Triple-Negative Breast Cancer Intrinsic FTSJ1 Favors Tumor Progression and Attenuates CD8+ T Cell Infiltration
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Public Database
2.2. Cell Culture
2.3. Plasmid and Lentivirus Preparation
2.4. Establishment of Stable TNBC Cell Lines
2.5. Cell Proliferation Analysis
2.6. Wound Healing Assay
2.7. Real-Time PCR
- CD276-F: 5′-CTGGCTTTCGTGTGCTGGAGAA-3′;
- CD276-R:5′-GCTGTCAGAGTGTTTCAGAGGC-3′;
- CD252 (TNFSF4)-F: 5′-CCTACATCTGCCTGCACTTCTC-3′;
- CD252 (TNFSF4)-R: 5′-TGATGACTGAGTTGTTCTGCACC-3′;
- TNFSF9-F: 5′-GGCTGGAGTCTACTATGTCTTCT-3′;
- TNFSF9-R: 5′-CGTGTCCTCTTTGTAGCTCAGG-3′;
- CD70-F: 5′-GCTTTGGTCCCATTGGTCG-3′;
- CD70-R: 5′-CGTCCCACCCAAGTGACTC-3′;
- CD274 (PD-L1)-F: 5′-TGGCATTTGCTGAACGCATTT-3′;
- CD274 (PD-L1)-R: 5′-TGCAGCCAGGTCTAATTGTTTT-3′;
- GAPDH-F: 5′-CTGGGCTACACTGAGCACC-3′;
- GAPDH-R: 5′-AAGTGGTCGTTGAG GGCAATG-3′.
2.8. Western Blot Analysis
2.9. T Cell Separation, Activation, and Culture
2.10. T Cell Cytotoxicity Assays
2.11. Xenograft Tumor Mouse Models
2.12. Flow Cytometry Analysis
2.12.1. Apoptosis
2.12.2. Co-Culture Assay
2.12.3. Xenografts Tumors Infiltrating Lymphocyte Analysis
2.13. Clinical Specimens
2.14. Immunohistochemical and Immunofluorescence Staining
2.15. Statistical Analysis
3. Results
3.1. FTSJ1 Is Associated with Poor Prognosis in TNBC
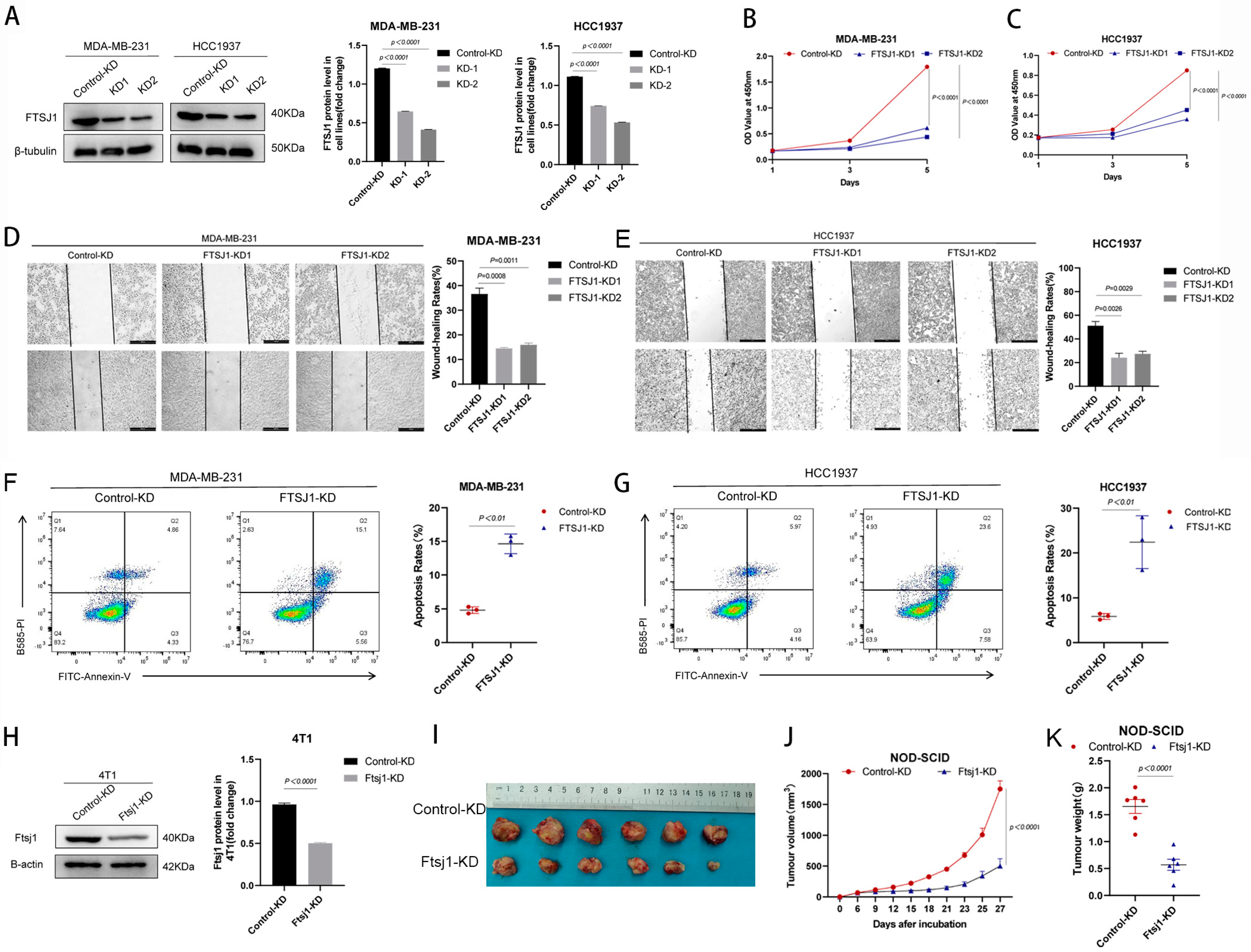
3.2. FTSJ1 Knockdown Suppresses TNBC Cell Proliferation, Migration and Promotes Apoptosis
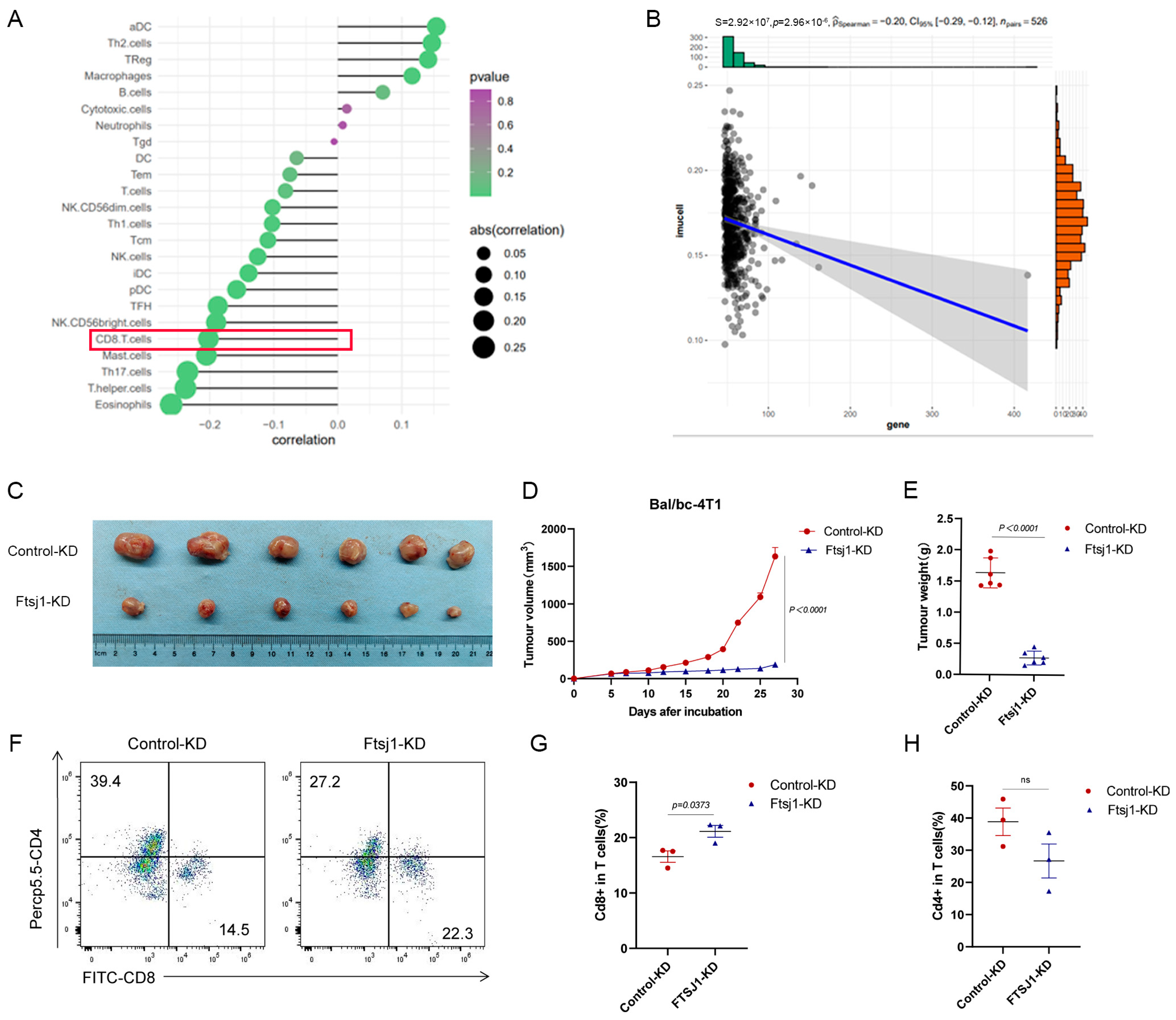
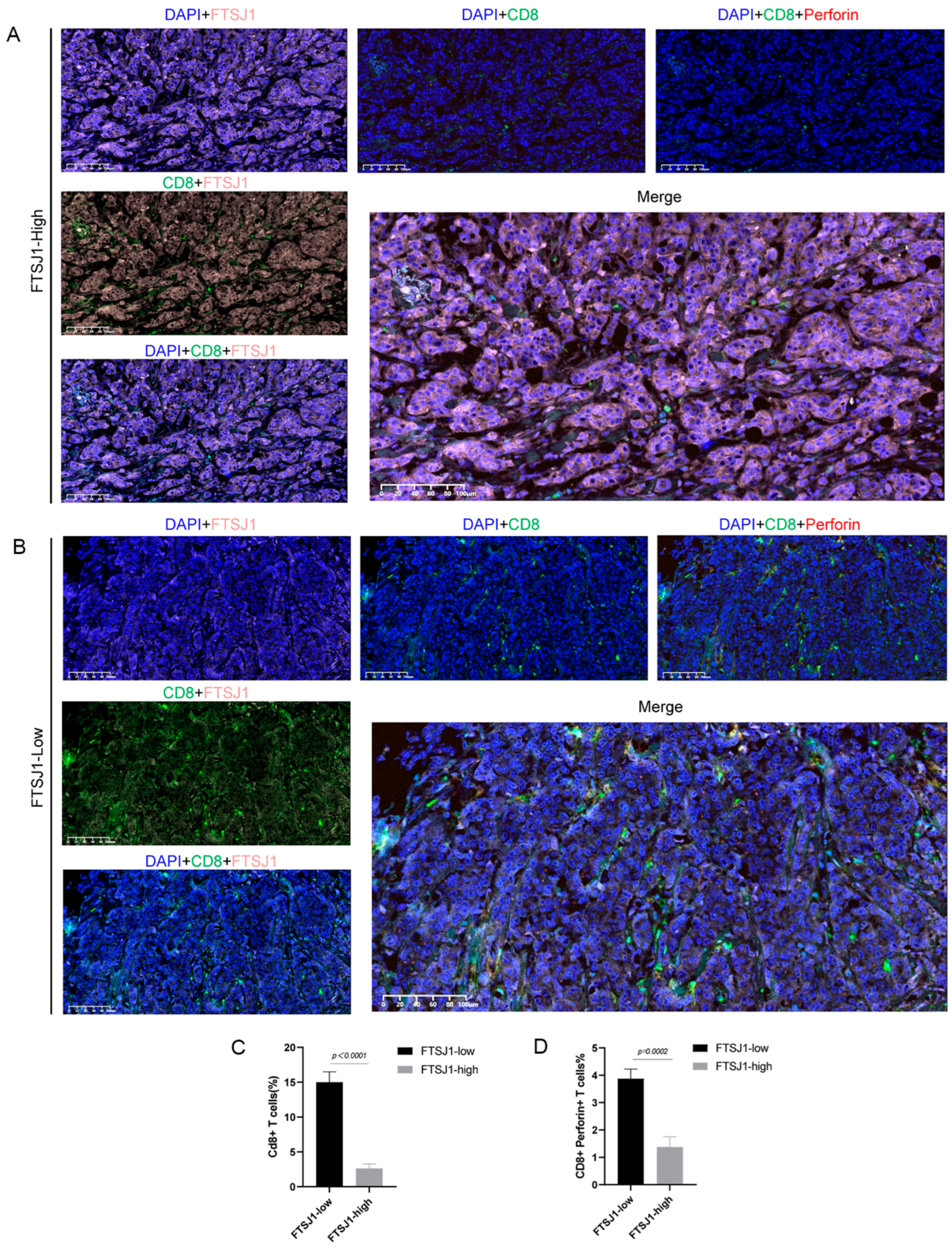
