RenseNet: A Deep Learning Network Incorporating Residual and Dense Blocks with Edge Conservative Module to Improve Small-Lesion Classification and Model Interpretation
Abstract
Simple Summary
Abstract
1. Introduction
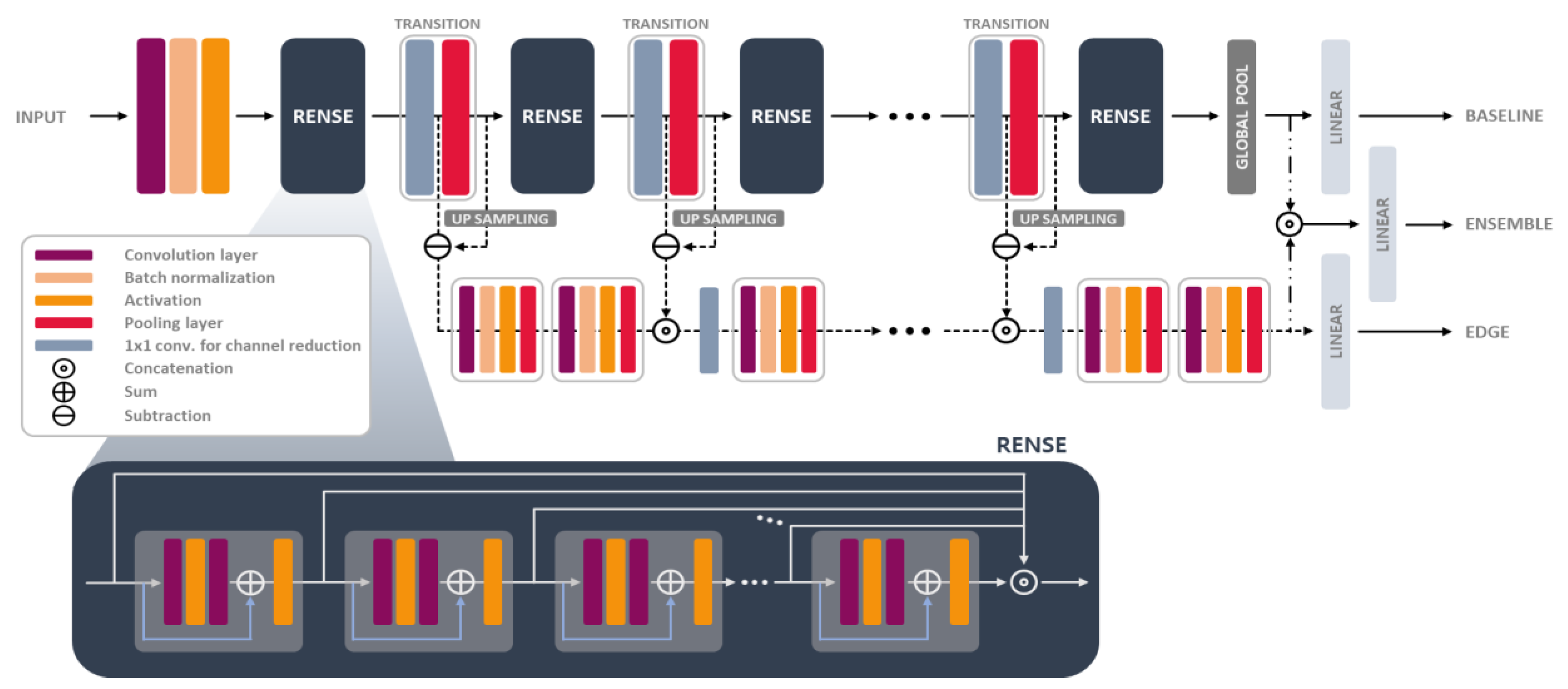
- A backbone network, which is composed of Rense blocks merged with residual and dense blocks, was proposed using mathematical analysis such that we can utilize the skip connections more efficiently and provide an insight for model structures.
