The PAX Genes: Roles in Development, Cancer, and Other Diseases
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Evolution of PAX-like Genes
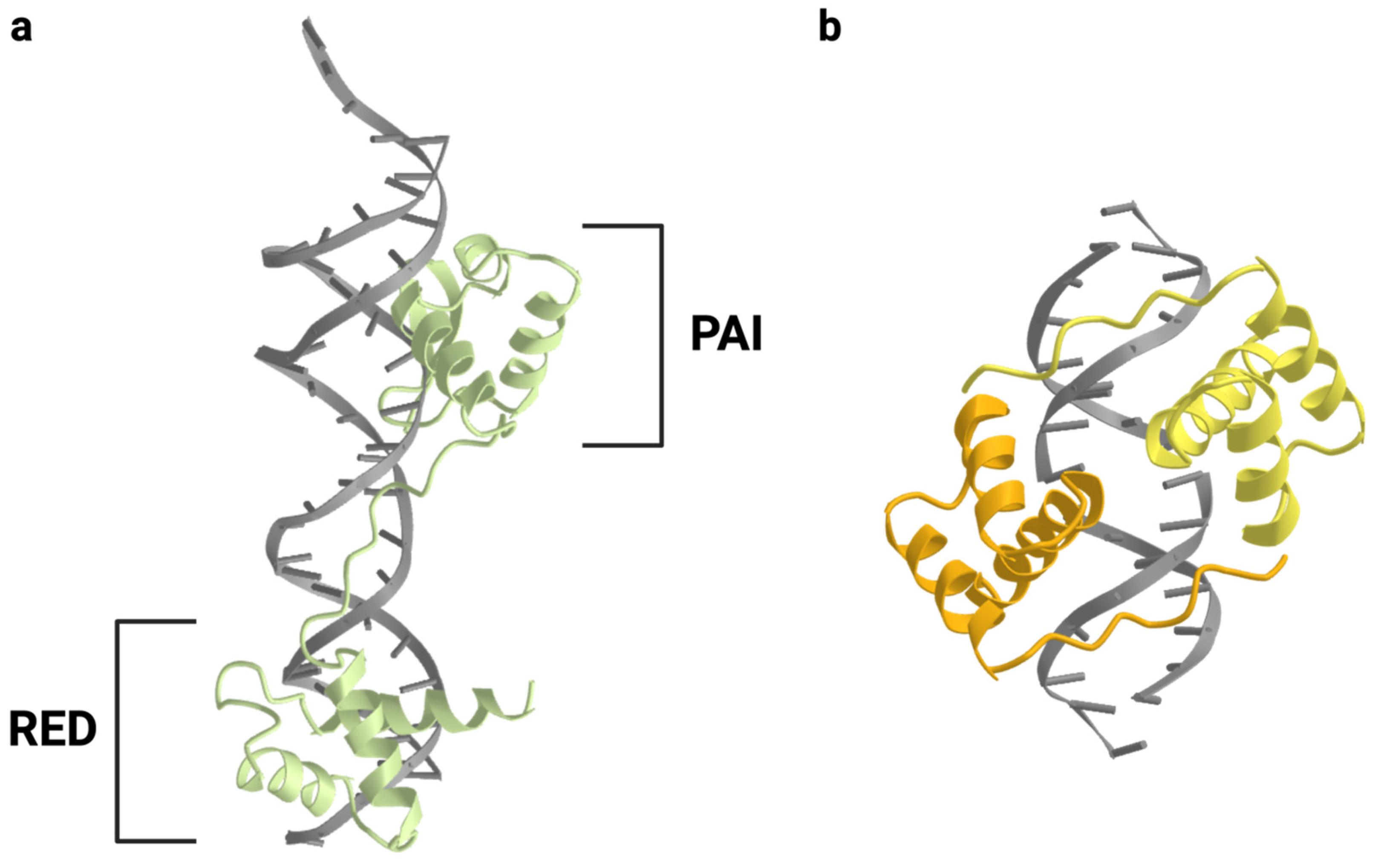
1.2. PAX Protein Structure
2. Regulation of PAX Family Gene Expression
2.1. Upstream Transcription Factors
2.2. miRNAs
2.3. Alternative Splicing and PAX Isoforms
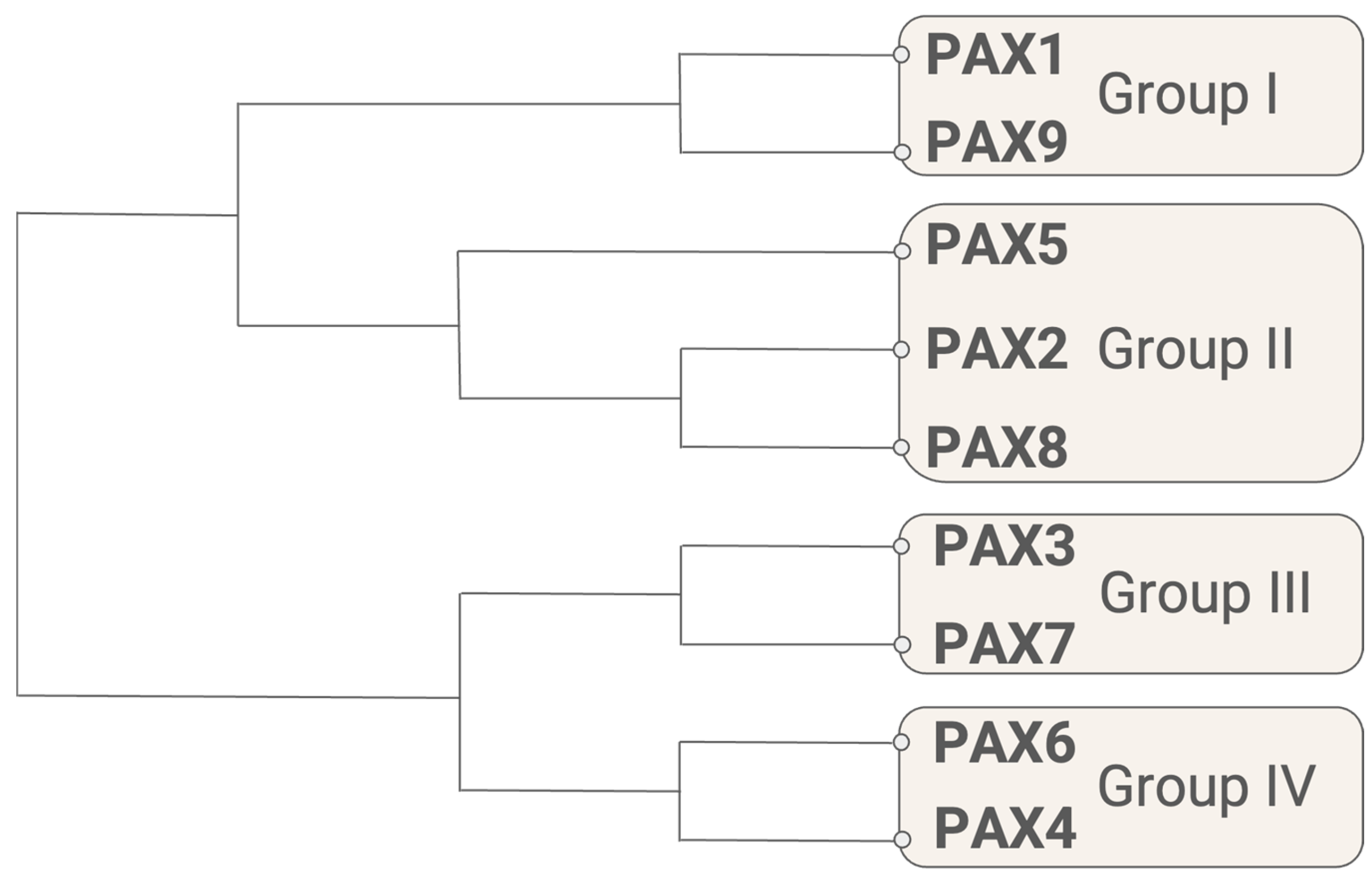
| Subgroup | Gene | Aliases | Developmental Role(s) | Associated Disorder(s) |
|---|---|---|---|---|
| Group I | PAX1 | HuP48 | Axial and appendicular skeleton, thymus, parathyroid gland | Otofaciocervical syndrome-2 (OTFCS2) |
| PAX9 | - | Axial and craniofacial skeleton, teeth | Selective tooth agenesis-3 (STHAG3) | |
| Group II | PAX2 | - | Kidney, CNS | Papillorenal syndrome (PAPRS), focal segmental glomerulosclerosis-7 (FSGS7) |
| PAX5 | BSAP | CNS, B cells | - | |
| PAX8 | - | Kidney, CNS, thyroid | Congenital hypothyroidism | |
| Group III | PAX3 | HuP2 | Skeletal muscle, neural crest, CNS | Waardenburg syndrome (Types 1 and 3), craniofacial-deafness-hand syndrome (CDHS) |
| PAX7 | HuP1, RMS2 | Skeletal muscle, neural crest, CNS | Congenital myopathy-19 (CMYP19) | |
| Group IV | PAX4 | - | GI endocrine cells | Maturity-onset diabetes of the young (Type 9, MODY9), diabetes mellitus (Type 2), ketosis-prone diabetes |
| PAX6 | MGDA, WAGR | GI endocrine cells, eye, CNS | Aniridia, anterior segment dysgenesis 5 (ASGD5), foveal hypoplasia 1 (FVH1), keratitis |
2.4. Post-Translational Modifications
2.5. Protein–Protein Interactions
3. PAX Genes during Development
3.1. PAX1 and PAX9 in Skeletal Development
3.2. PAX2 and PAX8 in Renal Development
3.3. PAX5 in B Cells
3.4. PAX8 in Thyroid Development
3.5. PAX3 and PAX7 in Myogenesis
3.6. PAX3 and PAX7 in Neural Crest Formation
3.7. PAX4 and PAX6 in Pancreatic Endocrine Cells
3.8. PAX6 in Ocular Development
4. PAX Genes in Human Disease
4.1. PAX1 in Otofaciocervical Syndrome
4.2. PAX9 in Tooth Agenesis
4.3. PAX2 in Renal and Ocular Disorders
4.4. PAX8 in Congenital Hypothyroidism
4.5. PAX3 in Waardenburg and Craniofacial-Deafness-Hand Syndromes
4.6. PAX7 in Congenital Myopathy
4.7. PAX4 in Diabetes
4.8. PAX6 in Ocular Disorders
5. PAX Genes and Cancer
| Gene | Associated Human Cancer | Observation | Reference |
|---|---|---|---|
| PAX1 | Colorectal carcinoma | Greater methylation at the PAX1 promoter and lower mRNA and protein expression levels of PAX1 in colorectal cancer vs. paired normal tissue samples | [125] |
| Cervical cancer | Greater methylation at the PAX1 promoter in patients with high-grade lesions as compared to patients with lower-grade lesions or normal cervical tissue | [126] | |
| PAX9 | Esophageal carcinoma | Inverse correlation between PAX9 protein expression level and malignancy of epithelial lesions | [127] |
| PAX2 | Ovarian cancer | Increased expression of PAX2 mRNA in low-grade and high-grade carcinoma samples as compared to normal ovarian surface epithelia. Increased PAX2 protein expression in low-grade carcinoma samples, compared to no PAX2 protein expression in high-grade carcinomas or normal ovarian surface epithelia | [128] |
| Wilms tumor | Higher PAX2 expression in Wilms tumor samples compared to normal adult kidney | [129,130] | |
| PAX5 | Non-Hodgkin lymphoma | A t(9;14)(p13;q32) chromosomal translocation generating the PAX5::IGH fusion gene, which brings the enhancer of the IGH gene in close proximity to the PAX5 gene and increases PAX5 transcription | [131,132,133] |
| Acute lymphoblastic leukemia | PAX5::ELN, PAX5::ETV6, PAX5::FOXP1, PAX5::PML gene fusions | [134,135,136,137] | |
| PAX8 | Follicular thyroid carcinoma | A t(2;3)(q13;p25) chromosomal translocation generating the PAX8::PPARg fusion protein, which acts as a dominant negative inhibitor of wild-type PPARg and activates transcription of some PAX8 and PPARg target genes | [138,139] |
| Wilms tumor | Higher PAX8 expression in Wilms tumor samples compared to normal adult kidney | [140] | |
| PAX3 | Alveolar rhabdomyosarcoma | A t(2;13)(q35;q14) chromosomal translocation generating the PAX3::FOXO1 fusion protein, which has a 10- to 100-fold increase in transcriptional activity compared to PAX3 at regulatory sites of target gene transcription | [141,142] |
| Melanoma | Increased expression of PAX3 in cutaneous melanoma as compared to benign lesions or normal skin | [143] | |
| PAX7 | Alveolar rhabdomyosarcoma | A t(1;13)(p36;q14) chromosomal translocation generating the PAX7::FOXO1 fusion protein | [144] |
| PAX4 | Insulinomas | Increased expression of PAX4 in insulinoma samples as compared to normal islets | [145] |
| PAX6 | Pancreatic carcinoma | Increased expression of PAX6 in pancreatic carcinoma tumors and cell lines as compared to normal adult pancreatic exocrine cells | [146] |
| Glioblastoma | Inverse correlation between PAX6 protein expression level and malignancy of astrocytic gliomas | [147] |
5.1. PAX Gene Expression in Cancer
5.2. PAX Gene Fusions in Cancer
5.3. PAX Genes as Favorable Prognostic Indicators
6. Therapeutic Strategies Targeting PAX Genes
6.1. Indirect Targeting of PAX Proteins
6.2. Direct Targeting of PAX Proteins
6.3. Additional Strategies
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kilchherr, F.; Baumgartner, S.; Bopp, D.; Frei, E.; Noll, M. Isolation of the paired gene of Drosophila and its spatial expression during early embryogenesis. Nature 1986, 321, 493–499. [Google Scholar] [CrossRef]
- McGinnis, W.; Garber, R.L.; Wirz, J.; Kuroiwa, A.; Gehring, W.J. A homologous protein-coding sequence in drosophila homeotic genes and its conservation in other metazoans. Cell 1984, 37, 403–408. [Google Scholar] [CrossRef]
- Deutsch, U.; Dressler, G.R.; Gruss, P. Pax 1, a member of a paired box homologous murine gene family, is expressed in segmented structures during development. Cell 1988, 53, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Burri, M.; Tromvoukis, Y.; Bopp, D.; Frigerio, G.; Noll, M. Conservation of the paired domain in metazoans and its structure in three isolated human genes. EMBO J. 1989, 8, 1183–1190. [Google Scholar] [CrossRef]
