Role of Post-Translational Modifications in Colorectal Cancer Metastasis
Abstract
Simple Summary
Abstract
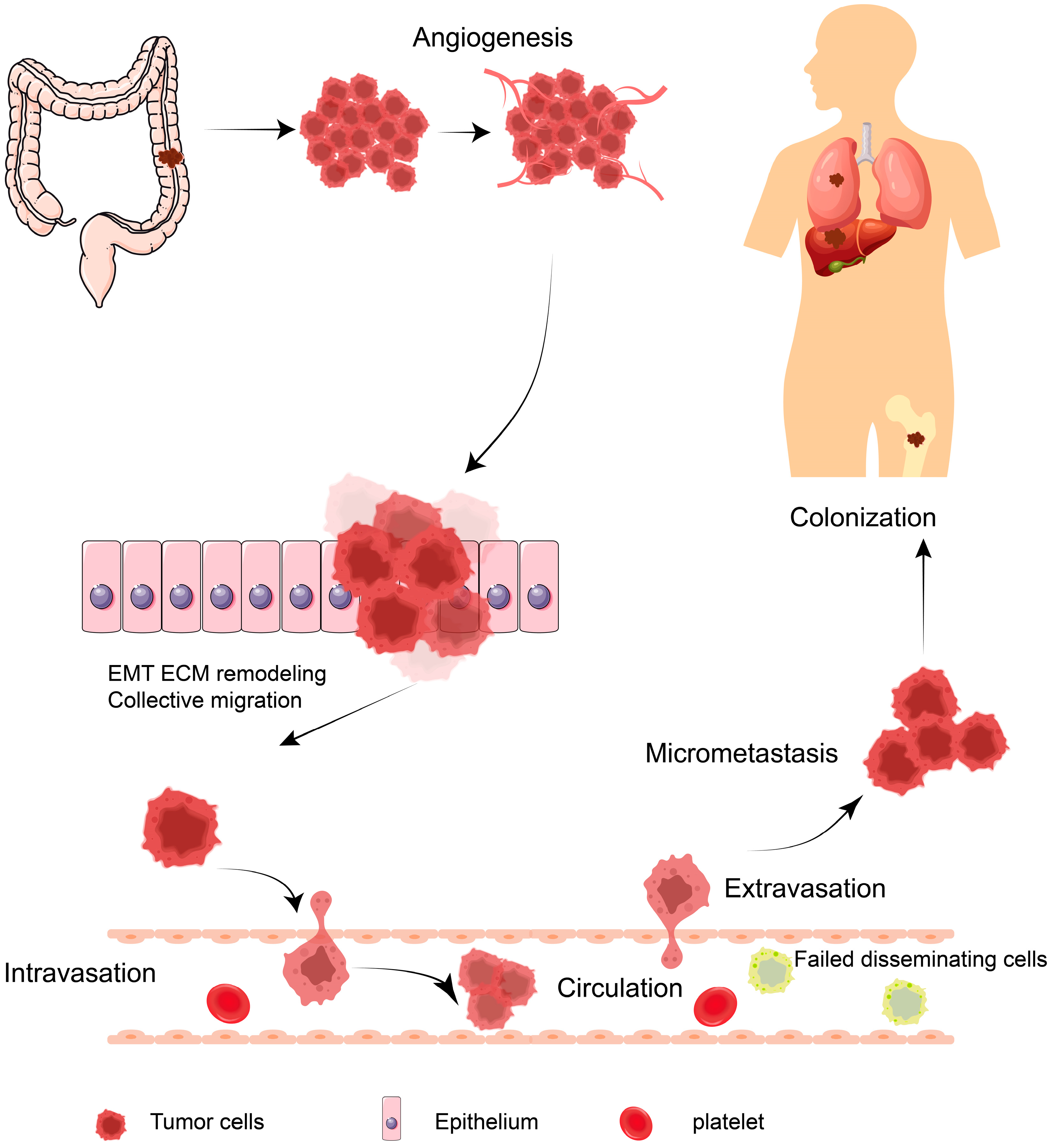
1. Introduction
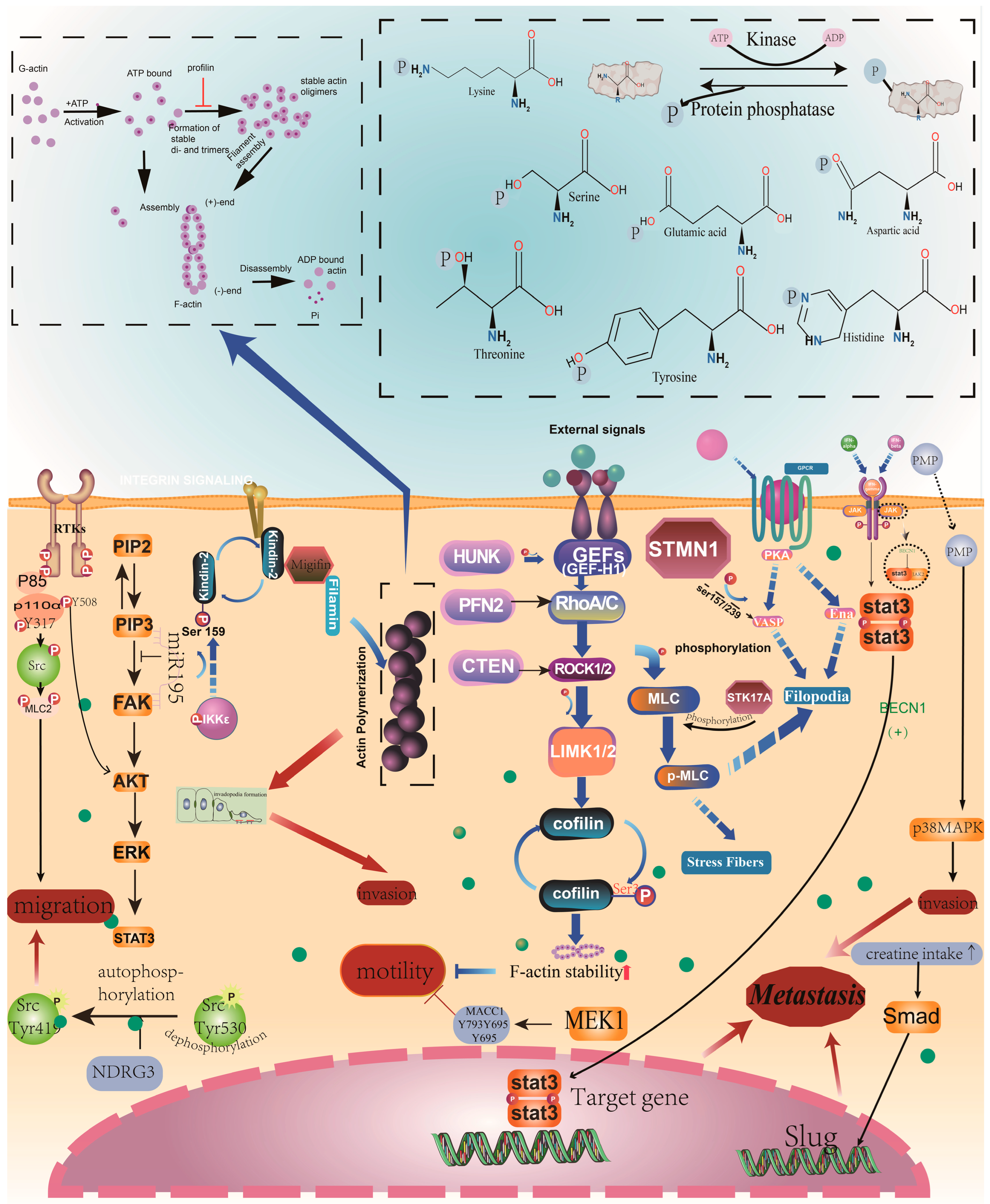
2. Phosphorylation and CRC Metastasis
2.1. Phosphorylation Regulates Cytoskeleton Rearrangement
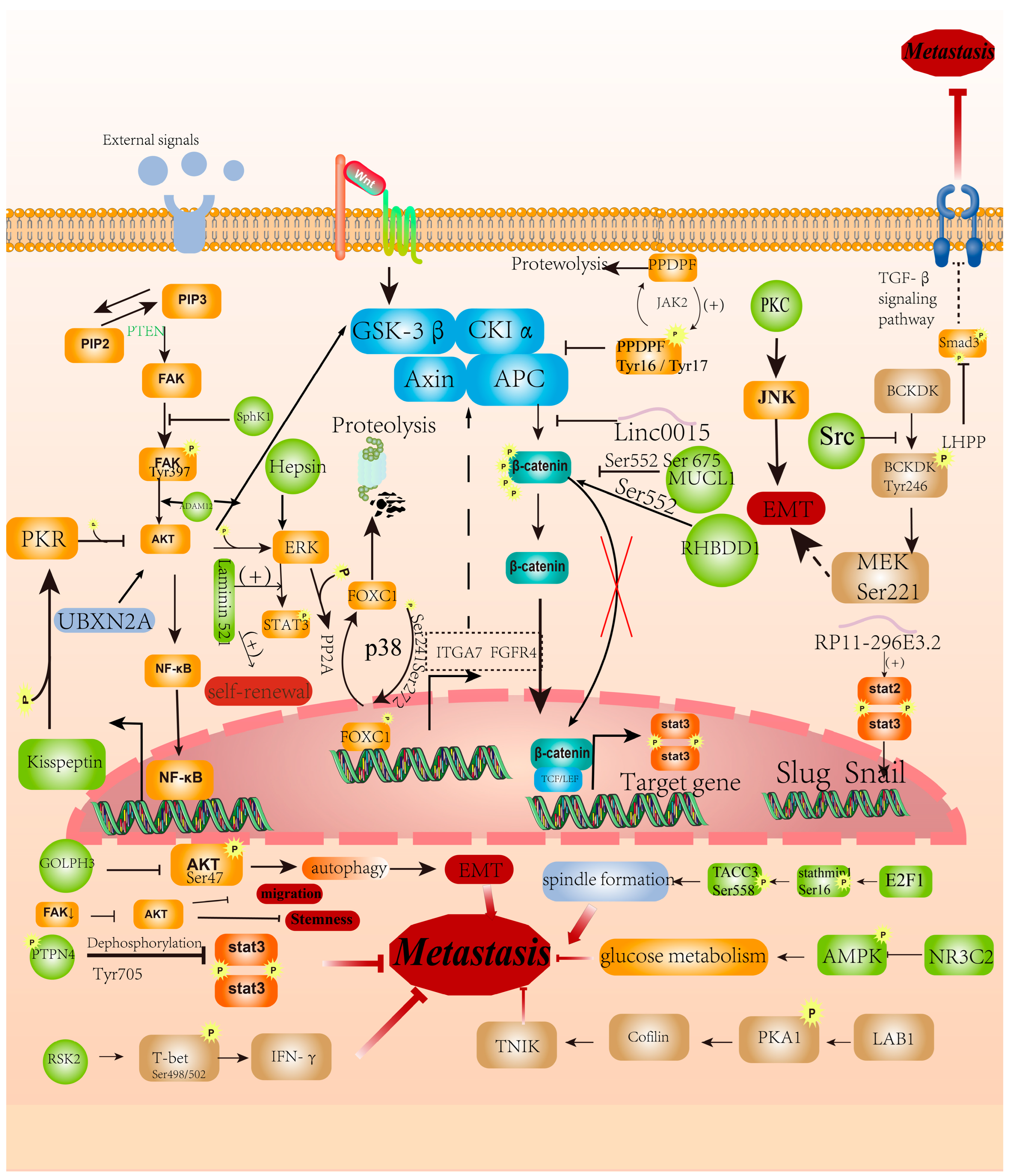
2.2. Phosphorylation Regulates EMT
2.3. Phosphorylation Regulates Invasion and Migration
2.4. Phosphorylation Regulates Other Characteristics of CRC Cells
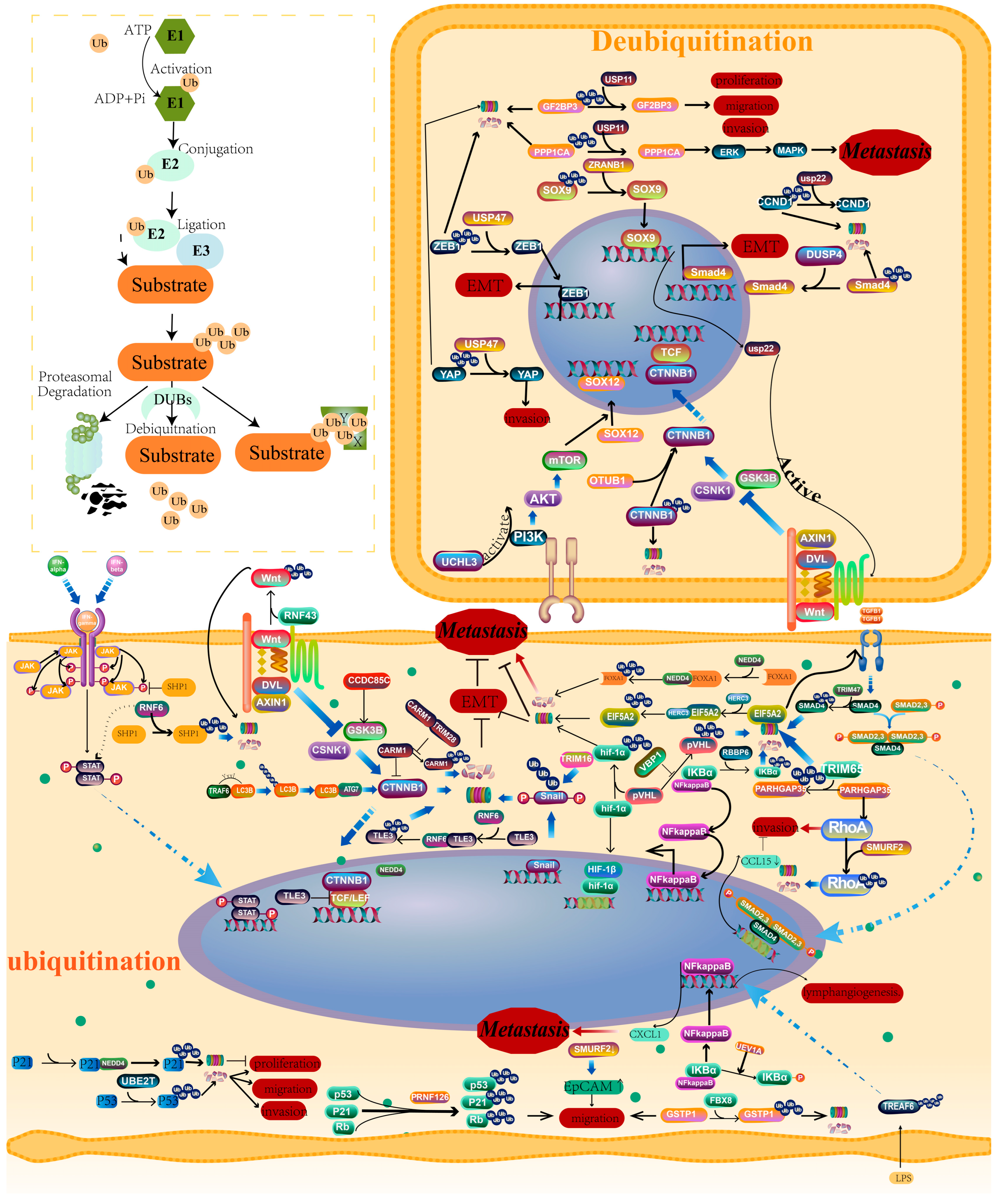
3. Ubiquitination/Deubiquitination and CRC Metastasis
3.1. Ubiquitination Regulates EMT
3.2. Ubiquitination Regulates Invasion and Migration
3.3. Ubiquitination Regulates Stemness
3.4. Ubiquitination Regulates Lymph Node Metastasis
3.5. Ubiquitin-Conjugating E2 Enzymes and CRC Metastasis
3.6. Deubiquitination and CRC Metastasis
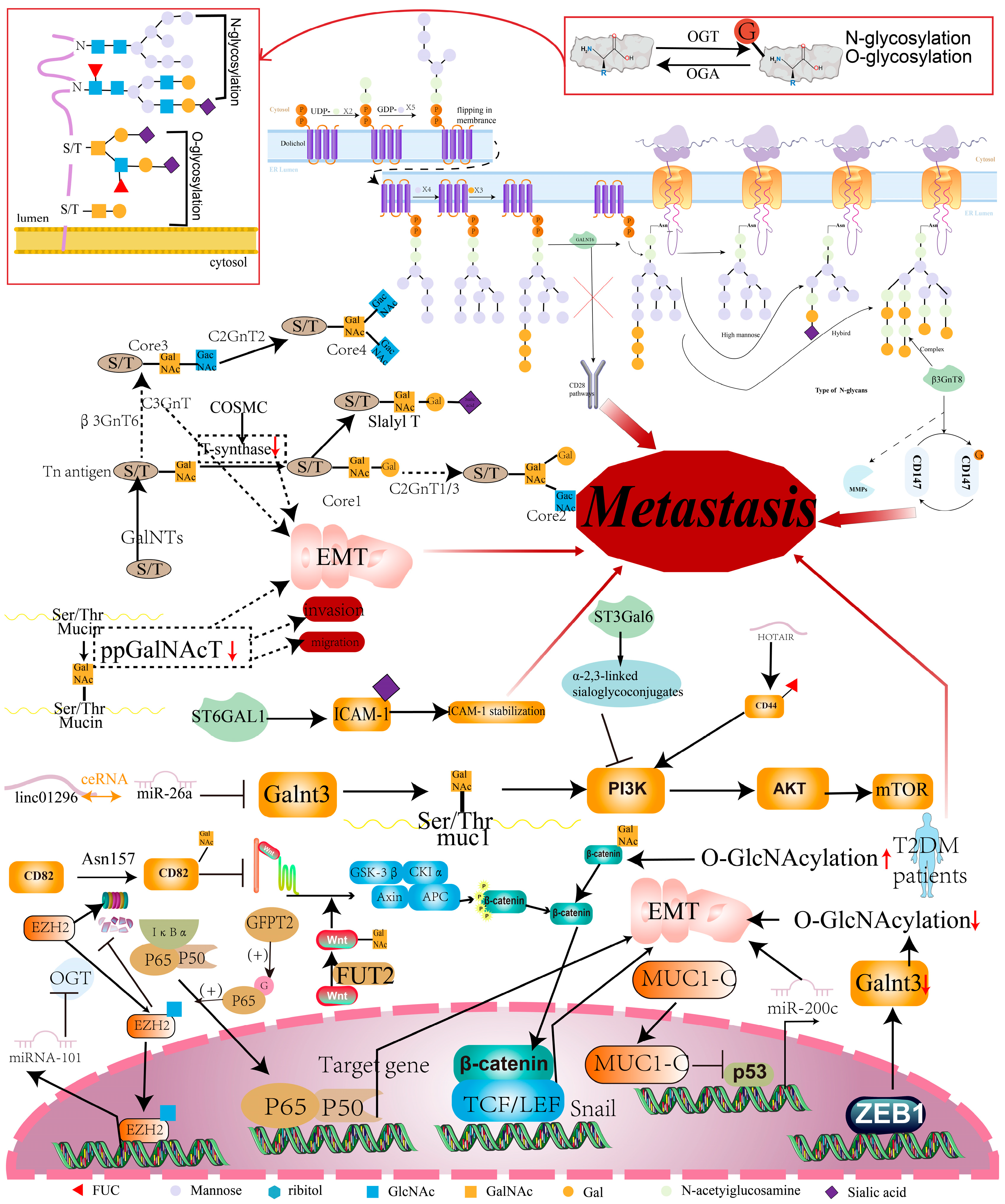
4. Glycosylation and CRC Metastasis
4.1. Glycosaminyltransferase and CRC Metastasis
4.2. Glycosylation of Key Proteins and CRC Metastasis
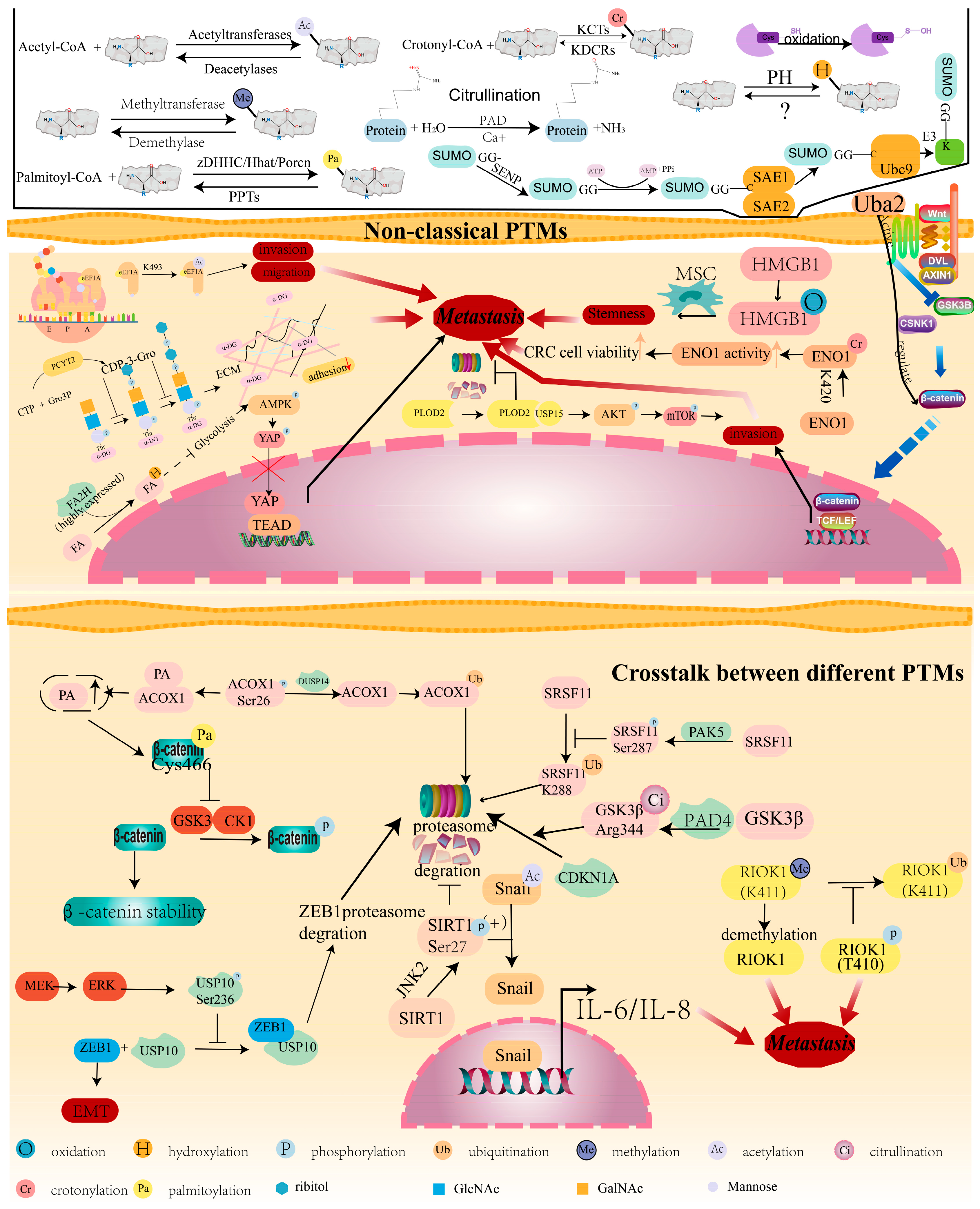
5. Non-Classical PTMs and CRC Metastasis
Crosstalk between Different PTMs Contributes to CRC Metastasis
6. Therapeutic Potential of Targeting PTMs to Inhibit CRC Metastasis
6.1. Drugs Targeting Phosphorylation
6.2. Drugs Targeting Glycosylation
6.3. Drugs Targeting Ubiquitination
7. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Jiang, Y.; Yuan, H.; Li, Z.; Ji, X.; Shen, Q.; Tuo, J.; Bi, J.; Li, H.; Xiang, Y. Global pattern and trends of colorectal cancer survival: A systematic review of population-based registration data. Cancer Biol. Med. 2021, 19, 175–186. [Google Scholar] [CrossRef]
- Dulskas, A.; Gaizauskas, V.; Kildusiene, I.; Samalavicius, N.E.; Smailyte, G. Improvement of Survival over Time for Colorectal Cancer Patients: A Population-Based Study. J. Clin. Med. 2020, 9, 4038. [Google Scholar] [CrossRef]
- Pretzsch, E.; Bosch, F.; Neumann, J.; Ganschow, P.; Bazhin, A.; Guba, M.; Werner, J.; Angele, M. Mechanisms of Metastasis in Colorectal Cancer and Metastatic Organotropism: Hematogenous versus Peritoneal Spread. J. Oncol. 2019, 2019, 7407190. [Google Scholar] [CrossRef]
- Chandra, R.; Karalis, J.D.; Liu, C.R.; Murimwa, G.Z.; Park, J.V.; Heid, C.A.; Reznik, S.I.; Huang, E.M.A.; Minna, J.D.; Brekken, R.A. The Colorectal Cancer Tumor Microenvironment and Its Impact on Liver and Lung Metastasis. Cancers 2021, 13, 6206. [Google Scholar] [CrossRef]
- Shasha, T.; Gruijs, M.; van Egmond, M. Mechanisms of colorectal liver metastasis development. Cell. Mol. Life Sci. CMLS 2022, 79, 607. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T.; Garneau-Tsodikova, S.; Gatto, G.J., Jr. Protein posttranslational modifications: The chemistry of proteome diversifications. Angew. Chem. Int. Ed. Engl. 2005, 44, 7342–7372. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Agar, J.N.; Amster, I.J.; Baker, M.S.; Bertozzi, C.R.; Boja, E.S.; Costello, C.E.; Cravatt, B.F.; Fenselau, C.; Garcia, B.A.; et al. How many human proteoforms are there? Nat. Chem. Biol. 2018, 14, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.D.; Gevaert, K.; De Smet, I. Protein Language: Post-Translational Modifications Talking to Each Other. Trends Plant Sci. 2018, 23, 1068–1080. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, L.; Liu, M.; Luo, J. Protein post-translational modifications in the regulation of cancer hallmarks. Cancer Gene Ther. 2022, 30, 529–547. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, F.; Zhang, X.; Lin, H.K.; Xu, C. Insights into the post-translational modification and its emerging role in shaping the tumor microenvironment. Signal Transduct. Target. Ther. 2021, 6, 422. [Google Scholar] [CrossRef]
- Senga, S.S.; Grose, R.P. Hallmarks of cancer-the new testament. Open Biol. 2021, 11, 200358. [Google Scholar] [CrossRef]
- Wu, Z.; Huang, R.; Yuan, L. Crosstalk of intracellular post-translational modifications in cancer. Arch. Biochem. Biophys. 2019, 676, 108138. [Google Scholar] [CrossRef]
- Pan, S.; Chen, R. Pathological implication of protein post-translational modifications in cancer. Mol. Asp. Med. 2022, 86, 101097. [Google Scholar] [CrossRef]
- Zou, Y.; Weng, J.; Rong, Y.; Tan, Y.; Chen, X.; Cai, J.; Lin, X. Mechanism of programmed cell death 1 ligand 1 promoting colorectal cancer metastasis by glycosylation modification. Chin. J. Exp. Surg. 2020, 37, 2097–2100. [Google Scholar]
- Liu, C.; Yang, Q.; Zhu, Q.; Lu, X.; Li, M.; Hou, T.; Li, Z.; Tang, M.; Li, Y.; Wang, H.; et al. CBP mediated DOT1L acetylation confers DOT1L stability and promotes cancer metastasis. Theranostics 2020, 10, 1758–1776. [Google Scholar] [CrossRef] [PubMed]
- Ohara, M.; Ohara, K.; Kumai, T.; Ohkuri, T.; Nagato, T.; Hirata-Nozaki, Y.; Kosaka, A.; Nagata, M.; Hayashi, R.; Harabuchi, S.; et al. Phosphorylated vimentin as an immunotherapeutic target against metastatic colorectal cancer. Cancer Immunol. Immunother. 2020, 69, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, C.; Yu, T.; Chen, T. Proteomic Analysis of Protein Ubiquitination Events in Human Primary and Metastatic Colon Adenocarcinoma Tissues. Front. Oncol. 2020, 10, 1684. [Google Scholar] [CrossRef] [PubMed]