3.3. FTSJ1 Knockdown Enhances Tumor-Infiltrating CD8+ T Cell
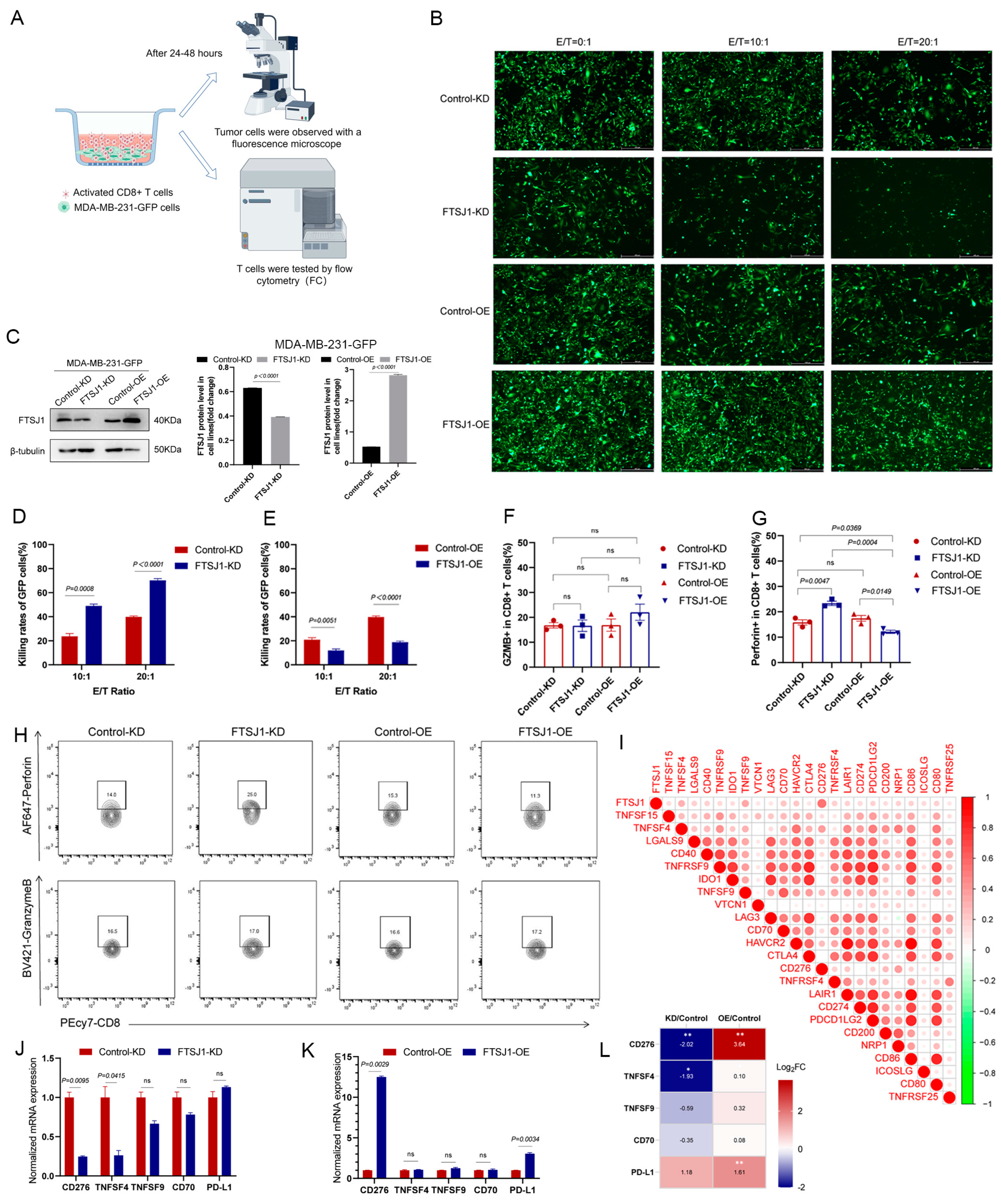
3.4. FTSJ1 Knockdown Increases the Sensitivity of TNBC to T Cell Killing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Breast Cancer. Available online: https://www.iarc.who.int/cancer-type/breast-cancer/ (accessed on 10 December 2023).
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-Negative Breast Cancer: Clinical Features and Patterns of Recurrence. Clin. Cancer Res. 2007, 13 Pt 1, 4429–4434. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Heppner, B.I.; Weber, K.E.; Budczies, J.; Huober, J.; Klauschen, F.; Furlanetto, J.; et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018, 19, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Gruosso, T.; Gigoux, M.; Manem, V.S.K.; Bertos, N.; Zuo, D.; Perlitch, I.; Saleh, S.M.I.; Zhao, H.; Souleimanova, M.; Johnson, R.M.; et al. Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J. Clin. Investig. 2019, 129, 1785–1800. [Google Scholar] [CrossRef] [PubMed]
- Marra, A.; Viale, G.; Curigliano, G. Recent advances in triple negative breast cancer: The immunotherapy era. BMC Med. 2019, 17, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Begley, U.; Dyavaiah, M.; Patil, A.; Rooney, J.P.; DiRenzo, D.; Young, C.M.; Conklin, D.S.; Zitomer, R.S.; Begley, T.J. Trm9-Catalyzed tRNA Modifications Link Translation to the DNA Damage Response. Mol. Cell 2007, 28, 860–870. [Google Scholar] [CrossRef] [PubMed]