- Paixão-Côrtes, V.R.; Salzano, F.M.; Bortolini, M.C. Origins and evolvability of the PAX family. Semin. Cell Dev. Biol. 2015, 44, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Balczarek, K.A.; Lai, Z.C.; Kumar, S. Evolution of functional diversification of the paired box (Pax) DNA-binding domains. Mol. Biol. Evol. 1997, 14, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.-i.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Breitling, R.; Gerber, J.K. Origin of the paired domain. Dev. Genes Evol. 2000, 210, 644–650. [Google Scholar] [CrossRef]
- Wang, Y.-T.; Pan, Y.-J.; Cho, C.-C.; Lin, B.-C.; Su, L.-H.; Huang, Y.-C.; Sun, C.-H. A Novel Pax-like Protein Involved in Transcriptional Activation of Cyst Wall Protein Genes in Giardia lamblia. J. Biol. Chem. 2010, 285, 32213–32226. [Google Scholar] [CrossRef] [PubMed]
- Bopp, D.; Burri, M.; Baumgartner, S.; Frigerio, G.; Noll, M. Conservation of a large protein domain in the segmentation gene paired and in functionally related genes of Drosophila. Cell 1986, 47, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, S.; Bopp, D.; Burri, M.; Noll, M. Structure of two genes at the gooseberry locus related to the paired gene and their spatial expression during Drosophila embryogenesis. Genes Dev. 1987, 1, 1247–1267. [Google Scholar] [CrossRef] [PubMed]
- Codutti, L.; van Ingen, H.; Vascotto, C.; Fogolari, F.; Corazza, A.; Tell, G.; Quadrifoglio, F.; Viglino, P.; Boelens, R.; Esposito, G. The solution structure of DNA-free Pax-8 paired box domain accounts for redox regulation of transcriptional activity in the pax protein family. J. Biol. Chem. 2008, 283, 33321–33328. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.E.; Rould, M.A.; Xu, W.; Epstein, J.A.; Maas, R.L.; Pabo, C.O. Crystal structure of the human Pax6 paired domain–DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev. 1999, 13, 1263–1275. [Google Scholar] [CrossRef]
- Lechner, M.S.; Dressler, G.R. Mapping of Pax-2 transcription activation domains. J. Biol. Chem. 1996, 271, 21088–21093. [Google Scholar] [CrossRef]
- Eberhard, D.; Jiménez, G.; Heavey, B.; Busslinger, M. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 2000, 19, 2292–2303. [Google Scholar] [CrossRef]
- Birrane, G.; Soni, A.; Ladias, J.A.A. Structural basis for DNA recognition by the human PAX3 homeodomain. Biochemistry 2009, 48, 1148–1155. [Google Scholar] [CrossRef]
- Corry, G.N.; Underhill, D.A. Pax3 target gene recognition occurs through distinct modes that are differentially affected by disease-associated mutations. Pigment. Cell Res. 2005, 18, 427–438. [Google Scholar] [CrossRef]
- Madej, T.; Lanczycki, C.J.; Zhang, D.; Thiessen, P.A.; Geer, R.C.; Marchler-Bauer, A.; Bryant, S.H. MMDB and VAST+: Tracking structural similarities between macromolecular complexes. Nucleic Acids Res. 2014, 42, D297–D303. [Google Scholar] [CrossRef]
- Curto, G.G.; Gard, C.; Ribes, V. Structures and properties of PAX linked regulatory networks architecting and pacing the emergence of neuronal diversity. Semin. Cell Dev. Biol. 2015, 44, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Goljanek-Whysall, K.; Sweetman, D.; Abu-Elmagd, M.; Chapnik, E.; Dalmay, T.; Hornstein, E.; Münsterberg, A. MicroRNA regulation of the paired-box transcription factor Pax3 confers robustness to developmental timing of myogenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 11936–11941. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Sarver, A.L.; Alamgir, S.; Subramanian, S. Downregulation of microRNAs miR-1, -206 and -29 stabilizes PAX3 and CCND2 expression in rhabdomyosarcoma. Lab. Investig. 2012, 92, 571–583. [Google Scholar] [CrossRef]
- Thompson, B.; Davidson, E.A.; Liu, W.; Nebert, D.W.; Bruford, E.A.; Zhao, H.; Dermitzakis, E.T.; Thompson, D.C.; Vasiliou, V. Overview of PAX gene family: Analysis of human tissue-specific variant expression and involvement in human disease. Hum. Genet. 2021, 140, 381–400. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Lawrence, E.J.; Strzelecki, D.; Rajput, P.; Xia, S.J.; Gottesman, D.M.; Barr, F.G. Co-expression of alternatively spliced forms of PAX3, PAX7, PAX3-FKHR and PAX7-FKHR with distinct DNA binding and transactivation properties in rhabdomyosarcoma. Int. J. Cancer 2005, 115, 85–92. [Google Scholar] [CrossRef]
- Vogan, K.J.; Underhill, D.A.; Gros, P. An alternative splicing event in the Pax-3 paired domain identifies the linker region as a key determinant of paired domain DNA-binding activity. Mol. Cell. Biol. 1996, 16, 6677–6686. [Google Scholar] [CrossRef]
- Pritchard, C.; Grosveld, G.; Hollenbach, A.D. Alternative splicing of Pax3 produces a transcriptionally inactive protein. Gene 2003, 305, 61–69. [Google Scholar] [CrossRef]
- Sasamoto, Y.; Hayashi, R.; Park, S.-J.; Saito-Adachi, M.; Suzuki, Y.; Kawasaki, S.; Quantock, A.J.; Nakai, K.; Tsujikawa, M.; Nishida, K. PAX6 Isoforms, along with Reprogramming Factors, Differentially Regulate the Induction of Cornea-specific Genes. Sci. Rep. 2016, 6, 20807. [Google Scholar] [CrossRef]
- Miller, P.J.; Dietz, K.N.; Hollenbach, A.D. Identification of serine 205 as a site of phosphorylation on Pax3 in proliferating but not differentiating primary myoblasts. Protein Sci. 2008, 17, 1979–1986. [Google Scholar] [CrossRef]
- Dietz, K.N.; Miller, P.J.; Hollenbach, A.D. Phosphorylation of serine 205 by the protein kinase CK2 persists on Pax3-FOXO1, but not Pax3, throughout early myogenic differentiation. Biochemistry 2009, 48, 11786–11795. [Google Scholar] [CrossRef]
- Cao, X.; Kambe, F.; Lu, X.; Kobayashi, N.; Ohmori, S.; Seo, H. Glutathionylation of two cysteine residues in paired domain regulates DNA binding activity of Pax-8. J. Biol. Chem. 2005, 280, 25901–25906. [Google Scholar] [CrossRef]
