Multiparametric Radiogenomic Model to Predict Survival in Patients with Glioblastoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Histopathological Data
2.2. Image Analysis
2.3. Statistical Analysis
3. Results
3.1. Clinical Characteristics of Patient Population
3.2. Training Model
3.3. External Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wesseling, P.; Capper, D. WHO 2016 Classification of Gliomas. Neuropathol. Appl. Neurobiol. 2018, 44, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Gladson, C.L.; Prayson, R.A.; Liu, W.M. The Pathobiology of Glioma Tumors. Annu. Rev. Pathol. Mech. Dis. 2010, 5, 33–50. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Bauchet, L.; Davis, F.G.; Deltour, I.; Fisher, J.L.; Langer, C.E.; Pekmezci, M.; Schwartzbaum, J.A.; Turner, M.C.; Walsh, K.M.; et al. The Epidemiology of Glioma in Adults: A State of the Science Review. Neuro Oncol. 2014, 16, 896–913. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Adamson, C.; Kanu, O.O.; Mehta, A.I.; Di, C.; Lin, N.; Mattox, A.K.; Bigner, D.D. Glioblastoma Multiforme: A Review of Where We Have Been and Where We Are Going. Expert. Opin. Investig. Drugs 2009, 18, 1061–1083. [Google Scholar] [CrossRef] [PubMed]
- Thakkar, J.P.; Dolecek, T.A.; Horbinski, C.; Ostrom, Q.T.; Lightner, D.D.; Barnholtz-Sloan, J.S.; Villano, J.L. Epidemiologic and Molecular Prognostic Review of Glioblastoma. Cancer Epidemiol. Biomark. Prev. 2014, 23, 1985–1996. [Google Scholar] [CrossRef]
- Szopa, W.; Burley, T.A.; Kramer-Marek, G.; Kaspera, W. Diagnostic and Therapeutic Biomarkers in Glioblastoma: Current Status and Future Perspectives. Biomed. Res. Int. 2017, 2017, 8013575. [Google Scholar] [CrossRef] [PubMed]
- Stark, A.M.; Van De Bergh, J.; Hedderich, J.; Mehdorn, H.M.; Nabavi, A. Glioblastoma: Clinical Characteristics, Prognostic Factors and Survival in 492 Patients. Clin. Neurol. Neurosurg. 2012, 114, 840–845. [Google Scholar] [CrossRef]
- Lamborn, K.R.; Chang, S.M.; Prados, M.D. Prognostic Factors for Survival of Patients with Glioblastoma: Recursive Partitioning Analysis. Neuro Oncol. 2004, 6, 227. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Felsberg, J.; Hartmann, C.; Berger, H.; Steinbach, J.P.; Schramm, J.; Westphal, M.; Schackert, G.; Simon, M.; Tonn, J.C.; et al. Molecular Predictors of Progression-Free and Overall Survival in Patients with Newly Diagnosed Glioblastoma: A Prospective Translational Study of the German Glioma Network. J. Clin. Oncol. 2009, 27, 5743–5750. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.F.; Ottaviani, D.; Tazare, J.; Gregson, J.; Kitchen, N.; Brandner, S.; Fersht, N.; Mulholland, P. Survival Outcomes and Prognostic Factors in Glioblastoma. Cancers 2022, 14, 3161. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro Oncol. 2021, 23, 1231. [Google Scholar] [CrossRef]
