google Scholar] [CrossRef]
- Nault, J.C.; Mallet, M.; Pilati, C.; Calderaro, J.; Bioulac-Sage, P.; Laurent, C.; Laurent, A.; Cherqui, D.; Balabaud, C.; Rossi, J.Z. High Frequency of Telomerase Reverse-Transcriptase Promoter Somatic Mutations in Hepatocellular Carcinoma and Preneoplastic Lesions. Nat. Commun. 2013, 4, 2577. [Google Scholar] [CrossRef]
- Pinyol, R.; Tovar, V.; Llovet, J.M. TERT Promoter Mutations: Gatekeeper and Driver of Hepatocellular Carcinoma. J. Hepatol. 2014, 61, 685–687. [Google Scholar] [CrossRef] [PubMed]
- Neiheisel, A.; Kaur, M.; Ma, N.; Havard, P.; Shenoy, A.K. Wnt Pathway Modulators in Cancer Therapeutics: An Update on Completed and Ongoing Clinical Trials. Int. J. Cancer 2022, 150, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Hori, Y.; Inoue, S.; Yamamoto, Y.; Iso, K.; Kamiyama, H.; Yamaguchi, A.; Kimura, T.; Uesugi, M.; Ito, J.; et al. E7386, a Selective Inhibitor of the Interaction between β-Catenin and CBP, Exerts Antitumor Activity in Tumor Models with Activated Canonical Wnt Signaling. Cancer Res. 2021, 81, 1052–1062. [Google Scholar] [CrossRef] [PubMed]
- Voskarides, K.; Giannopoulou, N. The Role of TP53 in Adaptation and Evolution. Cells 2023, 12, 512. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J. P53: 800 Million Years of Evolution and 40 Years of Discovery. Nat. Rev. Cancer 2020, 20, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Kruse, J.P.; Gu, W. Modes of P53 Regulation. Cell 2009, 137, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Ashcroft, M.; Kubbutat, M.H.G.; Vousden, K.H. Regulation of P53 Function and Stability by Phosphorylation. Mol. Cell. Biol. 1999, 19, 1751–1758. [Google Scholar] [CrossRef]
- Barboza, J.A.; Iwakuma, T.; Terzian, T.; El-Naggar, A.K.; Lozano, G. Mdm2 and Mdm4 Loss Regulates Distinct P53 Activities. Mol. Cancer Res. 2008, 6, 947–954. [Google Scholar] [CrossRef]
- Aubrey, B.J.; Kelly, G.L.; Janic, A.; Herold, M.J.; Strasser, A. How Does P53 Induce Apoptosis and How Does This Relate to P53-Mediated Tumour Suppression? Cell Death Differ. 2018, 25, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Rivlin, N.; Brosh, R.; Oren, M.; Rotter, V. Mutations in the P53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer 2011, 2, 466. [Google Scholar] [CrossRef] [PubMed]
- Hassin, O.; Oren, M. Drugging P53 in Cancer: One Protein, Many Targets. Nat. Rev. Drug Discov. 2023, 22, 127–144. [Google Scholar] [CrossRef] [PubMed]
- Nag, S.; Zhang, X.; Srivenugopal, K.S.; Wang, M.-H.; Wang, W.; Zhang, R. Targeting MDM2-P53 Interaction for Cancer Therapy: Are We There Yet? Curr. Med. Chem. 2014, 21, 553. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Guo, M.; Wei, H.; Chen, Y. Targeting P53 Pathways: Mechanisms, Structures, and Advances in Therapy. Signal Transduct. Target. Ther. 2023, 8, 92. [Google Scholar] [CrossRef] [PubMed]