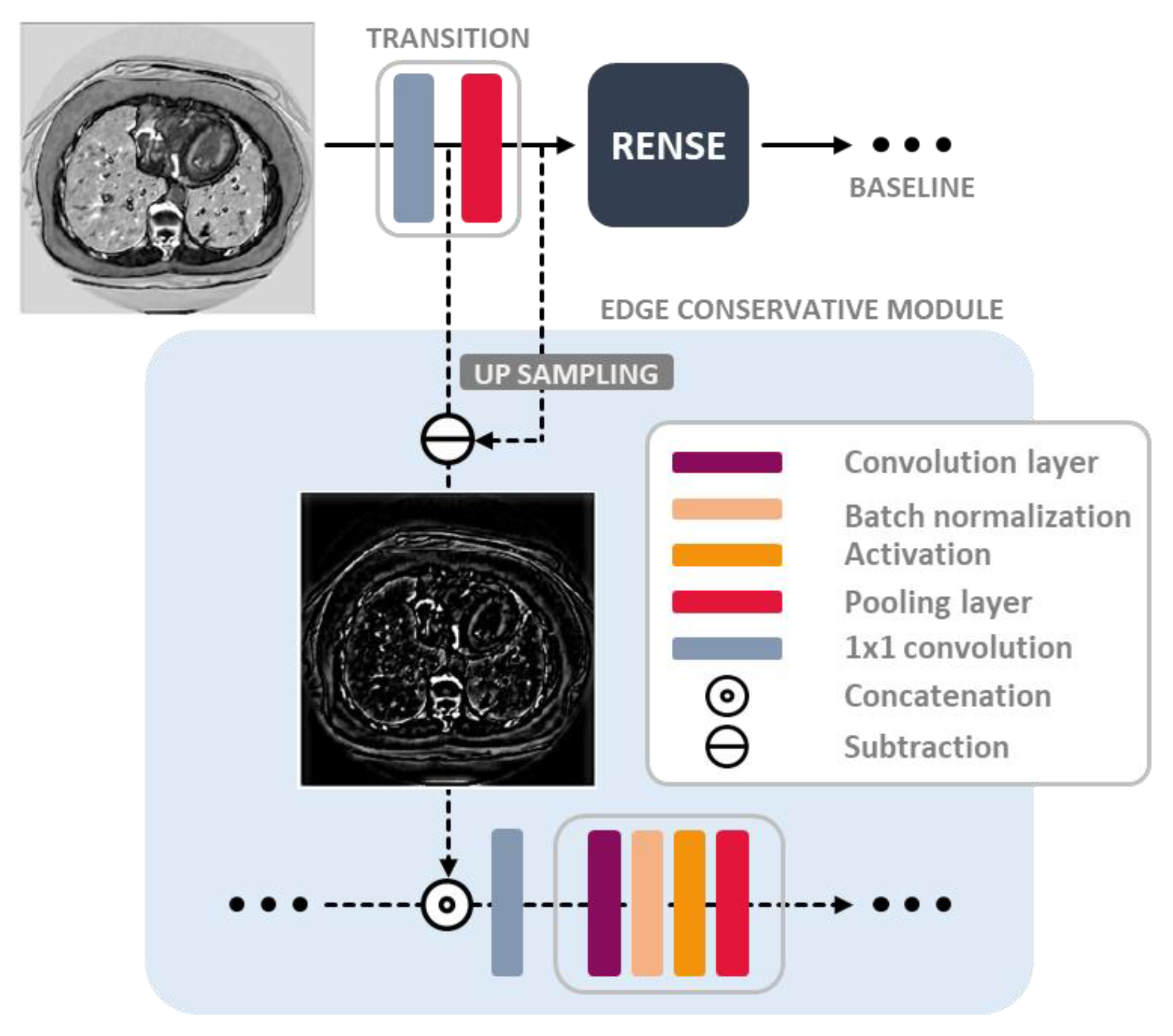
- The edge conservative module was designed in the compensation paths, by preserving the image detail lost from the subsampling (pooling) operation. Auxiliary features were then extracted in this module for better inference.