- Ozlu, N.; Akten, B.; Timm, W.; Haseley, N.; Steen, H.; Steen, J.A.J. Phosphoproteomics. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 255–276. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sun, Y.D.; Yu, G.Y.; Cui, J.R.; Lou, Z.; Zhang, H.; Huang, Y.; Bai, C.G.; Deng, L.L.; Liu, P.; et al. Integrated Omics of Metastatic Colorectal Cancer. Cancer Cell 2020, 38, 734–747.e739. [Google Scholar] [CrossRef] [PubMed]
- Li, X.C.; Wang, J.L. Mechanical tumor microenvironment and transduction: Cytoskeleton mediates cancer cell invasion and metastasis. Int. J. Biol. Sci. 2020, 16, 2014–2028. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Bao, Y.; Liu, C.; Zhu, Q.; Zhao, L.; Lu, X.; Zhu, Q.; Lv, Y.; Bai, F.; Wen, H.; et al. IKKε phosphorylates kindlin-2 to induce invadopodia formation and promote colorectal cancer metastasis. Theranostics 2020, 10, 2358–2373. [Google Scholar] [CrossRef] [PubMed]
- Short, S.P.; Thompson, J.J.; Bilotta, A.J.; Chen, X.; Revetta, F.L.; Washington, M.K.; Williams, C.S. Serine Threonine Kinase 17A Maintains the Epithelial State in Colorectal Cancer Cells. Mol. Cancer Res. 2019, 17, 882–894. [Google Scholar] [CrossRef]
- Moraes Sousa-Squiavinato, A.C.; Rocha, M.R.; Barcellos-de-Souza, P.; de Souza, W.F.; Morgado-Diaz, J.A. Cofilin-1 signaling mediates epithelial-mesenchymal transition by promoting actin cytoskeleton reorganization and cell-cell adhesion regulation in colorectal cancer cells. Biochim. Biophys. Acta-Mol. Cell Res. 2019, 1866, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.T.; Chung, M.C.M. Label-Free Quantitative Phosphoproteomics Reveals Regulation of Vasodilator-Stimulated Phosphoprotein upon Stathmin-1 Silencing in a Pair of Isogenic Colorectal Cancer Cell Lines. Proteomics 2018, 18, e1700242. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, W.; Yan, J.; Zhou, K.; Wan, B.; Shi, P.; Chen, Y.; He, S.; Li, D. Loss of profilin 2 contributes to enhanced epithelial-mesenchymal transition and metastasis of colorectal cancer. Int. J. Oncol. 2018, 53, 1118–1128. [Google Scholar] [CrossRef] [PubMed]
- Asiri, A.; Toss, M.S.; Raposo, T.P.; Akhlaq, M.; Thorpe, H.; Alfahed, A.; Asiri, A.; Ilyas, M. Cten promotes Epithelial-Mesenchymal Transition (EMT) in colorectal cancer through stabilisation of Src. Pathol. Int. 2019, 69, 381–391. [Google Scholar] [CrossRef]
- Han, X.; Jiang, S.; Gu, Y.; Ding, L.; Zhao, E.; Cao, D.; Wang, X.; Wen, Y.; Pan, Y.; Yan, X.; et al. HUNK inhibits epithelial-mesenchymal transition of CRC via direct phosphorylation of GEF-H1 and activating RhoA/LIMK-1/CFL-1. Cell Death Dis. 2023, 14, 327. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Du, G.S.; Qiu, Y.; Wang, W.S.; Peng, K.; Zhang, Z.C.; Li, X.S.; Xiao, W.D.; Yang, H. Knockdown on aPKC-ι inhibits epithelial-mesenchymal transition, migration and invasion of colorectal cancer cells through Rac1-JNK pathway. Exp. Mol. Pathol. 2019, 107, 57–67. [Google Scholar] [CrossRef]
- Liu, S.-Q.; Xu, C.-Y.; Wu, W.-H.; Fu, Z.-H.; He, S.-W.; Qin, M.-B.; Huang, J.-A. Sphingosine kinase 1 promotes the metastasis of colorectal cancer by inducing the epithelial-mesenchymal transition mediated by the FAK/AKT/MMPs axis. Int. J. Oncol. 2019, 54, 41–52. [Google Scholar] [CrossRef]
- Tian, Q.; Yuan, P.; Quan, C.; Li, M.; Xiao, J.; Zhang, L.; Lu, H.; Ma, T.; Zou, L.; Wang, F.; et al. Phosphorylation of BCKDK of BCAA catabolism at Y246 by Src promotes metastasis of colorectal cancer. Oncogene 2020, 39, 3980–3996. [Google Scholar] [CrossRef]
- Zhang, B.-D.; Li, Y.-R.; Ding, L.-D.; Wang, Y.-Y.; Liu, H.-Y.; Jia, B.-Q. Loss of PTPN4 activates STAT3 to promote the tumor growth in rectal cancer. Cancer Sci. 2019, 110, 2258–2272. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Miao, F.; Huang, R.; Liu, W.; Zhao, Y.; Jiao, T.; Lu, Y.; Wu, F.; Wang, X.; Wang, H.; et al. RHBDD1 promotes colorectal cancer metastasis through the Wnt signaling pathway and its downstream target ZEB1. J. Exp. Clin. Cancer Res. 2018, 37, 22. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.; Peng, C.; Zhang, Y.; Zykova, T.A.; Lee, M.H.; Lee, S.Y.; Rao, E.; Chen, H.; Ryu, J.; Wang, L.; et al. RSK2 phosphorylates T-bet to attenuate colon cancer metastasis and growth. Proc. Natl. Acad. Sci. USA 2017, 114, 12791–12796. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.H.; Park, Y.L.; Park, S.Y.; Joo, Y.E. A disintegrin and metalloprotease 12 contributes to colorectal cancer metastasis by regulating epithelial-mesenchymal transition. Int. J. Oncol. 2023, 62, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liao, Y.; Chen, C.; Sun, W.; Sun, X.; Liu, Y.; Xu, E.; Lai, M.; Zhang, H. p38-regulated FOXC1 stability is required for colorectal cancer metastasis. J. Pathol. 2020, 250, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, Z.; Li, X.; Chen, J.; Wang, G.; Tian, Z.; Qian, M.; Chen, Z.; Guo, H.; Tang, G.; et al. Forkhead box C1 promotes colorectal cancer metastasis through transactivating ITGA7 and FGFR4 expression. Oncogene 2018, 37, 5477–5491. [Google Scholar] [CrossRef] [PubMed]
- Abdulla, M.; Traiki, T.B.; Vaali-Mohammed, M.A.; El-Wetidy, M.S.; Alhassan, N.; Al-Khayal, K.; Zubaidi, A.; Al-Obeed, O.; Ahmad, R. Targeting MUCL1 protein inhibits cell proliferation and EMT by deregulating β-catenin and increases irinotecan sensitivity in colorectal cancer. Int. J. Oncol. 2022, 60, 1–12. [Google Scholar] [CrossRef]