- Yim, S.Y.; Lee, J.-S. An Overview of the Genomic Characterization of Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2021, 8, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Neuveut, C.; Wei, Y.; Buendia, M.A. Mechanisms of HBV-Related Hepatocarcinogenesis. J. Hepatol. 2010, 52, 594–604. [Google Scholar] [CrossRef]
- Liu, J.; Ma, Q.; Zhang, M.; Wang, X.; Zhang, D.; Li, W.; Wang, F.; Wu, E. Alterations of TP53 Are Associated with a Poor Outcome for Patients with Hepatocellular Carcinoma: Evidence from a Systematic Review and Meta-Analysis. Eur. J. Cancer 2012, 48, 2328–2338. [Google Scholar] [CrossRef]
- Zhang, W.W.; Li, L.; Li, D.; Liu, J.; Li, X.; Li, W.; Xu, X.; Zhang, M.J.; Chandler, L.A.; Lin, H.; et al. The First Approved Gene Therapy Product for Cancer Ad-P53 (Gendicine): 12 Years in the Clinic. Hum. Gene Ther. 2018, 29, 160–179. [Google Scholar] [CrossRef]
- Xia, Y.; Li, X.; Sun, W. Applications of Recombinant Adenovirus-P53 Gene Therapy for Cancers in the Clinic in China. Curr. Gene Ther. 2020, 20, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Chen, Y.; Zhang, Y.; Xu, M.; Guo, W.; Zhang, J.; Ma, G.; Liu, C.; Yang, J.; Wu, X. Recombinant Human Adenovirus P53 Combined with Transcatheter Arterial Chemoembolization for Liver Cancer: A Meta-Analysis. PLoS ONE 2023, 18, e0295323. [Google Scholar] [CrossRef]
- Yang, Z.X.; Wang, D.; Wang, G.; Zhang, Q.H.; Liu, J.M.; Peng, P.; Liu, X.H. Clinical Study of Recombinant Adenovirus-P53 Combined with Fractionated Stereotactic Radiotherapy for Hepatocellular Carcinoma. J. Cancer Res. Clin. Oncol. 2010, 136, 625–630. [Google Scholar] [CrossRef]
- Nishikawa, S.; Iwakuma, T. Drugs Targeting P53 Mutations with FDA Approval and in Clinical Trials. Cancers 2023, 15, 429. [Google Scholar] [CrossRef]
- Duffy, M.J.; Synnott, N.C.; O’Grady, S.; Crown, J. Targeting P53 for the Treatment of Cancer. Semin. Cancer Biol. 2022, 79, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M. Cyclin-Dependent Kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef] [PubMed]
- Łukasik, P.; Załuski, M.; Gutowska, I. Cyclin-Dependent Kinases (CDK) and Their Role in Diseases Development-Review. Int. J. Mol. Sci. 2021, 22, 2935. [Google Scholar] [CrossRef]
- Ding, L.; Cao, J.; Lin, W.; Chen, H.; Xiong, X.; Ao, H.; Yu, M.; Lin, J.; Cui, Q. The Roles of Cyclin-Dependent Kinases in Cell-Cycle Progression and Therapeutic Strategies in Human Breast Cancer. Int. J. Mol. Sci. 2020, 21, 1960. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Khoshbakht, T.; Hussen, B.M.; Dong, P.; Gassler, N.; Taheri, M.; Baniahmad, A.; Dilmaghani, N.A. A Review on the Role of Cyclin Dependent Kinases in Cancers. Cancer Cell Int. 2022, 22, 325. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, L.; Hei, R.; Li, X.; Cai, H.; Wu, X.; Zheng, Q.; Cai, C. CDK Inhibitors in Cancer Therapy, an Overview of Recent Development. Am. J. Cancer Res. 2021, 11, 1913. [Google Scholar]