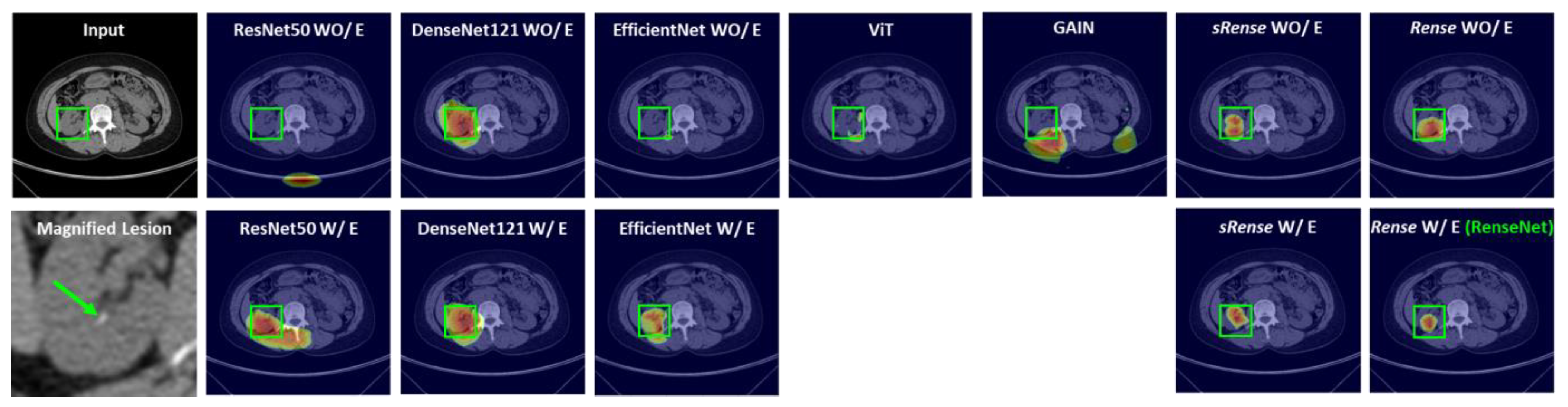
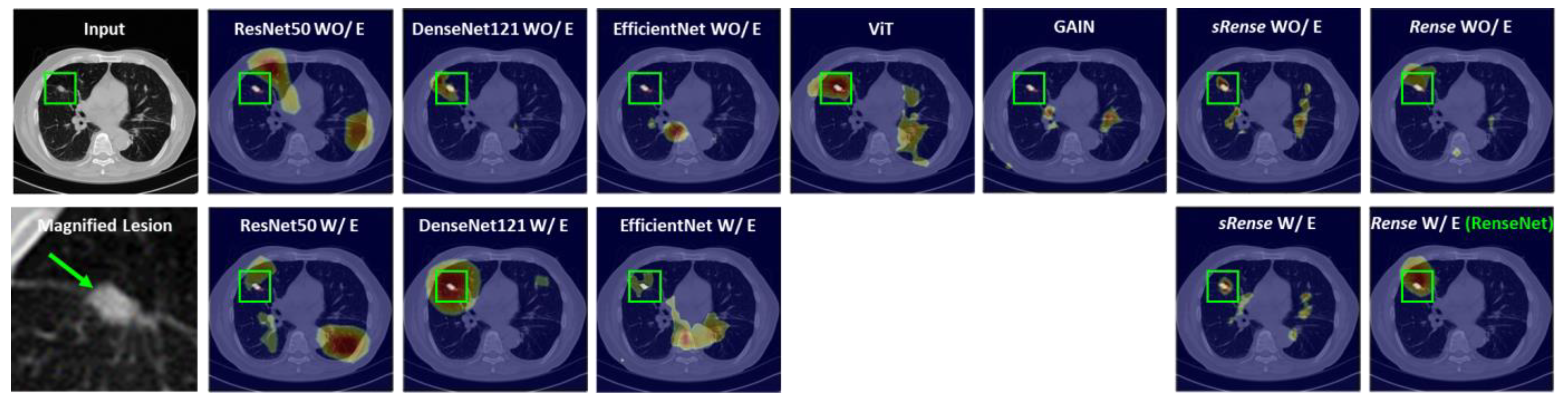
- The performance of the proposed model was evaluated using a public CT dataset of lung tumors and a private CT dataset of kidney stones, where there are often small-lesion scenarios. Validation results in terms of classification and Grad-CAM-based heatmaps show that the proposed model classifies small lesions better and can provide more trust to clinical users in their decision-making compared with cutting-edge techniques.