- Schäfer, B.W.; Czerny, T.; Bernasconi, M.; Genini, M.; Busslinger, M. Molecular cloning and characterization of a human PAX-7 cDNA expressed in normal and neoplastic myocytes. Nucleic Acids Res. 1994, 22, 4574–4582. [Google Scholar] [CrossRef]
- Verger, A.; Duterque-Coquillaud, M. When Ets transcription factors meet their partners. BioEssays 2002, 24, 362–370. [Google Scholar] [CrossRef]
- Patel, S.R.; Kim, D.; Levitan, I.; Dressler, G.R. The BRCT-Domain Containing Protein PTIP Links PAX2 to a Histone H3, Lysine 4 Methyltransferase Complex. Dev. Cell 2007, 13, 580–592. [Google Scholar] [CrossRef]
- Lee, T.I.; Young, R.A. Transcriptional regulation and its misregulation in disease. Cell 2013, 152, 1237–1251. [Google Scholar] [CrossRef]
- Bodmer, R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development 1993, 118, 719–729. [Google Scholar] [CrossRef]
- Lints, T.J.; Parsons, L.M.; Hartley, L.; Lyons, I.; Harvey, R.P. Nkx-2.5: A novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 1993, 119, 969. [Google Scholar] [CrossRef]
- He, A.; Kong, S.W.; Ma, Q.; Pu, W.T. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl. Acad. Sci. USA 2011, 108, 5632–5637. [Google Scholar] [CrossRef] [PubMed]
- Bouveret, R.; Waardenberg, A.J.; Schonrock, N.; Ramialison, M.; Doan, T.; de Jong, D.; Bondue, A.; Kaur, G.; Mohamed, S.; Fonoudi, H.; et al. NKX2-5 mutations causative for congenital heart disease retain functionality and are directed to hundreds of targets. eLife 2015, 4, e06942. [Google Scholar] [CrossRef] [PubMed]
- Walther, C.; Gruss, P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development 1991, 113, 1435–1449. [Google Scholar] [CrossRef] [PubMed]
- Wolf, L.V.; Yang, Y.; Wang, J.; Xie, Q.; Braunger, B.; Tamm, E.R.; Zavadil, J.; Cvekl, A. Identification of pax6-dependent gene regulatory networks in the mouse lens. PLoS ONE 2009, 4, e4159. [Google Scholar] [CrossRef]
- Grindley, J.C.; Davidson, D.R.; Hill, R.E. The role of Pax-6 in eye and nasal development. Development 1995, 121, 1433–1442. [Google Scholar] [CrossRef]
- Zhu, J.; Ordway, A.J.; Weber, L.; Buddika, K.; Kumar, J.P. Polycomb group (PcG) proteins and Pax6 cooperate to inhibit in vivo reprogramming of the developing Drosophila eye. Development 2018, 145, dev160754. [Google Scholar] [CrossRef]
- Neubüser, A.; Koseki, H.; Balling, R. Characterization and developmental expression of Pax9, a paired-box-containing gene related to Pax1. Dev. Biol. 1995, 170, 701–716. [Google Scholar] [CrossRef]
- Wallin, J.; Wilting, J.; Koseki, H.; Fritsch, R.; Christ, B.; Balling, R. The role of Pax-1 in axial skeleton development. Development 1994, 120, 1109–1121. [Google Scholar] [CrossRef]
- Wilm, B.; Dahl, E.; Peters, H.; Balling, R.; Imai, K. Targeted disruption of Pax1 defines its null phenotype and proves haploinsufficiency. Proc. Natl. Acad. Sci. USA 1998, 95, 8692–8697. [Google Scholar] [CrossRef]
- Kokubu, C.; Wilm, B.; Kokubu, T.; Wahl, M.; Rodrigo, I.; Sakai, N.; Santagati, F.; Hayashizaki, Y.; Suzuki, M.; Yamamura, K.-I.; et al. Undulated short-tail deletion mutation in the mouse ablates Pax1 and leads to ectopic activation of neighboring Nkx2-2 in domains that normally express Pax1. Genetics 2003, 165, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Peters, H.; Wilm, B.; Sakai, N.; Imai, K.; Maas, R.; Balling, R. Pax1 and Pax9 synergistically regulate vertebral column development. Development 1999, 126, 5399–5408. [Google Scholar] [CrossRef]
- Peters, H.; Neubüser, A.; Kratochwil, K.; Balling, R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998, 12, 2735–2747. [Google Scholar] [CrossRef]
- Li, R.; Chen, Z.; Yu, Q.; Weng, M.; Chen, Z. The Function and Regulatory Network of Pax9 Gene in Palate Development. J. Dent. Res. 2019, 98, 277–287. [Google Scholar] [CrossRef]
- Dressler, G.R.; Deutsch, U.; Chowdhury, K.; Nornes, H.O.; Gruss, P. Pax2, a new murine paired-box-containing gene and its expression in the developing excretory system. Development 1990, 109, 787–795. [Google Scholar] [CrossRef]
- Keller, S.A.; Jones, J.M.; Boyle, A.; Barrow, L.L.; Killen, P.D.; Green, D.G.; Kapousta, N.V.; Hitchcock, P.F.; Swank, R.T.; Meisler, M.H. Kidney and Retinal Defects (Krd), a Transgene-Induced Mutation with a Deletion of Mouse Chromosome 19 That Includes the Pax2 Locus. Genomics 1994, 23, 309–320. [Google Scholar] [CrossRef]
- Torres, M.; Gómez-Pardo, E.; Dressler, G.R.; Gruss, P. Pax-2 controls multiple steps of urogenital development. Development 1995, 121, 4057–4065. [Google Scholar] [CrossRef]
- Carroll, T.J.; Vize, P.D. Synergism between Pax-8 and lim-1 in Embryonic Kidney Development. Dev. Biol. 1999, 214, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Buisson, I.; Le Bouffant, R.; Futel, M.; Riou, J.-F.; Umbhauer, M. Pax8 and Pax2 are specifically required at different steps of Xenopus pronephros development. Dev. Biol. 2015, 397, 175–190. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, A.; Chowdhury, K.; Gruss, P. Follicular cells of the thyroid gland require Pax8 gene function. Nat. Genet. 1998, 19, 87–90. [Google Scholar] [CrossRef]
- Bouchard, M.; Souabni, A.; Mandler, M.; Neubüser, A.; Busslinger, M. Nephric lineage specification by Pax2 and Pax8. Genes Dev. 2002, 16, 2958–2970. [Google Scholar] [CrossRef]
- Cai, Y.; Brophy, P.D.; Levitan, I.; Stifani, S.; Dressler, G.R. Groucho suppresses Pax2 transactivation by inhibition of JNK-mediated phosphorylation. EMBO J. 2003, 22, 5522–5529. [Google Scholar] [CrossRef]
- Patel, S.R.; Ranghini, E.; Dressler, G.R. Mechanisms of Gene Activation and Repression by Pax Proteins in the Developing Kidney. Pediatr. Nephrol. 2014, 29, 589–595. [Google Scholar] [CrossRef][Green Version]
- Holmes, M.L.; Pridans, C.; Nutt, S.L. The regulation of the B-cell gene expression programme by Pax5. Immunol. Cell Biol. 2008, 86, 47–53. [Google Scholar] [CrossRef]
- Urbánek, P.; Wang, Z.Q.; Fetka, I.; Wagner, E.F.; Busslinger, M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 1994, 79, 901–912. [Google Scholar] [CrossRef]