- Syn, N.L.; Lim, P.L.; Kong, L.R.; Wang, L.; Wong, A.L.A.; Lim, C.M.; Loh, T.K.S.; Siemeister, G.; Goh, B.C.; Hsieh, W.S. Pan-CDK Inhibition Augments Cisplatin Lethality in Nasopharyngeal Carcinoma Cell Lines and Xenograft Models. Signal Transduct. Target. Ther. 2018, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Jessen, B.A.; Lee, L.; Koudriakova, T.; Haines, M.; Lundgren, K.; Price, S.; Nonomiya, J.; Lewis, C.; Stevens, G.J. Peripheral White Blood Cell Toxicity Induced by Broad Spectrum Cyclin-Dependent Kinase Inhibitors. J. Appl. Toxicol. 2007, 27, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Massard, C.; Soria, J.C.; Anthoney, D.A.; Proctor, A.; Scaburri, A.; Pacciarini, M.A.; Laffranchi, B.; Pellizzoni, C.; Kroemer, G.; Armand, J.P.; et al. A First in Man, Phase I Dose-Escalation Study of PHA-793887, an Inhibitor of Multiple Cyclin-Dependent Kinases (CDK2, 1 and 4) Reveals Unexpected Hepatotoxicity in Patients with Solid Tumors. Cell Cycle 2011, 10, 963–970. [Google Scholar] [CrossRef] [PubMed]
- DiPippo, A.J.; Patel, N.K.; Barnett, C.M. Cyclin-Dependent Kinase Inhibitors for the Treatment of Breast Cancer: Past, Present, and Future. Pharmacotherapy 2016, 36, 652–667. [Google Scholar] [CrossRef]
- Kwapisz, D. Cyclin-Dependent Kinase 4/6 Inhibitors in Breast Cancer: Palbociclib, Ribociclib, and Abemaciclib. Breast Cancer Res. Treat. 2017, 166, 41–54. [Google Scholar] [CrossRef]
- Beaver, J.A.; Amiri-Kordestani, L.; Charlab, R.; Chen, W.; Palmby, T.; Tilley, A.; Zirkelbach, J.F.; Yu, J.; Liu, Q.; Zhao, L.; et al. FDA Approval: Palbociclib for the Treatment of Postmenopausal Patients with Estrogen Receptor-Positive, HER2-Negative Metastatic Breast Cancer. Clin. Cancer Res. 2015, 21, 4760–4766. [Google Scholar] [CrossRef]
- Shah, A.; Bloomquist, E.; Tang, S.; Fu, W.; Bi, Y.; Liu, Q.; Yu, J.; Zhao, P.; Palmby, T.R.; Goldberg, K.B.; et al. FDA Approval: Ribociclib for the Treatment of Postmenopausal Women with Hormone Receptor-Positive, HER2-Negative Advanced or Metastatic Breast Cancer. Clin. Cancer Res. 2018, 24, 2999–3004. [Google Scholar] [CrossRef]
- Royce, M.; Osgood, C.; Mulkey, F.; Bloomquist, E.; Pierce, W.F.; Roy, A.; Kalavar, S.; Ghosh, S.; Philip, R.; Rizvi, F.; et al. FDA Approval Summary: Abemaciclib with Endocrine Therapy for High-Risk Early Breast Cancer. J. Clin. Oncol. 2022, 40, 1155–1162. [Google Scholar] [CrossRef]