2. Related Works
3. Materials and Methods
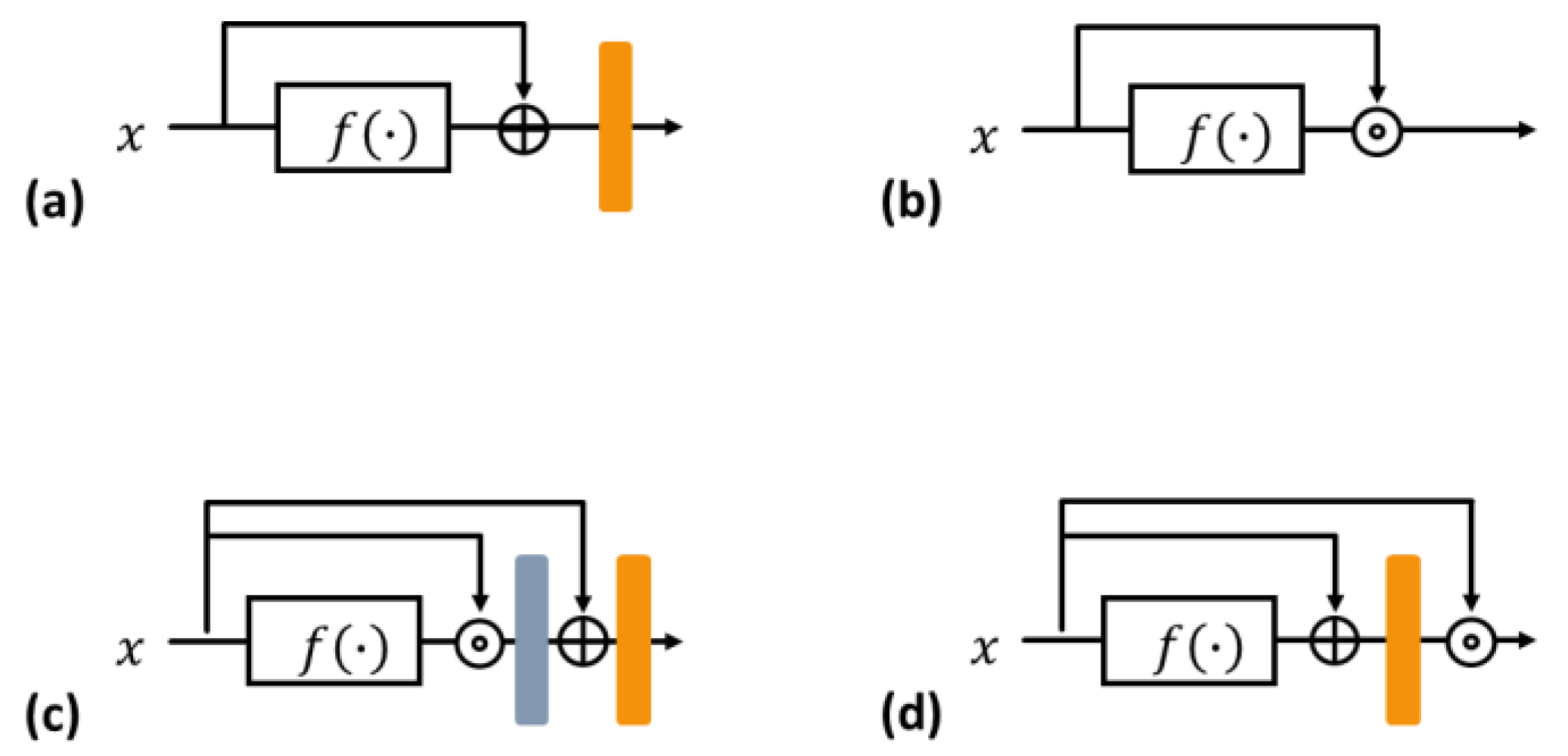
3.1. Residual and Dense Blocks
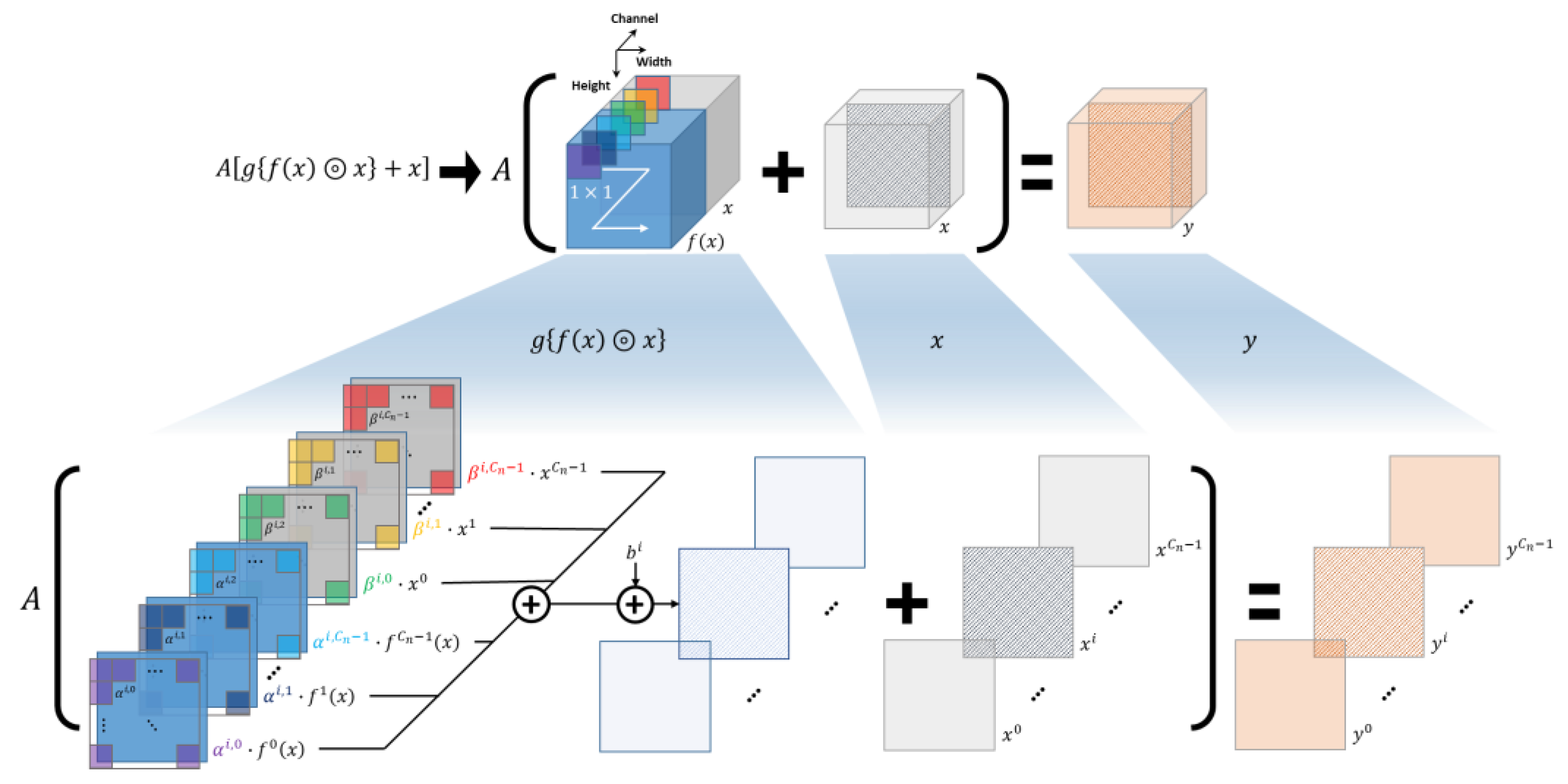
3.2. Rense Block
3.3. Edge Conservative Module
3.4. RenseNet Details
3.5. Dataset and Preparation
3.6. Grad-CAM-Based Heatmap
4. Results
4.1. Kidney Stone Dataset
4.2. Lung Tumor Dataset
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shen, L.; Zhao, W.; Xing, L. Patient-specific reconstruction of volumetric computed tomography images from a single projection view via deep learning. Nat. Biomed. Eng. 2019, 3, 880–888. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.; Badiei Khuzani, M.; Vasudevan, V.; Huang, C.; Ren, H.; Xiao, R.; Jia, X.; Xing, L. Machine learning techniques for biomedical image segmentation: An overview of technical aspects and introduction to state-of-art applications. Med. Phys. 2020, 47, e148–e167. [Google Scholar] [CrossRef]
- Seo, H.; Huang, C.; Bassenne, M.; Xiao, R.; Xing, L. Modified U-Net (mU-Net) with incorporation of object-dependent high level features for improved liver and liver-tumor segmentation in CT images. IEEE Trans. Med. Imaging 2019, 39, 1316–1325. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.; Bassenne, M.; Xing, L. Closing the gap between deep neural network modeling and biomedical decision-making metrics in segmentation via adaptive loss functions. IEEE Trans. Med. Imaging 2020, 40, 585–593. [Google Scholar] [CrossRef]
- Seo, H.; Yu, L.; Ren, H.; Li, X.; Shen, L.; Xing, L. Deep neural network with consistency regularization of multi-output channels for improved tumor detection and delineation. IEEE Trans. Med. Imaging 2021, 40, 3369–3378. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, H.; Shim, E.; Hwang, B.Y.; Kim, Y.; Lee, J.W.; Seo, H. Deep Learning-Based Automatic Segmentation of Mandible and Maxilla in Multi-Center CT Images. Appl. Sci. 2022, 12, 1358. [Google Scholar] [CrossRef]
- Gohel, P.; Singh, P.; Mohanty, M. Explainable AI: Current status and future directions. arXiv 2021, arXiv:2107.07045. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization. arXiv 2016, arXiv:1610.02391. [Google Scholar]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning Deep Features for Discriminative Localization. arXiv 2015, arXiv:1512.04150. [Google Scholar]
- Chattopadhay, A.; Sarkar, A.; Howlader, P.; Balasubramanian, V.N. Grad-cam++: Generalized gradient-based visual explanations for deep convolutional networks. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 839–847. [Google Scholar]