- Begley, U.; Sosa, M.S.; Avivar-Valderas, A.; Patil, A.; Endres, L.; Estrada, Y.; Chan, C.T.; Su, D.; Dedon, P.C.; Aguirre-Ghiso, J.A.; et al. A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-α. EMBO Mol. Med. 2013, 5, 366–383. [Google Scholar] [CrossRef]
- Rapino, F.; Delaunay, S.; Rambow, F.; Zhou, Z.; Tharun, L.; De Tullio, P.; Sin, O.; Shostak, K.; Schmitz, S.; Piepers, J.; et al. Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature 2018, 558, 605–609. [Google Scholar] [CrossRef]
- Delaunay, S.; Rapino, F.; Tharun, L.; Zhou, Z.; Heukamp, L.; Termathe, M.; Shostak, K.; Klevernic, I.; Florin, A.; Desmecht, H.; et al. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J. Exp. Med. 2016, 213, 2503–2523. [Google Scholar] [CrossRef]
- Dai, L.; Xing, L.; Gong, P.; Zhang, K.; Gao, X.; Zheng, Z.; Zhou, J.; Guo, Y.; Guo, S.; Zhang, F. Positive association of the FTSJ1 gene polymorphisms with nonsyndromic X-linked mental retardation in young Chinese male subjects. J. Hum. Genet. 2008, 53, 592–597. [Google Scholar] [CrossRef]
- Nagayoshi, Y.; Chujo, T.; Hirata, S.; Nakatsuka, H.; Chen, C.-W.; Takakura, M.; Miyauchi, K.; Ikeuchi, Y.; Carlyle, B.C.; Kitchen, R.R.; et al. Loss of Ftsj1 perturbs codon-specific translation efficiency in the brain and is associated with X-linked intellectual disability. Sci. Adv. 2021, 7, eabf3072. [Google Scholar] [CrossRef]
- Takano, K.; Nakagawa, E.; Inoue, K.; Kamada, F.; Kure, S.; Goto, Y.I.; Japanese Mental Retardation Consortium. A loss-of-function mutation in the FTSJ1 gene causes nonsyndromic X-linked mental retardation in a Japanese family. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2008, 147B, 479–484. [Google Scholar] [CrossRef]