- Wiestler, B.; Capper, D.; Holland-Letz, T.; Korshunov, A.; Von Deimling, A.; Pfister, S.M.; Platten, M.; Weller, M.; Wick, W. ATRX Loss Refines the Classification of Anaplastic Gliomas and Identifies a Subgroup of IDH Mutant Astrocytic Tumors with Better Prognosis. Acta Neuropathol. 2013, 126, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.E.; Mazor, T.; Hong, C.; Barnes, M.; Aihara, K.; McLean, C.Y.; Fouse, S.D.; Yamamoto, S.; Ueda, H.; Tatsuno, K.; et al. Mutational Analysis Reveals the Origin and Therapy-Driven Evolution of Recurrent Glioma. Science 2014, 343, 189. [Google Scholar] [CrossRef] [PubMed]
- Wick, W.; Platten, M.; Meisner, C.; Felsberg, J.; Tabatabai, G.; Simon, M.; Nikkhah, G.; Papsdorf, K.; Steinbach, J.P.; Sabel, M.; et al. Temozolomide Chemotherapy Alone versus Radiotherapy Alone for Malignant Astrocytoma in the Elderly: The NOA-08 Randomised, Phase 3 Trial. Lancet Oncol. 2012, 13, 707–715. [Google Scholar] [CrossRef]
- Jiao, Y.; Killela, P.J.; Reitman, Z.J.; Rasheed, B.A.; Heaphy, C.M.; de Wilde, R.F.; Rodriguez, F.J.; Rosemberg, S.; Oba-Shinjo, S.M.; Marie, S.K.N.; et al. Frequent ATRX, CIC, FUBP1 and IDH1 Mutations Refine the Classification of Malignant Gliomas. Oncotarget 2012, 3, 709. [Google Scholar] [CrossRef]
- Heimberger, A.B.; Hlatky, R.; Suki, D.; Yang, D.; Weinberg, J.; Gilbert, M.; Sawaya, R.; Aldape, K. Prognostic Effect of Epidermal Growth Factor Receptor and EGFRvIII in Glioblastoma Multiforme Patients. Clin. Cancer Res. 2005, 11, 1462–1466. [Google Scholar] [CrossRef]
- Huang, P.H.; Xu, A.M.; White, F.M. Oncogenic EGFR Signaling Networks in Glioma. Sci. Signal 2009, 2, re6. [Google Scholar] [CrossRef]
- Burger, P.C.; Kleihues, P. Cytologic Composition of the Untreated Glioblastoma with Implications for Evaluation of Needle Biopsies. Cancer 1989, 63, 2014–2023. [Google Scholar] [CrossRef]
- Vaquero, J.; Martínez, R.; Manrique, M. Stereotactic Biopsy for Brain Tumors: Is It Always Necessary? Surg. Neurol. 2000, 53, 432–438. [Google Scholar] [CrossRef]
- Bruner, J.M.; Inouye, L.; Fuller, G.N.; Langford, L.A. Diagnostic Discrepancies and Their Clinical Impact in a Neuropathology Referral Practice. Cancer 1997, 79, 796–803. [Google Scholar] [CrossRef]
- McGirt, M.J.; Villavicencio, A.T.; Bulsara, K.R.; Friedman, A.H. MRI-Guided Stereotactic Biopsy in the Diagnosis of Glioma: Comparison of Biopsy and Surgical Resection Specimen. Surg. Neurol. 2003, 59, 279–283. [Google Scholar] [CrossRef]
- Reithmeier, T.; Lopez, W.O.; Doostkam, S.; MacHein, M.R.; Pinsker, M.O.; Trippel, M.; Nikkhah, G. Intraindividual Comparison of Histopathological Diagnosis Obtained by Stereotactic Serial Biopsy to Open Surgical Resection Specimen in Patients with Intracranial Tumours. Clin. Neurol. Neurosurg. 2013, 115, 1955–1960. [Google Scholar] [CrossRef]
- Aftab, K.; Aamir, F.B.; Mallick, S.; Mubarak, F.; Pope, W.B.; Mikkelsen, T.; Rock, J.P.; Enam, S.A. Radiomics for Precision Medicine in Glioblastoma. J. Neurooncol. 2022, 156, 217–231. [Google Scholar] [CrossRef]