- Hou, B.; Li, W.; Xia, P.; Zhao, F.; Liu, Z.; Zeng, Q.; Wang, S.; Chang, D. LHPP suppresses colorectal cancer cell migration and invasion in vitro and in vivo by inhibiting Smad3 phosphorylation in the TGF-beta pathway. Cell Death Discov. 2021, 7, 273. [Google Scholar] [CrossRef]
- Kim, J.N.; Kim, T.H.; Yoon, J.H.; Cho, S.G. Kisspeptin Inhibits Colorectal Cancer Cell Invasiveness by Activating PKR and PP2A. Anticancer Res. 2018, 38, 5791–5798. [Google Scholar] [CrossRef] [PubMed]
- Zaragoza-Huesca, D.; Nieto-Olivares, A.; Garcia-Molina, F.; Ricote, G.; Montenegro, S.; Sanchez-Canovas, M.; Garrido-Rodriguez, P.; Penas-Martinez, J.; Vicente, V.; Martinez, F.; et al. Implication of Hepsin from Primary Tumor in the Prognosis of Colorectal Cancer Patients. Cancers 2022, 14, 3106. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, V.; Fournier-Level, A.; Cooper, H.M.; Murray, M.J. Loss of Neogenin1 in human colorectal carcinoma cells causes a partial EMT and wound-healing response. Sci. Rep. 2019, 9, 4110. [Google Scholar] [CrossRef] [PubMed]
- Sane, S.; Srinivasan, R.; Potts, R.A.; Eikanger, M.; Zagirova, D.; Freeling, J.; Reihe, C.A.; Antony, R.M.; Gupta, B.K.; Lynch, D.; et al. UBXN2A suppresses the Rictor-mTORC2 signaling pathway, an established tumorigenic pathway in human colorectal cancer. Oncogene 2023, 42, 1763–1776. [Google Scholar] [CrossRef] [PubMed]
- Yue, B.; Liu, C.; Sun, H.; Liu, M.; Song, C.; Cui, R.; Qiu, S.; Zhong, M. A Positive Feed-Forward Loop between LncRNA-CYTOR and Wnt/β-Catenin Signaling Promotes Metastasis of Colon Cancer. Mol. Ther. 2018, 26, 1287–1298. [Google Scholar] [CrossRef]
- Shi, Q.; He, Y.; He, S.; Li, J.; Xia, J.; Chen, T.; Huo, L.; Ling, Y.; Liu, Q.; Zang, W.; et al. RP11-296E3.2 acts as an important molecular chaperone for YBX1 and promotes colorectal cancer proliferation and metastasis by activating STAT3. J. Transl. Med. 2023, 21, 418. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, Z.; Yan, H.; Wang, W.; Wu, Z.; Zhang, F.; Zhang, Q.; Shi, G.; Du, J.; Cai, H.; et al. Creatine promotes cancer metastasis through activation of Smad2/3. Cell Metab. 2021, 33, 1111–1123.e1114. [Google Scholar] [CrossRef]
- Hu, F.; Li, G.; Huang, C.; Hou, Z.; Yang, X.; Luo, X.; Feng, Y.; Wang, G.; Hu, J.; Cao, Z. The autophagy-independent role of BECN1 in colorectal cancer metastasis through regulating STAT3 signaling pathway activation. Cell Death Dis. 2020, 11, 304. [Google Scholar] [CrossRef]
- Li, T.; Sun, R.; Lu, M.; Chang, J.; Meng, X.; Wu, H. NDRG3 facilitates colorectal cancer metastasis through activating Src phosphorylation. Onco Targets Ther. 2018, 11, 2843–2852. [Google Scholar] [CrossRef]
- Wang, Y.; Mu, L.; Huang, M. MicroRNA-195 suppresses rectal cancer growth and metastasis via regulation of the PI3K/AKT signaling pathway. Mol. Med. Rep. 2019, 20, 4449–4458. [Google Scholar] [CrossRef]
- Kobelt, D.; Perez-Hernandez, D.; Fleuter, C.; Dahlmann, M.; Zincke, F.; Smith, J.; Migotti, R.; Popp, O.; Burock, S.; Walther, W.; et al. The newly identified MEK1 tyrosine phosphorylation target MACC1 is druggable by approved MEK1 inhibitors to restrict colorectal cancer metastasis. Oncogene 2021, 40, 5286–5301. [Google Scholar] [CrossRef]
- Wang, T.; Sun, L.; Chen, C.; Zhang, Y.; He, B.; Zhang, Y.; Wang, Z.; Xue, H.; Hao, Y. Phosphorylation at tyrosine 317 and 508 are crucial for PIK3CA/p110α to promote CRC tumorigenesis. Cell Biosci. 2023, 13, 164. [Google Scholar] [CrossRef]
- Kassassir, H.; Papiewska-Pająk, I.; Kryczka, J.; Boncela, J.; Kowalska, M.A. Platelet-derived microparticles stimulate the invasiveness of colorectal cancer cells via the p38MAPK-MMP-2/MMP-9 axis. Cell Commun. Signal 2023, 21, 51. [Google Scholar] [CrossRef] [PubMed]
- La Vecchia, S.; Sebastián, C. Metabolic pathways regulating colorectal cancer initiation and progression. Semin. Cell Dev. Biol. 2020, 98, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.Y.; Tu, T.; Zhu, J.; Hu, A.P.; Song, J.W.; Huang, J.Q.; Yang, Y.; Zhu, Z.; Chen, Y. Golgi phosphoprotein 3 induces autophagy and epithelial-mesenchymal transition to promote metastasis in colon cancer. Cell Death Discov. 2022, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Lin, M.; Chen, S.; Liu, H.; Zhu, M.; Hu, Y.; Han, S.; Wang, Y.; Sun, L.; Zhu, F.; et al. E2F1 promotes cell cycle progression by stabilizing spindle fiber in colorectal cancer cells. Cell. Mol. Biol. Lett. 2022, 27, 90. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lei, W.; Li, Z.; Wang, X.; Zhou, L. NR3C2 inhibits the proliferation of colorectal cancer via regulating glucose metabolism and phosphorylating AMPK. J. Cell. Mol. Med. 2023, 27, 1069–1082. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Shembrey, C.; Smith, J.; Paquet-Fifield, S.; Behrenbruch, C.; Beyit, L.M.; Thomson, B.N.J.; Heriot, A.G.; Cao, Y.; Hollande, F. Laminin 521 enhances self-renewal via STAT3 activation and promotes tumor progression in colorectal cancer. Cancer Lett. 2020, 476, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhang, W.; Liu, C. FAK downregulation suppresses stem-like properties and migration of human colorectal cancer cells. PLoS ONE 2023, 18, e0284871. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhang, T.; Hua, D. Ubiquitination and SUMOylation: Protein homeostasis control over cancer. Epigenomics 2022, 14, 43–58. [Google Scholar] [CrossRef]