- von Halbach, V.B.U.; Venz, S.; Nwakor, S.; Hentschker, C.; Hammer, E.; Junker, H.; Kuss, A.W.; von Halbach, O.B.U.; Jensen, L.R. Deficiency in FTSJ1 Affects Neuronal Plasticity in the Hippocampal Formation of Mice. Biology 2022, 11, 1011. [Google Scholar] [CrossRef]
- He, Q.H.; Yang, L.; Gao, K.; Ding, P.; Chen, Q.; Xiong, J.; Yang, W.; Song, Y.; Wang, L.; Wang, Y.; et al. FTSJ1 regulates tRNA 2′-O-methyladenosine modification and suppresses the malignancy of NSCLC via inhibiting DRAM1 expression. Cell Death Dis. 2020, 11, 348. [Google Scholar] [CrossRef] [PubMed]
- Holzer, K.; Ori, A.; Cooke, A.; Dauch, D.; Drucker, E.; Riemenschneider, P.; Andres-Pons, A.; DiGuilio, A.L.; Mackmull, M.-T.; Baßler, J.; et al. Nucleoporin Nup155 is part of the p53 network in liver cancer. Nat. Commun. 2019, 10, 2147. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Li, C.; Tang, Z.; Zhang, W.; Ye, Z.; Liu, F. GEPIA2021: Integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. 2021, 49, W242–W246. [Google Scholar] [CrossRef]
- Ahmadzadeh, M.; Johnson, L.A.; Heemskerk, B.; Wunderlich, J.R.; Dudley, M.E.; White, D.E.; Rosenberg, S.A. Tumor antigen–specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 2009, 114, 1537–1544. [Google Scholar] [CrossRef]
- Whitford, P.; George, W.; Campbell, A.M. Flow cytometric analysis of tumour infiltrating lymphocyte activation and tumour cell MHC Class I and II expression in breast cancer patients. Cancer Lett. 1992, 61, 157–164. [Google Scholar] [CrossRef]