- Rolink, A.G.; Nutt, S.L.; Melchers, F.; Busslinger, M. Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature 1999, 401, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Fuxa, M.; Busslinger, M. Reporter gene insertions reveal a strictly B lymphoid-specific expression pattern of Pax5 in support of its B cell identity function. J. Immunol. 2007, 178, 3031–3037. [Google Scholar] [CrossRef]
- He, T.; Hong, S.Y.; Huang, L.; Xue, W.; Yu, Z.; Kwon, H.; Kirk, M.; Ding, S.-j.; Su, K.; Zhang, Z. Histone acetyltransferase p300 acetylates Pax5 and strongly enhances Pax5-mediated transcriptional activity. J. Biol. Chem. 2011, 286, 14137–14145. [Google Scholar] [CrossRef]
- Nilsson, M.; Williams, D. On the Origin of Cells and Derivation of Thyroid Cancer: C Cell Story Revisited. Eur. Thyroid. J. 2016, 5, 79–93. [Google Scholar] [CrossRef]
- Plachov, D.; Chowdhury, K.; Walther, C.; Simon, D.; Guenet, J.L.; Gruss, P. Pax8, a murine paired box gene expressed in the developing excretory system and thyroid gland. Development 1990, 110, 643–651. [Google Scholar] [CrossRef]
- Antonica, F.; Kasprzyk, D.F.; Opitz, R.; Iacovino, M.; Liao, X.-H.; Dumitrescu, A.M.; Refetoff, S.; Peremans, K.; Manto, M.; Kyba, M.; et al. Generation of functional thyroid from embryonic stem cells. Nature 2012, 491, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Pasca di Magliano, M.; Di Lauro, R.; Zannini, M. Pax8 has a key role in thyroid cell differentiation. Proc. Natl. Acad. Sci. USA 2000, 97, 13144–13149. [Google Scholar] [CrossRef] [PubMed]
- Bober, E.; Franz, T.; Arnold, H.-H.; Gruss, P.; Tremblay, P. Pax-3 is required for the development of limb muscles: A possible role for the migration of dermomyotomal muscle progenitor cells. Development 1994, 120, 603–612. [Google Scholar] [CrossRef]
- Franz, T.; Kothary, R.; Surani, M.A.; Halata, Z.; Grim, M. The Splotch mutation interferes with muscle development in the limbs. Anat. Embryol. 1993, 187, 153–160. [Google Scholar] [CrossRef]
- Borycki, A.-G.; Li, J.; Jin, F.; Emerson, C.P.; Epstein, J.A. Pax3 functions in cell survival and in pax7 regulation. Development 1999, 126, 1665–1674. [Google Scholar] [CrossRef]
- Seale, P.; Sabourin, L.A.; Girgis-Gabardo, A.; Mansouri, A.; Gruss, P.; Rudnicki, M.A. Pax7 Is Required for the Specification of Myogenic Satellite Cells. Cell 2000, 102, 777–786. [Google Scholar] [CrossRef]
- Relaix, F.; Rocancourt, D.; Mansouri, A.; Buckingham, M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 2005, 435, 948–953. [Google Scholar] [CrossRef]
- Xi, M.; Lui, F. Neuroanatomy, Neural Crest. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Goulding, M.D.; Chalepakis, G.; Deutsch, U.; Erselius, J.R.; Gruss, P. Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 1991, 10, 1135–1147. [Google Scholar] [CrossRef]
- Chi, N.; Epstein, J.A. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002, 18, 41–47. [Google Scholar] [CrossRef]
- Lang, D.; Chen, F.; Milewski, R.; Li, J.; Lu, M.M.; Epstein, J.A. Pax3 is required for enteric ganglia formation and functions with Sox10 to modulate expression of c-ret. J. Clin. Investig. 2000, 106, 963–971. [Google Scholar] [CrossRef]
- Mansouri, A.; Stoykova, A.; Torres, M.; Gruss, P. Dysgenesis of cephalic neural crest derivatives in Pax7−/− mutant mice. Development 1996, 122, 831–838. [Google Scholar] [CrossRef]
- Sosa-Pineda, B.; Chowdhury, K.; Torres, M.; Oliver, G.; Gruss, P. The Pax4 gene is essential for differentiation of insulin-producing beta cells in the mammalian pancreas. Nature 1997, 386, 399–402. [Google Scholar] [CrossRef]
- Prado, C.L.; Pugh-Bernard, A.E.; Elghazi, L.; Sosa-Pineda, B.; Sussel, L. Ghrelin cells replace insulin-producing β cells in two mouse models of pancreas development. Proc. Natl. Acad. Sci. USA 2004, 101, 2924–2929. [Google Scholar] [CrossRef]
- St-Onge, L.; Sosa-Pineda, B.; Chowdhury, K.; Mansouri, A.; Gruss, P. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature 1997, 387, 406–409. [Google Scholar] [CrossRef] [PubMed]
- Swisa, A.; Avrahami, D.; Eden, N.; Zhang, J.; Feleke, E.; Dahan, T.; Cohen-Tayar, Y.; Stolovich-Rain, M.; Kaestner, K.H.; Glaser, B.; et al. PAX6 maintains β cell identity by repressing genes of alternative islet cell types. J. Clin. Investig. 2016, 127, 230–243. [Google Scholar] [CrossRef]
- Graw, J. Chapter Ten–Eye Development. In Current Topics in Developmental Biology; Koopman, P., Ed.; Organogenesis in Development; Academic Press: Cambridge, MA, USA, 2010; Volume 90, pp. 343–386. [Google Scholar]
- Hyde, R.R. An Eyeless Mutant in DROSOPHILA HYDEI. Genetics 1922, 7, 319–334. [Google Scholar] [CrossRef]
- Quiring, R.; Walldorf, U.; Kloter, U.; Gehring, W.J. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science 1994, 265, 785–789. [Google Scholar] [CrossRef]
- Hogan, B.L.M.; Hirst, E.M.A.; Horsburgh, G.; Hetherington, C.M. Small eye (Sey): A mouse model for the genetic analysis of craniofacial abnormalities. Development 1988, 103, 115–119. [Google Scholar] [CrossRef]
- Favor, J.; Peters, H.; Hermann, T.; Schmahl, W.; Chatterjee, B.; Neuhäuser-Klaus, A.; Sandulache, R. Molecular characterization of Pax6(2Neu) through Pax6(10Neu): An extension of the Pax6 allelic series and the identification of two possible hypomorph alleles in the mouse Mus musculus. Genetics 2001, 159, 1689–1700. [Google Scholar] [CrossRef]
- Epstein, J.A.; Glaser, T.; Cai, J.; Jepeal, L.; Walton, D.S.; Maas, R.L. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994, 8, 2022–2034. [Google Scholar] [CrossRef] [PubMed]