- Zhang, Z.; Sie, B.; Chang, A.; Leng, Y.; Nardone, C.; Timms, R.T.; Elledge, S.J. Elucidation of E3 ubiquitin ligase specificity through proteome-wide internal degron mapping. Mol. Cell 2023, 83, 3377–3392.e3376. [Google Scholar] [CrossRef]
- Yue, M.; Yun, Z.; Li, S.; Yan, G.; Kang, Z. NEDD4 triggers FOXA1 ubiquitination and promotes colon cancer progression under microRNA-340-5p suppression and ATF1 upregulation. RNA Biol. 2021, 18, 1981–1995. [Google Scholar] [CrossRef]
- Zhang, S.; Yu, C.R.; Yang, X.; Hong, H.J.J.; Lu, J.Y.; Hu, W.J.; Hao, X.H.; Li, S.C.; Aikemu, B.; Yang, G.; et al. N-myc downstream-regulated gene 1 inhibits the proliferation of colorectal cancer through emulative antagonizing NEDD4-mediated ubiquitylation of p21. J. Exp. Clin. Cancer Res. 2019, 38, 490. [Google Scholar] [CrossRef]
- Zhang, Z.; He, G.; Lv, Y.; Liu, Y.; Niu, Z.; Feng, Q.; Hu, R.; Xu, J. HERC3 regulates epithelial-mesenchymal transition by directly ubiquitination degradation EIF5A2 and inhibits metastasis of colorectal cancer. Cell Death Dis. 2022, 13, 74. [Google Scholar] [CrossRef]
- Liang, Q.; Ma, D.; Zhu, X.; Wang, Z.; Sun, T.-T.; Shen, C.; Yan, T.; Tian, X.; Yu, T.; Guo, F.; et al. RING-Finger Protein 6 Amplification Activates JAK/STAT3 Pathway by Modifying SHP-1 Ubiquitylation and Associates with Poor Outcome in Colorectal Cancer. Clin. Cancer Res. 2018, 24, 1473–1485. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, Y.; Wong, C.C.; Zhang, J.; Dong, Y.; Li, X.; Kang, W.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. RNF6 Promotes Colorectal Cancer by Activating the Wnt/beta-Catenin Pathway via Ubiquitination of TLE3. Cancer Res. 2018, 78, 1958–1971. [Google Scholar] [CrossRef]
- Wang, S.; Wang, T.; Wang, L.; Zhong, L.; Li, K. Overexpression of RNF126 Promotes the Development of Colorectal Cancer via Enhancing p53 Ubiquitination and Degradation. Onco Targets Ther. 2020, 13, 10917–10929. [Google Scholar] [CrossRef]
- Neumeyer, V.; Grandl, M.; Dietl, A.; Brutau-Abia, A.; Allgaeuer, M.; Kalali, B.; Zhang, Y.; Pan, K.-F.; Steiger, K.; Vieth, M.; et al. Loss of endogenous RNF43 function enhances proliferation and tumour growth of intestinal and gastric cells. Carcinogenesis 2019, 40, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Choi, D.K.; Min, J.S.; Kang, I.; Kim, J.C.; Kim, S.; Ahn, J.K. VBP1 represses cancer metastasis by enhancing HIF-1α degradation induced by pVHL. FEBS J. 2018, 285, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Li, Y.; Zhang, X.; Wu, H.; Wang, Q.; Cai, J.; Cui, Y.; Liu, H.; Lan, P.; Wang, J.; et al. Ubiquitin ligase TRIM65 promotes colorectal cancer metastasis by targeting ARHGAP35 for protein degradation. Oncogene 2019, 38, 6429–6444. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Tang, C.; Tang, M.; Zhang, Q.; Gao, Y.; Ge, Z. TRIM47 is up-regulated in colorectal cancer, promoting ubiquitination and degradation of SMAD4. J. Exp. Clin. Cancer Res. 2019, 38, 159. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Hu, J.; Ye, Z.; Fan, Y.; Li, Y.; Wang, G.; Wang, L.; Wang, Z. TRIM28 protects CARM1 from proteasome-mediated degradation to prevent colorectal cancer metastasis. Sci. Bull. 2019, 64, 986–997. [Google Scholar] [CrossRef] [PubMed]
- Ruan, L.; Liu, W.; Yang, Y.; Chu, Z.; Yang, C.; Yang, T.; Sun, J. TRIM16 overexpression inhibits the metastasis of colorectal cancer through mediating Snail degradation. Exp. Cell Res. 2021, 406, 112735. [Google Scholar] [CrossRef] [PubMed]
- Guangwei, Z.; Zhibin, C.; Qin, W.; Chunlin, L.; Penghang, L.; Ruofan, H.; Hui, C.; Hoffman, R.M.; Jianxin, Y. TRAF6 regulates the signaling pathway influencing colorectal cancer function through ubiquitination mechanisms. Cancer Sci. 2022, 113, 1393–1405. [Google Scholar] [CrossRef]
- Wu, H.; Lu, X.-X.; Wang, J.-R.; Yang, T.-Y.; Li, X.-M.; He, X.-S.; Li, Y.; Ye, W.-L.; Wu, Y.; Gan, W.-J.; et al. TRAF6 inhibits colorectal cancer metastasis through regulating selective autophagic CTNNB1/beta-catenin degradation and is targeted for GSK3B/GSK3 beta-mediated phosphorylation and degradation. Autophagy 2019, 15, 1506–1522. [Google Scholar] [CrossRef]
- Xiao, C.; Wu, G.; Zhou, Z.; Zhang, X.; Wang, Y.; Song, G.; Ding, E.; Sun, X.; Zhong, L.; Li, S.; et al. RBBP6, a RING finger-domain E3 ubiquitin ligase, induces epithelial-mesenchymal transition and promotes metastasis of colorectal cancer. Cell Death Dis. 2019, 10, 833. [Google Scholar] [CrossRef]
- Wang, F.; Xu, H.; Yan, Y.; Wu, P.; Wu, J.; Zhu, X.; Li, J.; Sun, J.; Zhou, K.; Ren, X.; et al. FBX8 degrades GSTP1 through ubiquitination to suppress colorectal cancer progression. Cell Death Dis. 2019, 10, 351. [Google Scholar] [CrossRef]
- Liu, H.; Wang, K.; Fu, H.; Song, J. Low expression of the ubiquitin ligase FBXW7 correlates with poor prognosis of patients with colorectal cancer. Int. J. Clin. Exp. Pathol. 2018, 11, 413–419. [Google Scholar]