- Chin, Y.; Janseens, J.; Vandepitte, J.; Vandenbrande, J.; Opdebeek, L.; Raus, J. Phenotypic analysis of tumor-infiltrating lymphocytes from human breast cancer. Anticancer Res. 1992, 12, 1463–1466. [Google Scholar]
- Savas, P.; Salgado, R.; Denkert, C.; Sotiriou, C.; Darcy, P.K.; Smyth, M.J.; Loi, S. Clinical relevance of host immunity in breast cancer: From TILs to the clinic. Nat. Rev. Clin. Oncol. 2016, 13, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Loibl, S.; Noske, A.; Roller, M.; Müller, B.M.; Komor, M.; Budczies, J.; Darb-Esfahani, S.; Kronenwett, R.; Hanusch, C.; et al. Tumor-Associated Lymphocytes as an Independent Predictor of Response to Neoadjuvant Chemotherapy in Breast Cancer. J. Clin. Oncol. 2010, 28, 105–113. [Google Scholar] [CrossRef]
- Loi, S.; Sirtaine, N.; Piette, F.; Salgado, R.; Viale, G.; Van Eenoo, F.; Rouas, G.; Francis, P.; Crown, J.P.; Hitre, E.; et al. Prognostic and Predictive Value of Tumor-Infiltrating Lymphocytes in a Phase III Randomized Adjuvant Breast Cancer Trial in Node-Positive Breast Cancer Comparing the Addition of Docetaxel to Doxorubicin With Doxorubicin-Based Chemotherapy: BIG 02-98. J. Clin. Oncol. 2013, 31, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.R.; Provenzano, E.; Dawson, S.-J.; Blows, F.M.; Liu, B.; Shah, M.; Earl, H.M.; Poole, C.J.; Hiller, L.; Dunn, J.A.; et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann. Oncol. 2014, 25, 1536–1543. [Google Scholar] [CrossRef] [PubMed]