- Pohl, E.; Aykut, A.; Beleggia, F.; Karaca, E.; Durmaz, B.; Keupp, K.; Arslan, E.; Onay, M.P.; Yigit, G.; Özkinay, F.; et al. A hypofunctional PAX1 mutation causes autosomal recessively inherited otofaciocervical syndrome. Hum. Genet. 2013, 132, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Paganini, I.; Sestini, R.; Capone, G.l.; Putignano, A.l.; Contini, E.; Giotti, I.; Gensini, F.; Marozza, A.; Barilaro, A.; Porfirio, B.; et al. A novel PAX1 null homozygous mutation in autosomal recessive otofaciocervical syndrome associated with severe combined immunodeficiency. Clin. Genet. 2017, 92, 664–668. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.J.; Das Bhowmik, A.; Bhat, V.; Satidevi Vineeth, V.; Vasudevamurthy, R.; Dalal, A. Autosomal recessive otofaciocervical syndrome type 2 with novel homozygous small insertion in PAX1 gene. Am. J. Med. Genet. Part A 2018, 176, 1200–1206. [Google Scholar] [CrossRef]
- Stockton, D.W.; Das, P.; Goldenberg, M.; D’Souza, R.N.; Patel, P.I. Mutation of PAX9 is associated with oligodontia. Nat. Genet. 2000, 24, 18–19. [Google Scholar] [CrossRef]
- Das, P.; Stockton, D.W.; Bauer, C.; Shaffer, L.G.; D’Souza, R.N.; Wright, T.J.; Patel, P.I. Haploinsufficiency of PAX9 is associated with autosomal dominant hypodontia. Hum. Genet. 2002, 110, 371–376. [Google Scholar] [CrossRef]
- Lammi, L.; Halonen, K.; Pirinen, S.; Thesleff, I.; Arte, S.; Nieminen, P. A missense mutation in PAX9 in a family with distinct phenotype of oligodontia. Eur. J. Hum. Genet. 2003, 11, 866–871. [Google Scholar] [CrossRef]
- Zhao, J.-l.; Chen, Y.-x.; Bao, L.; Xia, Q.-j.; Wu, T.-j.; Zhou, L. Novel mutations of PAX9 gene in Chinese patients with oligodontia. Zhonghua Kou Qiang Yi Xue Za Zhi 2005, 40, 266–270. [Google Scholar]
- Kapadia, H.; Frazier-Bowers, S.; Ogawa, T.; D’Souza, R.N. Molecular characterization of a novel PAX9 missense mutation causing posterior tooth agenesis. Eur. J. Hum. Genet. 2006, 14, 403–409. [Google Scholar] [CrossRef]
- Wang, Y.; Groppe, J.C.; Wu, J.; Ogawa, T.; Mues, G.; D’Souza, R.N.; Kapadia, H. Pathogenic mechanisms of tooth agenesis linked to paired domain mutations in human PAX9. Hum. Mol. Genet. 2009, 18, 2863–2874. [Google Scholar] [CrossRef]
- Schimmenti, L.A. Renal coloboma syndrome. Eur. J. Hum. Genet. 2011, 19, 1207–1212. [Google Scholar] [CrossRef]
- Amiel, J.; Audollent, S.; Joly, D.; Dureau, P.; Salomon, R.; Tellier, A.-L.; Augé, J.; Bouissou, F.; Antignac, C.; Gubler, M.-C.; et al. PAX2 mutations in renal–coloboma syndrome: Mutational hotspot and germline mosaicism. Eur. J. Hum. Genet. 2000, 8, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Porteous, S.; Torban, E.; Cho, N.-P.; Cunliffe, H.; Chua, L.; McNoe, L.; Ward, T.; Souza, C.; Gus, P.; Giugliani, R.; et al. Primary renal hypoplasiain humans and mice with PAX2 mutations: Evidence of increased apoptosis in fetal kidneys of Pax21Neu +/– mutant mice. Hum. Mol. Genet. 2000, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Barua, M.; Stellacci, E.; Stella, L.; Weins, A.; Genovese, G.; Muto, V.; Caputo, V.; Toka, H.R.; Charoonratana, V.T.; Tartaglia, M.; et al. Mutations in PAX2 Associate with Adult-Onset FSGS. J. Am. Soc. Nephrol. 2014, 25, 1942. [Google Scholar] [CrossRef] [PubMed]
- Macchia, P.E.; Lapi, P.; Krude, H.; Pirro, M.T.; Missero, C.; Chiovato, L.; Souabni, A.; Baserga, M.; Tassi, V.; Pinchera, A.; et al. PAX8 mutations associated with congenital hypothyroidism caused by thyroid dysgenesis. Nat. Genet. 1998, 19, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Meeus, L.; Gilbert, B.; Rydlewski, C.; Parma, J.; Roussie, A.L.; Abramowicz, M.; Vilain, C.; Christophe, D.; Costagliola, S.; Vassart, G. Characterization of a Novel Loss of Function Mutation of PAX8 in a Familial Case of Congenital Hypothyroidism with In-Place, Normal-Sized Thyroid. J. Clin. Endocrinol. Metab. 2004, 89, 4285–4291. [Google Scholar] [CrossRef]
- Tell, G.; Pellizzari, L.; Esposito, G.; Pucillo, C.; Macchia, P.E.; Di Lauro, R.; Damante, G. Structural defects of a Pax8 mutant that give rise to congenital hypothyroidism. Biochem. J. 1999, 341 Pt 1, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Pingault, V.; Ente, D.; Dastot-Le Moal, F.; Goossens, M.; Marlin, S.; Bondurand, N. Review and update of mutations causing Waardenburg syndrome. Hum. Mutat. 2010, 31, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, C.T.; Hoth, C.F.; Amos, J.A.; da-Silva, E.O.; Milunsky, A. An exonic mutation in the HuP2 paired domain gene causes Waardenburg’s syndrome. Nature 1992, 355, 637–638. [Google Scholar] [CrossRef]
- Tassabehji, M.; Read, A.P.; Newton, V.E.; Harris, R.; Balling, R.; Gruss, P.; Strachan, T. Waardenburg’s syndrome patients have mutations in the human homologue of the Pax-3 paired box gene. Nature 1992, 355, 635–636. [Google Scholar] [CrossRef] [PubMed]
- Zlotogora, J.; Lerer, I.; Bar-David, S.; Ergaz, Z.; Abeliovich, D. Homozygosity for Waardenburg syndrome. Am. J. Hum. Genet. 1995, 56, 1173–1178. [Google Scholar]
- Wildhardt, G.; Winterpacht, A.; Hilbert, K.; Menger, H.; Zabel, B. Two different PAX3 gene mutations causing Waardenburg syndrome type I. Mol. Cell. Probes 1996, 10, 229–231. [Google Scholar] [CrossRef]
- Hoth, C.F.; Milunsky, A.; Lipsky, N.; Sheffer, R.; Clarren, S.K.; Baldwin, C.T. Mutations in the paired domain of the human PAX3 gene cause Klein-Waardenburg syndrome (WS-III) as well as Waardenburg syndrome type I (WS-I). Am. J. Hum. Genet. 1993, 52, 455–462. [Google Scholar]
- Fortin, A.S.; Underhill, D.A.; Gros, P. Reciprocal Effect of Waardenburg Syndrome Mutations on DNA Binding by the Pax-3 Paired Domain and Homeodomain. Hum. Mol. Genet. 1997, 6, 1781–1790. [Google Scholar] [CrossRef]
- Gad, A.; Laurino, M.; Maravilla, K.R.; Matsushita, M.; Raskind, W.H. Sensorineural deafness, distinctive facial features, and abnormal cranial bones: A new variant of Waardenburg syndrome? Am. J. Med. Genet. Part A 2008, 146A, 1880–1885. [Google Scholar] [CrossRef]
- Asher, J.H., Jr.; Sommer, A.; Morell, R.; Friedman, T.B. Missense mutation in the paired domain of PAX3 causes craniofacial-deafness-hand syndrome. Hum. Mutat. 1996, 7, 30–35. [Google Scholar] [CrossRef]