- Nuechterlein, N.; Li, B.; Feroze, A.; Holland, E.C.; Shapiro, L.; Haynor, D.; Fink, J.; Cimino, P.J.; Allen, P.G. Radiogenomic Modeling Predicts Survival-Associated Prognostic Groups in Glioblastoma. Neurooncol. Adv. 2021, 3, vdab004. [Google Scholar] [CrossRef]
- Sun, Q.; Chen, Y.; Liang, C.; Zhao, Y.; Lv, X.; Zou, Y.; Yan, K.; Zheng, H.; Liang, D.; Li, Z.C. Biologic Pathways Underlying Prognostic Radiomics Phenotypes from Paired Mri and Rna Sequencing in Glioblastoma. Radiology 2021, 301, 654–663. [Google Scholar] [CrossRef]
- Ellingson, B.M.; Bendszus, M.; Boxerman, J.; Barboriak, D.; Erickson, B.J.; Smits, M.; Nelson, S.J.; Gerstner, E.; Alexander, B.; Goldmacher, G.; et al. Editor’s Choice: Consensus Recommendations for a Standardized Brain Tumor Imaging Protocol in Clinical Trials. Neuro Oncol. 2015, 17, 1188. [Google Scholar] [CrossRef]
- Soni, N.; Priya, S.; Bathla, G.; Soni, X.N.; Priya, X.S.; Bathla, X.G. Texture Analysis in Cerebral Gliomas: A Review of the Literature. Am. J. Neuroradiol. 2019, 40, 928–934. [Google Scholar] [CrossRef] [PubMed]
- Amadasun, M.; King, R. Textural Features Corresponding to Textural Properties. Inst. Electr. Electron. Eng. Trans. Syst. Man. Cybern. 1989, 19, 1264–1274. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression Shrinkage and Selection via the Lasso. J. R. Stat. Soc. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- DeLong, E.R.; Delong, D.M.; Clarke-Pearson, D.L. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, C.; Hentschel, B.; Simon, M.; Westphal, M.; Schackert, G.; Tonn, J.C.; Loeffler, M.; Reifenberger, G.; Pietsch, T.; Von Deimling, A.; et al. Long-Term Survival in Primary Glioblastoma with versus without Isocitrate Dehydrogenase Mutations. Clin. Cancer Res. 2013, 19, 5146–5157. [Google Scholar] [CrossRef]
- Zhou, M.; Hall, L.; Goldgof, D.; Russo, R.; Balagurunathan, Y.; Gillies, R.; Gatenby, R. Radiologically Defined Ecological Dynamics and Clinical Outcomes in Glioblastoma Multiforme: Preliminary Results. Transl. Oncol. 2014, 7, 5–13. [Google Scholar] [CrossRef]
- Heydari, M. Prognosis of Glioblastoma Multiforme Using Textural Properties on MRI. ProQuest Dissertations and Theses, University of Alberta. 2009. Available online: https://ui.adsabs.harvard.edu/abs/2009MsT..........2H/abstract (accessed on 25 August 2023).
- Chaddad, A.; Tanougast, C. Extracted Magnetic Resonance Texture Features Discriminate between Phenotypes and Are Associated with Overall Survival in Glioblastoma Multiforme Patients. Med. Biol. Eng. Comput. 2016, 54, 1707–1718. [Google Scholar] [CrossRef]
- Li, Z.C.; Li, Q.; Sun, Q.; Luo, R.; Chen, Y. Identifying a Radiomics Imaging Signature for Prediction of Overall Survival in Glioblastoma Multiforme. In Proceedings of the BMEiCON 2017-10th Biomedical Engineering International Conference, Hokkaido, Japan, 31 August–2 September 2017; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2017; Volume 2017, pp. 1–4. [Google Scholar]