- Wu, Y.; Liu, B.; Lin, W.; Zhao, R.; Han, W.; Xie, J. AAMP promotes colorectal cancer metastasis by suppressing SMURF2-mediated ubiquitination and degradation of RhoA. Mol. Ther.-Oncolytics 2021, 23, 515–530. [Google Scholar] [CrossRef]
- Sato, N.; Sakai, N.; Furukawa, K.; Takayashiki, T.; Kuboki, S.; Takano, S.; Ohira, G.; Miyauchi, H.; Matsubara, H.; Ohtsuka, M. Tumor-suppressive role of Smad ubiquitination regulatory factor 2 in patients with colorectal cancer. Sci. Rep. 2022, 12, 5495. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Shen, J.; Tao, Y.; Dong, D.; Lu, S.; Li, L.; Sun, D.; Fan, M.; Xu, C.; Shen, W.; et al. CCDC85C suppresses colorectal cancer cells proliferation and metastasis through activating GSK-3β and promoting β-catenin degradation. Cell. Signal. 2023, 109, 110799. [Google Scholar] [CrossRef]
- Wu, M.; Li, X.; Huang, W.; Chen, Y.; Wang, B.; Liu, X. Ubiquitin-conjugating enzyme E2T(UBE2T) promotes colorectal cancer progression by facilitating ubiquitination and degradation of p53. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101493. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Neufeld, H.; Torlakovic, E.; Xiao, W. Uev1A-Ubc13 promotes colorectal cancer metastasis through regulating CXCL1 expression via NF-κB activation. Oncotarget 2018, 9, 15952–15967. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-Y.; Zhang, C.-M.; Dai, Y.-B.; Lin, J.-G.; Lin, N.; Huang, Z.-X.; Xu, T.-W. USP11 facilitates colorectal cancer proliferation and metastasis by regulating IGF2BP3 stability. Am. J. Transl. Res. 2021, 13, 480–496. [Google Scholar] [PubMed]
- Sun, H.; Ou, B.; Zhao, S.; Liu, X.; Song, L.; Liu, X.; Wang, R.; Peng, Z. USP11 promotes growth and metastasis of colorectal cancer via PPP1CA-mediated activation of ERK/MAPK signaling pathway. eBioMedicine 2019, 48, 236–247. [Google Scholar] [CrossRef]
- Miao, D.; Wang, Y.; Jia, Y.; Tong, J.; Jiang, S.; Liu, L. ZRANB1 enhances stem-cell-like features and accelerates tumor progression by regulating Sox9-mediated USP22/Wnt/beta-catenin pathway in colorectal cancer. Cell. Signal. 2022, 90, 110200. [Google Scholar] [CrossRef]
- Gennaro, V.J.; Stanek, T.J.; Peck, A.R.; Sun, Y.; Wang, F.; Qie, S.; Knudsen, K.E.; Rui, H.; Butt, T.; Diehl, J.A.; et al. Control of CCND1 ubiquitylation by the catalytic SAGA subunit USP22 is essential for cell cycle progression through G1 in cancer cells. Proc. Natl. Acad. Sci. USA 2018, 115, E9298–E9307. [Google Scholar] [CrossRef]
- Ye, D.-x.; Wang, S.-s.; Huang, Y.; Wang, X.-j.; Chi, P. USP43 directly regulates ZEB1 protein, mediating proliferation and metastasis of colorectal cancer. J. Cancer 2021, 12, 404–416. [Google Scholar] [CrossRef]
- Pan, B.; Yang, Y.; Li, J.; Wang, Y.; Fang, C.; Yu, F.-X.; Xu, Y. USP47-mediated deubiquitination and stabilization of YAP contributes to the progression of colorectal cancer. Protein Cell 2020, 11, 138–143. [Google Scholar] [CrossRef]
- Li, J.; Zheng, Y.; Li, X.; Dong, X.; Chen, W.; Guan, Z.; Zhang, C. UCHL3 promotes proliferation of colorectal cancer cells by regulating SOX12 via AKT/mTOR signaling pathway. Am. J. Transl. Res. 2020, 12, 6445–6454. [Google Scholar]
- Ye, D.; Wang, S.; Wang, X.; Lin, Y.; Huang, Y.; Chi, P. Overexpression of OTU domain-containing ubiquitin aldehyde-binding protein 1 exacerbates colorectal cancer malignancy by inhibiting protein degradation of β-Catenin via Ubiquitin-proteasome pathway. Bioengineered 2022, 13, 9106–9116. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Chen, B.; Ke, D.; Chen, X. DUSP4 directly deubiquitinates and stabilizes Smad4 protein, promoting proliferation and metastasis of colorectal cancer cells. Aging 2020, 12, 17634–17646. [Google Scholar] [CrossRef]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749. [Google Scholar] [CrossRef]
- Eichler, J. Protein glycosylation. Curr. Biol. 2019, 29, R229–R231. [Google Scholar] [CrossRef]
- Stowell, S.R.; Ju, T.; Cummings, R.D. Protein glycosylation in cancer. Annu. Rev. Pathol. 2015, 10, 473–510. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Chen, L.; Gao, H.; Zhen, T.; Li, H.; Liang, J.; Zhang, F.; Shi, H.; Han, A. GALNT6 suppresses progression of colorectal cancer. Am. J. Cancer Res. 2018, 8, 2419–2435. [Google Scholar] [PubMed]
- Jiang, Z.; Zhang, H.; Liu, C.; Yin, J.; Tong, S.; Lv, J.; Wei, S.; Wu, S. beta 3GnT8 Promotes Colorectal Cancer Cells Invasion via CD147/MMP2/Galectin3 Axis. Front. Physiol. 2018, 9, 588. [Google Scholar] [CrossRef]
- Yan, X.; Lu, J.; Zou, X.; Zhang, S.; Cui, Y.; Zhou, L.; Liu, F.; Shan, A.; Lu, J.; Zheng, M.; et al. The polypeptide N-acetylgalactosaminyltransferase 4 exhibits stage-dependent expression in colorectal cancer and affects tumorigenesis, invasion and differentiation. FEBS J. 2018, 285, 3041–3055. [Google Scholar] [CrossRef]
- Balcik-Ercin, P.; Cetin, M.; Yalim-Camci, I.; Odabas, G.; Tokay, N.; Sayan, A.E.; Yagci, T. Genome-wide analysis of endogenously expressed ZEB2 binding sites reveals inverse correlations between ZEB2 and GalNAc-transferase GALNT3 in human tumors. Cell. Oncol. 2018, 41, 379–393. [Google Scholar] [CrossRef]
- Dong, X.; Jiang, Y.; Liu, J.; Liu, Z.; Gao, T.; An, G.; Wen, T. T-Synthase Deficiency Enhances Oncogenic Features in Human Colorectal Cancer Cells via Activation of Epithelial-Mesenchymal Transition. BioMed Res. Int. 2018, 2018, 9532389. [Google Scholar] [CrossRef]