- Feichtinger, R.G.; Mucha, B.E.; Hengel, H.; Orfi, Z.; Makowski, C.; Dort, J.; D’Anjou, G.; Nguyen, T.T.M.; Buchert, R.; Juenger, H.; et al. Biallelic variants in the transcription factor PAX7 are a new genetic cause of myopathy. Genet. Med. 2019, 21, 2521–2531. [Google Scholar] [CrossRef]
- Shimajiri, Y.; Sanke, T.; Furuta, H.; Hanabusa, T.; Nakagawa, T.; Fujitani, Y.; Kajimoto, Y.; Takasu, N.; Nanjo, K. A Missense Mutation of Pax4 Gene (R121W) Is Associated With Type 2 Diabetes in Japanese. Diabetes 2001, 50, 2864–2869. [Google Scholar] [CrossRef]
- Plengvidhya, N.; Kooptiwut, S.; Songtawee, N.; Doi, A.; Furuta, H.; Nishi, M.; Nanjo, K.; Tantibhedhyangkul, W.; Boonyasrisawat, W.; Yenchitsomanus, P.-T.; et al. PAX4 Mutations in Thais with Maturity Onset Diabetes of the Young. J. Clin. Endocrinol. Metab. 2007, 92, 2821–2826. [Google Scholar] [CrossRef]
- Mauvais-Jarvis, F.; Smith, S.B.; Le May, C.; Leal, S.M.; Gautier, J.-F.; Molokhia, M.; Riveline, J.-P.; Rajan, A.S.; Kevorkian, J.-P.; Zhang, S.; et al. PAX4 gene variations predispose to ketosis-prone diabetes. Hum. Mol. Genet. 2004, 13, 3151–3159. [Google Scholar] [CrossRef]
- Robinson, D.O.; Howarth, R.J.; Williamson, K.A.; van Heyningen, V.; Beal, S.J.; Crolla, J.A. Genetic analysis of chromosome 11p13 and the PAX6 gene in a series of 125 cases referred with aniridia. Am. J. Med. Genet. Part A 2008, 146A, 558–569. [Google Scholar] [CrossRef]
- Singh, S.; Chao, L.Y.; Mishra, R.; Davies, J.; Saunders, G.F. Missense mutation at the C-terminus of PAX6 negatively modulates homeodomain function. Hum. Mol. Genet. 2001, 10, 911–918. [Google Scholar] [CrossRef]
- Jordan, T.; Hanson, I.; Zaletayev, D.; Hodgson, S.; Prosser, J.; Seawright, A.; Hastie, N.; van Heyningen, V. The human PAX6 gene is mutated in two patients with aniridia. Nat. Genet. 1992, 1, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Solomon, B.D.; Pineda-Alvarez, D.E.; Balog, J.Z.; Hadley, D.; Gropman, A.L.; Nandagopal, R.; Han, J.C.; Hahn, J.S.; Blain, D.; Brooks, B.; et al. Compound heterozygosity for mutations in PAX6 in a patient with complex brain anomaly, neonatal diabetes mellitus, and microophthalmia. Am. J. Med. Genet. Part A 2009, 149A, 2543–2546. [Google Scholar] [CrossRef] [PubMed]
- Mirzayans, F.; Pearce, W.G.; MacDonald, I.M.; Walter, M.A. Mutation of the PAX6 gene in patients with autosomal dominant keratitis. Am. J. Hum. Genet. 1995, 57, 539–548. [Google Scholar] [PubMed]
- Hanson, I.M.; Fletcher, J.M.; Jordan, T.; Brown, A.; Taylor, D.; Adams, R.J.; Punnett, H.H.; van Heyningen, V. Mutations at the PAX6 locus are found in heterogeneous anterior segment malformations including Peters’ anomaly. Nat. Genet. 1994, 6, 168–173. [Google Scholar] [CrossRef]
- Robson, E.J.D.; He, S.-J.; Eccles, M.R. A PANorama of PAX genes in cancer and development. Nat. Rev. Cancer 2006, 6, 52–62. [Google Scholar] [CrossRef]
- Huang, J.; Tan, Z.-R.; Yu, J.; Li, H.; Lv, Q.-L.; Shao, Y.-Y.; Zhou, H.-H. DNA hypermethylated status and gene expression of PAX1/SOX1 in patients with colorectal carcinoma. OncoTargets Ther. 2017, 10, 4739–4751. [Google Scholar] [CrossRef]
- Kan, Y.-Y.; Liou, Y.-L.; Wang, H.-J.; Chen, C.-Y.; Sung, L.-C.; Chang, C.-F.; Liao, C.-I. PAX1 methylation as a potential biomarker for cervical cancer screening. Int. J. Gynecol. Cancer 2014, 24, 928–934. [Google Scholar] [CrossRef]
- Gerber, J.-K.; Richter, T.; Kremmer, E.; Adamski, J.; Höfler, H.; Balling, R.; Peters, H. Progressive loss of PAX9 expression correlates with increasing malignancy of dysplastic and cancerous epithelium of the human oesophagus. J. Pathol. 2002, 197, 293–297. [Google Scholar] [CrossRef]
- Tung, C.S.; Mok, S.C.; Tsang, Y.T.M.; Zu, Z.; Song, H.; Liu, J.; Deavers, M.T.; Malpica, A.; Wolf, J.K.; Lu, K.H.; et al. PAX2 expression in low malignant potential ovarian tumors and low-grade ovarian serous carcinomas. Mod. Pathol. 2009, 22, 1243–1250. [Google Scholar] [CrossRef]
- Ryan, G.; Steele-Perkins, V.; Morris, J.F.; Rauscher, F.J., III; Dressler, G.R. Repression of Pax-2 by WT1 during normal kidney development. Development 1995, 121, 867–875. [Google Scholar] [CrossRef] [PubMed]
- Dressler, G.R.; Douglass, E.C. Pax-2 is a DNA-binding protein expressed in embryonic kidney and Wilms tumor. Proc. Natl. Acad. Sci. USA 1992, 89, 1179–1183. [Google Scholar] [CrossRef]
- Busslinger, M.; Klix, N.; Pfeffer, P.; Graninger, P.G.; Kozmik, Z. Deregulation of PAX-5 by translocation of the Emu enhancer of the IgH locus adjacent to two alternative PAX-5 promoters in a diffuse large-cell lymphoma. Proc. Natl. Acad. Sci. USA 1996, 93, 6129–6134. [Google Scholar] [CrossRef] [PubMed]
- Iida, S.; Rao, P.H.; Nallasivam, P.; Hibshoosh, H.; Butler, M.; Louie, D.C.; Dyomin, V.; Ohno, H.; Chaganti, R.S.; Dalla-Favera, R. The t(9;14)(p13;q32) chromosomal translocation associated with lymphoplasmacytoid lymphoma involves the PAX-5 gene. Blood 1996, 88, 4110–4117. [Google Scholar] [CrossRef] [PubMed]
- Morrison, A.M.; Jäger, U.; Chott, A.; Schebesta, M.; Haas, O.A.; Busslinger, M. Deregulated PAX-5 transcription from a translocated IgH promoter in marginal zone lymphoma. Blood 1998, 92, 3865–3878. [Google Scholar] [CrossRef]
- Bousquet, M.; Broccardo, C.; Quelen, C.; Meggetto, F.; Kuhlein, E.; Delsol, G.; Dastugue, N.; Brousset, P. A novel PAX5-ELN fusion protein identified in B-cell acute lymphoblastic leukemia acts as a dominant negative on wild-type PAX5. Blood 2007, 109, 3417–3423. [Google Scholar] [CrossRef] [PubMed]
- Cazzaniga, G.; Daniotti, M.; Tosi, S.; Giudici, G.; Aloisi, A.; Pogliani, E.; Kearney, L.; Biondi, A. The paired box domain gene PAX5 is fused to ETV6/TEL in an acute lymphoblastic leukemia case. Cancer Res. 2001, 61, 4666–4670. [Google Scholar]
- Mullighan, C.G.; Goorha, S.; Radtke, I.; Miller, C.B.; Coustan-Smith, E.; Dalton, J.D.; Girtman, K.; Mathew, S.; Ma, J.; Pounds, S.B.; et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007, 446, 758–764. [Google Scholar] [CrossRef] [PubMed]