- Loi, S.; Michiels, S.; Salgado, R.; Sirtaine, N.; Jose, V.; Fumagalli, D.; Kellokumpu-Lehtinen, P.-L.; Bono, P.; Kataja, V.; Desmedt, C.; et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: Results from the FinHER trial. Ann. Oncol. 2014, 25, 1544–1550. [Google Scholar] [CrossRef]
- Adams, S.; Gray, R.J.; Demaria, S.; Goldstein, L.; Perez, E.A.; Shulman, L.N.; Martino, S.; Wang, M.; Jones, V.E.; Saphner, T.J.; et al. Prognostic Value of Tumor-Infiltrating Lymphocytes in Triple-Negative Breast Cancers from Two Phase III Randomized Adjuvant Breast Cancer Trials: ECOG 2197 and ECOG. J. Clin. Oncol. 2014, 32, 2959–2966. [Google Scholar] [CrossRef]
- Salgado, R.; Denkert, C.; Campbell, C.; Savas, P.; Nuciforo, P.; Aura, C.; de Azambuja, E.; Eidtmann, H.; Ellis, C.E.; Baselga, J.; et al. Tumor-Infiltrating Lymphocytes and Associations with Pathological Complete Response and Event-Free Survival in HER2-Positive Early-Stage Breast Cancer Treated with Lapatinib and Trastuzumab: A Secondary Analysis of the NeoALTTO Trial. JAMA Oncol. 2015, 1, 448–454. [Google Scholar] [CrossRef] [PubMed]
- So, J.Y.; Ohm, J.; Lipkowitz, S.; Yang, L. Triple negative breast cancer (TNBC): Non-genetic tumor heterogeneity and immune microenvironment: Emerging treatment options. Pharmacol. Ther. 2022, 237, 108253. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Sharma, P.; Allison, J.P. Immune Checkpoint Targeting in Cancer Therapy: Toward Combination Strategies with Curative Potential. Cell 2015, 161, 205–214. [Google Scholar] [CrossRef]