- Liu, L.; Pan, Y.; Ren, X.; Zeng, Z.; Sun, J.; Zhou, K.; Liang, Y.; Wang, F.; Yan, Y.; Liao, W.; et al. GFPT2 promotes metastasis and forms a positive feedback loop with p65 in colorectal cancer. Am. J. Cancer Res. 2020, 10, 2510. [Google Scholar]
- Iwai, T.; Kudo, T.; Kawamoto, R.; Kubota, T.; Togayachi, A.; Hiruma, T.; Okada, T.; Kawamoto, T.; Morozumi, K.; Narimatsu, H. Core 3 synthase is down-regulated in colon carcinoma and profoundly suppresses the metastatic potential of carcinoma cells. Proc. Natl. Acad. Sci. USA 2005, 102, 4572–4577. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Wei, X.; Shang, Y.; Pan, Q.; Yang, M.; Tian, Y.; He, Y.; Peng, Z.; Chen, L.; Chen, W.; et al. Core 3 mucin-type O-glycan restoration in colorectal cancer cells promotes MUC1/p53/miR-200c-dependent epithelial identity. Oncogene 2017, 36, 6391–6407. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Pan, S.; Xiao, Y.; Liu, Q.; Xu, J.; Jia, L. LINC01296/miR-26a/GALNT3 axis contributes to colorectal cancer progression by regulating O-glycosylated MUC1 via PI3K/AKT pathway. J. Exp. Clin. Cancer Res. 2018, 37, 316. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, S.; Zou, X.; Lu, J.; Yang, X.; Xu, Z.; Shan, A.; Jia, W.; Liu, F.; Yan, X.; et al. The beta-galactoside alpha 2,6-sialyltranferase 1 (ST6GAL1) inhibits the colorectal cancer metastasis by stabilizing intercellular adhesion molecule-1 via sialylation. Cancer Manag. Res. 2019, 11, 6185–6199. [Google Scholar] [CrossRef]
- Liu, P.; Liu, J.Y.; Ding, M.Y.; Liu, Y.J.; Zhang, Y.; Chen, X.M.; Zhou, Z.X. FUT2 promotes the tumorigenicity and metastasis of colorectal cancer cells via the Wnt/β-catenin pathway. Int. J. Oncol. 2023, 62, 1–12. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, J.; Dong, X.; Hu, X.; Jiang, Y.; Li, L.; Du, T.; Yang, L.; Wen, T.; An, G.; et al. Tn antigen promotes human colorectal cancer metastasis via H-Ras mediated epithelial-mesenchymal transition activation. J. Cell. Mol. Med. 2019, 23, 2083–2092. [Google Scholar] [CrossRef]
- Harosh-Davidovich, S.B.; Khalaila, I. O-GlcNAcylation affects β-catenin and E-cadherin expression, cell motility and tumorigenicity of colorectal cancer. Exp. Cell Res. 2018, 364, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Shan, Y.; Ma, J.; Pan, Y.; Zhou, H.; Jiang, L.; Jia, L. LncRNA ST3Gal6-AS1/ST3Gal6 axis mediates colorectal cancer progression by regulating α-2,3 sialylation via PI3K/Akt signaling. Int. J. Cancer 2019, 145, 450–460. [Google Scholar] [CrossRef]
- Jiang, M.; Xu, B.; Li, X.; Shang, Y.; Chu, Y.; Wang, W.; Chen, D.; Wu, N.; Hu, S.; Zhang, S.; et al. O-GlcNAcylation promotes colorectal cancer metastasis via the miR-101-O-GlcNAc/EZH2 regulatory feedback circuit. Oncogene 2019, 38, 301–316. [Google Scholar] [CrossRef]
- Naka, Y.; Okada, T.; Nakagawa, T.; Kobayashi, E.; Kawasaki, Y.; Tanaka, Y.; Tawa, H.; Hirata, Y.; Kawakami, K.; Kakimoto, K.; et al. Enhancement of O-linked N-acetylglucosamine modification promotes metastasis in patients with colorectal cancer and concurrent type 2 diabetes mellitus. Oncol. Lett. 2020, 20, 1171–1178. [Google Scholar] [CrossRef]
- Pan, S.; Liu, Y.; Liu, Q.; Xiao, Y.; Liu, B.; Ren, X.; Qi, X.; Zhou, H.; Zeng, C.; Jia, L. HOTAIR/miR-326/FUT6 axis facilitates colorectal cancer progression through regulating fucosylation of CD44 via PI3K/AKT/mTOR pathway. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 750–760. [Google Scholar] [CrossRef]
- Zhu, L.; Chen, Y.; Du, H.; Cong, Y.; Yan, W.; Ma, K.; Huang, X. N-glycosylation of CD82 at Asn157 is required for suppressing migration and invasion by reversing EMT via Wnt/?-catenin pathway in colon cancer. Biochem. Biophys. Res. Commun. 2022, 629, 121–127. [Google Scholar] [CrossRef]
- Umezawa, F.; Natsume, M.; Fukusada, S.; Nakajima, K.; Yamasaki, F.; Kawashima, H.; Kuo, C.-W.; Khoo, K.-H.; Shimura, T.; Yagi, H.; et al. Cancer Malignancy Is Correlated with Upregulation of PCYT2-Mediated Glycerol Phosphate Modification of alpha-Dystroglycan. Int. J. Mol. Sci. 2022, 23, 6662. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yang, X.; Huang, X.; Yao, Y.; Wei, X.; Yang, S.; Zhou, D.; Zhang, W.; Long, Z.; Xu, X.; et al. 2-Hydroxylation of Fatty Acids Represses Colorectal Tumorigenesis and Metastasis via the YAP Transcriptional Axis. Cancer Res. 2021, 81, 289–302. [Google Scholar] [CrossRef]
- Kishi, S.; Fujiwara-Tani, R.; Honoki, K.; Sasaki, R.; Mori, S.; Ohmori, H.; Sasaki, T.; Miyagawa, Y.; Kawahara, I.; Kido, A.; et al. Oxidized high mobility group B-1 enhances metastability of colorectal cancer via modification of mesenchymal stem/stromal cells. Cancer Sci. 2022, 113, 2904–2915. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.-Y.; Cao, J.; Gao, L.-J.; Zhang, F.-P.; Shen, J.; Zhou, L.; Shi, J.-Y.; Feng, Y.-L.; Yan, Z.; Wang, D.-P.; et al. Upregulation of a enolase (ENO1) crotonylation in colorectal cancer and its promoting effect on cancer cell metastasis. Biochem. Biophys. Res. Commun. 2021, 578, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Sun, X.; Li, J.; He, P.; Liu, W.; Meng, X. Knockdown of Uba2 inhibits colorectal cancer cell invasion and migration through downregulation of the Wnt/-catenin signaling pathway. J. Cell. Biochem. 2018, 119, 6914–6925. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, Y.; Liu, B.; Yang, X.; Wang, Z.; Han, M.; Li, H.; Luo, J.; Yao, H. Dynamic regulation of eEF1A1 acetylation affects colorectal carcinogenesis. Biol. Chem. 2023, 404, 585–599. [Google Scholar] [CrossRef]