- Digiacomo, G.; Fumarola, C.; La Monica, S.; Bonelli, M.A.; Cretella, D.; Alfieri, R.; Cavazzoni, A.; Galetti, M.; Bertolini, P.; Missale, G.; et al. Simultaneous Combination of the CDK4/6 Inhibitor Palbociclib with Regorafenib Induces Enhanced Anti-Tumor Effects in Hepatocarcinoma Cell Lines. Front. Oncol. 2020, 10, 563249. [Google Scholar] [CrossRef]
- Limousin, W.; Laurent-Puig, P.; Ziol, M.; Ganne-Carrié, N.; Nahon, P.; Ait-Omar, A.; Seror, O.; Sidali, S.; Campani, C.; Blanc, P.; et al. Molecular-Based Targeted Therapies in Patients with Hepatocellular Carcinoma and Hepato-Cholangiocarcinoma Refractory to Atezolizumab/Bevacizumab. J. Hepatol. 2023, 79, 1450–1458. [Google Scholar] [CrossRef]
- Wang, H.; Liao, P.; Zeng, S.X.; Lu, H. Co-Targeting P53-R249S and CDK4 Synergistically Suppresses Survival of Hepatocellular Carcinoma Cells. Cancer Biol. Ther. 2020, 21, 269–277. [Google Scholar] [CrossRef]
- Jindal, A.; Thadi, A.; Shailubhai, K. Hepatocellular Carcinoma: Etiology and Current and Future Drugs. J. Clin. Exp. Hepatol. 2019, 9, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Digiacomo, G.; Fumarola, C.; La Monica, S.; Bonelli, M.; Cavazzoni, A.; Galetti, M.; Terenziani, R.; Eltayeb, K.; Volta, F.; Zoppi, S.; et al. CDK4/6 Inhibitors Improve the Anti-Tumor Efficacy of Lenvatinib in Hepatocarcinoma Cells. Front. Oncol. 2022, 12, 942341. [Google Scholar] [CrossRef]
- Robertson, I.B.; Rifkin, D.B. Unchaining the Beast; Insights from Structural and Evolutionary Studies on TGFβ Secretion, Sequestration, and Activation. Cytokine Growth Factor Rev. 2013, 24, 355–372. [Google Scholar] [CrossRef]
- Budi, E.H.; Duan, D.; Derynck, R. Transforming Growth Factor-β Receptors and Smads: Regulatory Complexity and Functional Versatility. Trends Cell Biol. 2017, 27, 658–672. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Budi, E.H. Specificity, Versatility, and Control of TGF-β Family Signaling. Sci. Signal. 2019, 12, eaav5183. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ren, J.; Ten Dijke, P. Targeting TGFβ Signal Transduction for Cancer Therapy. Signal Transduct. Target. Ther. 2021, 6, 8. [Google Scholar] [CrossRef]
- Oberhammer, F.A.; Pavelka, M.; Sharma, S.; Tiefenbacher, R.; Purchio, A.F.; Bursch, W.; Schulte-Hermann, R. Induction of Apoptosis in Cultured Hepatocytes and in Regressing Liver by Transforming Growth Factor Beta 1. Proc. Natl. Acad. Sci. USA 1992, 89, 5408–5412. [Google Scholar] [CrossRef]
- Sánchez, A.; Álvarez, A.M.; Benito, M.; Fabregat, I. Apoptosis Induced by Transforming Growth Factor-Beta in Fetal Hepatocyte Primary Cultures: Involvement of Reactive Oxygen Intermediates. J. Biol. Chem. 1996, 271, 7416–7422. [Google Scholar] [CrossRef]
- Polyak, K.; Kato, J.Y.; Solomon, M.J.; Sherr, C.J.; Massague, J.; Roberts, J.M.; Koff, A. P27Kip1, a Cyclin-Cdk Inhibitor, Links Transforming Growth Factor-Beta and Contact Inhibition to Cell Cycle Arrest. Genes Dev. 1994, 8, 9–22. [Google Scholar] [CrossRef]