- Ditmer, A.; Zhang, B.; Shujaat, T.; Pavlina, A.; Luibrand, N.; Gaskill-Shipley, M.; Vagal, A. Diagnostic Accuracy of MRI Texture Analysis for Grading Gliomas. J. Neurooncol. 2018, 140, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Jiang, J.; Zhang, S.; Shi, J.; Xu, K.; Shen, N.; Zhang, J.; Li, L.; Zhao, L.; Zhang, J.; et al. Radiomics Based on Multicontrast MRI Can Precisely Differentiate among Glioma Subtypes and Predict Tumour-Proliferative Behaviour. Eur. Radiol. 2019, 29, 1986–1996. [Google Scholar] [CrossRef]
- Yang, D.; Rao, G.; Martinez, J.; Veeraraghavan, A.; Rao, A. Evaluation of Tumor-derived MRI-texture Features for Discrimination of Molecular Subtypes and Prediction of 12month Survival Status in Glioblastoma. Int. J. Med. Phys. Res. Pract. 2015, 42, 6725–6735. [Google Scholar] [CrossRef]
- Drabycz, S.; Roldan, G.; de Robles, P.; Adler, D.; McIntyre, J.B.; Magliocco, A.M.; Cairncross, J.G.; Mitchell, J.R. An Analysis of Image Texture, Tumor Location, and MGMT Promoter Methylation in Glioblastoma using Magnetic Resonance Imaging. NeuroImage 2010, 49, 1398–1405. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kim, S.H.; Cho, J.; Kim, J.W.; Chang, J.H.; Kim, D.S.; Lee, K.S.; Suh, C.O. MGMT Gene Promoter Methylation as a Potent Prognostic Factor in Glioblastoma Treated with Temozolomide-Based Chemoradiotherapy: A Single-Institution Study. Int. J. Radiat. Oncol. Biol. Phys. 2012, 84, 661–667. [Google Scholar] [CrossRef]
- Binabaj, M.M.; Bahrami, A.; ShahidSales, S.; Joodi, M.; Joudi Mashhad, M.; Hassanian, S.M.; Anvari, K.; Avan, A. The Prognostic Value of MGMT Promoter Methylation in Glioblastoma: A Meta-Analysis of Clinical Trials. J. Cell Physiol. 2018, 233, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.-C.; Gorlia, T.; Hamou, M.-F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.; Niu, L.; Zhang, Y.; Wang, X.; Ma, K.; Yin, H.; Dai, J.; Zhou, W.; Pan, Y. Defining Optimal Cutoff Value of MGMT Promoter Methylation by ROC Analysis for Clinical Setting in Glioblastoma Patients. J. Neurooncol. 2017, 133, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Tixier, F.; Um, H.; Bermudez, D.; Iyer, A.; Apte, A.; Graham, M.S.; Nevel, K.S.; Deasy, J.O.; Young, R.J.; Veeraraghavan, H. Preoperative MRI-Radiomics Features Improve Prediction of Survival in Glioblastoma Patients over MGMT Methylation Status Alone. Oncotarget 2019, 10, 660–672. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, K.; Das, B.; Singh, A.K.; Misra, A.; Misra, S.; Misra, S.S. Prognostic Significance of Epidermal Growth Factor Receptor in Patients of Glioblastoma Multiforme. J. Clin. Diagn. Res. 2017, 11, EC05–EC08. [Google Scholar] [CrossRef]