- Lan, J.; Zhang, S.; Zheng, L.; Long, X.; Chen, J.; Liu, X.; Zhou, M.; Zhou, J. PLOD2 promotes colorectal cancer progression by stabilizing USP15 to activate the AKT/mTOR signaling pathway. Cancer Sci. 2023, 114, 3190–3202. [Google Scholar] [CrossRef]
- Pieroni, L.; Iavarone, F.; Olianas, A.; Greco, V.; Desiderio, C.; Martelli, C.; Manconi, B.; Sanna, M.T.; Messana, I.; Castagnola, M.; et al. Enrichments of post-translational modifications in proteomic studies. J. Sep. Sci. 2020, 43, 313–336. [Google Scholar] [CrossRef] [PubMed]
- Geffen, Y.; Anand, S.; Akiyama, Y.; Yaron, T.M.; Song, Y.; Johnson, J.L.; Govindan, A.; Babur, Ö.; Li, Y.; Huntsman, E.; et al. Pan-cancer analysis of post-translational modifications reveals shared patterns of protein regulation. Cell 2023, 186, 3945–3967.e3926. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-H.; Kim, S.-J.; Fang, X.; Song, N.-Y.; Kim, D.-H.; Suh, J.; Na, H.-K.; Kim, K.-O.; Baek, J.-H.; Surh, Y.-J. JNK-mediated Ser27 phosphorylation and stabilization of SIRT1 promote growth and progression of colon cancer through deacetylation-dependent activation of Snail. Mol. Oncol. 2022, 16, 1555–1571. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Huang, H.; Qiu, X.; Ding, Z.; Feng, X.; Zhu, Y.; Zhuo, H.; Hou, J.; Zhao, J.; Cai, W.; et al. Targeting posttranslational modifications of RIOK1 inhibits the progression of colorectal and gastric cancers. eLife 2018, 7, e29511. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Chang, S.; Xiao, S.; Peng, Y.; Gao, Y.; Hu, F.; Liang, J.; Xu, Y.; Du, K.; Chen, Y.; et al. PAD4-dependent citrullination of nuclear translocation of GSK3 beta promotes colorectal cancer progression via the degradation of nuclear CDKN1A. Neoplasia 2022, 33, 100835. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.J.; Huo, F.C.; Kang, M.J.; Liu, B.W.; Wu, M.D.; Pei, D.S. Alternative splicing of HSPA12A pre-RNA by SRSF11 contributes to metastasis potential of colorectal cancer. Clin. Transl. Med. 2022, 12, e1113. [Google Scholar] [CrossRef]
- Sun, L.; Yu, J.; Guinney, J.; Qin, B.; Sinicrope, F.A. USP10 Regulates ZEB1 Ubiquitination and Protein Stability to Inhibit ZEB1-Mediated Colorectal Cancer Metastasis. Mol. Cancer Res. 2023, 21, 578–590. [Google Scholar] [CrossRef]
- Zhang, Q.; Yang, X.; Wu, J.; Ye, S.; Gong, J.; Cheng, W.M.; Luo, Z.; Yu, J.; Liu, Y.; Zeng, W.; et al. Reprogramming of palmitic acid induced by dephosphorylation of ACOX1 promotes β-catenin palmitoylation to drive colorectal cancer progression. Cell Discov. 2023, 9, 26. [Google Scholar] [CrossRef]
- Zhu, G.; Jin, L.; Sun, W.; Wang, S.; Liu, N. Proteomics of post-translational modifications in colorectal cancer: Discovery of new biomarkers. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188735. [Google Scholar] [CrossRef]
- Tan, X.; Xu, M.; Liu, F.; Xu, M.; Yao, Y.; Tang, D. Antimetastasis Effect of Astragalus membranaceus-Curcuma zedoaria via -Catenin Mediated CXCR4 and EMT Signaling Pathway in HCT116. Evid. Based Complement. Alternat. Med. 2019, 2019, 9692350. [Google Scholar] [CrossRef]
- Han, Y.H.; Kee, J.Y.; Hong, S.H. Rosmarinic Acid Activates AMPK to Inhibit Metastasis of Colorectal Cancer. Front. Pharmacol. 2018, 9, 68. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, P.; Yan, R.; Wang, Q.; Fang, E.; Wu, H.; Li, S.; Tan, H.; Zhou, X.; Ma, X.; et al. MnTE-2-PyP Attenuates TGF-beta-Induced Epithelial-Mesenchymal Transition of Colorectal Cancer Cells by Inhibiting the Smad2/3 Signaling Pathway. Oxid. Med. Cell. Longev. 2019, 2019, 8639791. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.T.; Lin, H.; Wang, C.S.; Chang, C.H.; Lin, A.M.; Yang, J.C.; Lo, Y.L. Improving the anticancer effect of afatinib and microRNA by using lipid polymeric nanoparticles conjugated with dual pH-responsive and targeting peptides. J. Nanobiotechnol. 2019, 17, 89. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Shen, W.; Zhang, F.; Fan, H.; Xu, C.; Li, L.; Tan, J.; Miao, Y.; Zhang, H.; Yang, Y.; et al. alpha-Hederin inhibits interleukin 6-induced epithelial-to-mesenchymal transition associated with disruption of JAK2/STAT3 signaling in colon cancer cells. Biomed. Pharmacother. 2018, 101, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.Z.; Huang, D.L.; Tang, Z.R.; Zhang, J.H.; Xiong, T.; Zhou, C.; Zhang, N.X.; Fu, R.; Cheng, Y.X.; Wu, Z.Q. Pharmacological targeting of Axin2 suppresses cell growth and metastasis in colorectal cancer. Br. J. Pharmacol. 2023, 180, 3071–3091. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Shi, Z.; Zhang, S.; Li, X.; To, S.K.Y.; Peng, Y.; Liu, J.; Chen, S.; Hu, H.; Wong, A.S.T.; et al. The Ginsenoside Compound K Suppresses Stem-Cell-like Properties and Colorectal Cancer Metastasis by Targeting Hypoxia-Driven Nur77-Akt Feed-Forward Signaling. Cancers 2022, 15, 24. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Lee, S.K.; Park, J.; Jung, M.J.; An, S.E.; Yang, H.J.; Chung, W.Y. Buddlejasaponin IV induces apoptotic cell death by activating the mitochondrial-dependent apoptotic pathway and reducing α(2)β(1) integrin-mediated adhesion in HT-29 human colorectal cancer cells. Oncol. Rep. 2023, 49, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hegde, P.; Rajakumar, S.B.; Swamy, B.M.; Inamdar, S.R. A mitogenic lectin from Rhizoctonia bataticola arrests growth, inhibits metastasis, and induces apoptosis in human colon epithelial cancer cells. J. Cell. Biochem. 2018, 119, 5632–5645. [Google Scholar] [CrossRef]