- Giannelli, G.; Bergamini, C.; Fransvea, E.; Sgarra, C.; Antonaci, S. Laminin-5 with Transforming Growth Factor-Beta1 Induces Epithelial to Mesenchymal Transition in Hepatocellular Carcinoma. Gastroenterology 2005, 129, 1375–1383. [Google Scholar] [CrossRef]
- Malfettone, A.; Soukupova, J.; Bertran, E.; Crosas-Molist, E.; Lastra, R.; Fernando, J.; Koudelkova, P.; Rani, B.; Fabra, Á.; Serrano, T.; et al. Transforming Growth Factor-β-Induced Plasticity Causes a Migratory Stemness Phenotype in Hepatocellular Carcinoma. Cancer Lett. 2017, 392, 39–50. [Google Scholar] [CrossRef]
- Gotzmann, J.; Fischer, A.N.M.; Zojer, M.; Mikula, M.; Proell, V.; Huber, H.; Jechlinger, M.; Waerner, T.; Weith, A.; Beug, H.; et al. A Crucial Function of PDGF in TGF-Beta-Mediated Cancer Progression of Hepatocytes. Oncogene 2006, 25, 3170–3185. [Google Scholar] [CrossRef]
- Soukupova, J.; Malfettone, A.; Hyroššová, P.; Hernández-Alvarez, M.I.; Peñuelas-Haro, I.; Bertran, E.; Junza, A.; Capellades, J.; Giannelli, G.; Yanes, O.; et al. Role of the Transforming Growth Factor-β in Regulating Hepatocellular Carcinoma Oxidative Metabolism. Sci. Rep. 2017, 7, 12486. [Google Scholar] [CrossRef]
- Kim, J.; Hong, S.J.; Lim, E.K.; Yu, Y.S.; Kim, S.W.; Roh, J.H.; Do, I.G.; Joh, J.W.; Kim, D.S. Expression of Nicotinamide N-Methyltransferase in Hepatocellular Carcinoma Is Associated with Poor Prognosis. J. Exp. Clin. Cancer Res. 2009, 28, 20. [Google Scholar] [CrossRef]
- Campagna, R.; Vignini, A. NAD+ Homeostasis and NAD+-Consuming Enzymes: Implications for Vascular Health. Antioxidants 2023, 12, 376. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; You, S.; Zhang, S.; Hu, Q.; Wang, F.; Chi, X.; Zhao, W.; Xie, C.; Zhang, C.; Yu, Y.; et al. Elevated N-Methyltransferase Expression Induced by Hepatic Stellate Cells Contributes to the Metastasis of Hepatocellular Carcinoma via Regulation of the CD44v3 Isoform. Mol. Oncol. 2019, 13, 1993–2009. [Google Scholar] [CrossRef] [PubMed]
- Caballero-Díaz, D.; Bertran, E.; Peñuelas-Haro, I.; Moreno-Càceres, J.; Malfettone, A.; López-Luque, J.; Addante, A.; Herrera, B.; Sánchez, A.; Alay, A.; et al. Clathrin Switches Transforming Growth Factor-β Role to pro-Tumorigenic in Liver Cancer. J. Hepatol. 2020, 72, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Dzieran, J.; Gu, X.; Marhenke, S.; Vogel, A.; Machida, K.; Weiss, T.S.; Ruemmele, P.; Kollmar, O.; Hoffmann, P.; et al. Smad7 Regulates Compensatory Hepatocyte Proliferation in Damaged Mouse Liver and Positively Relates to Better Clinical Outcome in Human Hepatocellular Carcinoma. Clin. Sci. 2015, 128, 761–774. [Google Scholar] [CrossRef]