- Drake, C.G.; Lipson, E.J.; Brahmer, J.R. Breathing new life into immunotherapy: Review of melanoma, lung and kidney cancer. Nat. Rev. Clin. Oncol. 2014, 11, 24–37. [Google Scholar] [CrossRef]
- Schmid, P.; Rugo, H.S.; Adams, S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Henschel, V.; Molinero, L.; Chui, S.Y.; et al. Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): Updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2020, 21, 44–59. [Google Scholar] [CrossRef]
- Harlin, H.; Kuna, T.V.; Peterson, A.C.; Meng, Y.; Gajewski, T.F. Tumor progression despite massive influx of activated CD8+ T cells in a patient with malignant melanoma ascites. Cancer Immunol. Immunother. 2006, 55, 1185–1197. [Google Scholar] [CrossRef]
- Li, K.-K.; Adams, D.H. Antitumor CD8+T cells in hepatocellular carcinoma: Present but exhausted. Hepatology 2014, 59, 1232–1234. [Google Scholar] [CrossRef]
- Sowell, R.T.; Kaech, S.M. Probing the Diversity of T Cell Dysfunction in Cancer. Cell 2016, 166, 1362–1364. [Google Scholar] [CrossRef]
- Chapoval, A.I.; Ni, J.; Lau, J.S.; Wilcox, R.S.; Flies, D.B.; Liu, D.; Dong, H.; Sica, G.L.; Zhu, G.; Tamada, K.; et al. B7-H3: A costimulatory molecule for T cell activation and IFN-gamma production. Nat. Immunol. 2001, 2, 269–274. [Google Scholar] [CrossRef]
- Flem-Karlsen, K.; Fodstad, Ø.; Tan, M.; Nunes-Xavier, C.E. B7-H3 in Cancer—Beyond Immune Regulation. Trends Cancer 2018, 4, 401–404. [Google Scholar] [CrossRef]