- Ramaswamy, H.G. Ablation-cam: Visual explanations for deep convolutional network via gradient-free localization. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Snowmass, CO, USA, 1–5 March 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 983–991. [Google Scholar]
- Fu, R.; Hu, Q.; Dong, X.; Guo, Y.; Gao, Y.; Li, B. Axiom-based grad-cam: Towards accurate visualization and explanation of cnns. arXiv 2020, arXiv:2008.02312. [Google Scholar]
- Binder, A.; Montavon, G.; Lapuschkin, S.; Müller, K.-R.; Samek, W. Layer-wise relevance propagation for neural networks with local renormalization layers. In Proceedings of the 25th International Conference on Artificial Neural Networks, Barcelona, Spain, 6–9 September 2016; Springer: Cham, Switzerland, 2016; pp. 63–71. [Google Scholar]
- Choi, S.J.; Kim, E.S.; Choi, K. Prediction of the histology of colorectal neoplasm in white light colonoscopic images using deep learning algorithms. Sci. Rep. 2021, 11, 5311. [Google Scholar] [CrossRef]
- Ganatra, N. A Comprehensive Study of Applying Object Detection Methods for Medical Image Analysis. In Proceedings of the 2021 8th International Conference on Computing for Sustainable Global Development (INDIACom), New Delhi, India, 17–19 March 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 821–826. [Google Scholar]
- Kitanovski, I.; Trojacanec, K.; Dimitrovski, I.; Loskovska, S. Modality classification using texture features. In International Conference on ICT Innovations; Springer: Berlin/Heidelberg, Germany, 2011; pp. 189–198. [Google Scholar]
- Riaz, F.; Silva, F.B.; Ribeiro, M.D.; Coimbra, M.T. Invariant gabor texture descriptors for classification of gastroenterology images. IEEE Trans. Biomed. Eng. 2012, 59, 2893–2904. [Google Scholar] [CrossRef] [PubMed]
- Manivannan, S.; Wang, R.; Trucco, E. Inter-cluster features for medical image classification. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Boston, MA, USA, 14–18 September 2014; Springer: Cham, Switzerland, 2014; pp. 345–352. [Google Scholar]
- Khan, S.; Yong, S.-P.; Deng, J.D. Ensemble classification with modified sift descriptor for medical image modality. In Proceedings of the 2015 International Conference on Image and Vision Computing New Zealand (IVCNZ), Auckland, New Zealand, 23–24 November 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 1–6. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 4700–4708. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; PMLR: New York, NY, USA, 2019; pp. 6105–6114. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16×16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Müftüoğlu, Z.; Kizrak, M.A.; Yildlnm, T. Differential privacy practice on diagnosis of COVID-19 radiology imaging using EfficientNet. In Proceedings of the 2020 International Conference on INnovations in Intelligent SysTems and Applications (INISTA), Novi Sad, Serbia, 24–26 August 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–6. [Google Scholar]
- Wang, H.; Ji, Y.; Song, K.; Sun, M.; Lv, P.; Zhang, T. ViT-P: Classification of Genitourinary Syndrome of Menopause from OCT Images Based on Vision Transformer Models. IEEE Trans. Instrum. Meas. 2021, 70, 1–14. [Google Scholar] [CrossRef]
- Chen, W.F.; Ou, H.Y.; Lin, H.Y.; Wei, C.P.; Liao, C.C.; Cheng, Y.F.; Pan, C.T. Development of Novel Residual-Dense-Attention (RDA) U-Net Network Architecture for Hepatocellular Carcinoma Segmentation. Diagnostics 2022, 12, 1916. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Jha, D.; Chanda, S.; Pal, U.; Johansen, H.D.; Johansen, D.; Riegler, M.A.; Ali, S.; Halvorsen, P. MSRF-Net: A multi-scale residual fusion network for biomedical image segmentation. IEEE J. Biomed. Health Inform. 2021, 26, 2252–2263. [Google Scholar] [CrossRef] [PubMed]
- Hénaff, O.J.; Simoncelli, E.P. Geodesics of learned representations. arXiv 2015, arXiv:1511.06394. [Google Scholar]
- Lustig, M.; Donoho, D.L.; Santos, J.M.; Pauly, J.M. Compressed sensing MRI. IEEE Signal Process. Mag. 2008, 25, 72–82. [Google Scholar] [CrossRef]
- Simpson, A.L.; Antonelli, M.; Bakas, S.; Bilello, M.; Farahani, K.; Van Ginneken, B.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; et al. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv 2019, arXiv:1902.09063. [Google Scholar]
- Li, K.; Wu, Z.; Peng, K.-C.; Ernst, J.; Fu, Y. Guided attention inference network. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 42, 2996–3010. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.; So, S.; Yun, S.; Lee, S.; Barg, J. Spatial Feature Conservation Networks (SFCNs) for Dilated Convolutions to Improve Breast Cancer Segmentation from DCE-MRI. In International Workshop on Applications of Medical AI; Springer: Cham, Switzerland, 2022; pp. 118–127. [Google Scholar]
| Kidney Stone | ||||||||
|---|---|---|---|---|---|---|---|---|
| Metrics | ResNet50 | DenseNet121 | EfficientNet [23] | ViT [24] | GAIN [32] | sRense Block | Rense Block | |
| Accuracy | WO/E | 96.98 | 95.26 | 95.69 | 75.86 | 70.37 | 83.19 | 97.80 |