- Nebral, K.; König, M.; Harder, L.; Siebert, R.; Haas, O.A.; Strehl, S. Identification of PML as novel PAX5 fusion partner in childhood acute lymphoblastic leukaemia. Br. J. Haematol. 2007, 139, 269–274. [Google Scholar] [CrossRef]
- Kroll, T.G.; Sarraf, P.; Pecciarini, L.; Chen, C.J.; Mueller, E.; Spiegelman, B.M.; Fletcher, J.A. PAX8-PPARgamma1 fusion oncogene in human thyroid carcinoma [corrected]. Science 2000, 289, 1357–1360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, J.; Grachtchouk, V.; Qin, T.; Lumeng, C.N.; Sartor, M.A.; Koenig, R.J. Genomic binding of PAX8-PPARG fusion protein regulates cancer-related pathways and alters the immune landscape of thyroid cancer. Oncotarget 2017, 8, 5761–5773. [Google Scholar] [CrossRef]
- Poleev, A.; Fickenscher, H.; Mundlos, S.; Winterpacht, A.; Zabel, B.; Fidler, A.; Gruss, P.; Plachov, D. PAX8, a human paired box gene: Isolation and expression in developing thyroid, kidney and Wilms’ tumors. Development 1992, 116, 611–623. [Google Scholar] [CrossRef]
- Galili, N.; Davis, R.J.; Fredericks, W.J.; Mukhopadhyay, S.; Rauscher, F.J.; Emanuel, B.S.; Rovera, G.; Barr, F.G. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat. Genet. 1993, 5, 230–235. [Google Scholar] [CrossRef]
- Fredericks, W.J.; Galili, N.; Mukhopadhyay, S.; Rovera, G.; Bennicelli, J.; Barr, F.G.; Rauscher, F.J. The PAX3-FKHR fusion protein created by the t(2;13) translocation in alveolar rhabdomyosarcomas is a more potent transcriptional activator than PAX3. Mol. Cell. Biol. 1995, 15, 1522–1535. [Google Scholar] [CrossRef]
- Scholl, F.A.; Kamarashev, J.; Murmann, O.V.; Geertsen, R.; Dummer, R.; Schäfer, B.W. PAX3 is expressed in human melanomas and contributes to tumor cell survival. Cancer Res. 2001, 61, 823–826. [Google Scholar]
- Davis, R.J.; D’Cruz, C.M.; Lovell, M.A.; Biegel, J.A.; Barr, F.G. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994, 54, 2869–2872. [Google Scholar]
- Miyamoto, T.; Kakizawa, T.; Ichikawa, K.; Nishio, S.; Kajikawa, S.; Hashizume, K. Expression of dominant negative form of PAX4 in human insulinoma. Biochem. Biophys. Res. Commun. 2001, 282, 34–40. [Google Scholar] [CrossRef]
- Lang, D.; Mascarenhas, J.B.; Powell, S.K.; Halegoua, J.; Nelson, M.; Ruggeri, B.A. PAX6 is expressed in pancreatic adenocarcinoma and is downregulated during induction of terminal differentiation. Mol. Carcinog. 2008, 47, 148–156. [Google Scholar] [CrossRef]
- Zhou, Y.-H.; Tan, F.; Hess, K.R.; Yung, W.K.A. The expression of PAX6, PTEN, vascular endothelial growth factor, and epidermal growth factor receptor in gliomas: Relationship to tumor grade and survival. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2003, 9, 3369–3375. [Google Scholar]
- Liu, Y.; Cui, S.; Li, W.; Zhao, Y.; Yan, X.; Xu, J. PAX3 is a biomarker and prognostic factor in melanoma: Database mining. Oncol. Lett. 2019, 17, 4985–4993. [Google Scholar] [CrossRef]
- Watanabe, A.; Takeda, K.; Ploplis, B.; Tachibana, M. Epistatic relationship between Waardenburg Syndrome genes MITF and PAX3. Nat. Genet. 1998, 18, 283–286. [Google Scholar] [CrossRef]
- Smith, M.P.; Brunton, H.; Rowling, E.J.; Ferguson, J.; Arozarena, I.; Miskolczi, Z.; Lee, J.L.; Girotti, M.R.; Marais, R.; Levesque, M.P.; et al. Inhibiting Drivers of Non-mutational Drug Tolerance Is a Salvage Strategy for Targeted Melanoma Therapy. Cancer Cell 2016, 29, 270–284. [Google Scholar] [CrossRef] [PubMed]
- Li, H.G.; Wang, Q.; Li, H.M.; Kumar, S.; Parker, C.; Slevin, M.; Kumar, P. PAX3 and PAX3-FKHR promote rhabdomyosarcoma cell survival through downregulation of PTEN. Cancer Lett. 2007, 253, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Sefidbakht, S.; Khorsand, A.; Omidi, S.; Mohsenpourian, S.; Mirzaian, E. Expression of PAX2 and PAX8 in Wilms Tumor: A Tissue Microarray-based Immunohistochemical Study. Iran. J. Pathol. 2021, 16, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Dehbi, M.; Pelletier, J. PAX8-mediated activation of the wt1 tumor suppressor gene. EMBO J. 1996, 15, 4297–4306. [Google Scholar] [CrossRef] [PubMed]
- Dressler, G.R. Pax-2, kidney development, and oncogenesis. Med. Pediatr. Oncol. 1996, 27, 440–444. [Google Scholar] [CrossRef]
- Ohno, H.; Ueda, C.; Akasaka, T. The t(9;14)(p13;q32) translocation in B-cell non-Hodgkin’s lymphoma. Leuk. Lymphoma 2000, 36, 435–445. [Google Scholar] [CrossRef]
- Fazio, G.; Bresolin, S.; Silvestri, D.; Quadri, M.; Saitta, C.; Vendramini, E.; Buldini, B.; Palmi, C.; Bardini, M.; Grioni, A.; et al. PAX5 fusion genes are frequent in poor risk childhood acute lymphoblastic leukaemia and can be targeted with BIBF1120. eBioMedicine 2022, 83, 104224. [Google Scholar] [CrossRef]
- Poppe, B.; De Paepe, P.; Michaux, L.; Dastugue, N.; Bastard, C.; Herens, C.; Moreau, E.; Cavazzini, F.; Yigit, N.; Van Limbergen, H.; et al. PAX5/IGH rearrangement is a recurrent finding in a subset of aggressive B-NHL with complex chromosomal rearrangements. Genes Chromosomes Cancer 2005, 44, 218–223. [Google Scholar] [CrossRef]
- Skapek, S.X.; Anderson, J.; Barr, F.G.; Bridge, J.A.; Gastier-Foster, J.M.; Parham, D.M.; Rudzinski, E.R.; Triche, T.; Hawkins, D.S. PAX-FOXO1 fusion status drives unfavorable outcome for children with rhabdomyosarcoma: A children’s oncology group report: PAX-FOXO1 Influences Survival in RMS. Pediatr. Blood Cancer 2013, 60, 1411–1417. [Google Scholar] [CrossRef]
- Bennicelli, J.L.; Edwards, R.H.; Barr, F.G. Mechanism for transcriptional gain of function resulting from chromosomal translocation in alveolar rhabdomyosarcoma. Proc. Natl. Acad. Sci. USA 1996, 93, 5455–5459. [Google Scholar] [CrossRef]
- Gryder, B.E.; Yohe, M.E.; Chou, H.-C.; Zhang, X.; Marques, J.; Wachtel, M.; Schaefer, B.; Sen, N.; Song, Y.; Gualtieri, A.; et al. PAX3–FOXO1 Establishes Myogenic Super Enhancers and Confers BET Bromodomain Vulnerability. Cancer Discov. 2017, 7, 884–899. [Google Scholar] [CrossRef]