- Loureiro, L.R.; Sousa, D.P.; Ferreira, D.; Chai, W.; Lima, L.; Pereira, C.; Lopes, C.B.; Correia, V.G.; Silva, L.M.; Li, C.; et al. Novel monoclonal antibody L2A5 specifically targeting sialyl-Tn and short glycans terminated by alpha-2-6 sialic acids. Sci. Rep. 2018, 8, 12196. [Google Scholar] [CrossRef]
- Martinez-Iglesias, O.; Casas-Pais, A.; Castosa, R.; Diaz-Diaz, A.; Roca-Lema, D.; Concha, A.; Cortes, A.; Gago, F.; Figueroa, A. Hakin-1, a New Specific Small-Molecule Inhibitor for the E3 Ubiquitin-Ligase Hakai, Inhibits Carcinoma Growth and Progression. Cancers 2020, 12, 1340. [Google Scholar] [CrossRef]
- Malki, A.; Abu ElRuz, R.; Gupta, I.; Allouch, A.; Vranic, S.; Al Moustafa, A.E. Molecular Mechanisms of Colon Cancer Progression and Metastasis: Recent Insights and Advancements. Int. J. Mol. Sci. 2021, 22, 130. [Google Scholar] [CrossRef]
- Liebl, M.C.; Hofmann, T.G. The Role of p53 Signaling in Colorectal Cancer. Cancers 2021, 13, 2125. [Google Scholar] [CrossRef]
- Ren, S.; Shao, Y.; Zhao, X.; Hong, C.S.; Wang, F.; Lu, X.; Li, J.; Ye, G.; Yan, M.; Zhuang, Z.; et al. Integration of Metabolomics and Transcriptomics Reveals Major Metabolic Pathways and Potential Biomarker Involved in Prostate Cancer. Mol. Cell Proteom. 2016, 15, 154–163. [Google Scholar] [CrossRef]
- He, X.F.; Hu, X.; Wen, G.J.; Wang, Z.; Lin, W.J. O-GlcNAcylation in cancer development and immunotherapy. Cancer Lett. 2023, 566, 216258. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.M.; Li, C.W.; Lai, Y.J.; Hung, M.C. Posttranslational Modifications of PD-L1 and Their Applications in Cancer Therapy. Cancer Res. 2018, 78, 6349–6353. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, S.; Kaur, K.; Huang, R.; Zhang, Q.; Kaur, P.; Yazdani, H.O.; Bilal, M.U.; Zheng, J.; Zheng, L.; Wang, X.S. MicroRNAs in colorectal cancer: Role in metastasis and clinical perspectives. World J. Gastroenterol. 2014, 20, 17011–17019. [Google Scholar] [CrossRef]
- Fang, Y.; Wu, F.; Shang, G.; Yin, C. SCARA5 in bone marrow stromal cell-derived exosomes inhibits colorectal cancer progression by inactivating the PI3K/Akt pathway. Genomics 2023, 115, 110636. [Google Scholar] [CrossRef] [PubMed]
- Noberini, R.; Robusti, G.; Bonaldi, T. Mass spectrometry-based characterization of histones in clinical samples: Applications, progress, and challenges. FEBS J. 2022, 289, 1191–1213. [Google Scholar] [CrossRef]
| E3 | Substrate | Inhibit/Promote Metastasis | Cellular Function | Molecular Pathway | References |
|---|---|---|---|---|---|
| NEDD4 | F0XA1 | promote | EMT, migration, invasion Invasion | N/A | [62] |
| NEDD4 | P21 | promote | EMT, migration, invasion | N/A | [63] |
| HERC3 | EIF5A2 | inhibit | metastasis | TGF-/smad2/3 | [64] |
| RNF6 | STAT3 | promote | metastasis | JAK-STAT3 | [65] |
| RNF6 | TLE3 | promote | EMT | WNT/β-catenin | [66] |
| RNF126 | P53 | promote | migration | P21/Rb | [67] |
| RNF43 | Fzd | inhibit | stemness | WNT | [68] |
| pVHL | HIF-1α | inhibit | EMT, metastasis | N/A | [69] |
| TRIM65 | ARHGAP35 | promote | Migration, metastasis | N/A | [70] |
| TRIM47 | SMAD4 | promote | invasion | CCL15-CCR1 | [71] |
| TRIM28 | CARM 1 | inhibit | EMT | WNT/β-catenin | [72] |
| TRIM16 | snail | inhibit | EMT, migration, invasion | N/A | [73] |
| TRAF6 | N/A | promote | Migration, invasion | NF-κB- VEGF-C | [74] |
| TRAF6 | LC3B | inhibit | EMT, metastasis | WNT/β-catenin | [75] |
| RBBP6 | N/A | promote | EMT, metastasis | NF-κB pathway | [76] |
| Fbx8 | GSTP1 | Inhibit | Invasion | N/A | [77] |
| FBXW7 | N/A | promote | metastasis | N/A | [78] |
| SMURF2 | RhoA | Inhibit | migration and invasion | N/A | [79] |
| SMURF2 | EpCAM | Inhibit | stemness | N/A | [80] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, N.; Liu, J.; Hai, S.; Liu, Y.; Zhao, H.; Liu, W. Role of Post-Translational Modifications in Colorectal Cancer Metastasis. Cancers 2024, 16, 652. https://doi.org/10.3390/cancers16030652
Peng N, Liu J, Hai S, Liu Y, Zhao H, Liu W. Role of Post-Translational Modifications in Colorectal Cancer Metastasis. Cancers. 2024; 16(3):652. https://doi.org/10.3390/cancers16030652
Chicago/Turabian StylePeng, Na, Jingwei Liu, Shuangshuang Hai, Yihong Liu, Haibo Zhao, and Weixin Liu. 2024. "Role of Post-Translational Modifications in Colorectal Cancer Metastasis" Cancers 16, no. 3: 652. https://doi.org/10.3390/cancers16030652
APA StylePeng, N., Liu, J., Hai, S., Liu, Y., Zhao, H., & Liu, W. (2024). Role of Post-Translational Modifications in Colorectal Cancer Metastasis. Cancers, 16(3), 652. https://doi.org/10.3390/cancers16030652