| W/E | 97.41 | 96.98 | 96.12 | - | - | 92.24 | 97.84 | |
| Precision | WO/E | 96.36 | 91.23 | 91.38 | 75.00 | 39.98 | 66.67 | 96.41 |
| W/E | 96.43 | 98.11 | 98.04 | - | - | 93.48 | 96.49 | |
| Sensitivity | WO/E | 91.38 | 89.53 | 91.38 | 5.17 | 81.25 | 65.52 | 94.75 |
| W/E | 93.10 | 89.66 | 86.21 | - | - | 74.14 | 94.83 | |
| Specificity | WO/E | 96.82 | 97.09 | 97.05 | 95.37 | 85.94 | 89.08 | 97.13 |
| W/E | 98.86 | 99.41 | 98.85 | - | - | 98.28 | 99.43 | |
| F1 score | WO/E | 93.80 | 90.37 | 91.38 | 9.67 | 53.59 | 66.09 | 95.57 |
| W/E | 94.74 | 93.69 | 91.75 | - | - | 82.69 | 95.65 | |
| Kidney Stone | ||||||||
|---|---|---|---|---|---|---|---|---|
| Metrics | ResNet50 | DenseNet121 | EfficientNet [23] | ViT [24] | GAIN [32] | sRense Block | Rense Block | |
| Accuracy | WO/E | 0.0061 | 0.0004 | 0.0102 | 0.0037 | 0.0032 | 0.0023 | 0.0127 |
| W/E | 0.0069 | 0.0108 | 0.0037 | 0.0048 | 0.0153 | |||
| Precision | WO/E | 0.0007 | 0.0158 | 0.0094 | 0.0136 | 0.0157 | 0.0076 | 0.0057 |
| W/E | 0.0052 | 0.0057 | 0.0064 | 0.0053 | 0.0071 | |||
| Sensitivity | WO/E | 0.0036 | 0.0069 | 0.0073 | 0.0037 | 0.0035 | 0.0058 | 0.0027 |
| W/E | 0.0032 | 0.0115 | 0.0161 | 0.0014 | 0.0024 | |||
| Specificity | WO/E | 0.0128 | 0.0111 | 0.0090 | 0.0104 | 0.0127 | 0.0140 | 0.0090 |
| W/E | 0.0142 | 0.0110 | 0.0086 | 0.0167 | 0.0029 | |||
| F1 score | WO/E | 0.0037 | 0.0045 | 0.0119 | 0.0065 | 0.0146 | 0.0067 | 0.0071 |
| W/E | 0.0145 | 0.0087 | 0.0064 | 0.0003 | 0.0039 | |||
| Lung Tumor | ||||||||
|---|---|---|---|---|---|---|---|---|
| Metrics | ResNet50 | DenseNet121 | EfficientNet [23] | ViT [24] | GAIN [32] | sRense Block | RENSE Block | |
| Accuracy | WO/E | 66.67 | 76.98 | 68.45 | 71.63 | 65.43 | 71.43 | 77.01 |
| W/E | 75.59 | 77.58 | 73.61 | - | - | 73.41 | 79.58 | |
| Precision | WO/E | 71.72 | 71.04 | 82.31 | 65.37 | 20.81 | 69.37 | 76.73 |
| W/E | 80.54 | 79.39 | 84.68 | - | - | 70.69 | 84.97 | |
| Sensitivity | WO/E | 56.89 | 91.23 | 48.41 | 91.40 | 90.58 | 78.87 | 83.55 |
| W/E | 58.41 | 84.00 | 58.84 | - | - | 82.04 | 84.23 | |
| Specificity | WO/E | 77.63 | 62.01 | 78.68 | 51.45 | 57.07 | 65.29 | 89.56 |
| W/E | 94.04 | 94.40 | 76.36 | - | - | 66.12 | 94.85 | |
| F1 score | WO/E | 63.45 | 79.88 | 60.96 | 76.22 | 33.84 | 73.82 | 79.99 |
| W/E | 67.71 | 81.63 | 69.43 | - | - | 75.94 | 84.59 | |
| DSC | WO/E | 25.49 | 40.86 | 22.84 | 28.01 | 26.65 | 42.33 | 49.13 |
| W/E | 31.77 | 52.27 | 28.20 | - | - | 58.97 | 63.52 | |
| Lung Tumor | ||||||||
|---|---|---|---|---|---|---|---|---|