- Maeda, N.; Yoshimura, K.; Yamamoto, S.; Kuramasu, A.; Inoue, M.; Suzuki, N.; Watanabe, Y.; Maeda, Y.; Kamei, R.; Tsunedomi, R.; et al. Expression of B7-H3, a Potential Factor of Tumor Immune Evasion in Combination with the Number of Regulatory T Cells, Affects Against Recurrence-Free Survival in Breast Cancer Patients. Ann. Surg. Oncol. 2014, 21, 546–554. [Google Scholar] [CrossRef]
- Arigami, T.; Narita, N.; Mizuno, R.; Nguyen, L.B.; Ye, X.M.; Chung, A.; Giuliano, A.E.; Hoon, D.S.B.M. B7–H3 Ligand Expression by Primary Breast Cancer and Associated with Regional Nodal Metastasis. Ann. Surg. 2010, 252, 1044–1051. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Liu, Q.; Zhong, S.; Wei, R.; Luo, J.-L. Triple-Negative Breast Cancer Intrinsic FTSJ1 Favors Tumor Progression and Attenuates CD8+ T Cell Infiltration. Cancers 2024, 16, 597. https://doi.org/10.3390/cancers16030597
Sun Y, Liu Q, Zhong S, Wei R, Luo J-L. Triple-Negative Breast Cancer Intrinsic FTSJ1 Favors Tumor Progression and Attenuates CD8+ T Cell Infiltration. Cancers. 2024; 16(3):597. https://doi.org/10.3390/cancers16030597
Chicago/Turabian StyleSun, Yangqing, Qingqing Liu, Shangwei Zhong, Rui Wei, and Jun-Li Luo. 2024. "Triple-Negative Breast Cancer Intrinsic FTSJ1 Favors Tumor Progression and Attenuates CD8+ T Cell Infiltration" Cancers 16, no. 3: 597. https://doi.org/10.3390/cancers16030597
APA StyleSun, Y., Liu, Q., Zhong, S., Wei, R., & Luo, J.-L. (2024). Triple-Negative Breast Cancer Intrinsic FTSJ1 Favors Tumor Progression and Attenuates CD8+ T Cell Infiltration. Cancers, 16(3), 597. https://doi.org/10.3390/cancers16030597