- Böhm, M.; Wachtel, M.; Marques, J.G.; Streiff, N.; Laubscher, D.; Nanni, P.; Mamchaoui, K.; Santoro, R.; Schäfer, B.W. Helicase CHD4 is an epigenetic coregulator of PAX3-FOXO1 in alveolar rhabdomyosarcoma. J. Clin. Investig. 2016, 126, 4237–4249. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.J.; Barr, F.G. Fusion genes resulting from alternative chromosomal translocations are overexpressed by gene-specific mechanisms in alveolar rhabdomyosarcoma. Proc. Natl. Acad. Sci. USA 1997, 94, 8047–8051. [Google Scholar] [CrossRef] [PubMed]
- Raman, P.; Koenig, R.J. PAX8-PPARγ fusion protein in thyroid carcinoma. Nat. Rev. Endocrinol. 2014, 10, 616–623. [Google Scholar] [CrossRef]
- Au, A.Y.M.; McBride, C.; Wilhelm, K.G.; Koenig, R.J.; Speller, B.; Cheung, L.; Messina, M.; Wentworth, J.; Tasevski, V.; Learoyd, D.; et al. PAX8-peroxisome proliferator-activated receptor gamma (PPARgamma) disrupts normal PAX8 or PPARgamma transcriptional function and stimulates follicular thyroid cell growth. Endocrinology 2006, 147, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Hassan, Z.K.; Hafez, M.M.; Kamel, M.M.; Zekri, A.R.N. Human Papillomavirus Genotypes and Methylation of CADM1, PAX1, MAL and ADCYAP1 Genes in Epithelial Ovarian Cancer Patients. Asian Pac. J. Cancer Prev. 2017, 18, 169–176. [Google Scholar] [CrossRef]
- Zhou, Y.-H.; Wu, X.; Tan, F.; Shi, Y.-X.; Glass, T.; Liu, T.J.; Wathen, K.; Hess, K.R.; Gumin, J.; Lang, F.; et al. PAX6 suppresses growth of human glioblastoma cells. J. Neurooncol. 2005, 71, 223–229. [Google Scholar] [CrossRef]
- Mascarenhas, J.B.; Young, K.P.; Littlejohn, E.L.; Yoo, B.K.; Salgia, R.; Lang, D. PAX6 Is Expressed in Pancreatic Cancer and Actively Participates in Cancer Progression through Activation of the MET Tyrosine Kinase Receptor Gene. J. Biol. Chem. 2009, 284, 27524–27532. [Google Scholar] [CrossRef]
- Liu, J.; Perumal, N.B.; Oldfield, C.J.; Su, E.W.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder in transcription factors. Biochemistry 2006, 45, 6873–6888. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Piovesan, D.; Del Conte, A.; Clementel, D.; Monzon, A.M.; Bevilacqua, M.; Aspromonte, M.C.; Iserte, J.A.; Orti, F.E.; Marino-Buslje, C.; Tosatto, S.C.E. MobiDB: 10 years of intrinsically disordered proteins. Nucleic Acids Res. 2023, 51, D438–D444. [Google Scholar] [CrossRef] [PubMed]
- Bradford, S.T.J.; Grimley, E.; Laszczyk, A.M.; Lee, P.H.; Patel, S.R.; Dressler, G.R. Identification of Pax protein inhibitors that suppress target gene expression and cancer cell proliferation. Cell Chem. Biol. 2022, 29, 412–422.e4. [Google Scholar] [CrossRef]
- Abraham, J.; Nishijo, K.; Huang, E.T.; Prajapati, S.I.; Walker, R.L.; Davis, S.; Rebeles, J.; Wiebush, H.; McCleish, A.T.; Hampton, S.T.; et al. Lineage of origin in rhabdomyosarcoma informs pharmacological response. Genes Dev. 2014, 28, 1578–1591. [Google Scholar] [CrossRef] [PubMed]
- Ghayad, S.E.; Rammal, G.; Sarkis, O.; Basma, H.; Ghamloush, F.; Fahs, A.; Karam, M.; Harajli, M.; Rabeh, W.; Mouawad, J.E.; et al. The histone deacetylase inhibitor Suberoylanilide Hydroxamic Acid (SAHA) as a therapeutic agent in rhabdomyosarcoma. Cancer Biol. Ther. 2019, 20, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Herrero Martín, D.; Boro, A.; Schäfer, B.W. Cell-Based Small-Molecule Compound Screen Identifies Fenretinide as Potential Therapeutic for Translocation-Positive Rhabdomyosarcoma. PLoS ONE 2013, 8, e55072. [Google Scholar] [CrossRef]
- Grimley, E.; Liao, C.; Ranghini, E.J.; Nikolovska-Coleska, Z.; Dressler, G.R. Inhibition of Pax2 Transcription Activation with a Small Molecule that Targets the DNA Binding Domain. ACS Chem. Biol. 2017, 12, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, K.; Shaw, T.; Song, Y.K.; Kouassi-Brou, M.; Molotkova, A.; Tiwari, P.B.; Chou, H.-C.; Wen, X.; Wei, J.S.; Deniz, E.; et al. Piperacetazine Directly Binds to the PAX3::FOXO1 Fusion Protein and Inhibits Its Transcriptional Activity. Cancer Res. Commun. 2023, 3, 2030–2043. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, C. Identification of a new class of PAX3-FKHR target promoters: A role of the Pax3 paired box DNA binding domain. Oncogene 2007, 26, 1595–1605. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Sinhari, A.; Jain, P.; Jadhav, H.R. A perspective on oligonucleotide therapy: Approaches to patient customization. Front. Pharmacol. 2022, 13, 1006304. [Google Scholar] [CrossRef]
- Mackall, C.L.; Rhee, E.H.; Read, E.J.; Khuu, H.M.; Leitman, S.F.; Bernstein, D.; Tesso, M.; Long, L.M.; Grindler, D.; Merino, M.; et al. A pilot study of consolidative immunotherapy in patients with high-risk pediatric sarcomas. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 4850–4858. [Google Scholar] [CrossRef]


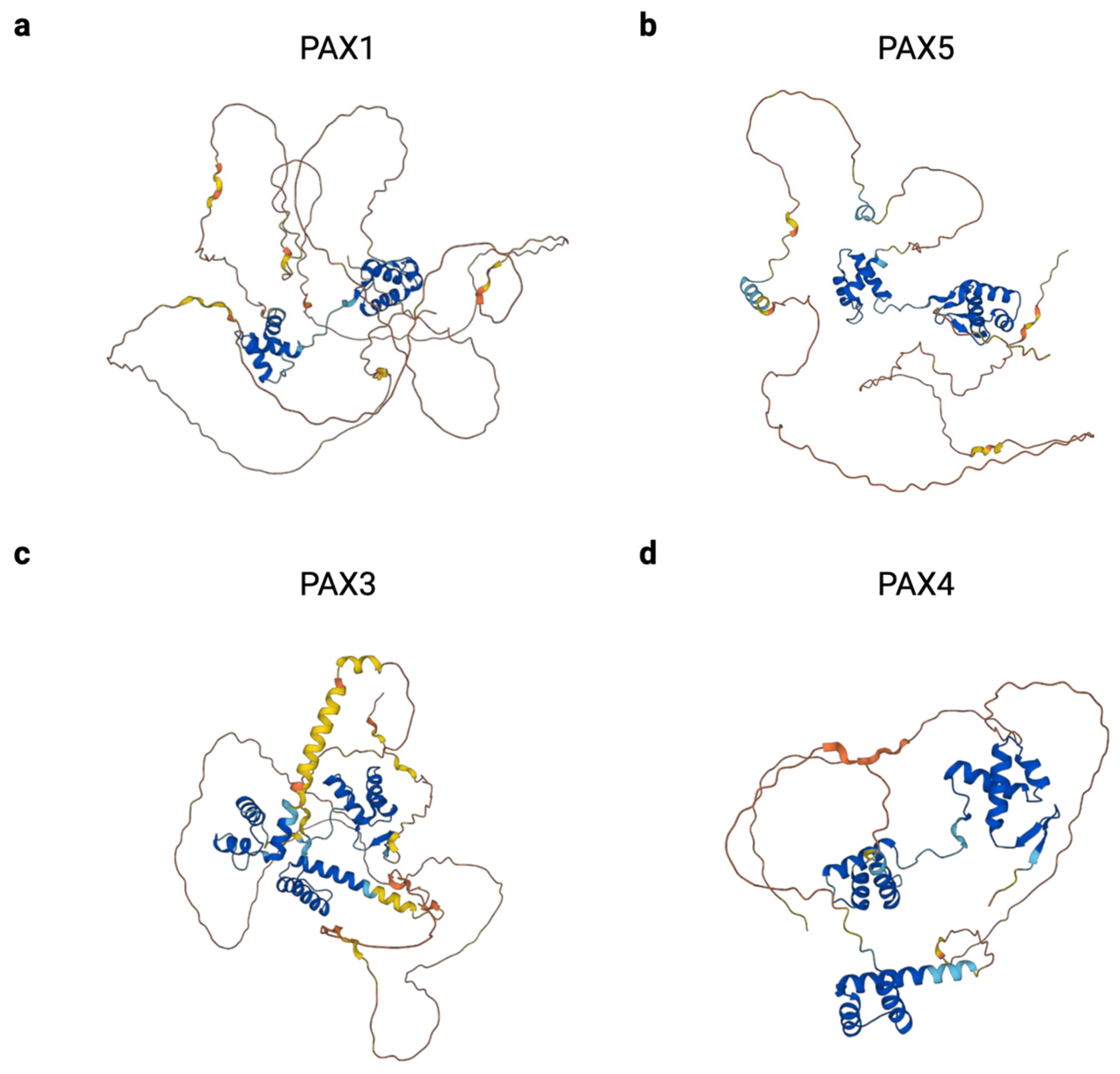
| Subgroup | Gene | Isoforms | Longest Isoform (# of Amino Acids) | Shortest Isoform (# of Amino Acids) |
|---|---|---|---|---|
| Group I | PAX1 | 1, 2 | 1 (534) | 2 (457) |
| PAX9 | - | 341 | - | |
| Group II | PAX2 | a–g (7) | e (432) | g (102) |
| PAX5 | 1–11 | 1 (391) | 6 (220) | |
| PAX8 | A, C, D, E (4) | A (450) | E (287) | |
| Group III | PAX3 | a, b, c, d, e, g, h, i | e (505) | b (206) |
| PAX7 | 1–3 | 1 (520) | 3 (505) | |
| Group IV | PAX4 | 1, 2 | 1 (351) | 2 (348) |
| PAX6 | a–o (15) | e (503) | o (221) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaw, T.; Barr, F.G.; Üren, A. The PAX Genes: Roles in Development, Cancer, and Other Diseases. Cancers 2024, 16, 1022. https://doi.org/10.3390/cancers16051022
Shaw T, Barr FG, Üren A. The PAX Genes: Roles in Development, Cancer, and Other Diseases. Cancers. 2024; 16(5):1022. https://doi.org/10.3390/cancers16051022
Chicago/Turabian StyleShaw, Taryn, Frederic G. Barr, and Aykut Üren. 2024. "The PAX Genes: Roles in Development, Cancer, and Other Diseases" Cancers 16, no. 5: 1022. https://doi.org/10.3390/cancers16051022
APA StyleShaw, T., Barr, F. G., & Üren, A. (2024). The PAX Genes: Roles in Development, Cancer, and Other Diseases. Cancers, 16(5), 1022. https://doi.org/10.3390/cancers16051022