- Xie, Y.; Tan, Y.; Yang, C.; Zhang, X.; Xu, C.; Qiao, X.; Xu, J.; Tian, S.; Fang, C.; Kang, C. Omics-Based Integrated Analysis Identified ATRX as a Biomarker Associated with Glioma Diagnosis and Prognosis. Cancer Biol. Med. 2019, 16, 784–796. [Google Scholar] [CrossRef]
- Hoebel, K.V.; Patel, J.B.; Beers, A.L.; Chang, K.; Singh, P.; Brown, J.M.; Pinho, M.C.; Batchelor, T.T.; Gerstner, E.R.; Rosen, B.R.; et al. Radiomics Repeatability Pitfalls in a Scan-Rescan MRI Study of Glioblastoma. Radiol. Artif. Intell. 2021, 3, e190199. [Google Scholar] [CrossRef]


| Total | Survival 18 Months (n = 42) | Survival < 18 Months (n = 74) | p Value | |
|---|---|---|---|---|
| Age (mean ± SD) | 59.6 ± 13.9 | 55.8 ± 14.6 | 61.7 ± 13.1 | 0.030 |
| Sex (M/F) | 62/54 | 21/21 | 41/33 | 0.575 |
| Tumor side (right/left) | 65/51 | 22/20 | 43/31 | 0.552 |
| Tumor location (n/%) | 0.266 | |||
| Frontal lobe | 45/39% | 16/14% | 29/25% | |
| Parietal lobe | 10/9% | 3/3% | 7/6% | |
| Temporal lobe | 48/41% | 16/14% | 32/27% | |
| Occipital lobe | 5/4% | 4/3% | 1/1% | |
| Cerebellum | 1/1% | 1/1% | 0/0% | |
| Basal ganglia | 7/6% | 2/2% | 5/4% | |
| MGMT (methylated/non-methylated) | 53/63 | 25/17 | 29/45 | 0.035 |
| * ATRX (wildtype/mutated) | 85/18 | 29/9 | 56/9 | 0.221 |
| ** EGFR (amplified/non-amplified) | 74/41 | 25/17 | 49/24 | 0.414 |
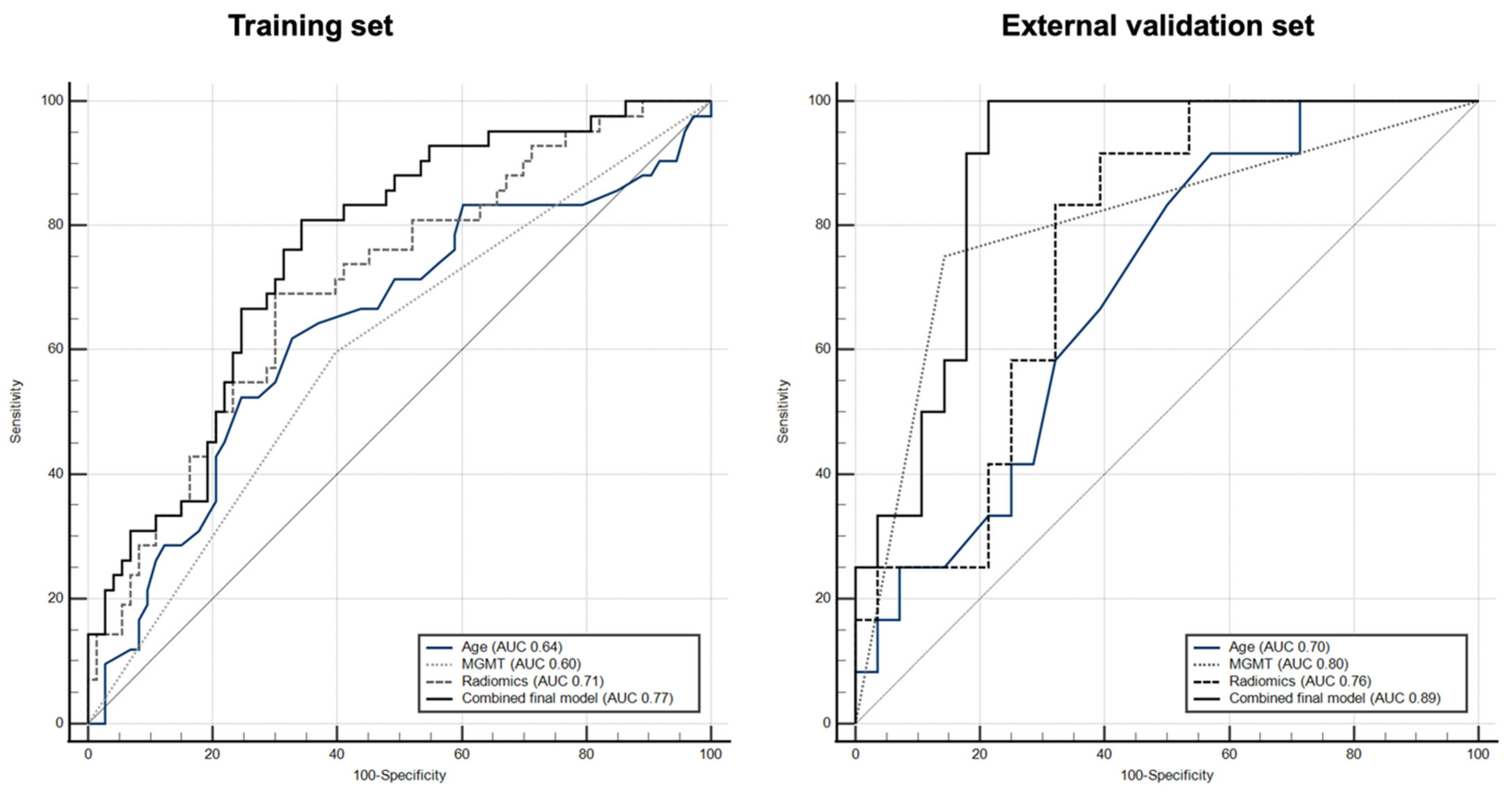
| Training Set (n = 116) | External Validation Set (n = 40) | |
|---|---|---|
| AUC/Sensitivity/Specificity | AUC/Sensitivity/Specificity | |
| Age | 0.64/61.9/67.6 | 0.70/91.7/42.9 |
| MGMT status | 0.60/59.5/60.8 | 0.80/75.0/85.7 |
| Radiomic model (combined 7 textures) | 0.71/69.0/70.3 | 0.76/91.7/60.7 |
| * Radiogenomic model | 0.77/81.0/66.0 | 0.89/100/78.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahmoudi, K.; Kim, D.H.; Tavakkol, E.; Kihira, S.; Bauer, A.; Tsankova, N.; Khan, F.; Hormigo, A.; Yedavalli, V.; Nael, K. Multiparametric Radiogenomic Model to Predict Survival in Patients with Glioblastoma. Cancers 2024, 16, 589. https://doi.org/10.3390/cancers16030589
Mahmoudi K, Kim DH, Tavakkol E, Kihira S, Bauer A, Tsankova N, Khan F, Hormigo A, Yedavalli V, Nael K. Multiparametric Radiogenomic Model to Predict Survival in Patients with Glioblastoma. Cancers. 2024; 16(3):589. https://doi.org/10.3390/cancers16030589
Chicago/Turabian StyleMahmoudi, Keon, Daniel H. Kim, Elham Tavakkol, Shingo Kihira, Adam Bauer, Nadejda Tsankova, Fahad Khan, Adilia Hormigo, Vivek Yedavalli, and Kambiz Nael. 2024. "Multiparametric Radiogenomic Model to Predict Survival in Patients with Glioblastoma" Cancers 16, no. 3: 589. https://doi.org/10.3390/cancers16030589
APA StyleMahmoudi, K., Kim, D. H., Tavakkol, E., Kihira, S., Bauer, A., Tsankova, N., Khan, F., Hormigo, A., Yedavalli, V., & Nael, K. (2024). Multiparametric Radiogenomic Model to Predict Survival in Patients with Glioblastoma. Cancers, 16(3), 589. https://doi.org/10.3390/cancers16030589