- Badawi, M.; Kim, J.; Dauki, A.; Sutaria, D.; Motiwala, T.; Reyes, R.; Wani, N.; Kolli, S.; Jiang, J.; Coss, C.C.; et al. CD44 Positive and Sorafenib Insensitive Hepatocellular Carcinomas Respond to the ATP-Competitive MTOR Inhibitor INK128. Oncotarget 2018, 9, 26032–26045. [Google Scholar] [CrossRef] [PubMed]
- Dzieran, J.; Fabian, J.; Feng, T.; Coulouarn, C.; Ilkavets, I.; Kyselova, A.; Breuhahn, K.; Dooley, S.; Meindl-Beinker, N.M. Comparative Analysis of TGF-β/Smad Signaling Dependent Cytostasis in Human Hepatocellular Carcinoma Cell Lines. PLoS ONE 2013, 8, e72252. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J. Recent Advances in the Development of TGF-β Signaling Inhibitors for Anticancer Therapy. J. Cancer Prev. 2020, 25, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Chung, C.L.; Hu, T.H.; Chen, J.J.; Liu, P.F.; Chen, C.L. Recent Progress in TGF-β Inhibitors for Cancer Therapy. Biomed. Pharmacother. 2021, 134, 111046. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.X.; Kang, Y.K.; Yen, C.J.; Finn, R.S.; Galle, P.R.; Llovet, J.M.; Assenat, E.; Brandi, G.; Pracht, M.; Lim, H.Y.; et al. Ramucirumab after Sorafenib in Patients with Advanced Hepatocellular Carcinoma and Increased α-Fetoprotein Concentrations (REACH-2): A Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet. Oncol. 2019, 20, 282–296. [Google Scholar] [CrossRef]
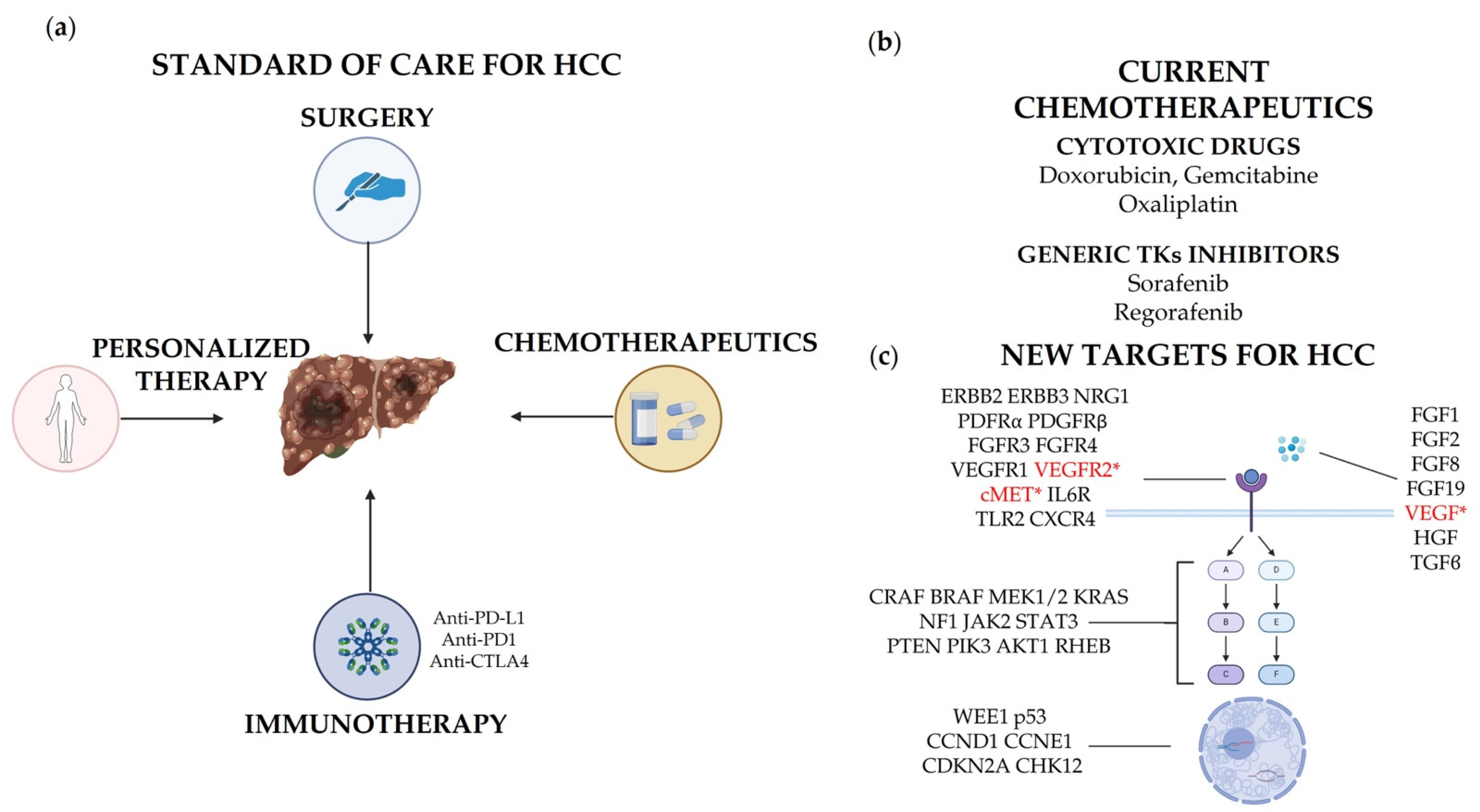
| Drug | Commercial Name | Target | Format | Reference | Drug Associated in Combination Therapy |
|---|---|---|---|---|---|
| Atezolizumab | Tecentriq™ | PD-L1 (CD274) | Fc optimized (no ADCC or CDC), humanized | [18] | Bevacizumab |
| Durvalumab | Imfinzi™ | PD-L1 | Monoclonal | [19] | Tremelimumab |
| Pembrolizumab | Keytruda® | PD-1 | Humanized, IgG4 Ab | [20] | N.A. |
| Nivolumab | Opdivo™ | PD-1 | Human, IgG4 Ab | [21] | Ipilimumab |
| Ipilimumab | Yervoy™ | CTLA4 | Human, IgG1 Ab | [21] | Nivolumab |
| Tremelimumab | Imjudo® | CTLA4 | Human, IgG2 Ab | [19] | Durvalumab |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pessino, G.; Scotti, C.; Maggi, M.; Immuno-HUB Consortium. Hepatocellular Carcinoma: Old and Emerging Therapeutic Targets. Cancers 2024, 16, 901. https://doi.org/10.3390/cancers16050901
Pessino G, Scotti C, Maggi M, Immuno-HUB Consortium. Hepatocellular Carcinoma: Old and Emerging Therapeutic Targets. Cancers. 2024; 16(5):901. https://doi.org/10.3390/cancers16050901
Chicago/Turabian StylePessino, Greta, Claudia Scotti, Maristella Maggi, and Immuno-HUB Consortium. 2024. "Hepatocellular Carcinoma: Old and Emerging Therapeutic Targets" Cancers 16, no. 5: 901. https://doi.org/10.3390/cancers16050901
APA StylePessino, G., Scotti, C., Maggi, M., & Immuno-HUB Consortium. (2024). Hepatocellular Carcinoma: Old and Emerging Therapeutic Targets. Cancers, 16(5), 901. https://doi.org/10.3390/cancers16050901