| Metrics | ResNet50 | DenseNet121 | EfficientNet [23] | ViT [24] | GAIN [32] | sRense Block | Rense Block | |
| Accuracy | WO/E | 0.0124 | 0.0017 | 0.0052 | 0.0039 | 0.0152 | 0.0175 | 0.0076 |
| W/E | 0.0144 | 0.0078 | 0.0029 | 0.0186 | 0.0208 | |||
| Precision | WO/E | 0.0028 | 0.0084 | 0.0070 | 0.0116 | 0.0070 | 0.0174 | 0.0109 |
| W/E | 0.0084 | 0.0202 | 0.0197 | 0.0149 | 0.0001 | |||
| Sensitivity | WO/E | 0.0170 | 0.0146 | 0.0153 | 0.0197 | 0.0061 | 0.0008 | 0.0175 |
| W/E | 0.0168 | 0.0092 | 0.0160 | 0.0022 | 0.0093 | |||
| Specificity | WO/E | 0.0042 | 0.0018 | 0.0112 | 0.0210 | 0.0171 | 0.0185 | 0.0109 |
| W/E | 0.0051 | 0.0154 | 0.0202 | 0.0138 | 0.0089 | |||
| F1 score | WO/E | 0.0125 | 0.0101 | 0.0140 | 0.0024 | 0.0204 | 0.0051 | 0.0099 |
| W/E | 0.0083 | 0.0057 | 0.0159 | 0.0053 | 0.0195 | |||
| DSC | WO/E | 2.7190 | 3.0903 | 3.8134 | 1.7193 | 2.7462 | 2.2437 | 1.7087 |
| W/E | 1.6711 | 2.4934 | 3.4014 | 1.9083 | 3.3200 | 1.6607 | 2.9846 | |
| ResNet50 | DenseNet121 | EfficientNet [23] | ViT [24] | GAIN [32] | sRense Block | Rense Block | ||
|---|---|---|---|---|---|---|---|---|
| Total Number of Parameters [106] | WO/E | 23.51 | 6.95 | 63.58 | 85.64 | 103.45 | 0.13 | 0.15 |
| W/E | 41.91 | 24.66 | 91.39 | - | - | 0.41 | 0.43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, H.; Lee, S.; Yun, S.; Leem, S.; So, S.; Han, D.H. RenseNet: A Deep Learning Network Incorporating Residual and Dense Blocks with Edge Conservative Module to Improve Small-Lesion Classification and Model Interpretation. Cancers 2024, 16, 570. https://doi.org/10.3390/cancers16030570
Seo H, Lee S, Yun S, Leem S, So S, Han DH. RenseNet: A Deep Learning Network Incorporating Residual and Dense Blocks with Edge Conservative Module to Improve Small-Lesion Classification and Model Interpretation. Cancers. 2024; 16(3):570. https://doi.org/10.3390/cancers16030570
Chicago/Turabian StyleSeo, Hyunseok, Seokjun Lee, Sojin Yun, Saebom Leem, Seohee So, and Deok Hyun Han. 2024. "RenseNet: A Deep Learning Network Incorporating Residual and Dense Blocks with Edge Conservative Module to Improve Small-Lesion Classification and Model Interpretation" Cancers 16, no. 3: 570. https://doi.org/10.3390/cancers16030570
APA StyleSeo, H., Lee, S., Yun, S., Leem, S., So, S., & Han, D. H. (2024). RenseNet: A Deep Learning Network Incorporating Residual and Dense Blocks with Edge Conservative Module to Improve Small-Lesion Classification and Model Interpretation. Cancers, 16(3), 570. https://doi.org/10.3390/cancers16030570






